Abstract
Recent findings revealed rare copy number variants and missense changes in the X-linked gene PTCHD1 in autism spectrum disorder (ASD) and intellectual disability (ID). Here, we aim to explore the contribution of common PTCHD1 variants in ASD and gain additional evidence for the role of rare variants of this gene in ASD and ID. A two-stage case–control association study investigated 28 tag single nucleotide polymorphisms (SNPs) in 994 ASD cases and 1035 controls from four European populations. Mutation screening was performed in 673 individuals who included 240 ASD cases, 183 ID patients and 250 controls. The case–control association study showed a significant association with rs7052177 (P=6.13E-4) in the ASD discovery sample that was replicated in an independent sample (P=0.03). A Mantel-Haenszel meta-analysis for rs7052177T considering the four European populations showed an odds ratio of 0.58 (P=7E-05). This SNP is predicted to be located in a transcription factor binding site. No rare missense PTCHD1 variants were found in our ASD cohort and only one was identified in the ID sample. A duplication (27 bp) in the promoter region, absent from 590 controls, was found in three ASD patients (Fisher exact test, P=0.024). A gene reporter assay showed a significant decrease in the transcriptional activity (26%) driven by this variant. Moreover, we found that the longest allele of a trinucleotide repeat located upstream from PTCHD1 was associated with ASD (P=0.003, permP=0.0186). Our results further support the involvement of PTCHD1 in ASD, suggesting that both common and rare variants contribute to the disorder.
Introduction
Autism spectrum disorder (ASD) is a severe neurodevelopmental disorder characterized by impairments in reciprocal social interaction, verbal and non-verbal communication and stereotyped patterns of behavior. The prevalence for ASD is estimated to be about 0.5–1%, and it is approximately four times more frequent in males than in females.1, 2, 3 Family and twin studies in recent decades have provided strong evidence showing that ASD is one of the most heritable neuropsychiatric disorders. Sibling recurrence risk is approximately 20%, and concordance among twins may range from 76 to 88% for monozygotic twins and from 0 to 31% for dizygotic twins, depending on phenotypic criteria.4, 5 The genetic model for idiopathic autism is complex. Recent studies suggest that at least 1000 genes contribute to the disorder, with a combination of common variants of small to moderate effect and rare variants with potential larger effect sizes.6, 7
Common single nucleotide polymorphisms (SNPs) that act in an additive manner are estimated to explain about 17% of the variance in liability to ASD, whereas copy number variants explain approximately 10% of the autism phenotype.8, 9, 10 Exome and genome sequencing studies have provided strong evidence for the involvement of rare variants in autism, suggesting novel candidate genes in the disorder.11, 12, 13, 14 However, most of the underlying genetic factors remain still unknown, and none of the reported mutations accounts globally for more than 1% of ASD cases.15 The observed sex bias in ASD has suggested in the past a major role for X-linked genes in the disorder, although none of the main linkage studies detected loci on this chromosome,16 apart from modest evidence of linkage at Xq26 and Xq12.17, 18 However, genes mapping on the X chromosome remain strong candidates for ASD. Indeed, a considerable number of genes with neuronal function map on chromosome X, and their expression in the brain has been reported to be 1.1–1.2 higher than that of autosomal genes.19, 20, 21 The relevant role of X-linked genes in brain function is also suggested by the high number of genes responsible for intellectual disability (ID) that have been mapped on this chromosome. A considerable proportion of autism cases (around 30%) also present ID. Recently, it has been suggested that truncating variants may have a predominant role in those cases of autism associated with ID.13 Interestingly, several of these truncated genes, such as NLGN4X, TMLHE or DRP2, among others, map on chromosome X.13, 22, 23
Recent studies have pointed to the X-linked PTCHD1 gene as one of the most interesting candidates for involvement in ASD and ID. PTCHD1 maps on chromosome Xp22.11 and encodes the patched domain containing protein 1 (PTCHD1), which is mainly expressed in the developing brain and in adult brain tissues, with the highest expression in the cerebellum.24 PTCHD1 has been suggested to be implicated in Hedgehog (Hh) signalling, inhibiting Gli-dependent transcription in a similar way as its homologues PTCH1 and PTCH2.24 Deletions spanning the gene have been reported in ASD and ID patients.24, 25, 26, 27, 28, 29 Furthermore, a mutation screening performed in 900 ASD and 225 ID patients revealed seven missense variants in eight families (six ASD and two ID) that were absent from a control sample of 700 individuals.24 All were transmitted from the healthy mothers to the affected male probands, compatible with an X-linked inheritance model. Recently, a systematic clinical description of 23 individuals with truncating variants or deletions involving PTCHD1 suggested that these are highly penetrant genetic factors that may cause infantile hypotonia, motor coordination problems, subtle dysmorphic features and a wide range of neurodevelopmental conditions, including ID, ASD and attention deficit hyperactivity disorder.30
Here, we performed the first analysis of PTCHD1 common variants in ASD through a two-stage case–control association study of 28 SNPs in 994 ASD cases and 1035 controls. Moreover, we aimed to gain additional evidence for the role of rare variants of PTCHD1 in ASD and ID by performing a mutational screening of this gene in a Spanish sample of 240 ASD cases, 183 ID patients and 250 controls. The results of the present study provide new insights into the involvement of PTCHD1 in autism.
Materials and methods
Subjects
The mutational screening of PTCHD1 was performed in 240 ASD patients (216 males, 24 females) and 183 ID subjects (all males). Known ID syndromes were excluded by clinical evaluation. The control sample consisted of 250 healthy subjects (225 males and 25 females), and was increased to 590 subjects for the mutational analysis of the promoter region (535 males and 55 females). All individuals were European and Caucasian.
The sample used in the case–control association study comprised 994 ASD cases and 1035 controls from four European populations: Spanish, Dutch, German and Italian (Table 1). The discovery sample consisted of 595 ASD patients (270 Spanish, 247 Dutch and 78 German) and 671 gender-matched controls (320 Spanish, 269 Dutch and 82 German). Positive findings were replicated in an independent sample of 399 cases (230 Italian, 45 Spanish and 124 German) and 364 gender-matched controls (175 Italian, 58 Spanish and 131 German).
Table 1. European ASD samples used in the two-stage case–control association study.
| Discovery (28 SNPs) | Replication (1 SNP) | Pooled sample (1 SNP) | ||||
|---|---|---|---|---|---|---|
| Cases (%M) | Controls (%M) | Cases (%M) | Controls (%M) | Cases (%M) | Controls (%M) | |
| Spanish | 270 (88) | 320 (88) | 45 (91) | 58 (91) | 315 (88) | 378 (88) |
| Dutch | 247 (78) | 269 (78) | — | — | 247 (78) | 269 (78) |
| German | 78 (89) | 82 (89) | 124 (90) | 131 (90) | 202 (90) | 213 (90) |
| Italian | — | — | 230 (82) | 175 (81) | 230 (82) | 175 (81) |
| Total | 595 (84) | 671 (84) | 399 (86) | 364 (86) | 994 (85) | 1035 (85) |
%M, indicates the percentage of male individuals.
Abbreviation: SNP, single nucleotide polymorphism.
All ASD patients met ICD-10 or DSM-IV TR criteria for autism, Asperger disorder or pervasive developmental disorder not otherwise specified (PDD-NOS), assessed using ADI-R (Autism-Diagnostic Interview-Revised)31 and when possible also the ADOS (Autism Diagnostic Observation Schedule).32 Cytogenetic abnormalities and positive Fragile X test were considered as exclusion criteria in our ASD and ID samples. The study was approved by the relevant ethics committee in each center and written informed consent was obtained from all parents/guardians or, where possible, by affected individuals.
Genomic DNA was extracted from peripheral blood samples using standard salting-out methods, or alternatively from saliva samples using the Oragene DNA Extraction kit (DNA Genotek, Ontario, Canada), following the manufacturer's protocol. The DNA concentration of all samples was measured on a NanoDrop spectrophotometer (NanoDrop Technologies, LLC, Wilmington, DE, USA) or determined on a Gemini XPS fluorometer (Molecular Devices, Sunnyvale, CA, USA) using the PicoGreen dsDNA Quantification Kit (Molecular Probes, Eugene, OR, USA).
Selection of tag SNPs in the PTCHD1 gene and genotyping
The selection of tag SNPs was performed by considering patterns of linkage disequilibrium (LD) of PTCHD1 in the CEU panel using the HapMap project data (www.hapmap.org; phases 1, 2, 3; release 28). The covered region included the whole gene plus 5 kb upstream and 3 kb downstream from the coding region (Figure 1). A total of 29 tag SNPs were selected using the Tagger implementation in HaploView v4.2,33 according to the following criteria: r2≥0.8 and minor allele frequency≥0.05. The discovery sample was genotyped using the VeraCode technology (Illumina, San Diego, CA, USA) at the Spanish National Genotyping Center (CeGen). Five HapMap trios were genotyped and used as controls for genotyping quality and for genotype clustering. Only 1 SNP from the 29 SNPs initially genotyped failed (rs2363130, chrX.hg19.g.23367895C>A). The sample success rate was 98.3%. All polymorphisms were assessed for Hardy–Weinberg Equilibrium (threshold set at P<0.01 in our control sample) and checked for minor allele frequency (values>0.05) (Supplementary Table 1). The LD patterns and r2 values were calculated in our genotyping data using Haploview v4.2 (Supplementary Figure 1).33 The SNP included in the replication study was genotyped at the Santiago de Compostela node of the National Genotyping Center using the iPlex-Sequenom technology (Sequenom, San Diego, CA, USA).
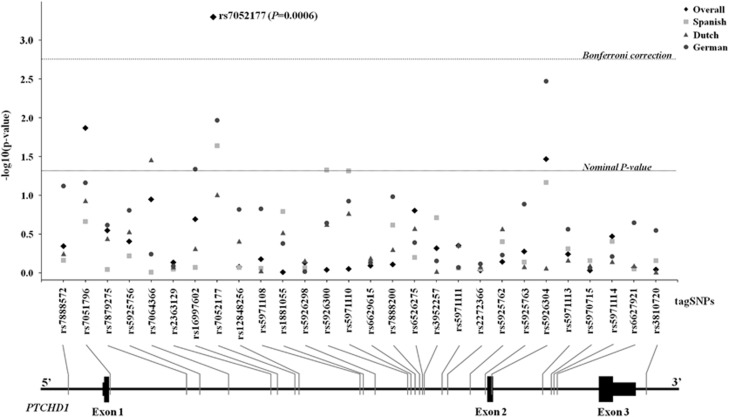
Figure 1.
Results from the case–control association study in the discovery sample (595 ASD cases and 671 controls). The x axis shows the PTCHD1 tag SNPs and their location along the gene. The y axis indicates the significance of the association as –log (P-value). The dashed lines represent the thresholds for statistical significance: Nominal P-value (P=0.05) and Bonferroni correction (P=0.0018). The P-values for the whole sample (◊) and the three populations under study (Spanish (□), Dutch (Δ) and German (o)) are indicated.
Mutation screening of the PTCHD1 gene
In this study, the coding regions and splice sites of the PTCHD1 gene were screened for rare variants. A 403 bp region upstream from the transcription start site (TSS) of the gene was also included in the mutation screening. Primers and PCR conditions are described in Supplementary Table 2. Purified PCR fragments were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit and an ABI PRISM 3730 automatic sequencer (Applied Biosystems, Foster City, CA, USA). The GRCh37/hg19 assembly of the UCSC Human Genome Browser (genome.ucsc.edu) was used for genome reference positions and all rare variants were submitted to the Leiden Open Variation Database (databases.lovd.nl/shared/genes). Exons were numbered according to the reference sequence NG_021300.1. We describe a novel trinucleotide repeat located 80 bp upstream from the TSS of PTCHD1 (chrX.hg19.g.23352905GCC(10_14)), that we have reported to the European Genome-phenome Archive (EGA-EBI, www.ebi.ac.uk/ega/home). The corresponding microsatellite alleles were assigned using sequencing data from 240 ASD patients and 585 controls.
Promoter cloning and gene reporter assay
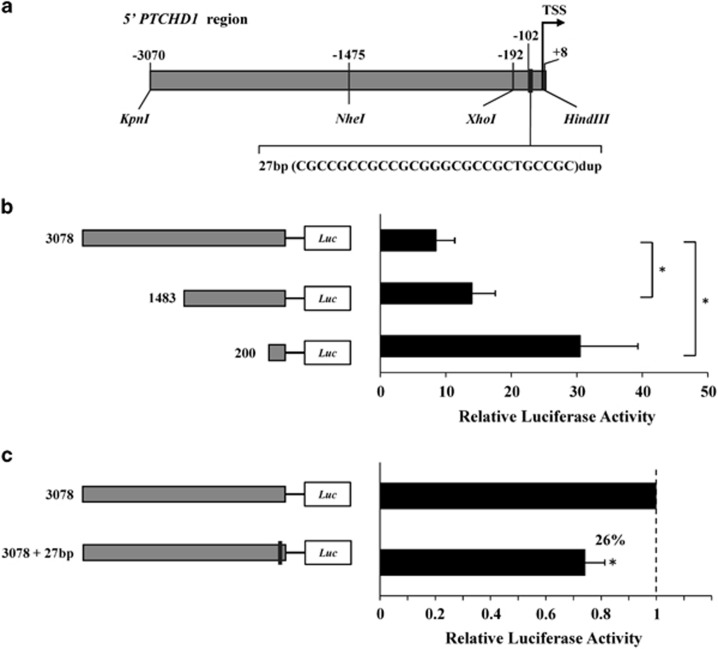
The duplication identified in the promoter region of PTCHD1 (chrXhg19.g.23352883_23352909dup) was assessed using a luciferase reporter assay to evaluate its influence on gene expression. Two fragments located upstream from PTCHD1 were synthesized: the wild-type (WT) promoter region (3078 bp, chrX.hg19.g.23349914-23352992) and the mutant, containing the 27-bp duplication (3078 bp+27 bp) (GENEWIZ Inc., South Plainfield, NJ, USA) (Figure 2a). Both fragments were cloned into a pGL3-Basic vector (Promega, Madison, WI, USA) using the unique digestion sites KpnI and HindIII. Two additional constructs were obtained by removing the most upstream fragments, resulting in shorter promoter regions of 1483 bp and 200 bp (Figure 2b). Constructs, sequences, plasmids and conditions are available upon request.
Figure 2.
Luciferase-based expression analysis of the PTCHD1 promoter. (a) Schematic representation of the upstream region of the PTCHD1 gene studied. Three constructs were generated by digestion with different restriction enzymes (3078, 1483 and 200 bp). For each segment, we prepared the WT construct and the counterpart with the duplication found in our autistic patients. Expression plasmids were co-transfected into SH-SY5Y cells with a Renilla plasmid for normalization. (b) Promoter activity based on Relative Luciferase Activity (RLA) for each of the three WT promoter regions. (c) Comparison of the luciferase expression for the longest WT construct and its mutated counterpart. In this graph, the RLA for the WT construct was scaled to 1.
We used the human SH-SY5Y neuroblastoma cell line grown in Dulbecco's Modified Eagle Medium and Nutrient Mixture F-12 (Ham's, Life Technologies, Paisley, UK) (1:1) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin (GIBCO, Carlsbad, CA, USA) in a 5% CO2 humidified atmosphere at 37 °C. The cell line was grown in 60-mm-diameter plates at a concentration of 2 × 106 cells/well, co-transfected with 1.2 μg Renilla plasmid (Promega) and an equimolar amount of each pGL3 construct under study, using the Amaxa Cell Line Nucleofector Kit V (Lonza, Basel, Switzerland) according to the manufacturer's instructions. Transfection for each reporter construct was conducted in triplicate and repeated twice (six biological replicates). Luciferase activities were measured in duplicate 24 h after transfection using the Dual Luciferase Reporter Assay System (Promega). The luminescence of 20 μl of lysis was measured using the Modulus Microplate Multimode Reader (Turner Biosystems Inc., Sunnyvale, CA, USA). Luciferase efficiencies were normalized to the Renilla luciferase activity to obtain a measure of ‘relative luciferase activity'.
Match software,34 which uses transcription factor binding sites from the Transfac Public database (www.gene-regulation.com), was used to predict the presence of transcription factor binding sites in the region.
Statistical analysis
The analysis of Hardy–Weinberg Equilibrium and minor allele frequency in the association study was performed using the SNPassoc R package.35 Twenty-eight markers were assessed in the discovery sample for the case–control association study under the additive model using the PLINK package.36 The analysis was performed for each population separately and also in all populations combined. Multiple testing was corrected using the Bonferroni correction considering 28 tests (P<0.0018). Haplotype-based analysis was performed using the Haploview v4.2 software.33 PLINK was also used in the replication study and in the pooled study of rs7052177 (chrX.hg19.g.23374882T>G) that combined the discovery and replication samples.36
An analysis of minimal statistical power was performed with the CaTS Power Calculator software (sph.umich.edu/csg/abecasis/CaTS),37 assuming an odds ratio (OR) of 1.3, estimated disease prevalence of 0.006,1, 2, 3 significance level of 0.05 (α) and minor allele frequency of 0.1, under the additive model. A meta-analysis study was performed for rs7052177 using genotype data from both the discovery and the replication sets. The study was performed using the rmeta R package (www.cran.r-project.org/web/packages/rmeta/index.html) and the PLINK package.36 The Q-statistic was assessed to ensure no heterogeneity among studies. Then the pooled OR was estimated using a fixed-effects model.
To assess the possible functional effect of the associated SNP rs7052177, the Transfac matrix database (v.7) (www.gene-regulation.com/) was used to predict possible changes in transcription factor affinity driven by the risk allele.
A case–control association study was performed for the trinucleotide repeat at 5' of PTCHD1 (chrX.hg19.g.23352905GCC(10_14)) using UNPHASED v3.0.38 Allele frequencies were estimated for 240 cases and 585 controls (Supplementary Table 3).
Statistical analysis of the reporter gene experiments in the promoter region of PTCHD1 was performed as follows: To compare the ‘relative luciferase activity' for different segments of the promoter considered in our study, normality was rejected using the Kolmogorov–Smirnov test, and non-parametric tests were applied. Statistical significance for the comparisons among the different WT constructs was calculated with the Kruskal–Wallis test, using Bonferroni's correction for multiple comparisons (P<0.017). The Wilcoxon signed-rank test was used to compare each WT construct against the corresponding mutant. Statistical significance was considered at a P-value <0.05.
Results
Case–control association study
Twenty-eight tag SNPs capturing the genetic variability of PTCHD1 were assessed for association with ASD using a discovery sample of 595 cases and 671 controls from three European populations (Table 1). All markers were analyzed under the additive model. The results are summarized in Figure 1, showing nominal associations in individual populations (Spanish: rs7052177, P=0.026; Dutch: rs7064366 chrX.hg19.g.23366722A>G, P=0.038; German: rs7052177, P=0.0123 and rs5926304 chrX.hg19.g.23397964C>T, P=0.004) (Supplementary Table 1). When the whole sample was considered, the most significant results were achieved for rs7051796 chrX.hg19.g.23353433G>C (P=0.015), rs7052177 (P=6.1E-04) and rs5926304 (P=0.038). The only marker that remained significant after applying multiple testing corrections was rs7052177 (threshold for Bonferroni correction, P<0.0018). The pairwise LD map of PTCHD1 was calculated in the discovery sample, which defined five LD blocks (Supplementary Figure 1). The haplotype analysis of these blocks failed to detect any association (all P>0.05, data not shown). The associated marker rs7052177 was not included in any of these LD blocks, being a singleton SNP.
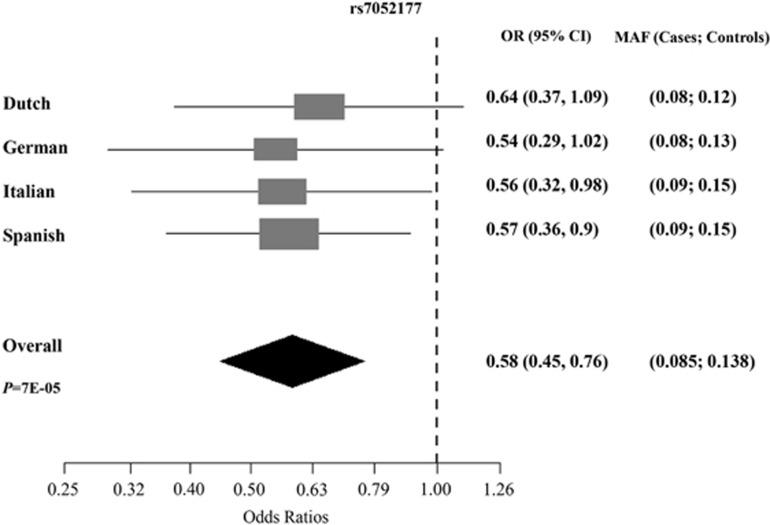
Subsequently, we sought to replicate the association identified in the discovery sample for rs7052177 in an additional European sample of 399 ASD cases and 364 controls (details of the samples used are shown in Table 1). We obtained significant association in the replication study (P=0.03). The minimal statistical power for the discovery sample to identify a variant of 1.3 OR under the additive model was 0.73, whereas for the replication study it was 0.49. We also performed the analysis for rs7052177 by combining the discovery and replication samples (994 ASD cases and 1035 controls); this achieved a significant result (P=6.8E-05) even after applying permutations (permP=9.9E-05). The association was driven by the Spanish (P=0.016) and Italian (P=0.04) samples and a trend in the German (P=0.055) and Dutch (P=0.102) samples (Table 2). Finally, when we considered the male and female samples separately, association was found in both groups (P=6.1E-04 and P=0.042, respectively) (Table 2). A meta-analysis study was performed for rs7052177 using the four European samples. No heterogeneity was detected (Q-statistic, P=0.98). Meta-analysis under the fixed effects model (Mantel-Haenszel) showed an OR of 0.58 (95% CI= 0.45–0.76) (P=7E-05). The meta-analysis Forest plot is shown in Figure 3 where OR estimations showed the same trend in all populations under study.
Table 2. Results of the case–control association study of rs7052177 showing the association of the major T allele with ASD.
| Pooled analysis (discovery+replication) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Population | Gender | All | |||||||
| Discovery | Replication | Spanish | Dutch | German | Italian | Males | Females | ||
| rs7052177 | 0.0006 | 0.0297 | 0.0164 | 0.1025 | 0.0559 | 0.0405 | 0.0006 | 0.0422 | 0.000068(permP=9.9E-05) |
permP, Permuted P-value obtained after 10 000 permutations.
Figure 3.
Forest plot for the OR (95% CI) of marker rs7052177 (allele T). ORs and minor allele frequency are shown for each European population under study and in the combined sample (Mantel-Haenszel meta-analysis). The overall P-value (P) for the meta-analysis is also indicated.
Mutation screening of the PTCHD1 gene
We screened the coding regions of PTCHD1 for rare variants in a sample of 673 individuals: 240 ASD patients, 183 ID patients and 250 controls. The results revealed two missense changes, one in an ID patient (chrX.hg19.g.23353144G>A, p.(Ser51Asn)) and one in a control individual (chrX.hg19.g.23412280T>C, p.(Val882Ala)), but none in the ASD sample (Table 3). The analysis of the promoter region identified several variants, most of them indels. The most interesting variant from this study was a 27-bp duplication upstream from the TSS (chrX.hg19.g.23352883_23352909dup), which was found in 3 out of 240 ASD probands and was absent from a control sample of 590 individuals (Fisher exact test, P=0.024). However, another 27-bp duplication (chrX.hg19.g.23352878_23352904dup) that partially overlaps the previous one was identified in a control individual (Table 3).
Table 3. Rare variants of the X-linked PTCHD1 gene in the ASD, ID and control samples from the present study and from the study of Noor et al. 24 .
| Present study | Noor et al.24 | EVSa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variantb | Location | dbSNP ID | Amino acidc | ASD (n=240) | ID (n=183) | Controls (n=250d) | ASD (n=900) | ID (n=225) | Controls (n=700) | Controls (n=1872) | PolyPhen2 | SIFTe |
| g.23352872_23352904del | promoter | 1(M) | ||||||||||
| g.23352872_23352924del | promoter | 1(M) | ||||||||||
| g.23352878_23352904dup | promoter | 1(M) | ||||||||||
| g.23352882_23352914del | promoter | 1(M) | ||||||||||
| g.23352882_23352921del | promoter | 1(F) | ||||||||||
| g.23352883_23352909dup | promoter | 3(M) | ||||||||||
| g.23352898_23352907del | promoter | 1(F) | ||||||||||
| g.23352938C>G | promoter | 1(M) | ||||||||||
| g.23353109C>G | Exon 1 | p.(=) | 1(M) | — | ||||||||
| g.23353144G>A | Exon 1 | p.(S51N) | 1(M) | — | B | B | ||||||
| g.23353209C>T | Exon 1 | rs373105249 | p.(L73F) | 1(M) | B | B | ||||||
| g.23353237G>A | Exon 1 | rs371935424 | p.(R82H) | 1 (M) | B | B | ||||||
| g.23397704A>C | Intron 1 | 1(M) | 1(M) | — | ||||||||
| g.23397873A>G | Exon 2 | rs147324438 | p.(I173V) | 2(M) | 2 (M) | P | B | |||||
| g.23397939G>A | Exon 2 | p.(V195I) | 1(M) | — | B | B | ||||||
| g.23398046G>A | Exon 2 | p.(=) | 1(M) | — | ||||||||
| g.23398107C>T | Exon 2 | rs368662150 | p.(P251S) | 1 (M) | B | B | ||||||
| g.23398108C>T | Exon 2 | p.(P251L) | 1(F) | — | P | B | ||||||
| g.23398364_23398365delinsTA | Exon 2 | p.(ML336-7II) | 1(M) | — | M336I: B; L337I: P | M336I: B; L337I: B | ||||||
| g.23410711A>G | Exon 3 | p.(H359R) | 1(M) | — | P | B | ||||||
| g.23411044C>A | Exon 3 | p.(A470D) | 1(M) | — | B | B | ||||||
| g.23411071A>G | Exon 3 | p.(E479G) | 1(M) | — | B | B | ||||||
| g.23411126C>A | Exon 3 | rs35880456 | p.(N497K) | 1(F) | — | P | B | |||||
| g.23412280T>C | Exon 3 | p.(V882A) | 1(M) | — | P | P | ||||||
| Total missense variants | 0/240 | 1/183 | 1/250 | 6/900 | 2/225 | 2/700 | ||||||
| Total missense variants (only males) | 0/216 | 1/183 | 1/225 | 6/723 | 2/225 | 0/531 | 4/1872 | |||||
Abbreviations: ASD, autism spectrum disorder; B, predicted benign; dbSNP, database of single nucleotide polymorphism; F, female; ID, intellectual disability; M, male; P, predicted pathogenic.
The exome variant server database (EVS) (evs.gs.washington.edu/EVS/) was used to include additional controls in this study. Only male individuals from the European-American EVS sample were included.
The GRCh37/hg19 assembly of the UCSC Human Genome Browser (genome.ucsc.edu) was used for genome reference positions.
NG_021300.1 was used as reference sequence for exon numbering and for amino acid positions
Additional controls were included for the promoter region up to 590 individuals.
Amino acid substitution predictor (SIFT, Sorting Intolerant from Tolerant).
Interestingly, we also found an as yet unannotated polymorphic GCC trinucleotide repeat in the promoter region, located 80 bp upstream from the TSS (chrX.hg19.g.23352905GCC(10_14)). We performed a case–control association study in 240 ASD patients and 585 controls. The results showed association in the global test (P=0.037), and the longest allele of this microsatellite (14 repeats) was significantly associated with ASD (P=0.003, permP=0.0186) (Supplementary Table 3).
Luciferase-based analysis of the 27-bp duplication effect on the PTCHD1 promoter
The 27-bp duplication found in three autistic individuals was functionally studied to assess its effect on PTCHD1 promoter activity. An initial analysis performed in silico with TRANSFAC34 predicted three transcription factor binding sites along the promoter region for the following proteins: ZNF354C (zinc finger protein 354C), ZBTB14 (zinc finger and BTB domain-containing protein 14) and TEAD2 (TEA domain family member 2). TEAD2 is predicted to bind the region found duplicated in our study. A luciferase reporter assay was performed to evaluate the impact of this duplication on gene expression. We generated three luciferase-expressing constructs containing a progressively smaller genomic region immediately upstream from the PTCHD1 gene (−3070 to +8, −1475 to +8 and −192 to +8, with respect to the TSS) (Figure 2b). The ability of the three WT constructs to drive transcription was studied in the SH-SY5Y neuroblastoma cell line. The highest promoter activity was detected with the shortest construct (200 bp) (Figure 2b). We investigated the effect of the 27-bp duplication by comparing luciferase expression for each of the three different WT constructs with the corresponding construct containing the 27-bp duplication (Figure 2c). Our results showed a 26% decrease in the reporter gene activity for the longest construct (P=0.031), whereas there were no significant differences for the two shorter constructs.
Discussion
Here, we present additional evidence for the involvement of the PTCHD1 gene in ASD. Until now, only truncating variants and deletions of this gene have been clearly associated with ASD and ID.
We performed a two-stage case–control association study to explore the contribution of common variants of PTCHD1 to ASD in a European sample of 994 patients and 1035 controls. We identified an association with rs7052177 that was subsequently replicated in an independent sample. This marker was associated in the Spanish and Italian populations and presented a trend for association in the Dutch and German samples (Figure 3 and Table 2). Furthermore, the SNP showed association in both the male and female samples. Rs7052177 is a singleton SNP located in the first intron of the PTCHD1 gene that falls within a predicted binding site for several members of the STAT protein family, which regulate many aspects of growth, survival and cell differentiation. The transcription factors STAT5A, STAT5B and STAT3 are predicted to bind their target preferentially when the rs7052177T allele, associated with ASD in our study, is present. This suggests that this common variant may have a functional impact on the regulation of the PTCHD1 gene.
Although several reports have confirmed a clear involvement of rare deletions or coding indels of PTCHD1 in ASD and ID,24, 25, 27, 30 it remains unclear whether rare missense variants of this gene may also contribute to the etiology of these disorders. In our study, we screened a Spanish sample of 240 ASD and 183 ID patients for variants in the PTCHD1 gene. Unlike the previous work of Noor et al.,24 which showed a significant enrichment of PTCHD1 missense variants in an ASD male sample, we failed to identify neither non-synonymous nor truncating variants in our ASD cohort. The results showed a missense change (p.(Ser51Asn)) predicted to be benign in one ID patient and also a missense change (p.(Val882Ala)) predicted to be damaging in one control individual. Overall, our study failed to find additional evidence for the involvement of rare missense variants of PTCHD1 in ASD, although we only investigated a small sample. When we considered a broader sample combining the data from the present study with those of Noor et al.24 and the European-American sample from the Exome Variant Server (evs.gs.washington.edu/EVS/), the enrichment of PTCHD1 rare missense variants in the male autism sample was significant (Fisher exact test, P=0.044), although these missense variants in ASD probands were predicted to be mostly benign (Table 3).
The most relevant result of our mutation screening was a 27-bp duplication in the promoter region of the PTCHD1 gene, which was present in three unrelated ASD probands and absent from a Spanish control sample of 590 individuals. Interestingly, one of these three patients has a dizygotic twin diagnosed with ASD, who is also a carrier of the duplication. The variant identified in these patients spans from −102 to −76 from the TSS. This region is predicted to harbor binding sites for the transcription factor TEAD2. This transcription factor belongs to a highly conserved protein family involved in the control of apoptosis and cell proliferation.39, 40, 41 TEAD2 is expressed in early stages of brain development,42, 43 and has been suggested to play a role in neural development.44, 45
In order to evaluate the possible effect of this duplication on the regulation of PTCHD1, we performed a gene reporter assay in the SH-SY5Y human neuroblastoma cell line. The study showed a significant decrease in gene expression (26%) when the longest promoter construct with the duplication was considered (Figure 2). The presence of additional TEAD2-binding sites in the promoter bearing the duplication may interfere with the correct assembly of the transcription initiation complex and could be responsible for the reduction in gene expression reported in this study. However, no differences were found between the WT and the mutated promoter regions for the two shorter constructs. We speculate that upstream regulatory elements in the longest construct interact with basal promoter elements. Interestingly, Noor et al.24 reported an enrichment of deletions in the PTCHD1 upstream region in male ASD patients compared with male controls. These deletions were suggested to disrupt putative enhancer elements.
We also identified a previously undescribed trinucleotide repeat, located at −80 bp from the TSS of PTCHD1, that showed a significant association with ASD driven by its longest allele. Expansion of tandem repeats is frequently associated with changes in gene expression and in some cases can be responsible for disease.46, 47, 48 Interestingly, the region found duplicated in the three ASD probands and the long microsatellite allele found associated are slightly overlapping and may exert the same effect on promoter activity.
In summary, our results further support the involvement of PTCHD1 in autism and suggest that regulatory elements of the gene may play an important role. We describe: (i) a common intronic variant (rs7052177) significantly associated with ASD; (ii) a rare 27-bp duplication in the promoter region of three ASD patients that alters gene expression and (iii) an additional association with ASD of the longest allele of a microsatellite in the PTCHD1 promoter region.
Recent findings indicate that disrupting variants in PTCHD1 are highly penetrant and responsible for an X-linked condition associated with several neurodevelopmental features.30 Given our results, we speculate that other variants of modest effect in the gene, located in regulatory regions, may contribute to the susceptibility to ASD.
Further studies are warranted to clarify the actual role of missense variants of this gene in the disease and to corroborate our findings.
Acknowledgments
We are grateful to all the patients, their families and clinical collaborators (Montse Causi and Ayhesa Ruiz). We would like to thank Magda Monfort and Maria Torres for technical support at the Spanish National Genotyping Center (CeGen) in Barcelona and Santiago de Compostela, respectively, and Gemma Marfany and Concepció Arenas for helpful suggestions on functional and statistical analyses, respectively. BT was supported by AGAUR (Generalitat de Catalunya), CT by the European Union (Marie Curie, PIEF-GA-2009-254930) and NF-C by the Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER). Financial support was received from ‘Fundació La Marató de TV3' (092010), ‘Fundación Alicia Koplowitz' (AKOPLOWITZ11_006), AGAUR (2014SGR932, 2014SGR603), and the Spanish ‘Ministerio de Economía y Competitividad' (SAF2012-33484). Additional funding was received from the University of Bologna (RFO), Karakter Child and Adolescent Psychiatry (to JB), the Netherlands Organisation for Scientific Research (NWO grant # 91610024 to NR), and Saarland University (grant # T 2042103-01 to CMF).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Elsabbagh M, Divan G, Koh YJ et al: Global prevalence of autism and other pervasive developmental disorders. Autism Res 2012; 5: 160–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Fombonne E: Pervasive developmental disorders in preschool children: confirmation of high prevalence. Am J Psychiatry 2005; 162: 1133–1141. [DOI] [PubMed] [Google Scholar]
- Fombonne E: Epidemiology of pervasive developmental disorders. Pediatr Res 2009; 65: 591–598. [DOI] [PubMed] [Google Scholar]
- Ozonoff S, Young GS, Carter A et al: Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics 2011; 128: e488–e495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald A, Hoekstra RA: Autism spectrum disorders and autistic traits: a decade of new twin studies. Am J Med Genet B Neuropsychiatr Genet 2011; 156B: 255–274. [DOI] [PubMed] [Google Scholar]
- Berg JM, Geschwind DH: Autism genetics: searching for specificity and convergence. Genome Biol 2012; 13: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persico AM, Napolioni V: Autism genetics. Behav Brain Res 2013; 251: 95–112. [DOI] [PubMed] [Google Scholar]
- Klei L, Sanders SJ, Murtha MT et al: Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism 2012; 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM et al: Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45: 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Scherer SW: Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev 2012; 22: 229–237. [DOI] [PubMed] [Google Scholar]
- Krumm N, O'Roak BJ, Shendure J, Eichler EE: A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci 2014; 37: 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Chahrour MH, Coulter ME et al: Using whole-exome sequencing to identify inherited causes of autism. Neuron 2013; 77: 259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma C, Torrico B, Hervas A et al: Exome sequencing in multiplex autism families suggests a major role for heterozygous truncating mutations. Mol Psychiatry 2013; 19: 784–790. [DOI] [PubMed] [Google Scholar]
- An JY, Cristino AS, Zhao Q et al: Towards a molecular characterization of autism spectrum disorders: an exome sequencing and systems approach. Transl Psychiatry 2014; 4: e394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeste SS, Geschwind DH: Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol 2014; 10: 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams BS, Geschwind DH: Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet 2008; 9: 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Nyholt DR, Magnussen P et al: A genomewide screen for autism susceptibility loci. Am J Hum Genet 2001; 69: 327–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auranen M, Vanhala R, Varilo T et al: A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25-27. Am J Hum Genet 2002; 71: 777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM: High expression of the mammalian X chromosome in brain. Brain Res 2006; 1126: 46–49. [DOI] [PubMed] [Google Scholar]
- Kemkemer C, Kohn M, Kehrer-Sawatzki H, Fundele RH, Hameister H: Enrichment of brain-related genes on the mammalian X chromosome is ancient and predates the divergence of synapsid and sauropsid lineages. Chromosome Res 2009; 17: 811–820. [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM: Dosage compensation of the active X chromosome in mammals. Nat Genet 2006; 38: 47–53. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M et al: X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet 2004; 74: 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava C, Lamari F, Heron D et al: Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE. Transl Psychiatry 2012; 2: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Whibley A, Marshall CR et al: Disruption at the PTCHD1 Locus on Xp22.11 in Autism spectrum disorder and intellectual disability. Sci Transl Med 2010; 2: 49ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB et al: Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 2008; 82: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L et al: Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010; 466: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whibley AC, Plagnol V, Tarpey PS et al: Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am J Hum Genet 2010; 87: 173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filges I, Rothlisberger B, Blattner A et al: Deletion in Xp22.11: PTCHD1 is a candidate gene for X-linked intellectual disability with or without autism. Clin Genet 2011; 79: 79–85. [DOI] [PubMed] [Google Scholar]
- Marshall CR, Scherer SW: Detection and characterization of copy number variation in autism spectrum disorder. Methods Mol Biol 2012; 838: 115–135. [DOI] [PubMed] [Google Scholar]
- Chaudhry A, Noor A, Degagne B et al: Phenotypic spectrum associated with PTCHD1 deletions and truncating mutations includes intellectual disability and autism spectrum disorder. Clin Genet e-pub ahead of print 14 August 2014. doi:10.1111/cge.12482. [DOI] [PubMed]
- Lord C, Rutter M, Le Couteur A: Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 1994; 24: 659–685. [DOI] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L et al: The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord 2000; 30: 205–223. [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265. [DOI] [PubMed] [Google Scholar]
- Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E: MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 2003; 31: 3576–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JR, Armengol L, Sole X et al: SNPassoc: an R package to perform whole genome association studies. Bioinformatics 2007; 23: 644–645. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR et al: Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 2006; 38: 209–213. [DOI] [PubMed] [Google Scholar]
- Dudbridge F: Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 2003; 25: 115–121. [DOI] [PubMed] [Google Scholar]
- Mahoney WM Jr., Hong JH, Yaffe MB, Farrance IK: The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J 2005; 388: 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML: TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev 2001; 15: 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tumaneng K, Guan KL: The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol 2011; 13: 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin P, Hwang JJ, Martial JA, Dolle P, Davidson I: A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J Biol Chem 1996; 271: 21775–21785. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, Cullinan EB, Latham KE, DePamphilis ML: Transcription factor mTEAD-2 is selectively expressed at the beginning of zygotic gene expression in the mouse. Development 1997; 124: 1963–1973. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, Kohn MJ, Liu C, DePamphilis ML: Transcription factor TEAD2 is involved in neural tube closure. Genesis 2007; 45: 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H: Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol 2008; 28: 3177–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszturi E, Kiraly O, Csapo Z et al: Association between the 120-bp duplication of the dopamine D4 receptor gene and attention deficit hyperactivity disorder: genetic and molecular analyses. Am J Med Genet B Neuropsychiatr Genet 2007; 144B: 231–236. [DOI] [PubMed] [Google Scholar]
- Paredes UM, Quinn JP, D'Souza UM: Allele-specific transcriptional activity of the variable number of tandem repeats in 5' region of the DRD4 gene is stimulus specific in human neuronal cells. Genes Brain Behav 2013; 12: 282–287. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Hagerman RJ, Hagerman PJ, Leehey MA: Fragile-X syndrome and fragile X-associated tremor/ataxia syndrome: two faces of FMR1. Lancet Neurol 2007; 6: 45–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.