Abstract
Wine is the most popular alcoholic beverage around the world and because of its importance in society has been widely studied. Understanding what drives its flavor has been a quest for decades but much is still unknown and will be determined at least in part by individual taste preferences. Recently studies in the genetics of taste have uncovered the role of different genes in the determination of food preferences giving new insight on its physiology. In this context we have performed a genome-wide association study on red and white wine liking using three isolated populations collected in Italy, and replicated our results on two additional populations coming from the Netherland and Central Asia for a total of 3885 samples. We have found a significant association (P=2.1 × 10−8) between white wine liking and rs9276975:C>T a polymorphism in the HLA-DOA gene encoding a non-canonical MHC II molecule, which regulates other MHC II molecules. The same association was also found with red wine liking (P=8.3 × 10−6). Sex-separated analysis have also revealed that the effect of HLA-DOA is twice as large in women as compared to men suggesting an interaction between this polymorphism and gender. Our results are one of the first examples of genome-wide association between liking of a commonly consumed food and gene variants. Moreover, our results suggest a role of the MHC system in the determination of food preferences opening new insight in this field in general.
Introduction
Wine is probably the oldest produced beverage in human history dating its first apparition in Georgia 7000 BC. It is extremely important for many cultures not only for its economy but also from the social and religious point of view. For these reasons it is also the most common and consumed type of alcoholic beverage throughout the world.1, 2 In addition, several studies indicate that modest wine consumption can have some beneficial health effects, including protection against cardiovascular and Alzheimer's disease.3, 4, 5, 6
There has been extensive research aiming to improve both its production and its flavor. The taste of wine depends on many different sensory perceptions ranging from taste and olfaction to texture.7 Wine is a mixture of thousands of molecules that can contribute to its final taste,8 and its composition varies greatly depending on the grapes used, climate and production method.9 Although the flavor of some of these compounds is very well known, it is really difficult to understand what their role in final taste is.7, 10, 11
The relevant role of genetic factors both on alcohol consumption and dependence has been clearly demonstrated by genome-wide association study (GWAS) studies.12, 13, 14, 15, 16 Moreover, studies on the genetic variations in bitter taste receptors have also shown that variations in TAS2R16 and TAS2R38 genes can influence alcohol intake17, 18, 19 but not dependence.19
On the contrary, very little is known about the possible contribution of genetic factors to alcohol preferences in humans. Recently, it has been demonstrated that ethanol flavor perception (but not taste) is associated to variants in the olfactory receptor (OR) OR7D4 gene and to the SCNN1D, a gene encoding a subunit of the salt taste receptor.20 Moreover, Mc Rae et al have demonstrated that a non-synonymous variant of OR2J3, an OR gene, impairs the ability to detect cis-3-hexen-1-ol, a key component in many different foods including wine.21 Finally, we conducted a study on numerous taste-related genes, which allowed us to associate white wine and vodka liking to variations on the TAS1R2 gene in Central Asia populations, suggesting a general role in ethanol perception for this sweet receptor gene.22
Nevertheless, the molecular bases underlying food liking in general and more specifically to wine is still poorly understood. For these reasons, we have conducted the first GWAS on red and white wine liking on five isolated populations coming from different geographic areas.
Materials and methods
Study populations
Samples have been collected in various populations from Europe and Central Asia. More specifically, our study includes 381 individuals come from INGI-CARL a population coming from Carlantino, a small village located in Puglia (Southern Italy); 744 from INGI-FVG which refers to six villages all situated in the Friuli Venezia Region in Northern-Eastern Italy; and finally 1115 from INGI-VB a population coming from the Val Borbera Valley in Northern-Western Italy. In total, 1261 samples were from the Erasmus Rucphen Family (ERF) study, a cross-sectional cohort including 3000 living descendants of 22 couples who had at least six children baptized in the community church around 1850–1900. Finally, Silk Road (SR) is a cohort of ~1000 individuals resulting from the sampling of 20 communities coming from five nations (Armenia, Azerbaijan, Georgia, Uzbekistan, Tajikistan and Kazakhstan) located along the silk road, in particular, 335 have been used for this study.
Wine-liking ascertainment
Red and white wine liking was ascertained through a questionnaire where each participant was asked to rate his liking for each type of wine on a scale from 1 (dislike extremely) to 9 (like extremely).23 To assess individual liking in the SR population a five-point scale coupled with smiley faces was used. This scale is commonly used in case of linguistic barriers or when working with illiterate people as was the case of the SR population.24 Given the differences in the two scales data have been standardized by dividing each score for the number of categories of the used scale, so 9 for the European populations and 5 for the SR study. This approach is the same as the ‘Simple proportion method' described in Colman et al (1977)25 and similar to the formulas used in Preston and Colman (2000)26 and Dawes (2002).27 The only difference in a regression would be noticed on the intercept; however, as, to perform meta-analysis, we are interested only in betas and association is performed within each group, this difference is negligible.
Genotyping and Imputation
Genotyping was carried out as previously described.28, 29, 30 In brief, INGI-CARL, INGI-FVG and INGI-VB have been genotyped with Illumina 370 k high-density SNP array, whereas SR has been genotyped with Illumina 700 k high-density SNP arrays. Genotype imputation on the INGI cohorts and SR was conducted after standard QC using SHAPEIT231 for the phasing step and IMPUTE232 for the imputation using the 1000 Genomes phase I v3 reference set.33 ERF has been genotyped with different genotyping platform: Illumina 318 k, 350 k, 610 k and Affymetrics 200 k. Genotypes were pooled together after QC, phased and imputed to the 1000Genomes data set phase I v333 using MaCH and minimac.34 After imputation, we excluded from the statistical analyses SNPs with MAF<0.01 or Info<0.4 for all populations but ERF for which R2<0.3 was used instead.
Association analysis
Association analysis was conducted using mixed model linear regression, whereas the standardized wine liking was used as the dependent variable and the SNP dosages as the independent variable. Sex and age were used as covariates. The kinship matrix based on all available genotyped SNPs was used as the random effect. For ERF the kinship matrix was estimated on 14.4 k SNPs common to all different genotyping platform used.35 The GenABEL R package was used to eliminate the effect of relatedness from the trait. Corrected environmental residuals were estimated according to the formula: trait~sex+age+genomic kinship using the GRAMMAR+ method36 as implemented in the GenABEL 1.7–2. MixABEL35 was used for running the linear regressions between the estimated residuals and all the imputed SNPs. Only SNPs that passed post imputation quality control were used for the association analysis. SNPs that did not pass quality control for more than one population were discarded as well. For the association analysis we used a two-step approach.37 For the genome-wide discovery step, association analysis was conducted separately for each INGI cohort and results have been pooled together using the inverse-variance weighting method. After association analysis all SNPs, which showed P<1 × 10−5, were selected to be used for the replication step using ERF and SR. We considered SNPs with P<5 × 10−8 as significant at the replication step. All meta-analysis have been conducted using in house R scripts. Power analysis was conducted using the ‘Genetic Power Calculator' software.38
Data availability
Genotypes for the cohorts used in this study have been deposited in the EGA database https://www.ebi.ac.uk/ega/home. In particular, under the following accession numbers: INGI-CARL EGAS00001001005, INGI-FVG EGAS00001001006, INGI-VB EGAS00001001007, ERF EGAS00001001134 and SR EGAS00001001008.
Results
Table 1 describes the each cohort's characteristics and trait distribution. The mean liking rating was very similar across the Italian populations, whereas lower liking was observed in SR and ERF. In particular, ERF shows the lowest liking as compared with the other populations. Although means were different, SDs were consistent across all populations showing that the standardization procedure was effective in making the measures comparable.
Table 1. Cohort demographics and trait distribution.
| Population | Number of samples red/white wine | Mean age (SD) | Percentage of women | Red wine liking (SD) | White wine liking (SD) |
|---|---|---|---|---|---|
| INGI-CARL | 398/381 | 52.56 (17.26) | 58% | 0.76 (0.27) | 0.69 (0.29) |
| INGI-FVG | 751/744 | 50.85 (15.80) | 59% | 0.68 (0.25) | 0.63 (0.26) |
| INGI-VB | 1122/1115 | 53.23 (16.62) | 63% | 0.73 (0.25) | 0.63 (0.26) |
| ERF | 1246/1261 | 47.4 (13.23) | 55% | 0.53 (0.26) | 0.58 (0.26) |
| SR | 353/335 | 39.12 (15.86) | 59% | 0.64 (0.30) | 0.59 (0.30) |
For all cohorts, numerosity of the sample for the red and white wine, age and sex distribution and mean liking with SD for red and white wine is reported.
For the discovery step we performed genome-wide association analysis on red wine liking and white wine liking on the three Italian populations (2271 samples) and then pulled the results together using the inverse-variance weighting method. We then chose all SNPs with P<1 × 10−5 to use for the second step joint analysis.
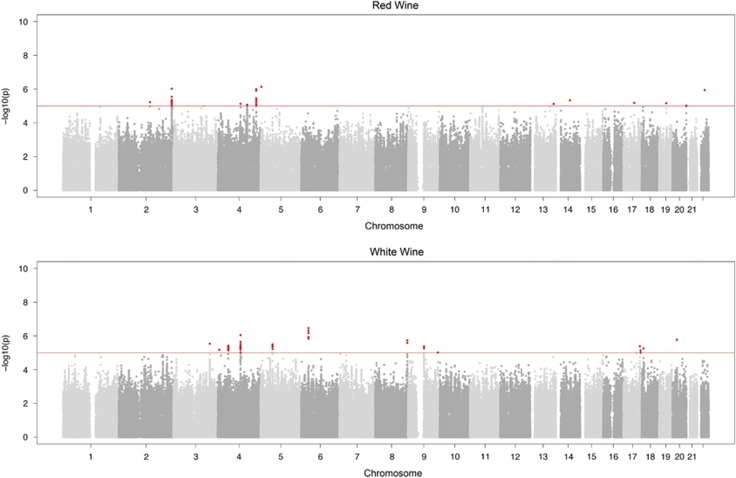
Figure 1 represents the Manhattan plot for the discovery step that formed the basis SNPs selected for replication. The selected SNPs were 42 for white wine and 72 for red wine. The selected variants were tested for association to wine preference in two additional cohorts: ERF using 1261 samples and SR using 335 samples. Joint analysis revealed a genome-wide significant association between white wine and rs9276975 (NC_000006.12:g.33005822C>T) (P=2.1 × 10−8) a SNP in the 3′-UTR region of the HLA-DOA gene. Given that the inverse-variance method for meta-analysis is sensible to differences in trait distributions and that we had to transform the SR scale to adapt it to the other populations, we performed also z-score based meta-analysis. The results were almost identical to the ones obtained with the inverse-variance method (regression coefficient 0.98), with the P-value for rs9276975:C>T equal to 1.5 × 10−8. This result shows that the transformation applied to the traits did not influence the final result. To verify that we did not have residual stratification in the replication step, we performed genome-wide meta-analysis on both analyzed traits. Lambda was 1.01 for red wine and 1.002 for white wine, QQ-plots are reported in the additional materials (Supplementary Figures S2 and S3). Finally, power analysis using the Genetic Power Calculator revealed that we have 0.54 power to detect association at α=5 × 10−8 under the same conditions of the described association (MAF=0.15 β=0.053). We also estimated that we have 0.8 power to detect association at α=5 × 10−8 at 0.01 explained variance regardless of the MAF.
Figure 1.
Manhattan Plot for the meta-analysis on the Italian Populations. The red line is set at P=1 × 10−5, whereas the red points represent the SNPs selected for replication. The top figure refers to red wine, whereas the lower one to white wine.
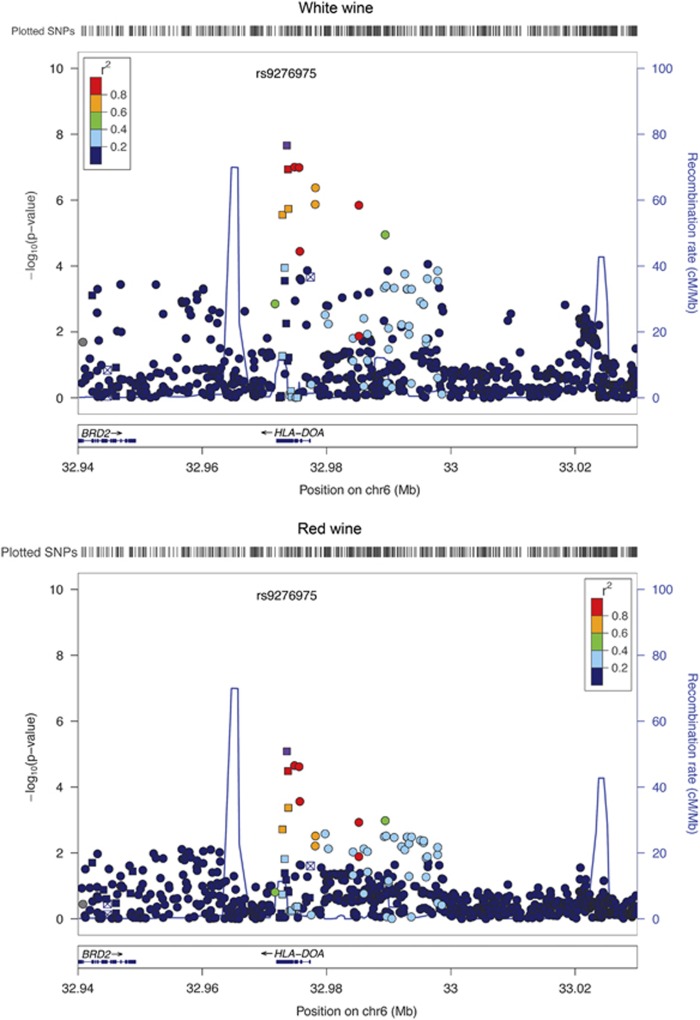
Figure 2 shows the regional association plot around rs9276975:C>T for white and red wine, whereas Supplementary Figure S1 shows a similar plot for the whole HLA locus. The association signal is clearly confined to the HLA-DOA locus not affecting the wider HLA region, which is in strong linkage disequilibrium. This is probably due to the fact that HLA-DOA is within two recombination hotspots that isolate it from the rest of the HLA locus.
Figure 2.
Regional association plot of the locus containing the HLA-DOA gene. The top figure refers to white wine, whereas the bottom one to red wine. Clearly, the two figures look alike, although in the case of red wine the association signal is much weaker. Different colors refer to the r2 with rs9276975 in the CEU population. Squares represent coding SNPs, whereas circles represent non-coding SNPs.
Table 2 reports the results for the significant SNP, whereas Supplementary Table S2 contains the results for all the SNPs selected in step one.
Table 2. Results for the association analyses for rs9276975 for white wine and red wine.
| rs9276975 effect allele T | Population | β | P | Effect allele frequency | Info/R2 |
|---|---|---|---|---|---|
| White wine | INGI-CARL | 0.0764 | 1.05 × 10−02 | 0.15 | 0.98 |
| INGI-FVG | 0.0503 | 9.29 × 10−03 | 0.15 | 0.97 | |
| INGI-VB | 0.0751 | 2.23 × 10−04 | 0.09 | 0.97 | |
| Discovery | 0.0647 | 3.47 × 10−07 | |||
| ERF | 0.0425 | 1.18 × 10−02 | 0.15 | 0.79 | |
| SR | 0.0286 | 3.55 × 10−01 | 0.17 | Typed | |
| Discovery+replication | 0.0539 | 2.18 × 10−08 | |||
| Red wine | INGI-CARL | 0.0451 | 9.91 × 10−02 | 0.15 | 0.98 |
| INGI-FVG | 0.0389 | 3.70 × 10−02 | 0.15 | 0.97 | |
| INGI-VB | 0.0423 | 2.64 × 10−02 | 0.09 | 0.97 | |
| Discovery | 0.0415 | 5.43 × 10−04 | |||
| ERF | 0.0471 | 7.54 × 10−03 | 0.15 | 0.79 | |
| SR | 0.0302 | 3.13 × 10−01 | 0.17 | Typed | |
| Discovery+replication | 0.0419 | 8.26 × 10−06 |
Beta represents the effect of the effect allele, P represents the P-value and Info/R2 is the quality of the imputation of the SNP measured as Info for INGI populations and SR and R2 for ERF.
To verify whether the associated SNP had an effect on food likings in general or only on wine, we ran association analysis between the two SNPs and 40 more food likings, which were available on the studied cohorts and collected similar to the collection of data for wine liking. This analysis did not show significant or close to significant results (Supplementary Table S4), suggesting that the described associations are specific to wine liking.
No significant association was found for any of the 72 SNPs used for the replication step of red wine as shown in Supplementary Table S3. Supplementary materials Supplementary Table S1 report the descriptive statistics for all GWAS and meta-analysis, whereas Supplementary Figures S2 and S3 show QQ-plots for the same analyses.
Given the possible genetic differences between man and women, the analyses have been conducted also on man and women separately limited to the rs9276975:C>T SNP. This analysis revealed that the association between white wine and HLA-DOA was much stronger in women than man (P=1 × 10−7 in women vs P=0.01 in man). Looking at the effect size, women exhibited twice the effect as compared with man (0.067 vs 0.033). Table 3 shows the results for the sex-specific analysis of white wine and red wine, which showed again a similar effect.
Table 3. Summary of results for the sex-separated analysis.
| rs9276975 effect allele T | Numerosity | Direction | β | P |
|---|---|---|---|---|
| White Wine | ||||
| Man | 1681 | ++++− | 0.03 | 1.48 × 10−2 |
| Women | 2193 | +++++ | 0.07 | 4.50 × 10−7 |
| Red Wine | ||||
| Man | 1656 | ++++− | 0.03 | 3.54 × 10−2 |
| Women | 2183 | +++++ | 0.05 | 6.08 × 10−5 |
Direction represents the direction of the effect in INGI-CARL, INGI-FVG, INGI-VB, ERF and SR populations.
Annotating the rs9276975:C>T with HaploReg v2,39 SNPNexus and Regulome DB did not give any insight on what might be its role in the observed association. Also, extending the annotation to the all the SNPs in strong LD (r2>0.8) with rs9276975:C>T in the CEU 1000 G population. Only rs1367731 (NC_000006.12:g.33017422C>T) and rs9276981 (NC_000006.12:g.33007922G>C) are suggested to have a possible functional role by Regulome DB with a score of 5 and 6, respectively, which corresponds to the lowest possible scores and to very weak evidence.
Discussion
In this study we present the first GWAS on wine liking showing a significant association with HLA-DOA, a non-canonical class II MHC molecule, in particular with white wine. Although it is true that in the MHC locus there are at least two clusters of OR, HLA-DOA is situated in between two recombination hotspots (Figure 2), which makes it independent from the main MHC locus containing the OR genes. The most associated SNP, rs9276975:C>T, is located in the 3′-UTR region of the gene, which would suggest more a regulatory role than a functional one. This fits particularly well if we consider HLA-DOA function in the general MHC class II system. In fact its role is to inhibit HLA-DM another non-canonical MHC class II molecule who has the double role of activating all other MHC class II proteins by de-coupling from the CLIP protein and then, by binding to them, is responsible for the specificity of their binding to the correct antigens.40 In other words HLA-DO by inhibiting HLA-DM prevents the activation of the MHC class II molecules in general. Unfortunately the bioinformatics analysis was not able to give us any insights on possible regulatory elements modified by SNPs in LD with rs9276975; however, we must consider that given the particular nature of our traits not much information is available on the interested tissues such as the olfactory epithelium for example.
MHC could be linked to wine liking directly through the olfactory epithelium cells. In fact numerous studies have linked MHC molecules to mate choice through scent in various species including humans.41, 42, 43 In mice, non-canonical MHC class I molecules are specifically expressed in a particular section of the vomeronasal organ (VNO),44 which are thought to modulate the response to MHC antigens.45 Given that humans do not have a VNO, it is unclear how they could be able to distinguish different HLA types. However, studies on VNO sham mice show that they can recognize MHC class II antigens as well as non-sham mice suggesting that, at least for this type of molecules, the VNO is not necessary.46 Another recent study conducted on monogamous birds with highly developed olfaction shows that it is MHC class II molecules and not class I, which determine mate recognition.47 In humans it has been demonstrated that different HLA types show preference or dislike for specific scents, suggesting that some volatile molecules responsible for these odors are mimicking some HLA-specific scent.48, 49 MHC class II molecules are responsible for immune response to external stimuli and it is thus a more suitable candidate than class I MHC for a possible olfactory function. One of the main theories behind MHC recognition is that people with different HLA profile have different composition in bacterial flora, which affects body odor and thus generates the HLA-specific scent.50 Extending this view to our findings, the association we detect seems to be specific to wine in which bacteria are an important part of the production process and could be responsible to the difference in liking. It is thus possible that HLA-DOA, by regulating MHC class II molecules, modulate also the perception of specific volatile compounds present in wine, which are produced by the bacteria used in wine production. It is also very suggestive that this effect is much stronger in white wine than in red wine in which the mouth-feel component of its flavor is more accentuated owing to the presence of more tannins.51 The fact that the effect of rs9276975:C>T on wine liking is twice as large in women than in men seems to support the olfactory hypothesis. Women have in fact been described as being more sensitive than men to specific odors52, 53 also in the specific case of MHC driven body odor recognition.54, 55
The precise mechanism of how MHC class II molecules actually interact with the olfactory epithelium and how the signal is then transferred to the brain is still unknown and further studies are needed to clarify this point.
We have previously reported rs4920566 (NC_000001.11:g.18853330 A>G) in the TAS1R2 gene as being associated to white wine liking. Checking in the results from the combined meta-analysis, this SNP resulted to be non significant (P-value=0.83). This lack of replication could be due to allelic heterogeneity between the SR populations and the European ones, and further studies are needed to clarify this point.
In conclusion, this study is one of the first that examines the genetics of liking for a commonly consumed food on a genome-wide scale. Our findings suggest that at least in the case of wine MHC has an important role in defining its liking probably through olfaction. This opens a new prospective not only in food liking and thus choice but also in understanding the mechanisms that link flavor to olfaction in general.
Ethical statement
All studies adhered to the tenets of the Declaration of Helsinki. The ERF study was approved by the Medical Ethics Committee of the Erasmus Medical Center in Rotterdam. Informed consent was obtained after explanation of the nature and possible consequences of the study.
All subjects in the INGI-CARL, INGI-FVG and SR studies provided written informed consent before participation. Approval for the research protocol was obtained from the ethical committee of IRCCS Burlo Garofolo Hospital.
The VB study, including the overall plan and the informed consent form was reviewed and approved by the institutional review boards of San Raffaele Hospital in Milan and by the ethical committee of the Regional Authorities of Piemonte.
Acknowledgments
We would like to thank all the participants in the study for their contribution and support. The SR study has been founded by the Region Friuli Venezia Giulia grant number 35\09 Linea 2 ‘Sulle tracce di Marco Polo: geni, gusto e loro implicazioni sulla salute lungo la Via della Seta'. The INGI-FVG study was founded through the Italian Ministry of health. We thank the inhabitants and the administrators of the Val Borbera for their participation in the study. A special thanks to Professor Clara Camaschella, Dr Silvia Bione, Dr Laura Crocco, Ms Maria Rosa Biglieri, Dr Diego Sabbi for help with the data collection. We thank Fondazione Compagnia di San Paolo, Torino, Fondazione Cariplo, Milano and Health Ministry (Progetto Finalizzato and Italian Centre for Disease Prevention and Control) for financial support. The ERF study as a part of EUROSPAN (European Special Populations Research Network) was supported by European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947) and also received funding from the European Community's Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F4-2007-201413 by the European Commission under the programme ‘Quality of Life and Management of the Living Resources' of 5th Framework Programme (no. QLG2-CT-2002-01254). High-throughput analysis of the ERF data was supported by joint grant from the Netherlands Organisation for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). We are grateful to all study participants and their relatives, general practitioners and neurologists for their contributions and to P Veraart for her help in genealogy, J Vergeer for the supervision of the laboratory work and P Snijders for his help in data collection. Statistical analyses were partly carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003 PI: Posthuma) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam. This research was financially supported by BBMRI-NL, a Research Infrastructure financed by the Dutch Government (NWO 184.021.007).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Sieri S, Agudo A, Kesse E et al: Patterns of alcohol consumption in 10 European countries participating in the European Prospective Investigation into Cancer and Nutrition (EPIC) project. Public Health Nutr 2002; 5: 1287–1296. [DOI] [PubMed] [Google Scholar]
- Schröder H, Ferrández O, Jimenez Conde J, Sánchez-Font A, Marrugat J: Cardiovascular risk profile and type of alcohol beverage consumption: a population-based study. Ann Nutr Metab 49: 100–106. [DOI] [PubMed] [Google Scholar]
- Rimm EB, Giovannucci EL, Willett WC et al: Prospective study of alcohol consumption and risk of coronary disease in men. Lancet 1991; 338: 464–468. [DOI] [PubMed] [Google Scholar]
- Marmot MG: Alcohol and coronary heart disease. Int J Epidemiol 2001; 30: 724–729. [DOI] [PubMed] [Google Scholar]
- Di Castelnuovo A, Rotondo S, Iacoviello L, Donati MB, De Gaetano G: Meta-analysis of wine and beer consumption in relation to vascular risk. Circulation 2002; 105: 2836–2844. [DOI] [PubMed] [Google Scholar]
- Tolstrup J, Jensen MK, Tjønneland A, Overvad K, Mukamal KJ, Grønbaek M: Prospective study of alcohol drinking patterns and coronary heart disease in women and men. BMJ 2006; 332: 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polásková P, Herszage J, Ebeler SE: Wine flavor: chemistry in a glass. Chem Soc Rev 2008; 37: 2478–2489. [DOI] [PubMed] [Google Scholar]
- Rapp A: Volatile flavour of wine: correlation between instrumental analysis and sensory perception. Nahrung 1998; 42: 351–363. [DOI] [PubMed] [Google Scholar]
- Moreno-Arribas MV, Polo MC: Winemaking biochemistry and microbiology: current knowledge and future trends. Crit Rev Food Sci Nutr 2005; 45: 265–286. [DOI] [PubMed] [Google Scholar]
- Ardö Y: Flavour formation by amino acid catabolism. Biotechnol Adv 24: 238–242. [DOI] [PubMed] [Google Scholar]
- Snowdon EM, Bowyer MC, Grbin PR, Bowyer PK: Mousy off-flavor: a review. J Agric Food Chem 2006; 54: 6465–6474. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Wang J-C, Wetherill L et al: A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum Genet 2013; 132: 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK et al: A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA 2010; 107: 5082–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X et al: Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res 2010; 34: 840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Vink JM et al: A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Res Hum Genet 2010; 13: 10–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Cichon S, Treutlein J et al: Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict Biol 2012; 17: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Wallace MR, Knopik VS, Herbstman DM, Bartoshuk LM, Duffy VB: Allelic variation in TAS2R bitter receptor genes associates with variation in sensations from and ingestive behaviors toward common bitter beverages in adults. Chem Senses 2011; 36: 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy VB, Davidson AC, Kidd JR et al: Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcohol Clin Exp Res 2004; 28: 1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Bertelsen S et al: Functional variants in TAS2R38 and TAS2R16 influence alcohol consumption in high-risk families of African-American origin. Alcohol Clin Exp Res 2007; 31: 209–215. [DOI] [PubMed] [Google Scholar]
- Knaapila A, Hwang L-D, Lysenko A et al: Genetic analysis of chemosensory traits in human twins. Chem Senses 2012; 37: 869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae JF, Mainland JD, Jaeger SR, Adipietro KA, Matsunami H, Newcomb RD: Genetic variation in the odorant receptor OR2J3 is associated with the ability to detect the ‘grassy' smelling odor, cis-3-hexen-1-ol. Chem Senses 2012; 37: 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirastu N, Robino A, Lanzara C et al: Genetics of Food Preferences: A First View from Silk Road Populations. J Food Sci 2012; 77: S413–S418. [DOI] [PubMed] [Google Scholar]
- JONES LV, PERYAM DR, THURSTONE LL: Development of a scale for measuring soldiers'food preferences b. J Food Sci 1955; 20: 512–520. [Google Scholar]
- Coetzee H, Taylor JRN: The use and adaptation of the paired-comparison method in the sensory evaluation of hamburger-type patties by illiterate/semi-literate consumers. Food Qual Prefer 1996; 7: 81–85. [Google Scholar]
- Colman AM, Norris CE: PCC. Comparing Rating Scales of Different Lengths: Equivalence of Scores From 5-Point and 7-Point Scales. Psychol Rep 1997; 80: 355–362. [Google Scholar]
- Preston CC, Colman AM: Optimal number of response categories in rating scales: reliability, validity, discriminating power, and respondent preferences. Acta Psychol (Amst) 2000; 104: 1–15. [DOI] [PubMed] [Google Scholar]
- J.G. D: Five point vs eleven point scales: does it make a difference to data characteristics? Australas J Mark Res 2002; 10: 39–47. [Google Scholar]
- Girotto G, Pirastu N, Sorice R et al: Hearing function and thresholds: a genome-wide association study in European isolated populations identifies new loci and pathways. J Med Genet 2011; 48: 369–374. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Heutink P, Mackay I et al: Linkage disequilibrium in young genetically isolated Dutch population. Eur J Hum Genet 2004; 12: 527–534. [DOI] [PubMed] [Google Scholar]
- Pardo LM, MacKay I, Oostra B, van Duijn CM, Aulchenko YS: The effect of genetic drift in a young genetically isolated population. Ann Hum Genet 2005; 69: 288–295. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Zagury J-F, Marchini J: Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 2013; 10: 5–6. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J: A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Auton A, Brooks LD et al: An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR: Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44: 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM: GenABEL: an R library for genome-wide association analysis. Bioinformatics 2007; 23: 1294–1296. [DOI] [PubMed] [Google Scholar]
- Belonogova NM, Svishcheva GR, van Duijn CM, Aulchenko YS, Axenovich TI: Region-based association analysis of human quantitative traits in related individuals. PLoS One 2013; 8: e65395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ: Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 2005; 6: 95–108. [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC: Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003; 19: 149–150. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M: HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40: D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze M-SED, Wucherpfennig KW: The mechanism of HLA-DM induced peptide exchange in the MHC class II antigen presentation pathway. Curr Opin Immunol 2012; 24: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind C, Seebeck T, Bettens F, Paepke AJ: MHC-dependent mate preferences in humans. Proc Biol Sci 1995; 260: 245–249. [DOI] [PubMed] [Google Scholar]
- Ober C, Weitkamp LR, Cox N, Dytch H, Kostyu D, Elias S: HLA and mate choice in humans. Am J Hum Genet 1997; 61: 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capittini C, Martinetti M, Cuccia M: MHC variation, mate choice and natural selection: the scent of evolution. Riv Biol 101: 463–480. [PubMed] [Google Scholar]
- Ishii T, Mombaerts P: Expression of nonclassical class I major histocompatibility genes defines a tripartite organization of the mouse vomeronasal system. J Neurosci 2008; 28: 2332–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinders-Zufall T, Ishii T, Mombaerts P, Zufall F, Boehm T: Structural requirements for the activation of vomeronasal sensory neurons by MHC peptides. Nat Neurosci 2009; 12: 1551–1558. [DOI] [PubMed] [Google Scholar]
- Wysocki CJ, Yamazaki K, Curran M, Wysocki LM, Beauchamp GK: Mice (Mus musculus) lacking a vomeronasal organ can discriminate MHC-determined odortypes. Horm Behav 2004; 46: 241–246. [DOI] [PubMed] [Google Scholar]
- Strandh M, Westerdahl H, Pontarp M et al: Major histocompatibility complex class II compatibility, but not class I, predicts mate choice in a bird with highly developed olfaction. Proc Biol Sci 2012; 279: 4457–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milinski M: Evidence for MHC-correlated perfume preferences in humans. Behav Ecol 2001; 12: 140–149. [Google Scholar]
- Hämmerli A, Schweisgut C, Kaegi M: Population genetic segmentation of MHC-correlated perfume preferences. Int J Cosmet Sci 2012; 34: 161–168. [DOI] [PubMed] [Google Scholar]
- Singh PB, Herbert J, Roser B, Arnott L, Tucker DK, Brown RE: Rearing rats in a germ-free environment eliminates their odors of individuality. J Chem Ecol 1990; 16: 1667–1682. [DOI] [PubMed] [Google Scholar]
- Dinnella C, Recchia A, Tuorila H, Monteleone E: Individual astringency responsiveness affects the acceptance of phenol-rich foods. Appetite 2011; 56: 633–642. [DOI] [PubMed] [Google Scholar]
- Koelega HS, Köster EP: Some experiments on sex differences in odor perception. Ann N Y Acad Sci 1974; 237: 234–246. [DOI] [PubMed] [Google Scholar]
- Zucco GM, Aiello L, Turuani L, Köster E: Odor-evoked autobiographical memories: age and gender differences along the life span. Chem Senses 2012; 37: 179–189. [DOI] [PubMed] [Google Scholar]
- Santos PSC, Schinemann JA, Gabardo J, Bicalho M, da G: New evidence that the MHC influences odor perception in humans: a study with 58 Southern Brazilian students. Horm Behav 2005; 47: 384–388. [DOI] [PubMed] [Google Scholar]
- Wedekind C, Füri S: Body odour preferences in men and women: do they aim for specific MHC combinations or simply heterozygosity? Proc Biol Sci 1997; 264: 1471–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotypes for the cohorts used in this study have been deposited in the EGA database https://www.ebi.ac.uk/ega/home. In particular, under the following accession numbers: INGI-CARL EGAS00001001005, INGI-FVG EGAS00001001006, INGI-VB EGAS00001001007, ERF EGAS00001001134 and SR EGAS00001001008.