Abstract
The assignment of causative genes to noncoding variants identified in genome-wide association studies (GWASs) is challenging. We show how combination of knowledge from gene and pathway databases and chromatin interaction data leads to reinterpretation of published quantitative trait loci for blood metabolites. We describe a previously unidentified link between the rs2403254 locus, which is associated with the ratio of 3-methyl-2-oxobutanoate and alpha-hydroxyisovalerate levels, and the distal LDHA gene. We confirmed that lactate dehydrogenase can catalyze the conversion between these metabolites in vitro, suggesting that it has a role in branched-chain amino acid metabolism. Examining datasets from the ENCODE project we found evidence that the locus and LDHA promoter physically interact, showing that LDHA expression is likely under control of distal regulatory elements. Importantly, this discovery demonstrates that bioinformatic workflows for data integration can have a vital role in the interpretation of GWAS results.
Introduction
Owing to recent advances in metabolomics technology and decreasing costs, genome-wide association studies (GWASs) have now been performed on a wide range of metabolites.1, 2, 3, 4 These studies have provided new insights into how biochemical pathways are affected by genetic polymorphisms and have increased our understanding of the pathogenesis of metabolic disease.3, 5 As variation in metabolite levels is often linked to changes in enzyme or transporter activity, functional annotation of the loci that are located in or close to enzyme-coding genes has been straightforward. However, for a substantial part of the loci identified in mGWAS, no obvious link between the metabolite and proximal enzyme-coding genes exists, and the association with the phenotype is much harder to explain.
We developed an automated workflow for mapping the results of GWASs on pathway databases to assist in their interpretation. We applied the workflow on the 37 loci that have been reported by Suhre et al.3 and were able to provide a new functional annotation of the rs2403254 (chr11.hg19:g.18325146C>T) single nucleotide polymorphism (SNP) which associates with the ratio of 3-methyl-2-oxobutanoate and alpha-hydroxyisovalerate levels in the blood. Reanalysis of this locus uncovered a functional link to the gene coding for the lactate dehydrogenase (LDH) A enzyme and in vitro analysis confirmed that LDH could convert 3-methyl-2-oxobutanoate into alpha-hydroxyisovalerate. In addition, we found a physical association between the rs2403254 locus and the LDHA promoter region in the chromatin interaction data from the ENCODE project.6, 7, 8 Combined, our data suggest that LDH interacts with branched-chain amino acid metabolism and is affected by genetic variation at a distal locus.
Materials and methods
A description of the protocol for measuring LDH activity and of the workflow for automated annotation of GWAS results and characterization of the rs2403254 locus is given in the Supplementary Material and Methods.
Results
Automated annotation of GWAS results
The automated workflow we developed was used to generate reports for each of the 37 SNPs published by Suhre et al.,3 containing the associated protein, enzyme, metabolic reaction, pathway, and disease phenotypes of each gene within a distance of 500 kb of the locus. Inspection of the results showed that for one of the 37 SNPs, rs2403254, there was an alternative candidate gene that provided a more likely explanation of the association. The rs2403254 SNP was associated with alpha-hydroxyisovalerate levels in the blood (P=1.0 × 10−20) and showed an even stronger correlation with the ratio of 3-methyl-2-oxobutanoate and alpha-hydroxyisovalerate levels (P=7.9 × 10−28). The rs2403254 SNP lies in an intronic region of the HPS5 gene and had therefore been assigned to HPS5 by Suhre and colleagues. However, from inspection of the results from our workflow, it followed that a plausible alternative candidate gene in the vicinity of the locus was LDHA, which codes for the A-isoform of the LDH enzyme. In addition, from information retrieved from the KEGG database,9 it followed that LDH has a broad substrate specificity and can catalyze the conversion of several keto and hydroxy acids, even though 3-methyl-2-oxobutanoate or alpha-hydroxyisovalerate were not listed as substrates (Supplementary Text 1).
The rs2403254 locus
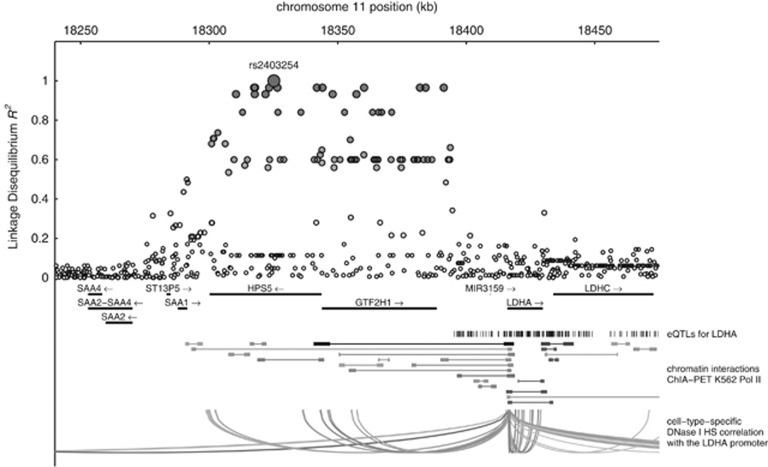
Closer examination of the locus showed that rs2403254 is located in a large linkage disequilibrium (LD) block, which lies approximately 20 kb upstream of LDHA (Figure 1). To investigate the presence of potential long-distance regulatory mechanisms, we first looked at expression quantitative trait loci (eQTLs) in lymphoblastoid cell lines that were associated with LDHA. These eQTLs were all located downstream of the LD block and were not in strong LD with rs2403254. In contrast, we found that rs2403254 is an eQTL for HPS5 (P=4.4 × 10−9) and GTF2H1 (P=8.4 × 10−15), most likely because it is in strong LD (R2>0.8) with several SNPs that lie close to the transcription start sites of these genes.
Figure 1.
Regional LD plot of the rs2403254 locus. The rs2403254 locus is located in a region of strong LD spanning over approximately 100 kb. Several eQTLs have been identified that associate with LDHA expression, but none lie in LD with rs2403254. In contrast, ChIA-PET data and cell type specific DHS correlations show that there are several chromatin interactions between the LDHA promoter and regulatory elements within the LD block.
Subsequently, we explored chromatin interactions and the presence of regulatory elements in the LD block by looking at the DNase I signal data from the ENCODE project.6, 7, 8 Interestingly, several regulatory regions were present within the LD block whose DNase I signal had a strong cross-cell-type correlation with the LDHA promoter (Figure 1; bottom). These potential distal interactions were supported by the Chromatin Interaction Analysis with Paired-End-Tag (ChIA-PET) sequencing data, which showed that there were significant chromatin interactions between the central region of the LD block and the region containing the LDHA promoter. Collectively, these data show that long-range regulation of LDHA by enhancers that are located more than 20 kbp upstream of its transcription start site is indeed plausible.
Experimental validation
Subsequently, we investigated whether LDH could catalyze the conversion between branched-chain alpha-keto and hydroxy acids. In previous studies LDH had already been shown to catalyze a broad range of substrates,10, 11 but because of the extremely low rates with which branched-chain alpha-keto and hydroxy acids were converted it was unclear whether this reaction was mediated by LDH or another dehydrogenase.12, 13 We therefore validated our finding by assaying the activity of LDH in the presence of NADH and the transaminated branched-chain keto acid products 3-methyl-2-oxobutanoate (valine), 4-methyl-2-oxopentanoate (leucine), 3-methyl-2-oxopentanoate (isoleucine) and pyruvate for a range of different substrate concentrations (Supplementary Figure 1). Results show that LDH was indeed able to convert 3-methyl-2-oxobutanoate, but it had a specificity constant kcat/Km that was approximately 3000 times lower than with pyruvate as substrate (Supplementary Table 1). In comparison, the kcat/Km of the other two branched-chain alpha-keto acids was around 10 times lower than with 3-methyl-2-oxobutanoate as substrate. Most probably 3-methyl-2-oxobutanoate is converted more efficiently by LDH because it is a smaller molecule, having a carbon chain of 4 atoms, whereas 3- and 4-methyl-2-oxopentanoate have a carbon chain of 5 atoms. Interestingly, the Km value for pyruvate and 3-methyl-2-oxopentanoate were very similar, while for the other two branched-chain alpha-keto acids it was three to four times larger.
Discussion
The discovery of the functional link between the rs2403254 locus and the LDHA gene has been made possible by the automated workflow we developed for annotating GWAS results, which allowed us to examine the set of loci reported by Suhre et al.3 in both a quick and thorough manner. In recent years, there has been an increasing interest in bioinformatics tools for the analysis, interpretation and integration of results from GWASs.14, 15 Suhre and colleagues have used the tool GRAIL from the Broad Institute16 in their study, which uses textual relationships between genes to prioritize candidate genes for a given locus. Nonetheless, using this method, the authors did not identify LDHA as a plausible candidate for the rs2403254 locus, most likely because alpha-hydroxyisovalerate is currently not present in pathway databases and its link to LDH has only been scarcely described in the literature. In fact, the locus was replicated in a recent meta-analysis on the same metabolomics platform,17 but HPS5 was still proposed as candidate gene.
In contrast, with our approach, we focused on integrating the knowledge present in several databases in order to produce succinct SNP reports containing relevant information about all neighboring genes. In the case of the rs2403254 locus, the SNP report showed that LDHA was the closest gene with a metabolic function and that LDH was documented in the KEGG pathway database as an enzyme that can catalyze multiple reactions. Subsequent investigation of the chemical structure of alpha-hydroxyisovalerate suggested that—in principle—its conversion could be catalyzed by LDH. This hypothesis was further reinforced by the observation that the same locus had a stronger association with the ratio of alpha-hydroxyisovalerate and 3-methyl-2-oxobutanoate levels and that alpha-hydroxyisovalerate is the product of 3-methyl-2-oxobutanoate after reduction of the alpha-carbonyl group.
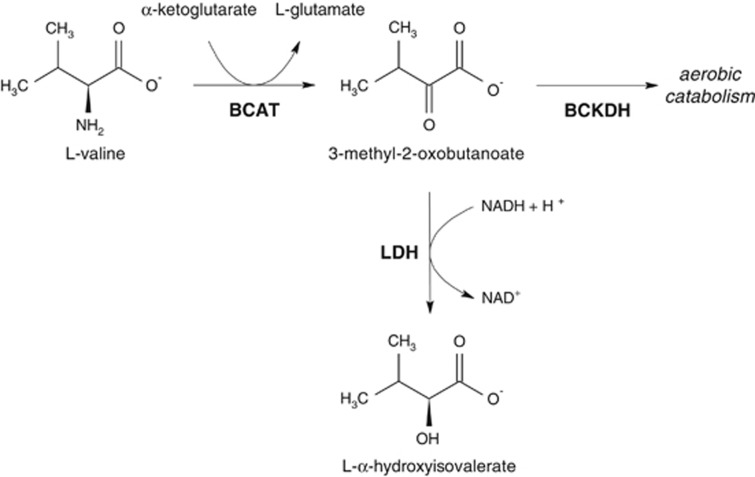
Our results suggest that there is a functional link between LDHA and alpha-hydroxyisovalerate levels and, more specifically, that LDH can compensate for large build-ups of branched-chain alpha-keto acids under hypoxic conditions. In fact, the first step of branched-chain amino acid catabolism involves the transamination of the amino group, which changes the oxidation level of the adjacent carbon atom. Under anaerobic conditions, the redox balance needs to be restored, which can be achieved through LDH by converting the alpha-keto acid into an alpha-hydroxycarboxylic acid (Figure 2). This process has been observed in babies who suffered from asphyxia during birth, where elevated levels of alpha-hydroxyisovalerate were found in the urine.18 Interestingly, also infants that suffer from Maple syrup urine disease, which is a defect in any of the genes coding for the components of the BCKDH enzyme complex, have elevated levels of alpha-hydroxyisovalerate in the urine.19 This can be explained by the fact that a blockage in the second step of the branched-chain amino acid degradation pathway causes a build-up of keto acid intermediates, which are then partly converted to hydroxycarboxylic acids via LDH (Figure 2).
Figure 2.
Visualization of the valine degradation pathway and the interaction with LDH. Abbreviations of enzyme names are shown in boldface BCAT: branched-chain aminotransferase; BCKDH: branched-chain alpha-keto acid dehydrogenase; LDH: lactate dehydrogenase. LDH can convert 3-methyl-2-oxobutanoate, which is the product of valine after transamination, into alpha-hydroxyisovalerate to balance the redox potential under hypoxic conditions. Under aerobic conditions, 3-methyl-2-oxobutanoate can be further degraded by the enzymes of branched-chain amino acid catabolism. In contrast, alpha-hydroxyisovalerate cannot be metabolized any further and is excreted in urine.
Interestingly, in the meta-analysis of Shin et al.,17 rs2403254 was found to have the strongest association with the ratio of 3-(4-hydroxyphenyl)lactate and alpha-hydroxyisovalerate levels. 3-(4-Hydroxyphenyl)lactate is the product of 4-hydroxyphenylpyruvate after reduction of the alpha-carbonyl group, which is the product of tyrosine after transamination. Given their structural differences, it is unlikely that 3-(4-hydroxyphenyl)lactate and alpha-hydroxyisovalerate can be converted into one another in one or two enzymatic steps. The association with rs2403254 is therefore probably due to other mechanisms, such as co-regulation of aromatic and branched-chain amino acid metabolism.
Importantly, our finding does not preclude the effects of rs2403254 on HPS5 or other genes. In fact, in our analysis, we found that rs2403254 is an expression QTL for HPS5 and GTF2H1, which shows that multiple genes can be affected by a single SNP. However, neither of these genes provide a biochemical explanation of the phenotype, namely, why rs2403254 is associated with the ratio of 3-methyl-2-oxobutanoate and alpha-hydroxyisovalerate levels. The absence of eQTLs for LDHA in the databases that we have enquired can be explained by the fact that distal eQTLs are more difficult to identify (because of, eg, tissue specificity20) and demonstrates the additional value of chromatin interaction data to establish SNP–gene interactions.
In conclusion, we have uncovered a novel functional link between LDH and the branched-chain keto acid intermediate of valine metabolism by reanalyzing published mGWAS results using automated workflows for integrating information from pathway databases.
Acknowledgments
We would like to thank Peter-Bram 't Hoen for critically reviewing the manuscript and Thomas Hankemeier for providing stock solutions of the branched-chain alpha-keto acids. This study has been funded by the European Network for Genetic and Genomic Epidemiology (ENGAGE) and by the Centre for Medical Systems Biology (CMSB) and Netherlands Consortium for Systems Biology (NCSB), both within the framework of the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Gieger C, Geistlinger L, Altmaier E et al: Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet 2008; 4: e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illig T, Gieger C, Zhai G et al: A genome-wide perspective of genetic variation in human metabolism. Nat Genet 2010; 42: 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Shin SY, Petersen AK et al: Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011; 477: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirkan A, van Duijn CM, Ugocsai P et al: Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet 2012; 8: e1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen J, Tukiainen T, Sarin AP et al: Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet 2012; 44: 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium Bernstein BE, Birney E An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK et al: Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 2012; 148: 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R et al: The accessible chromatin landscape of the human genome. Nature 2012; 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S: KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000; 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A, Schlegel HG: NAD-linked L(+)-lactate dehydrogenase from the strict aerobe Alcaligenes eutrophus. 1. Purification and properties. Eur J Biochem 1983; 130: 321–328. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Whitesides GM: L-Lactate dehydrogenase: substrate specificity and use as a catalyst in the synthesis of homochiral 2-hydroxy acids. J Am Chem Soc 1988; 110: 2959–2964. [DOI] [PubMed] [Google Scholar]
- Mamer OA, Reimer ML: On the mechanisms of the formation of L-alloisoleucine and the 2-hydroxy-3-methylvaleric acid stereoisomers from L-isoleucine in maple syrup urine disease patients and in normal humans. J Biol Chem 1992; 267: 22141–22147. [PubMed] [Google Scholar]
- Hoffer LJ, Taveroff A, Robitaille L, Mamer OA, Reimer ML: Alpha-keto and alpha-hydroxy branched-chain acid interrelationships in normal humans. J Nutr 1993; 123: 1513–1521. [DOI] [PubMed] [Google Scholar]
- Franke L, van Bakel H, Fokkens L, de Jong ED, Egmont-Petersen M, Wijmenga C: Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet 2006; 78: 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharuri H, Henneman P, Demirkan A et al: Automated workflow-based exploitation of pathway databases provides new insights into genetic associations of metabolite profiles. BMC Genomics 2013; 14: 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Plenge RM, Rossin EJ et al: Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet 2009; 5: e1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SY, Fauman EB, Petersen AK et al: An atlas of genetic influences on human blood metabolites. Nat Genet 2014; 46: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker V, Mills GA: Effects of birth asphyxia on urinary organic acid excretion. Biol Neonate 1992; 61: 162–172. [DOI] [PubMed] [Google Scholar]
- Chuang DT, Shih VE Maple syrup urine disease (branched-chain ketoaciduria). In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds): The Metabolic and Molecular Bases of Inherited Disease. New York, NY: McGraw-Hill, 2001,pp 1971–2005. [Google Scholar]
- Grundberg E, Small KS, Hedman AK et al: Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 2012; 44: 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.