Abstract
In most societies, surnames are passed down from fathers to sons, just like the Y chromosome. It follows that, theoretically, men sharing the same surnames would also be expected to share related Y chromosomes. Previous investigations have explored such relationships, but so far, the only detailed studies that have been conducted are on samples from the British Isles. In order to provide additional insights into the correlation between surnames and Y chromosomes, we focused on the Spanish population by analysing Y chromosomes from 2121 male volunteers representing 37 surnames. The results suggest that the degree of coancestry within Spanish surnames is highly dependent on surname frequency, in overall agreement with British but not Irish surname studies. Furthermore, a reanalysis of comparative data for all three populations showed that Irish surnames have much greater and older surname descent clusters than Spanish and British ones, suggesting that Irish surnames may have considerably earlier origins than Spanish or British ones. Overall, despite closer geographical ties between Ireland and Britain, our analysis points to substantial similarities in surname origin and development between Britain and Spain, while possibly hinting at unique demographic or social events shaping Irish surname foundation and development.
Introduction
In most human societies, surnames are paternally inherited, that is, they pass from a father to all of his children. The Y chromosome is also inherited paternally, but it is passed only from father to son. In theory then, males sharing the same surname should also share identical or closely related Y chromosomes.
Several studies over the past few years1, 2, 3, 4, 5, 6 have investigated the relationship between surnames and Y chromosomes, and have generally concluded that surnames are informative markers that can help disentangle hidden population structure and give insight into past historical events.
Up until now, all large-scale studies focusing on the relationship between Y chromosome and surname have been carried out on populations from Ireland and Britain.7, 8, 9 These studies reported a similar degree of correlation between surname and Y chromosome, the mean proportion of men belonging to groups of related Y chromosome haplotypes (indicated as descent clusters) being approximately 60% in both studies. However, the striking discrepancy in the correlation between Y chromosome sharing and surname frequency sets both works apart. In the British study, surnames with more than 5000 or so bearers at the national level showed relatively little or no Y chromosome coancestry, with an overall tendency towards more Y chromosome haplotype sharing as surnames became less frequent.7, 9 By contrast, common and extremely common Irish surnames displayed very strong Y chromosome coancestry levels.8 The authors concluded that the observed differences may have been caused by stronger drift in Ireland, perhaps brought about by the predominance of specific patrilineal clans or lineages, by smaller population sizes or by different demographic and historical events in Ireland and Britain.7, 8, 9
Given the reported discrepancy, it is not clear what the pattern might be in populations from other countries. To understand this further, we investigated Y chromosome variation and surnames in Spain, a Southern European population with a different historical, demographic and genetic background to the British Isles. Our results match the previous observations in British but not Irish surnames, suggesting perhaps that the inverse correlation between surname frequency and Y chromosome coancestry is a more general process, and that Irish surnames developed under more unusual and Irish-specific circumstances.
Materials and methods
Samples
A total of 1766 DNA samples from unrelated male volunteers each bearing one of 37 selected Spanish surnames and 355 Spanish controls were collected. Most of the samples were obtained by postal requests sent to potential participants randomly selected from the Spanish telephone directory. Volunteers performed self-collection of buccal cheek samples with a sterile cytology brush following the instructions included in the letter. Participants were asked to read and sign a written informed consent and to fill in a questionnaire with personal details such as name, place of birth of self, father, mother and paternal grandfather. Only samples with paternal grandfathers born in Spain were finally used. Buccal swabs and documents were sent back to CM-C by using a pre-stamped envelope.
The 37 Spanish surnames were selected to provide a wide geographical coverage, to represent surnames across the full frequency spectrum and from a sample of different surname types. Among the selected surnames, we defined five frequency classes: extremely frequent (surnames with more than 150 000 national bearers), moderately frequent (between 15 000 and 150 000 bearers), medium (between 5000 and 15 000 individuals), rare (between 3000 and 5000 bearers) and very rare (between 100 and 3000 bearers). If the medium and the rare categories are grouped together, each of these different frequency classes represents approximately 25% of the Spanish population. We also chose surnames with Castilian, Catalan and Basque origin and those that belong to different surname types or categories (Table 1). An additional set of 355 Spanish control subjects was also collected, genotyped and analysed. Controls were also recruited mainly via postal request; they all have different surnames, and were chosen randomly from the telephone directories of the same provinces where the surname samples were collected. The final number of controls from each province is proportional to the number of subjects used for the surname-based analysis from the selected provinces. All data regarding surname frequencies were taken from the Spanish National Statistics Institute (www.ine.es, year 2011).
Table 1. Surname information, haplogroup frequencies and gene diversities in 37 Spanish surnames and controls.
| Surname | Surname type | Origin | Frequency category | N | Total Population | E-M96 | G-M201 | I-M170 | J-M304 | R1a-1532 | R1b-M269 | T -M70 | Others | h Y-STRs | S.D. | Avg. h over STRs | SD | h Y-HGs | SD | % Men Clusters |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | − | − | − | 355 | 47 million | 8.5 | 1.7 | 6.6 | 7.7 | 3.9 | 67.9 | 2.3 | 1.4 | 1,000 | 0,001 | 0,584 | 0,299 | 0,538 | 0,041 | 0 |
| Fernandez | PA | CS | Extr. Freq. | 77 | 927 585 | 11.7 | 6.5 | 10.4 | 7.8 | 1.3 | 58.4 | 1.3 | 2.6 | 0,999 | 0,004 | 0,620 | 0,321 | 0,646 | 0,060 | 0 |
| Martinez | PA | CS | Extr. Freq. | 114 | 839 286 | 5.3 | 5.3 | 7 | 7.9 | - | 71.1 | 2.6 | 0.8 | 1,000 | 0,005 | 0,571 | 0,299 | 0,488 | 0,057 | 0 |
| Rubio | PC | CS | Frequent | 93 | 105 376 | 9.7 | 1.1 | 5.4 | 14 | − | 68.7 | 1.1 | − | 0,999 | 0,003 | 0,580 | 0,301 | 0,515 | 0,061 | 0 |
| Diez | PA | CS | Frequent | 108 | 64 715 | 18.5 | 6.5 | 1.9 | 8.3 | 1.9 | 59.3 | 2.8 | 0.8 | 1,000 | 0,006 | 0,610 | 0,319 | 0,615 | 0,048 | 0 |
| Pascual | PA | CS/CT | Frequent | 41 | 59 019 | 2.4 | − | − | 4.9 | − | 92.7 | − | − | 0,998 | 0,009 | 0,475 | 0,255 | 0,191 | 0,093 | 6.7 |
| Herrero | OC | CS | Frequent | 49 | 58 537 | 8.2 | 6.1 | 10.2 | 16.3 | − | 59.2 | − | − | 0,997 | 0,007 | 0,611 | 0,319 | 0,642 | 0,084 | 0 |
| Mateos | PA | CS | Frequent | 66 | 32 242 | 10.6 | 9.1 | 4.5 | 4.5 | 10.6 | 57.6 | − | 3.1 | 1,000 | 0,005 | 0,621 | 0,323 | 0,614 | 0,082 | 0 |
| Aguirre | TG | BQ | Frequent | 43 | 20 344 | 2.3 | 4.7 | 4.7 | 4.7 | 2.3 | 79 | 2.3 | − | 0,999 | 0,005 | 0,520 | 0,274 | 0,375 | 0,094 | 0 |
| Andreu | PA | CT | Frequent | 43 | 17 952 | 20.9 | 4.7 | 14 | 9.3 | − | 51.1 | − | − | 0,975 | 0,017 | 0,598 | 0,314 | 0,670 | 0,082 | 38.2 |
| Juarez | PA | CS | Frequent | 55 | 17 344 | 12.7 | − | 10.9 | 10.9 | 1.9 | 61.8 | 1.8 | − | 0,997 | 0,005 | 0,599 | 0,312 | 0,645 | 0,077 | 8.2 |
| Marques | OC | CS/CT | Frequent | 51 | 16 107 | 15.7 | 7.8 | 11.8 | − | 2 | 62.7 | − | − | 1,000 | 0,008 | 0,635 | 0,332 | 0,674 | 0,084 | 0 |
| Tirado | PC | CS | Medium | 39 | 11 878 | 15.4 | 2.6 | 5.1 | 15.4 | − | 56.3 | 2.6 | 2.6 | 0,998 | 0,009 | 0,618 | 0,325 | 0,722 | 0,076 | 6.7 |
| Alegre | PC | CS/CT | Medium | 46 | 9408 | 10.9 | 4.3 | 6.6 | 4.3 | − | 73.9 | − | − | 0,998 | 0,008 | 0,584 | 0,307 | 0,591 | 0,097 | 0 |
| Balaguer | TN | CT | Medium | 37 | 7588 | 10.8 | 27.1 | 2.7 | 8.1 | 2.7 | 48.6 | − | − | 0,972 | 0,025 | 0,622 | 0,328 | 0,658 | 0,087 | 18.5 |
| Quevedo | TN | CS | Medium | 42 | 7571 | 16.7 | − | 4.8 | 21.4 | − | 57.1 | − | − | 0,973 | 0,012 | 0,578 | 0,302 | 0,634 | 0,070 | 50.0 |
| Ibarra | TG | BQ | Medium | 42 | 7405 | 4.8 | − | 7.1 | 4.8 | 2.4 | 78.5 | − | 2.4 | 0,993 | 0,007 | 0,495 | 0,262 | 0,384 | 0,095 | 14.3 |
| Castell | TG | CT | Medium | 68 | 5592 | 1.5 | 1.5 | 5.9 | 41.2 | − | 49.9 | − | − | 0,911 | 0,031 | 0,557 | 0,291 | 0,589 | 0,037 | 75.0 |
| Cadenas | OC/PC | CS | Rare | 41 | 4607 | − | − | 4.9 | 2.4 | − | 92.7 | − | − | 0,934 | 0,025 | 0,333 | 0,184 | 0,142 | 0,072 | 68.3 |
| Moliner | OC | CT | Rare | 65 | 4237 | 18.5 | 1.5 | 21.5 | 3.1 | 3.1 | 52.3 | − | − | 0,988 | 0,010 | 0,556 | 0,293 | 0,617 | 0,085 | 34.3 |
| Huguet | PA | CT | Rare | 37 | 3951 | 5.4 | 8.1 | 2.7 | 5.4 | − | 78.4 | − | − | 0,960 | 0,019 | 0,492 | 0,262 | 0,386 | 0,100 | 48.6 |
| Valbuena | TN | CS | Rare | 57 | 3741 | 3.5 | 3.5 | − | 3.5 | 1.8 | 78.9 | 8.8 | − | 0,983 | 0,012 | 0,518 | 0,274 | 0,436 | 0,096 | 35.0 |
| Bengoechea | TG | BQ | Rare | 31 | 3672 | − | − | 3.2 | − | − | 96.8 | − | − | 0,914 | 0,027 | 0,395 | 0,215 | 0,065 | 0,059 | 80.6 |
| Villarroel | TN | CS | Rare | 61 | 3308 | 19.7 | − | 26.3 | 9.8 | 1.6 | 42.6 | − | − | 0,936 | 0,016 | 0,559 | 0,291 | 0,733 | 0,038 | 80.3 |
| Granell | PC/TG | CT | Rare | 53 | 3102 | 7.6 | 7.5 | 5.7 | − | − | 79.2 | − | − | 0,927 | 0,018 | 0,427 | 0,228 | 0,367 | 0,082 | 94.3 |
| Boluda | OC | CS | Very Rare | 20 | 2911 | 85 | − | − | − | − | 15 | − | − | 0,758 | 0,101 | 0,264 | 0,154 | 0,268 | 0,113 | 85.0 |
| Gordon | TN | CS | Very Rare | 43 | 2698 | 7 | − | 23.3 | 2.3 | 2.3 | 58.1 | 7 | − | 0,917 | 0,024 | 0,546 | 0,287 | 0,611 | 0,068 | 79.1 |
| Artola | TG | BQ | Very Rare | 33 | 2543 | − | − | − | − | − | 100 | − | − | 0,835 | 0,036 | 0,388 | 0,211 | 0,000 | 0,000 | 84.8 |
| Albiol | TN | CT | Very Rare | 45 | 2184 | − | − | − | 31.1 | − | 68.9 | − | − | 0,925 | 0,017 | 0,401 | 0,216 | 0,438 | 0,054 | 91.1 |
| Bibiloni | TN | CT | Very Rare | 28 | 1818 | − | − | 3.6 | 92.8 | − | 3.6 | − | − | 0,725 | 0,054 | 0,339 | 0,189 | 0,140 | 0,087 | 92.9 |
| Olaizola | TG | BQ | Very Rare | 40 | 1671 | − | − | − | − | − | 97.5 | − | 2.5 | 0,735 | 0,070 | 0,193 | 0,115 | 0,050 | 0,047 | 82.5 |
| Nortes | Unk. | CS | Very Rare | 18 | 1618 | − | − | − | − | − | 100 | − | − | 0,216 | 0,124 | 0,086 | 0,063 | 0,000 | 0,000 | 88.9 |
| Bennasar | PA | CT | Very Rare | 20 | 1437 | − | − | − | − | − | 100 | − | − | 0,742 | 0,096 | 0,112 | 0,076 | 0,000 | 0,000 | 95.0 |
| Ribalta | TG | CT | Very Rare | 44 | 1154 | − | − | 13.6 | 27.3 | − | 59.1 | − | − | 0,914 | 0,024 | 0,489 | 0,259 | 0,571 | 0,057 | 86.4 |
| Renau | PA/TN | CT | Very Rare | 53 | 1091 | − | 1.9 | 7.5 | 1.9 | − | 88.7 | − | − | 0,951 | 0,012 | 0,420 | 0,225 | 0,214 | 0,075 | 81.1 |
| Japon | TN | CS | Very Rare | 22 | 952 | − | − | − | 4.5 | − | 81.8 | − | 13.7 | 0,697 | 0,102 | 0,372 | 0,207 | 0,325 | 0,117 | 77.3 |
| Baigorri | TN | BQ | Very Rare | 22 | 882 | − | − | 22.7 | − | − | 77.3 | − | − | 0,784 | 0,066 | 0,430 | 0,236 | 0,368 | 0,100 | 95.5 |
| Ansotegui | PA | BQ | Very Rare | 19 | 660 | − | − | − | − | − | 100 | − | − | 0,714 | 0,073 | 0,201 | 0,123 | 0,000 | 0,000 | 78.9 |
| Total | − | − | − | 2121 | − | 8.9 | 3.2 | 7.2 | 9.8 | 1.6 | 67.3 | 1.3 | 0.8 | − | − | − | − | − | Avg. | 47.9 |
Abbreviations: N, sample size; h, gene diversity; PA, Patronymic; TN, Toponymic; TG, Topographic; OC, Occupational; PC, Personal Characteristics; Unk, Unknown origin. CS, Castilian origin; CT, Catalan; BQ, Basque. Extr. Freq., surnames with more than 150 000 carriers; Freq., 15 000 to 150 000; Medium, 5 000 to 15 000; Rare, 3000–5000; Very Rare, 100–3000. Gene diversity values for the SNP haplogroups were calculated using all SNPs genotyped. A table with the frequencies of all haplogroups, including the very rare ones clustered here under the ‘Others' column, is available in the Supplementary Material.
Only five spelling variants for the same surname were sampled and accounted for in this study: Ansotegui/Ansuategui, Babiloni/Bibiloni, Bengoechea/Bengochea, Castell/Castel/Castells and Nortes/Norte. Surname variants Bengochea, Castells, Castel and Norte were extremely uncommon and represented only by 3, 2, 1 and 1 samples, respectively.
Formal ethical approval for this project was obtained both from the Ethical Committee of the University of Santiago de Compostela and from the Ethical Committee of the Department of Health of the Autonomous Community of Galicia, Spain (‘Xunta de Galicia').
Laboratory methods
DNA extraction was performed using the ChargeSwitch Forensic DNA purification kit (Invitrogen, Eugene, OR, USA). Y chromosome STR genotyping was conducted using the Yfiler PCR Amplification kit (Applied Biosystems, Foster City, CA, USA),10 which co-amplifies a total of 17 Y chromosome STRs (DYS19, DYS385a,b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635 and GATAH4). Further information on the loci can be found on the YHRD and NIST webpages (www.yhrd.org; www.cstl.nist.gov/strbase/y_strs.htm). Twenty-two slowly mutating SNP markers (SRY-1532, M213, M9, M70, 92R7, M22, TAT, M173, P25, M269, M62, M172, M170, M26, M304, M201, M34, M81, M78, M35, M96 and M123) were grouped in three different multiplex sets, as previously described,11 and SNP results were used to assign Y chromosome haplogroups. Multiplex PCR amplification of DNA samples was performed using the Type-it Microsatellite PCR kit (QIAGEN, Dusseldorf, Germany). PCR reactions were carried out using 1ng of DNA as template, samples were run in an ABI PRISM 3730xl Genetic Analyzer, and the resulting genotypes were analysed with GeneMapper v3.2 (Applied Biosystems).
All new marker and haplotypic data contained in this study have been deposited in the YHRD database, reference access number YA004027 (www.yhrd.org), and it is also available in an excel table provided as supplementary material.
Statistical analysis
Basic descriptive statistics were calculated using the software package ARLEQUIN version 3.5.1.2.12 MDS analysis was based on the RST matrix of STR haplotypes using PROXSCAL in SPSS software version 11.5. These analyses were carried out using 15 STRs—DYS385 was not included owing to uncertainty in locus assignation, while DYS389II alleles were estimated after subtracting the number of repeats at the DYS389I locus.
Match probability scores were estimated for perfectly matching 17-STR haplotypes (including the DYS385 locus), and they were calculated for each individual and then averaged within surnames.
Median-joining networks were constructed using NETWORK, version 4.513 (www.fluxus-engineering.com/sharenet). All networks were constructed using 15 STR haplotypes (excluding the DYS385 locus) together with SNP markers. SNP markers were given a weight of 99, to reflect the virtual absence of back-mutations, while the more rapidly mutating STRs were given weights from 1 to 5 depending on their variance within haplogroups in our population, as described elsewhere.14
The criteria to identify and characterise descent clusters within surnames were defined following the exploration of our dataset. Thus, a numerical and neutral approach was used to define the descent clusters criteria, instead of establishing ad hoc rules based on the observation of the data as performed in the previous British and Irish studies.8, 9 We calculated the average number of mutational steps between all pairs of samples belonging to the same haplogroup within a surname. Assuming a monophyletic origin for the haplotypes within a surname, such estimates should provide a hint of the maximum number of mutational steps within a cluster, as well as being indicative of a plausible number for the maximum number of mutational steps without a linking haplotype. As R1b is by far the most common haplogroup in Spain (reaching 67% of the samples in this study), we considered that non-R1b descent clusters might better represent ‘natural' or ‘true' related groups, as they are probably less exposed to the introgression of identical by state chromosomes. This is expected to be even more so in the case of less frequent surnames. Therefore, we reasoned that the average number of mutational steps between haplotypes within non-R1b haplogroups in the very rare, rare and medium frequency surnames could be used as an indication of the maximum continuous number of steps that could be left ‘empty' or ‘unfilled' within a network when defining a descent cluster (the value calculated here was 2.08: a maximum of two mutational steps equalling to one ‘empty' or ‘unfilled' spot or haplotype). Following the same logic, we turned again to non-R1b haplogroups in medium, rare and very rare surnames, and considered as the maximum number of mutational steps between any two members of a cluster the upper 95% confidence interval of the mean number of mutational steps between haplotypes belonging to the same haplogroup within each surname (the value calculated here was 5.16). Thus, we decided that five would be the maximum number of mutational steps between any two members of a cluster. Additionally, two identical haplotypes were considered to be the minimum number of chromosomes to form a cluster. Finally, in cases in which more than one cluster could be identified or two clusters overlapped, the cluster with the largest number of total individuals was selected.
Times to the most recent common ancestor (TMRCAs) and their 95% confidence intervals were calculated for all haplotypes included in the different surname descent clusters using the Ytime package (available at www.ucl.ac.uk/tcga/software/), based on the ASD statistic (Average Squared Distance).15, 16 This approach requires the definition of a founder haplotype, and ASD is then calculated between all chromosomes in the cluster and the founder one. The founder haplotype of each cluster was calculated using the most common allele of each STR marker. The mutation rate used to estimate all TMRCAs was calculated by averaging the mutation rate values across the 15 STR markers used, as published in the Y Chromosome Haplotype Reference Database (www.yhrd.org, release 37; 0.002635 per locus per generation). A male generation time of 31 years was used,17, 18 and the mean age of all DNA sample donors (55 years) was subsequently added to the resulting TMRCA age estimate. It is also worth mentioning that TMRCA estimates are always subjected to errors inherent to time depth estimates from variable STR mutation rates. Note that Ytime can only be used when more than one different haplotype exists in a cluster. Thus, TMRCAs were only calculated for clusters harbouring more than one haplotype. The age of clusters represented by a single haplotype can be reasonably considered as not older than clusters composed by two haplotypes only. We considered the age of the oldest of such clusters (a total of 12 in the whole 15 STRs Spanish dataset) as being the most conservative way to provide an approximate estimate of the antiquity of single-haplotype clusters (273 years).
Comparative data
In order to rigorously compare the results obtained from Spain to published data from other populations, Y chromosome haplotype data on British and Irish surnames were taken from the literature,8, 9 and networks as well as descent clusters were ascertained as explained above. As STR data from the British and Irish study did not completely overlap with the Spanish one, only STRs genotyped in the three studies were used for all comparative calculations (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438 and DYS439). Owing to the reduced number of STR markers, descent cluster definition criteria for the comparative data were obviously somewhat stricter, and were established, as explained above, for samples belonging to rare or very rare surnames of the three populations (less than 5000 national carriers) and within infrequent haplogroups—R1b-M269 was excluded in all three populations, with the addition of I-M170 in Britain. There are only seven surnames in the Irish study that can be considered rare or very rare, and in those surnames there are only nine samples in total that do not belong to haplogroup R1b. For this reason, we removed the Irish data from our calculations. The combined estimates for the Spanish (20 rare and very rare surnames in total) and British (32 rare and very rare surnames in total) surnames were 1.73 (SD 0.9) average mutational steps between haplotypes belonging to uncommon haplogroups and 3.61 as the 95% upper distribution of the number of mutational steps between haplotypes. Following this, and visually inspecting the various networks in the comparative dataset, we allowed only one mutational step between haplotypes belonging to the same cluster (that is, no empty or unfilled haplotypes were accepted), and considered three as the maximum number of mutational steps between any two members of a cluster.
Results
The controls we used in this study showed haplogroup frequencies similar to previous Y chromosome studies in Spain,19, 20, 21 and most of the frequent and very frequent surnames showed haplogroup distributions similar to this control set (Table 1). For other less frequent surnames, in many cases, haplogroup composition was biased towards one or two haplogroups, indicating that sampling men with the same surname is far from random. This was the case for rare or very rare surnames, the most extreme cases being the very rare surnames Bennasar, Boluda, Bibiloni, Nortes, Artola, Ansotegui, Renau or Bengoechea (Table 1).
Both STR- and SNP-based gene diversity estimates were positively associated with surname frequency (Table 1; Spearman's r=0.896; P<0.0001 for STR haplotypes and r=0.749; P<0.0001 for haplogroups).
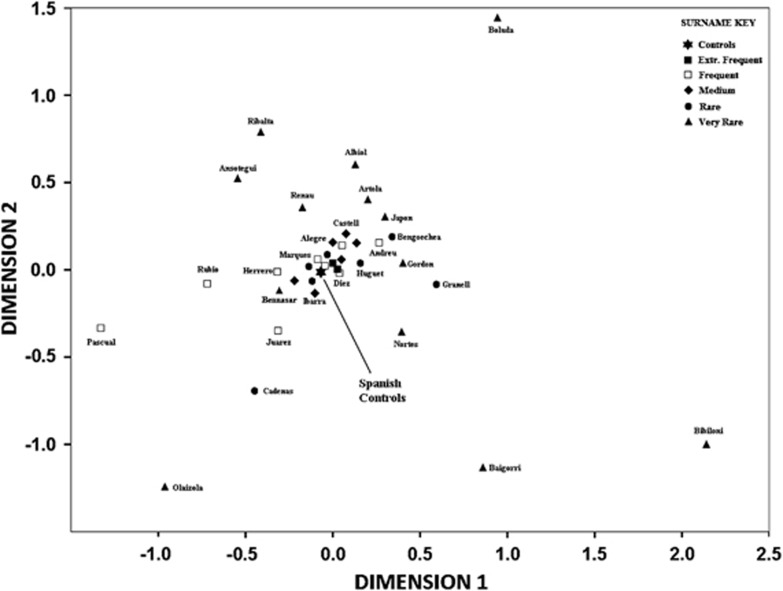
We used MDS analysis to explore Y-STR haplotype relationships among the different surnames and the control group (Figure 1). Surname frequency correlated with proximity to the centre of the plot and the Spanish controls (Figure 1). Indeed, except for two medium frequency surnames—Castell and Quevedo—all very frequent, frequent and medium frequency surnames were not significantly different from the control group (Fisher's exact test; P<0.05). On the other hand, many very rare surnames were not close to the other surnames or the controls, some of the rare and very rare surnames being outliers dominated entirely by one or two clusters of very similar haplotypes belonging to only one haplogroup (Figure 1).
Figure 1.
Multidimensional scaling plot based on Y-STR haplotypes. MDS analysis was based on an RST matrix of all surnames and controls.
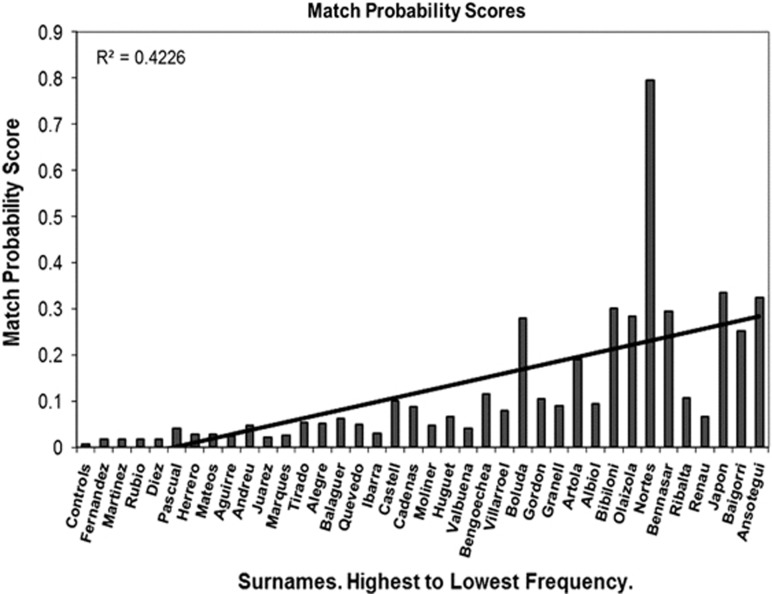
The match probability score determines the probability that two random men chosen from the same group (same surname in our case) will share an identical Y chromosome (17-STR haplotype; Figure 2). Although the probability scores were markedly different across surnames, the more frequent surnames tended to have low match probabilities, while the rare and very rare surnames tended to have higher match probability values (Spearman's Rank Correlation, r=0.906; P<0.0001).
Figure 2.
Match probability. Match probability scores plotted against surnames ranked by decreasing frequencies. A trend line and the R2 value are also shown.
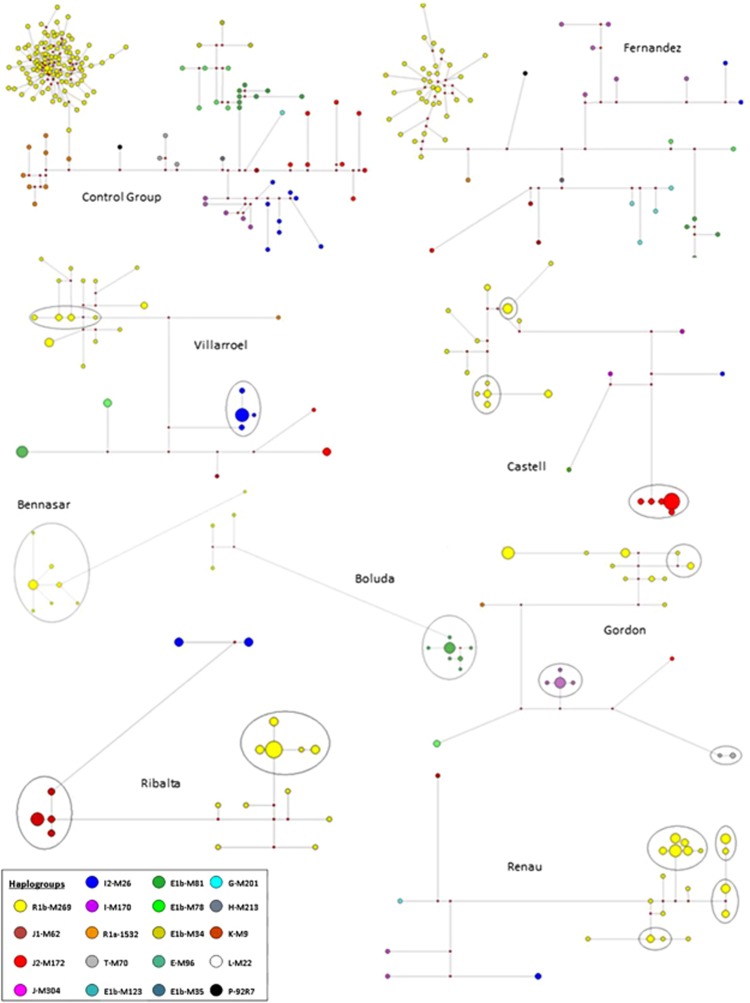
We identified clusters of related haplotypes using median-joining networks in the less frequent surnames, while networks of common or very common surnames show less haplotype clustering (Figure 3; Supplementary Material).
Figure 3.
Selected median-joining networks. A selection of median-joining networks representing Y haplogroup and Y STR diversity within surnames plus the control sample set. Each circle represents a distinct haplotype, with circle area proportional to frequency and with the smallest circle in each network representing n=1. The line length between haplotypes indicates their mutational divergence (SNP and STR mutational steps). The small red dots are median vectors (hypothesised and often ancestral sequences required to connect existing sequences within the network). Borders of descent clusters are displayed by the solid-line ellipses.
The haplotype network for the Spanish control group is almost entirely made up of different haplotypes occurring only once (singletons). Most of the frequent and very frequent surnames behave in quite a similar manner, showing remarkably high haplotype diversity, as well as a few of the less common ones (Supplementary Material), reflecting the low degree of coancestry intrinsic to these surnames. For example, surnames such as Fernandez, Martinez, Rubio, Diez, Herrero or Pascual are also almost entirely composed of singletons. Even less common surnames such as Marques, Aguirre, Ibarra, Juarez or Tirado are almost exclusively represented by unique haplotypes, and thus show a complete lack of descent clusters. It is worth underlining here that the proportion of the population represented by the most frequent surnames is extremely large in Spain, much larger than in Ireland or Britain.8, 9 For example, the 10 commonest surnames account for approximately 20% of the Spanish population (National Statistics Institute, www.ine.es), whereas these represent only 6.4 and 8.5% in Britain and Ireland, respectively.7, 22
Less common surnames display networks different from the controls or the frequent surnames, with most of their chromosomes being grouped in or around one single cluster of haplotypes. However, in some of the very rare surnames and in most, but not all, of the rare surnames, multiple clusters can be seen (usually three, four or more clusters) showing that, often, there is more than one ancestor to men sharing the same surname, even for rare ones (Supplementary Material).
Descent clusters and time depths
We defined a total of 84 descent clusters in the 37 surnames analysed in this study (Supplementary Material). Out of these 84 clusters, 40 contained more than one different haplotype, with the remaining ones being composed solely of identical haplotypes.
The average percentage of men included in descent clusters taking into account all sampled surnames was 47.9%, while the percentage for the control sample set was 0% (Table 1). The percentage of men in a descent cluster was also zero in 11 of the 13 most common surnames of the study, whereas a large fraction of the men belonging to rare or very rare surnames were included in descent clusters, with an average of 78% (Table 1). A strong correlation between surname frequency and the fraction of men included in descent clusters was observed (Spearman's Rank Correlation, r=0.859; P<0.0001).
TMRCAs for surname descent clusters
Ages of Spanish descent clusters ranged from 167 years for the Japon cluster to 1310 years for one of the Ribalta descent cluster. Only 8 clusters out of the 84 total descent clusters (9.5%) were estimated to be older than 800 years (Table 2), the earliest possible limit for hereditary surname establishment in Spain. We noted that 20 of the 40 descent clusters (50%) where the TMRCA was estimated—and potentially all of the 44 single-haplotype clusters—were younger than 450 years of age.
Table 2. Estimated ages of the Spanish surname descent clusters.
| MRCA | 95% CIs | TMRCAa | 95% CIsa | |||||
|---|---|---|---|---|---|---|---|---|
| Surname | Cluster No. | N | Time in gens. | Lower | Upper | Time in yrs. 31 y/gen | Lower | Upper |
| Albiol | 1 | 12 | 29.51 | 16.58 | 49.59 | 970 | 569 | 2087 |
| Albiol | 2 | 14 | 12.65 | 5.46 | 25.51 | 447 | 224 | 846 |
| Albiol | 3 | 15 | 35.42 | 21.72 | 54.23 | 1,153 | 728 | 2231 |
| Andreu | 4 | 8 | 12.65 | 4.51 | 30.43 | 447 | 195 | 998 |
| Artola | 5 | 10 | 5.06 | 1.12 | 15.93 | 212 | 90 | 549 |
| Artola | 6 | 9 | 5.62 | 1.25 | 18.58 | 229 | 94 | 631 |
| Baigorri | 7 | 13 | 11.68 | 4.5 | 24.96 | 417 | 195 | 829 |
| Bengoechea | 8 | 13 | 21.41 | 11.38 | 37.24 | 719 | 408 | 1705 |
| Bengoechea | 9 | 6 | 4.22 | 0.64 | 18.33 | 186 | 75 | 623 |
| Bennasar | 10 | 19 | 14.65 | 7.92 | 25.01 | 509 | 301 | 830 |
| Bibiloni | 11 | 15 | 8.43 | 3.36 | 18.92 | 316 | 159 | 642 |
| Boluda | 12 | 17 | 19.35 | 10.67 | 33.12 | 655 | 386 | 1577 |
| Cadenas | 13 | 22 | 34.5 | 22.79 | 49.94 | 1,124 | 762 | 2098 |
| Cadenas | 14 | 6 | 25.3 | 9.84 | 54.04 | 839 | 360 | 2225 |
| Castell | 15 | 21 | 16.87 | 9.45 | 27.93 | 578 | 348 | 921 |
| Castell | 16 | 7 | 14.46 | 4.79 | 35.03 | 503 | 203 | 1636 |
| Gordon | 17 | 10 | 7.59 | 2.29 | 21.25 | 290 | 126 | 714 |
| Gordon | 18 | 4 | 12.65 | 2.61 | 41.5 | 447 | 136 | 1837 |
| Gordon | 19 | 3 | 8.43 | 1.29 | 38.74 | 316 | 95 | 1751 |
| Granell | 20 | 29 | 20.06 | 13.16 | 29.81 | 677 | 463 | 979 |
| Granell | 21 | 7 | 25.3 | 10.31 | 52.8 | 839 | 375 | 2187 |
| Granell | 22 | 4 | 6.32 | 1.13 | 26.14 | 251 | 90 | 865 |
| Huguet | 23 | 12 | 23.19 | 12.39 | 41.06 | 774 | 439 | 1823 |
| Ibarra | 24 | 6 | 16.87 | 5.94 | 41.91 | 578 | 239 | 1849 |
| Japon | 25 | 14 | 3.61 | 0.78 | 11.38 | 167 | 79 | 408 |
| Moliner | 26 | 8 | 22.14 | 9.3 | 44.44 | 741 | 343 | 1928 |
| Olaizola | 27 | 33 | 18.4 | 11.88 | 27.77 | 625 | 423 | 916 |
| Quevedo | 28 | 4 | 12.65 | 2.72 | 42.34 | 447 | 139 | 1863 |
| Quevedo | 29 | 6 | 12.65 | 3.4 | 34.94 | 447 | 160 | 1633 |
| Quevedo | 30 | 4 | 6.32 | 1.01 | 29.26 | 251 | 86 | 962 |
| Renau | 31 | 23 | 18.7 | 11.39 | 29.96 | 635 | 408 | 984 |
| Renau | 32 | 7 | 7.23 | 1.65 | 24.99 | 279 | 106 | 830 |
| Renau | 33 | 7 | 21.68 | 8.43 | 46.28 | 727 | 316 | 1985 |
| Renau | 34 | 4 | 6.32 | 1.07 | 27.29 | 251 | 88 | 901 |
| Ribalta | 35 | 12 | 25.3 | 13.5 | 44.27 | 839 | 474 | 1922 |
| Ribalta | 36 | 20 | 40.48 | 27.01 | 58.27 | 1,310 | 892 | 2356 |
| Valbuena | 37 | 7 | 10.84 | 3.19 | 30.83 | 391 | 154 | 1011 |
| Valbuena | 38 | 5 | 5.06 | 0.92 | 22.77 | 212 | 83 | 761 |
| Villarroel | 39 | 16 | 11.07 | 4.75 | 22 | 398 | 202 | 737 |
| Villarroel | 40 | 9 | 28.11 | 14.55 | 52.08 | 926 | 506 | 2164 |
| Average | − | 11.5 | 16.07 | 7.52 | 34.02 | 553 | 288 | 1345 |
Abbreviations: N, sample size; gens., generations; yrs., years; CIs, confidence intervals.
Time in years plus 55 (mean age of all sample donors).
Comparative analysis of Spanish, British and Irish surnames
We constructed comparative median-joining networks and descent clusters for Spanish, British and Irish populations and estimated TMRCAs for surnames from all three populations with the 10 shared genotyped Y-STRs. These analyses were performed only for surnames with 15 or more samples (37 Spanish, 38 British and 21 Irish surnames; Table 3; Supplementary Material).
Table 3. Spanish, British and Irish comparative surname analysis.
| Spain | Britain | Ireland | |
|---|---|---|---|
| Total N | 1766 | 1654 | 953 |
| No. surnames analysed | 37 | 38 | 21 |
| No. clusters found | 79 | 80 | 41 |
| Avg. cluster size | 10.1 | 12.6 | 16.1 |
| % men in clusters | 45.1 | 59.2 | 67.7 |
| Avg. cluster age | 536 | 545 | 829 |
Proportion of men in clusters and average age of descent clusters.
Table 3 summarises the results obtained for all three populations using exactly the same conditions—Y chromosome STR markers, STR mutation rate, cluster definition criteria, TMRCA estimation (ASD) and male generation time (31 years). Irish surnames display much bigger descent clusters, with a greater number of individuals per cluster (average cluster size of 16.1) than Spanish (10.1) or British (12.6) surnames. Likewise, Irish surnames also show higher percentages of men within descent clusters (67.7%), as well as older clusters (829 years) than both the Spanish (45.1% and 536 years) and British surnames (59.2% and 545 years), even though the number of total descent clusters in Ireland is not larger than the one estimated for Spain or Britain (Table 3). Overall, British surnames adopt an intermediate position between the Irish—who have a higher percentage of men included in larger and older clusters—and the Spanish—who have lower percentages of men included in smaller and younger descent clusters—surnames, although as a whole they seem to be closer to the Spanish than to the Irish surnames in all the analyses performed. Tukey range test values after one-way ANOVA tests among all three populations for cluster size, proportion of men in clusters and cluster ages were all significant between Ireland and the other two populations (P values of 0.003, 0.007 and 0.009, respectively, between Ireland and Spain; and P values of 0.017, 0.037 and 0.011, respectively, between Ireland and Britain), whereas they were not significant between Britain and Spain (except for the proportion of men in clusters, P=0.043).
Discussion
The study of 37 surnames in the Spanish population reported here has shown that there is a remarkable correlation between Y chromosome haplotypes and many of the surnames in Spain. This is supported by several observations: (i) the gene diversity of men sharing a surname is in general low, lower than the gene diversity of the control populations; (ii) the probability of a man sharing an identical 17 STR Y chromosome haplotype with another man of the same surname (match probability) is greatly increased with respect to the general population (Figure 2); and (iii) descent clusters are present in most surnames, and absent in the controls, clearly suggesting that there is an association between Y chromosomes and surnames in Spain.
Correlation between surname frequency and degree of coancestry
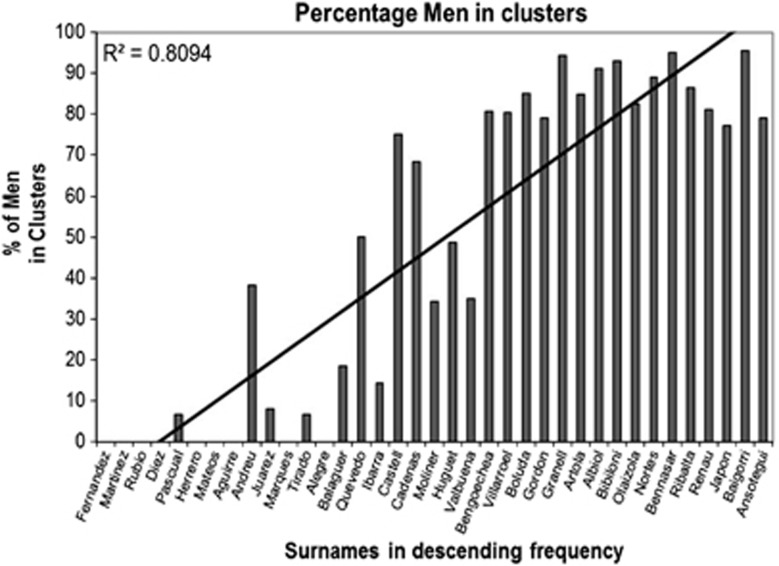
Our results also suggest that the degree of coancestry within surnames depends, to a large extent, on the frequency of the surname in the population. Frequent and very frequent surnames are characterised by high levels of gene diversity, entirely comparable with controls, whereas less common surnames show lower diversity values. Median-joining networks of common surnames are made up almost entirely of singletons, illustrating the low levels of coancestry present in those surnames. In fact, aside from a few exceptions, samples of moderately frequent, frequent and very frequent surnames (that is, surnames with more than 5000 Spanish bearers), would be almost indistinguishable from a Spanish control population (Table 1, Figure 4). Most of the individuals bearing rare and very rare surnames are grouped in clusters of related Y chromosomes (between 75 and 95% of samples within surnames), some of which are dominated by one or a few large clusters of haplotypes (Figure 3; Supplementary Material).
Figure 4.
Percentage of men in surname descent clusters. Proportion of haplotypes in descent clusters for each surname, ranked by decreasing frequencies.
Lack of correlation between surname type or origin and degree of coancestry
Surname frequency, not surname type, is correlated with Y chromosome coancestry in Spain. Although a larger number of surnames would increase power, we can make a few simple comparisons between different types of surnames (patronymic, toponymic, topographic, occupational, etc.). For example, we can clearly see that frequent and very frequent patronymic surnames—such as Fernandez, Martinez, Diez, Juarez, Mateos or Pascual—show high values of haplotype gene diversity, as well as low proportions of men included in clusters. However, rare or very rare surnames of patronymic origin such as Ansotegui, Huguet or Bennasar show low gene diversity values, as well as high proportions of men included in clusters (Table 1). Additionally, rare or very rare surnames of topographic origin—such as Artola, Ribalta, Olaizola, Bengoechea—or even a medium frequency surname—like Castell—show higher percentages of men in clusters and lower haplotype gene diversities than more frequent topographic surnames such as Aguirre or Ibarra. Occupational surnames also exhibit the same trend, with rare or very rare occupational surnames like Moliner and Boluda being less genetically diverse and displaying more individuals in clusters than frequent occupational names such as Herrero or Marques. Finally, a rare surname derived from a nickname or personal characteristic—Cadenas—also has higher percentages of men in clusters and lower haplotype gene diversity values than the frequent nickname surnames Alegre or Rubio.
Surnames from different provinces behaved accordingly to the number of bearers with no differences between surnames belonging to different regions (Castile, Catalonia or the Basque Country), the only observation being that Catalan and especially Basque surnames simply tend to be less frequent than Castilian ones.
All these examples suggest that surname type or surname origin have no bearing on the correlation between Y chromosome and surname, and that this association is only affected by surname frequency. Nevertheless, larger numbers of surnames belonging to all different surname types and regions are needed to fully test this relationship.
TMRCAs of descent clusters and the effect of genetic drift
The fact that roughly half of the descent clusters are younger than 450 years suggests that genetic drift has probably been at work since the inception of hereditary surnames in Spain, although we cannot rule out the possibility of sampling effects. Some extreme examples are represented by surnames such as Nortes or Artola (Table 2), in which large clusters have been dated to less than 200 years ago. This means that, owing to the fluctuation in lineage frequency caused by drift, many of the Y chromosome lineages that men had at the birth of each surname 700–800 years ago have probably gone extinct, whereas others have recently expanded. Therefore, the distribution of lineages within surnames that we see today might be quite different from the one present some 800 or even 400 years ago.
Some descent clusters (8 out of 40) are actually older than 800 years, and therefore discordant with surname establishment in Spain. All except two of them belong to common haplogroup R1b-M269. Probably, introgression of common similar haplotypes in these R1b-M269 clusters has inflated the age of the clusters belonging to this frequent haplogroup.
The study analysing the relationship between Y chromosomes and British surnames did not find a correlation between a surname's alleged origin and descent cluster data.9 When surname types likely of having a single origin were taken into account, the percentage of men in the dominant cluster did not suggest that purported single origin. To investigate this possibility in our Spanish dataset, we grouped surnames into two sets according to the likelihood of having a single origin. Ten toponymic surnames, derived from small villages or rare local place names and likely to have been founded only once (Albiol, Ansotegui, Baigorri, Balaguer, Gordon, Japon, Quevedo, Renau, Valbuena and Villarroel) were compared with 26 surnames that probably originated in more than one place at more than one time (patronymic, topographic, occupational, etc.). One surname of unknown origin, Nortes, was left out of this analysis.
When the percentage of men included in each surname's largest cluster—which is likely to have a single origin—was compared between both surname groups, no significant differences were found (unpaired t-test; t=0.743; P=0.463). This observation, together with the fact that many clusters appear to be, in general, much younger than the dates of origin of heritable surnames in Spain (Table 2) again suggests that drift has strongly influenced the history and development of most Spanish surnames—as previously proposed in relation to British surnames.9
Comparison to previous similar studies
Our results suggest that the ages of descent clusters in Spain are more similar to findings from Britain than those in Ireland (Table 3). Not only do Irish surnames display a greater proportion of men included in descent clusters than Spanish or British ones, but their clusters are also on average much larger, although the relative number of clusters does not seem to vary considerably between the three populations (Table 3). It therefore seems that it is greater Irish cluster size, and not greater cluster number, that explains the higher proportion of men included in Irish clusters. Moreover, the average estimated age of the Spanish descent clusters in this comparative study was 536 years, very close to the value of 545 years for British descent clusters, whereas the average age of the Irish descent clusters is much older, reaching 829 years (Table 3). This is consistent with the age of hereditary surnames in Ireland, as an older origin for Irish surnames than Spanish or British ones has been suggested. 9, 23 This disparity in descent cluster ages could simply reflect the different times of hereditary surname establishment between Ireland and the other two populations.8, 9, 23
Conclusions
In Spain, the correlation between Y chromosome type and surname is dependent on the frequency of a surname, in overall agreement with surnames in Britain,9 but not Ireland.8 As a whole, men bearing rare and very rare surnames share high levels of Y chromosome coancestry, whereas men with medium frequency, frequent and very frequent surnames do not. We also find no correlation between surname type and either Y chromosome coancestry or descent cluster formation. Drift has probably had a great impact in the history, development and current distribution of most Spanish surnames, as inferred from the nature and age of most descent clusters described in the Spanish surnames analysed in this study. Finally, as Irish descent clusters seem to be on average much older than British or Spanish ones, this study is in agreement with the historically well attested earlier time of surname establishment in Ireland than in Spain or Britain.23
Acknowledgments
We would like to thank all the DNA donors who volunteered to participate in this study. We would also like to thank Dr Chris Tyler-Smith, Dr Bruce Winney and especially Prof. Mark A. Jobling, for their extremely knowledgeable help as well as their comments and ideas. CM-C was supported by a Marie Curie Training and Mobility Research Fellowship from the European Commission of the EU. Part of the research leading to these results received funding from the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7/2007–2013/ under REA grant agreement no. 290344, and the grants from the ‘Ministerio de Ciencia e Innovación' (SAF2008-02971) and the Plan Galego IDT, Xunta de Galicia (EM 2012/045) given to AS. GBJB was supported by a BBSRC Doctoral Training Fellowship.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Hill EW, Jobling MA, Bradley DG: Y-chromosome variation and Irish origins. Nature 2000; 404: 351–352. [DOI] [PubMed] [Google Scholar]
- Moore LT, McEvoy B, Cape E, Simms K, Bradley DG: A Y-chromosome signature of hegemony in Gaelic Ireland. Am J Hum Genet 2006; 78: 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden GR, Balaresque P, King TE et al: Excavating past population structures by surname-based sampling: the genetic legacy of the Vikings in northwest England. Mol Biol Evol 2008; 25: 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winney B, Boumertit A, Day T et al: People of the British Isles: preliminary analysis of genotypes and surnames in a UK-control population. Eur J Hum Genet 2012; 20: 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gonzalez LJ, Martinez-Espin E, Alvarez JC et al: Surname and Y chromosome in Southern Europe: a case study with Colom/Colombo. Eur J Hum Genet 2012; 20: 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larmuseau MH, Vanoverbeke J, Van Geystelen A et al: Low historical rates of cuckoldry in a Western European human population traced by Y-chromosome and genealogical data. Proc Biol Sci 2013; 280: 20132400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TE, Ballereau SJ, Schürer KE, Jobling MA: Genetic signatures of coancestry within surnames. Curr Biol 2006; 160: 384–388. [DOI] [PubMed] [Google Scholar]
- McEvoy B, Bradley DG: Y-chromosomes and the extent of patrilineal ancestry in Irish surnames. Hum Genet 2006; 119: 212–219. [DOI] [PubMed] [Google Scholar]
- King TE, Jobling MA: Founders, drift, and infidelity: the relationship between Y chromosome diversity and patrilineal surnames. Mol Biol Evol 2009; 26: 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero JJ, Chang CW, Calandro LM et al: Development and validation of the AmpFlSTR Yfiler PCR amplification kit: a male specific, single amplification 17 Y-STR multiplex system. J Forensic Sci. 2006; 51: 64–75. [DOI] [PubMed] [Google Scholar]
- Brion M, Sobrino B, Blanco-Verea A, Lareu MV, Carracedo A: Hierarchical analysis of 30 Y-chromosome SNPs in European populations. Int J Legal Med 2005; 119: 10–15. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S: Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online 2005; 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A: Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Qamar R, Ayub Q, Mohyuddin A et al: Ychromosomal DNA variation in Pakistan. Am J Hum Genet 2002; 70: 1107–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW: An evaluation of genetic distances for use with microsatellite loci. Genetics 1995; 139: 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Thomas MG, Skorecki K et al: Multiple origins of Ashkenazi Levites: Y chromosome evidence for both Near Eastern and European ancestries. Am J Hum Genet 2003; 73: 768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Hrafnkelsson B, Gulcher JR, Ward R, Stefánsson K: A populationwide coalescent analysis of Icelandic matrilineal and patrilineal genealogies: evidence for a faster evolutionary rate of mtDNA lineages than Y chromosomes. Am J Hum Genet 2003; 72: 1370–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner JN: Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol 2005; 128: 415–423. [DOI] [PubMed] [Google Scholar]
- Flores C, Maca-Meyer N, González AM et al: Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography. Eur J Hum Genet 2004; 12: 855–863. [DOI] [PubMed] [Google Scholar]
- Cruciani F, La Fratta R, Trombetta B et al: Tracing past human male movements in northern/eastern Africa and western Eurasia: new clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol Biol Evol 2007; 24: 1300–1311. [DOI] [PubMed] [Google Scholar]
- Adams SM, Bosch E, Balaresque PL et al: The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula. Am J Hum Genet 2008; 83: 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telecom ÉireannIrish Telephone Directory 1998. Dublin, Ireland. [Google Scholar]
- MacLysaght E: The Surnames of Ireland. Dublin: Irish Academic Press, 1985. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.