Abstract
Background:
DNA hypomethylation of long interspersed nuclear elements-1 (LINEs-1) occurs during carcinogenesis, whereas information addressing LINE-1 methylation in Wilms tumor (WT) is limited. The main purpose of our study was to quantify LINE-1 methylation levels and evaluate their relationship with relative telomere length (TL) in WT.
Methods:
We investigated LINE-1 methylation and relative TL using bisulfite-polymerase chain reaction (PCR) pyrosequencing and quantitative PCR, respectively, in 20 WT tissues, 10 normal kidney tissues and a WT cell line. Significant changes were analyzed by t-tests.
Results:
LINE-1 methylation levels were significantly lower (P < 0.05) and relative TLs were significantly shorter (P < 0.05) in WT compared with normal kidney. There was a significant positive relationship between LINE-1 methylation and relative TL in WT (r = 0.671, P = 0.001). LINE-1 Methylation levels were significantly associated with global DNA methylation (r = 0.332, P < 0.01). In addition, relative TL was shortened and LINE-1 methylation was decreased in a WT cell line treated with the hypomethylating agent 5-aza-2′-deoxycytidine compared with untreated WT cell line.
Conclusion:
These results suggest that LINE-1 hypomethylation is common and may be linked to telomere shortening in WT.
Keywords: 5-aza-2’-deoxycytidine, Hypomethylation, Long Interspersed Nuclear Element-1, Relative Telomere Length, Wilms Tumor
INTRODUCTION
Long interspersed nuclear element-1 (LINE-1) sequences are highly repeated in human retrotransposons and constitute about 17% of the human genome. LINE-1 retrotransposition events lead to genomic deletions,[1] chromosome breaks and genomic instability.[2] LINE-1 retroelements may play roles in cell proliferation, differentiation, and tumor progression.[3] Targeting the LINE-1 would open the possibility of a promising gene therapy for a wide spectrum of human tumors.[4] LINE-1 transcription and retrotransposition are prevented in normal somatic cells by a variety of control mechanisms, including methylation of LINE-1, specifically LINE-1 promoters.[5] However, decreased methylation of LINE-1 sequences has been documented in many human cancers.[6] LINE-1 hypomethylation has the potential to act as molecular markers for detection and prognosis, but direct consequences of this epigenetic defect for cancer biology remain largely unknown.
Human telomeres consist of long (TTAGGG)n nucleotide repeats and an associated protein complex located at the chromosome ends. Telomeres play important roles in DNA replication and repair and are key factors in aging and cancer development.[7] Telomere shortening causes genome instability, resulting in loss of cell-cycle control, one of the hallmarks of cancer, and an increase in the rate of tumor initiation.[8] Mammalian telomeres can be elongated by telomerase or alternative lengthening of telomeres-related mechanisms. Moreover, several studies have implicated DNA methylation as an additional mechanism controlling telomere length (TL).[9,10] Dysfunctional telomeres can be a substrate for LINE-1 retrotransposition,[11,12] and LINE-1 is critical for telomere maintenance in telomerase positive tumor cells,[13] suggesting that LINE-1 methylation may affect TL. Although recent evidence has indicated an association between LINE-1 methylation and TL,[14,15,16] there have been few reports of this relationship in childhood solid tumors.
Concurrent telomere shortening and genomic instability have been observed in most embryonic tumors investigated, including Wilms tumor (WT).[17] WT is the most common renal tumor in childhood, affecting one in 10,000 children.[18] Approximately, 75% of cases occur in children younger than 5-year-old, with a peak incidence in 2 younger than 5r in childhood WTs are characterized by complex karyotypes showing various chromosomal instabilities, such as chromosome gains, deletions, and rearrangements.[19] Telomere-dependent mitotic instability and telomeric fusion as a chromosomal mechanism for the loss of bands 11p13 and 11p15 have both been detected in some WTs,[20] though the mechanisms responsible for these phenomena remain to be determined. DNA methylation may play a role in these processes through its effect on DNA instability and telomere function. Two apparently contradictory epigenetic events coexist in WT, namely global hypomethylation of DNA and hypermethylation. About 60% of WTs were found to show overall DNA hypomethylation, suggesting its involvement in oncogenesis and/or tumor progression.[21] LINE-1 methylation can act as a surrogate marker of global DNA methylation,[22] suggesting that LINE-1 hypomethylation might also occur in WT.
Based on the above reports, we hypothesized that LINE-1 hypomethylation may be significant and may also be related to TL in WT. In this study, we, therefore, compared the LINE-1 methylation and the relative average TL between WT and normal control tissues, using pyrosequencing and quantitative polymerase chain reaction (qPCR), respectively. In addition, we assessed the effect of genomic demethylation on TL in a WT cell line by treating it with a 5-aza-2′-deoxycytidine (5-aza-dC). The results of this study have important implications for understanding the pathogenesis of WT in children, as well as suggesting potential new strategies for the early diagnosis and individual treatment of WTs.
METHODS
Subjects
Twenty WT and ten normal kidney tissues were obtained from the Capital Institute of Pediatrics. The characteristics of the subjects are summarized in Table 1. This study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Ethical Committee of the Capital Institute of Pediatrics (No. SHERLL2013025). Normal tissues were obtained from patients after receiving the informed written consent of families or relatives at the hospital. The diagnosis of WT was based on pathology reports, the Institutional Review Board, and histological evaluations, according to the SIOP 2001 trials.[23] No patient had received chemotherapy or radiotherapy prior to surgery. Fresh tissue samples were stored at −80°C after resection.
Table 1.
Clinicopathologic characteristics of subjects (n)
| Characteristic | Normal control (n = 10) | WT patients (n = 20) |
|---|---|---|
| Age | ||
| ≤5 years | 9 | 17 |
| >5 years | 1 | 3 |
| Sex | ||
| Male | 6 | 12 |
| Female | 4 | 8 |
| Clinical stage | ||
| I + II | 15 | |
| III + IV | 5 |
WT: Wilms tumor
Cell lines
WT-CLS1 cells were obtained from Cell Lines Service GmbH (Eppelheim, Germany) and cultured in MG-80 medium (Cell Lines Service GmbH, Eppelheim, Germany) supplemented with 15% fetal bovine serum and 1% penicillin/streptomycin (Life Technologies, Carlsbad, CA, USA), and maintained at 37°C and 5% CO2.
DNA extraction and sodium bisulfite DNA modification
DNA was extracted using a commercial kit (Omega Bio-Tek, Inc., Norcross, GA, USA) according to the manufacturer's recommendations. One microgram of DNA (50 ng/μl) was subjected to bisulfite conversion using an EpiTect fast DNA bisulfite kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Final elution was performed with 30 μl of M-Elution Buffer (Qiagen, Hilden). Bisulfite-treated DNA was stored at −20°C.
DNA digestion and high performance liquid chromatography measurements of global genomic 5-methylcytosine content
Approximately, 20 μg DNA was dissolved in 150 μl of 1X TE; ribonuclease A (RNase A) was added to a final concentration of 100 μg/ml, RNase T1 was added to a final concentration of 2,000 units/ml, and the solution was incubated at 37°C for 2 h, followed by ethanol precipitation. After removal of RNA, DNA digested to nucleosides by treatment with deoxyribonuclease I and Nuclease P1 (Sigma-Aldrich, USA).
Global methylation levels of nucleosides were evaluated by measuring 5-methylcytosine (5 mC) content with high performance liquid chromatography (HPLC). 5 mC was measured as the proportion of total 2′-deoxycytidine-5′-monophosphate (5 mC + C). Deoxycytidine 5′-monophosphate (C) and 5-methyl-2′-deoxycytidine, 5′-monophosphate (5 mC) were purchased from USB (USA). The amounts of 5 mC and C were measured at 278 nm at a sensitivity of 0.005 absorbance-unit full scale following separation in an ESA C18 ((250 mm × 3.2 mm I.D., 5 μm, Chelmsford, USA) particle size 3 μm; USA) using 50 mmol/L ammonium orthophosphate (pH 4.1) as the mobile phase. All DNA samples were analyzed in duplicate, and the difference in 5 mC content between duplicate samples was <3%. The method was validated by determining the linearity (r2 ≥0.999). The coefficient of variation of intra- and inter-day precision were <5%. The assays were performed on each batch, but the case or control status of all specimens was unknown to the investigators. 5 mC content is reported as the ratio of 5 mC to the sum of C plus 5 mC. The percentage of 5 mC was calculated according to the following equation: %5 mC = ([5 mC]/[5 mC + C]) × 100.
Quantitative polymerase chain reaction and pyrosequencing assay for long interspersed nuclear element-1 methylation
The average of the amounts of cytosine in the three CpG sites was used as a measure of the overall LINE-1 methylation level in each specimen. LINE-1 methylation at three CpG sites (position 305–331 of GenBank accession no. ×58075) was measured using a commercially-available Pyromark kit (Qiagen, Valencia, CA, USA), following the manufacturer's instructions. Briefly, 1 μl of bisulfite-treated DNA was used in a 25 μl PCR reaction using Pyromark PCR kit (Qiagen, Valencia). The reverse PCR primer was biotinylated. A single PCR fragment spanning a part the genetic element was amplified and the degree of methylation was analyzed in a single pyrosequencing reaction using 20 μl of PCR product. The PCR product was purified using Streptavidin Sepharose High Performance Beads (Amersham Biosciences, Uppsala, Sweden). The sepharose beads containing immobilized PCR products were purified, washed, and denatured using 0.2 mol/L NaOH, and washed again using a vacuum prep tool (Pyrosequencing Inc., Westborough, MA). 12 μl of 0.3 μmol/L of pyrosequencing primer was annealed to the purified single-stranded PCR product, and pyrosequencing was done using PyroMark Q96 System (Qiagen, Valencia). The degree of methylation was expressed as the percentage of methylated cytosines divided by the sum of the methylated plus unmethylated cytosines. Bisulfite conversion was verified using non-CpG cytosine residues as built-in controls and complete conversion of cytosine at a non-CpG site confirmed successful bisulfite conversion.
Quantitative real-time polymerase chain reaction for relative telomere length analysis
Relative TL was measured using quantitative real-time polymerase chain reaction (qPCR) and the ratio of telomere repeat copy number to a single copy gene (hbg) copy number (T/S) was determined, based on the method reported by Cawthon.[24] The relative length of the telomeres was obtained by calculating the ratio (T/S) of telomere repeat and single gene copy products (hbg) for each individual, using the formula T/S = 2−ΔCT, where ΔCT = Ct telomere − Ct hbg. This ratio was then compared with the ratio for the reference DNA. All samples were run in triplicate and the average of the three telomere measurements was divided by the average of the three S measurements to calculate the average T/S ratio.
Chemical treatment
The demethylating effect of anticancer drug treatment was determined by treating WT-CLS1 cells were treated with 2.5 μmol/L 5-aza-dC (Sigma-Aldrich, USA) every 48 h for 12 days to achieve genomic demethylation using bisulfite-PCR pyrosequencing. Every control and treatment experiment was repeatedly performed for 3 wells in the same experiment.
Statistical analysis
Q–Q plots and one-sample Kolmogorov–Smirnov tests were used to determine if continuous parameters were normally distributed. Significant changes in the methylation status of LINE-1 and TLs in WTs were analyzed by unpaired or paired t-tests. Based on their fit to a normal distribution, correlations between LINE-1 and log10 TL were analyzed by Pearson's (parametric) tests. Correlations between LINE-1 methylation and log10 TL by Spearman's rank correlation test. All analyses were performed using the SPSS 16.0 software package (McGraw-Hill Inc., New York, NY, USA) and GraphPad Prism 5.01 (GraphPad Software Inc., La Jolla, CA, USA). All statistical tests were two-sided, and P < 0.05 was considered to indicate statistical significance.
RESULTS
Hypomethylation of long interspersed nuclear element-1 in Wilms tumors
We examined the methylation levels of the three promoter CpG islands in LINE-1 by pyrosequencing. Representative pyrograms are shown in Figure 1, and the results are summarized in Figure 2. The LINE-1 promoter was highly methylated with minimal variability in normal tissues (mean ± standard deviation [SD] methylation level: 64.03 ± 3.94%). The methylation levels were significantly lower in the corresponding tumor samples (mean: 56.5 ± 8.9%, P < 0.05), with 55% of the tumor samples showing hypomethylation (<57.15% =2 SDs below the normal mean).
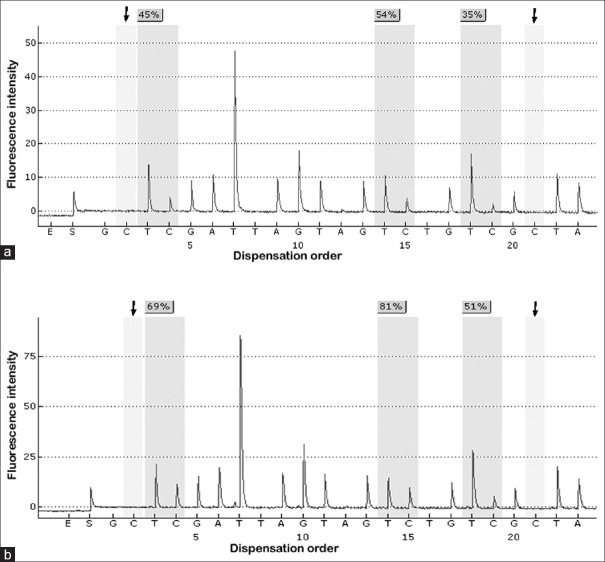
Figure 1.
Pyrosequencing of long interspersed nuclear element-1 methylation. (a) Long interspersed nuclear element-1 hypomethylated tumor (methylation level, 44.7%). (b) Long interspersed nuclear element-1 hypermethylated tumor (methylation level, 67%). The arrows indicate no residual C at the non-CpG site, confirming complete bisulfite conversion.
Figure 2.
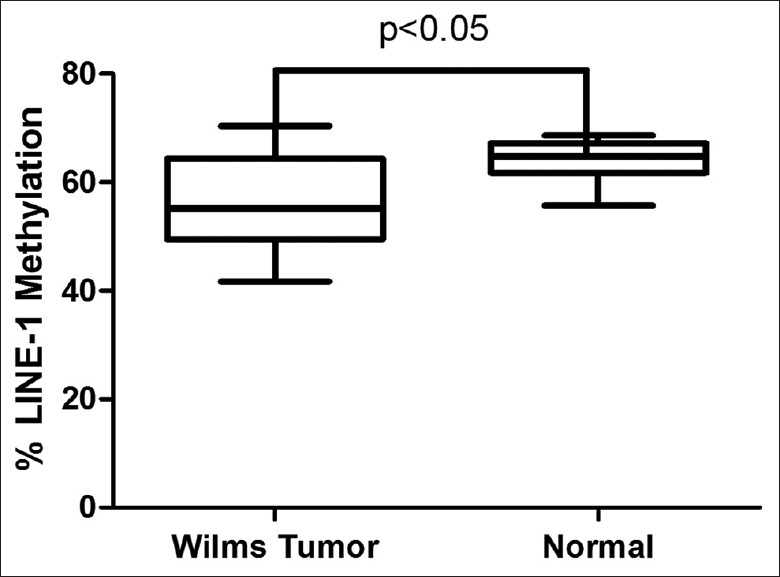
Box plot representation of long interspersed nuclear element-1 methylation values in Wilms tumor and normal control tissues. Hypomethylation of long interspersed nuclear element-1 was significantly increased in Wilms tumor tissues compared with normal controls. P values based on two-tailed unpaired t-tests.
Correlation between long interspersed nuclear element-1 methylation and global DNA methylation levels
The overall DNA 5 mC content was determined by HPLC on heat-denatured DNA digested to nucleosides. Using the HPLC assay, each global DNA methylation was tested on a panel of DNAs from WT, as well as normal kidney tissues samples, all of which had been tested for LINE-1 methylation levels by pyrosequencing. Linear regression analysis showed that the LINE-1 methylation was most closely associated with global DNA methylation as determined by HPLC [correlation coefficient, r = 0.332, P < 0.01, Figure 3].
Figure 3.
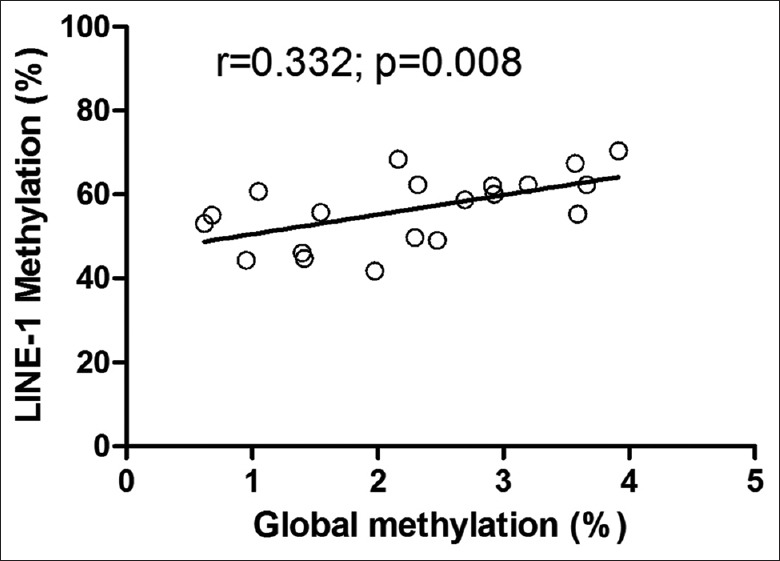
Correlation of long interspersed nuclear element-1 methylation levels by pyrosequencing measurements with high performance liquid chromatography-based global DNA methylation measurements for the samples.
Association between DNA methylation and relative telomere length
To determine if the changes in LINE-1 methylation between normal and cancer tissues were correlated with telomeres, we determined relative TLs by qPCR. Relative TLs have been shown to correlate well with absolute TL measured by Southern blotting.[25] The TL ratio was not distributed normally and was, therefore, transformed to a normal distribution using the normalized log10 TL values of individual ratios. Log10 TL levels in the ten normal kidney DNA samples ranged from −0.09 to 0.44 (mean 0.17 ± 0.18). Log10 TL levels in the 20 WT DNA samples ranged from −0.66 to 0.58 (mean − 0.15 ± 0.37), with 50% of the samples showing shortened log10 TL <−0.29 (2 SDs below the normal mean). The log10 TL for each WT sample was significantly shorter than that for normal kidney, according to Student's t-tests [Figure 4].
Figure 4.
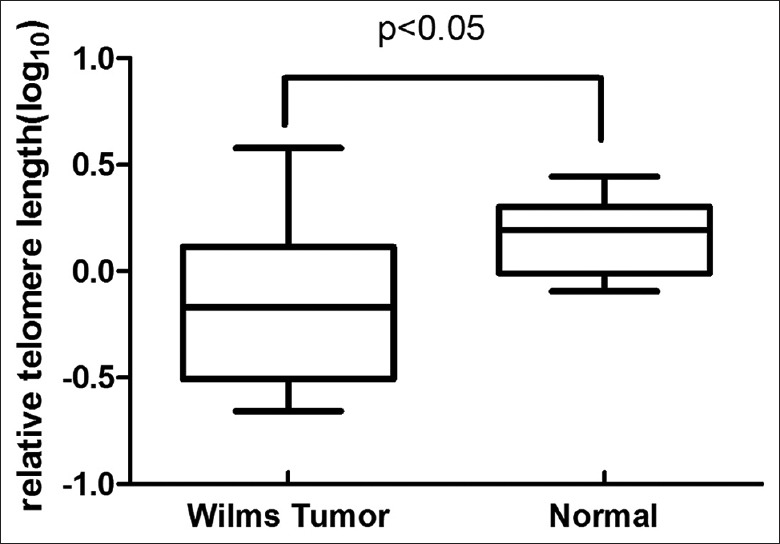
Box plot of relative telomere lengths in Wilms tumor and normal control tissues. There was significant shortening of relative telomere lengths in Wilms tumor compared with normal controls. P values based on two-tailed unpaired t-tests.
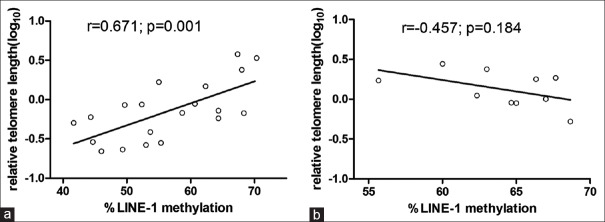
LINE-1 methylation and log10 TL level were positively correlated in WT DNA samples (r = 0.671, P = 0.001) [Figure 5a]. This suggests that LINE-1 hypomethylation in WT was associated with relatively shorter TL. In contrast, there was no significant between log10 TL and LINE-1 methylation (r = −0.457, P = 0.184) in unaffected controls [Figure 5b]. These nonsignificant results are in agreement with the previously-reported relationships in a panel of more than 20 different human cancer cell lines[9] and in unaffected dyskeratosis congenita relatives.[15]
Figure 5.
Correlation between relative telomere length and long interspersed nuclear element-1 methylation in Wilms tumor and normal tissues. (a) Wilms tumor tissues. (b) Normal tissues. r values represent Pearson's correlation coefficient between relative telomere length and long interspersed nuclear element-1 methylation.
Effects of 5-aza-2′-deoxycytidine on long interspersed nuclear element-1 methylation and telomere length
In order to ascertain whether or not the TL shortening occurred simultaneously with LINE-1 hypomethylation in the cells treated by demethylating reagent, we examined the effect of the demethylating agent 5-aza-dC in the WT cell line WT-CLS1. Treatment with 5-aza-dC resulted in LINE-1 hypomethylation and shortened TL compared with untreated cells [Table 2].
Table 2.
Effects of 5-aza-dC on LINE-1 methylation and relative telomere length in WT-CLS1 cells
| Cells | LINE-1 methylation (%) | Relative telomere length (T:S ratio) |
|---|---|---|
| Untreated (n = 3) | 62.00 ± 0.70 | 0.81 ± 0.03 |
| Treated (n = 3) | 25.90 ± 0.71* | 0.70 ± 0.02† |
*Significant variation compared with untreated cells (P < 0.05). †Significant variation compared with untreated cells (P < 0.001). P values based on paired t-tests. LINE-1: Long interspersed nuclear element-1; WT: Wilms tumor; 5-aza-dC: 5-aza-2’-deoxycytidine.
DISCUSSION
WT is considered to be associated with both genetic mutations and epigenetic abnormalities. Since the incidence of genetic mutations, such as WT1, WTX, and TP53 gene, is relatively low,[26] much attention has been paid to the role of epigenetic abnormalities in WT.[27] DNA methylation is a major epigenetic mechanism in carcinogenesis. Although many studies have focused on the relevance and clinical applications of DNA hypermethylation in WT, the level of LINE-1 methylation remains largely uninvestigated. In this study, we identified a significant reduction in LINE-1 methylation in WT compared with normal control tissues. We also found a positive association between LINE-1 methylation and TL in WT tissues. To the best of our knowledge, this study provides the first evidence of a relationship between LINE-1 methylation and TL in WT.
Weisenberger et al. evaluated methylation levels of Alu, LINE-1, and Sat2 using MethyLight in a panel of tumor DNA samples including WT.[22] However, they focused on the methods, and the role of LINE-1 methylation in WT was not reported in detail. In this study, we quantified the methylation levels of LINE-1 in WT and normal kidney tissues using pyrosequencing technology. This approach is considered more precise than the MethyLight and combined bisulfite and restriction analysis methods.[28] Our results show a statistically significant LINE-1 hypomethylation in WT compared with normal kidney tissues based on PCR pyrosequencing assay and indicate that LINE-1 hypomethylation is common in WT. These findings suggest that LINE-1 methylation might play an important role in WT development. LINE-1 hypomethylation is inversely associated with microsatellite instability, maybe various chromosomal instabilities in WT are associate with LINE-1 hypomethylation.[29] Further study needs to verify this hypothesis. Moreover, we found LINE-1 methylation was most closely associated with global DNA methylation, which is in agreement with previous finding.[22]
Telomere biology plays a crucial and complex role in the initiation and progression of cancer. Telomere shortening causes genome instability and increase in the rate of tumor initiation. Moreover, shortened telomere can active telomerase to enable telomere-length homeostasis and allow cancer cells to escape the antiproliferative barrier posed by short telomeres. In WT, as other tumors, shortened telomere may be the same mechanism. However, there is limited information on the occurrence of shortening telomeres. In this study, we identified a positive association between LINE-1 methylation and TL in WT tissues in agreement with previously-reported relationships in chronic lymphocytic leukemia.[14] This result supports existing findings of epigenetic regulation of TL.[9]
Possible explanations for the observed relationship include the following. One possibility is that LINE-1 may contain a disabled endonuclease able to use dysfunctional telomeres as an integration substrate.[11] There are similarities between the mechanisms of endonuclease-independent LINE-1 retrotransposition and telomerase.[12] In WT, as in other cancers, the telomerase gene is highly expressed and telomerase plays a major role in TL maintenance,[30] competitive inhibition of telomerase activity by LINE-1 retrotransposition due to hypomethylation may thus shorten TLs and cause genome instability. Alternatively, telomeres are located adjacent to gene-poor subtelomeric regions, which are also enriched in repetitive DNA.[16] Riethman et al. found that LINEs accounted for approximately 25% of the number of bases in the subtelomere, and there were very large strand- and subtelomere-specific biases in LINE, short interspersed element, long terminal repeat, and DNA repeat contents.[31] Previous investigations also found that methylation levels of subtelomeric regions were associated with TL and may be important for the epigenetic regulation of telomere dynamics.[10] The over-representation of LINEs in the subtelomeric region may thus account for the association between LINE-1 methylation and TL.
This study was limited by its relatively small sample size and limited clinical information, which precluded the analysis of specific subgroups. Further studies with larger sample sizes are necessary to elucidate the exact relationships between LINE-1 hypomethylation or shortened TL and tumor progression and prognosis in WT. However, despite the limited sample size, we still identified a strong significant association between LINE-1 methylation and TL in WT. This association might be relevant for cancer development. Hypomethylation promotes LINE-1 transcription to inactive telomere by DNA damage or other ways and promote tumorigenesis. On the other hand, inactivation of telomeres can be used as a substrate to activate LINE-1 transcription, promote DNA damage and other effects, and increase tumorigenesis. Further studies are needed to investigate the cause-effect relationship between hypomethylation of LINE-1 and shorter TL.
DNA methyltransferase inhibitors are being increasingly used in clinical treatment, and demethylation may have different results in different cancer cells. Vera et al. reported that treatment of human cancer cell lines with demethylating drugs resulted in hypomethylation of subtelomeric repeats and increased telomere recombination, which may in turn facilitate telomere elongation.[9] However, Zhang et al. reported that strongly repressed human telomerase reverse transcriptase expression decreased telomerase activity and remarkably shortened telomeres by demethylation following 5-aza-dC treatment.[32] Similarly, we found that demethylation with 5-aza-dC shortened TL in WT-CLS1 cell from a primary epithelial WT, though further studies are needed to clarify the mechanism in other WT cell lines. Demethylating agents are increasingly used in cancer treatment. It is, therefore, imperative to understand the mechanisms whereby cancer-related and/or drug-induced hypomethylation may initiate retrotransposition and shorten TL leading to subsequent genomic alterations.
In summary, the results of this study suggest that LINE-1 hypomethylation may be a common feature of WT. Moreover, the current findings show a positive significant relationship between LINE-1 methylation and TL in WT, suggesting that LINE-1 hypomethylation may be related to decrease TL. We also observed significant LINE-1 hypomethylation and shortened TLs in WT cells subjected to demethylation treatment, thus supporting the concept that epigenetic regulation may affect TL. Further work is required to clarify the molecular mechanisms involved in the relationship between DNA hypomethylation and TL, and to provide the basis for its clinical utilization in human cancer.
Financial support and sponsorship
National Natural Science Foundation of China (No. 81301773).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Yi Cui
REFERENCES
- 1.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–25. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 2.Kazazian HH, Jr, Goodier JL. LINE drive. Retrotransposition and genome instability. Cell. 2002;110:277–80. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- 3.Oricchio E, Sciamanna I, Beraldi R, Tolstonog GV, Schumann GG, Spadafora C. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 2007;26:4226–33. doi: 10.1038/sj.onc.1210214. [DOI] [PubMed] [Google Scholar]
- 4.Sinibaldi-Vallebona P, Lavia P, Garaci E, Spadafora C. A role for endogenous reverse transcriptase in tumorigenesis and as a target in differentiating cancer therapy. Genes Chromosomes Cancer. 2006;45:1–10. doi: 10.1002/gcc.20266. [DOI] [PubMed] [Google Scholar]
- 5.Piskareva O, Lackington W, Lemass D, Hendrick C, Doolan P, Barron N. The human L1 element: A potential biomarker in cancer prognosis, current status and future directions. Curr Mol Med. 2011;11:286–303. doi: 10.2174/156652411795677954. [DOI] [PubMed] [Google Scholar]
- 6.Bae JM, Shin SH, Kwon HJ, Park SY, Kook MC, Kim YW, et al. ALU and LINE-1 hypomethylations in multistep gastric carcinogenesis and their prognostic implications. Int J Cancer. 2012;131:1323–31. doi: 10.1002/ijc.27369. [DOI] [PubMed] [Google Scholar]
- 7.Harari Y, Romano GH, Ungar L, Kupiec M. Nature vs nurture: Interplay between the genetic control of telomere length and environmental factors. Cell Cycle. 2013;12:3465–70. doi: 10.4161/cc.26625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung AL, Deng W. Telomere dysfunction, genome instability and cancer. Front Biosci. 2008;13:2075–90. doi: 10.2741/2825. [DOI] [PubMed] [Google Scholar]
- 9.Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. Epigenetic regulation of telomeres in human cancer. Oncogene. 2008;27:6817–33. doi: 10.1038/onc.2008.289. [DOI] [PubMed] [Google Scholar]
- 10.Yehezkel S, Segev Y, Viegas-Péquignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17:2776–89. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- 11.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–12. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 12.Kopera HC, Moldovan JB, Morrish TA, Garcia-Perez JL, Moran JV. Similarities between long interspersed element-1 (LINE-1) reverse transcriptase and telomerase. Proc Natl Acad Sci U S A. 2011;108:20345–50. doi: 10.1073/pnas.1100275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aschacher T, Wolf B, Enzmann F, Kienzl P, Messner B, Sampl S, et al. LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines. Oncogene. 2015 doi: 10.1038/onc.2015.65. doi: 10.1038/onc.2015.65. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Hoxha M, Fabris S, Agnelli L, Bollati V, Cutrona G, Matis S, et al. Relevance of telomere/telomerase system impairment in early stage chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2014;53:612–21. doi: 10.1002/gcc.22171. [DOI] [PubMed] [Google Scholar]
- 15.Gadalla SM, Katki HA, Shebl FM, Giri N, Alter BP, Savage SA. The relationship between DNA methylation and telomere length in dyskeratosis congenita. Aging Cell. 2012;11:24–8. doi: 10.1111/j.1474-9726.2011.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong JY, De Vivo I, Lin X, Grashow R, Cavallari J, Christiani DC. The association between global DNA methylation and telomere length in a longitudinal study of boilermakers. Genet Epidemiol. 2014;38:254–64. doi: 10.1002/gepi.21796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalaby T, Hiyama E, Grotzer MA. Telomere maintenance as therapeutic target in embryonal tumours. Anticancer Agents Med Chem. 2010;10:196–212. doi: 10.2174/1871520611009030196. [DOI] [PubMed] [Google Scholar]
- 18.Stiller CA, Parkin DM. International variations in the incidence of childhood renal tumours. Br J Cancer. 1990;62:1026–30. doi: 10.1038/bjc.1990.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gratias EJ, Jennings LJ, Anderson JR, Dome JS, Grundy P, Perlman EJ. Gain of 1q is associated with inferior event-free and overall survival in patients with favorable histology Wilms tumor: A report from the Children's Oncology Group. Cancer. 2013;119:3887–94. doi: 10.1002/cncr.28239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewénius Y, Jin Y, Øra I, de Kraker J, Bras J, Frigyesi A, et al. Defective chromosome segregation and telomere dysfunction in aggressive Wilms’ tumors. Clin Cancer Res. 2007;13(22 Pt 1):6593–602. doi: 10.1158/1078-0432.CCR-07-1081. [DOI] [PubMed] [Google Scholar]
- 21.Ehrlich M, Jiang G, Fiala E, Dome JS, Yu MC, Long TI, et al. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene. 2002;21:6694–702. doi: 10.1038/sj.onc.1205890. [DOI] [PubMed] [Google Scholar]
- 22.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vujanic GM, Sandstedt B. The pathology of Wilms’ tumour (nephroblastoma): The International Society of Paediatric Oncology approach. J Clin Pathol. 2010;63:102–9. doi: 10.1136/jcp.2009.064600. [DOI] [PubMed] [Google Scholar]
- 24.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aviv A, Hunt SC, Lin J, Cao X, Kimura M, Blackburn E. Impartial comparative analysis of measurement of leukocyte telomere length/DNA content by Southern blots and qPCR. Nucleic Acids Res. 2011;39:e134. doi: 10.1093/nar/gkr634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Shen Y, Sun N, Jiang YP, Li ML, Sun L. Identification and analysis of mutations in WTX and WT1 genes in peripheral blood and tumor tissue of children with Wilms’ tumor. Chin Med J. 2012;125:1733–9. [PubMed] [Google Scholar]
- 27.Ohnishi K, Semi K, Yamada Y. Epigenetic regulation leading to induced pluripotency drives cancer development in vivo. Biochem Biophys Res Commun. 2014;455:10–5. doi: 10.1016/j.bbrc.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 28.Aparicio A, North B, Barske L, Wang X, Bollati V, Weisenberger D, et al. LINE-1 methylation in plasma DNA as a biomarker of activity of DNA methylation inhibitors in patients with solid tumors. Epigenetics. 2009;4:176–84. doi: 10.4161/epi.4.3.8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–73. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dome JS, Bockhold CA, Li SM, Baker SD, Green DM, Perlman EJ, et al. High telomerase RNA expression level is an adverse prognostic factor for favorable-histology Wilms’ tumor. J Clin Oncol. 2005;23:9138–45. doi: 10.1200/JCO.2005.00.562. [DOI] [PubMed] [Google Scholar]
- 31.Riethman H, Ambrosini A, Castaneda C, Finklestein J, Hu XL, Mudunuri U, et al. Mapping and initial analysis of human subtelomeric sequence assemblies. Genome Res. 2004;14:18–28. doi: 10.1101/gr.1245004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang ZX, Wang Y, Tao ZZ, Chen SM, Xiao BK, Zhou T. Subtelomeric demethylation deregulated hTERT expression, telomerase activity, and telomere length in four nasopharyngeal carcinoma cell lines. Cancer Biother Radiopharm. 2014;29:289–94. doi: 10.1089/cbr.2013.1581. [DOI] [PubMed] [Google Scholar]