Abstract
Background & objectives:
Epidemiological interventions and mosquito control are the available measures for dengue control. The former approach uses serotype and genetic information on the circulating virus strains. Dengue has been frequently reported from Nepal, but this information is mostly lacking. The present study was done to generate a comprehensive clinical and virological picture of a dengue outbreak in Nepal during 2013.
Methods:
A hospital-based study involving patients from five districts of Nepal was carried out. Demographic information, clinical details and dengue serological status were obtained. Viral RNA was characterized at the molecular level by reverse-transcription polymerase chain reaction (RT-PCR), nucleotide sequencing and phylogenetic analysis.
Results:
From among the 2340 laboratory-confirmed dengue cases during the study period, 198 patients consented for the study. Clinically they had fever (100%), headache (59.1%), rashes (18.2%), retro-orbital pain (30.3%), vomiting (15.1%), joint pain (28.8%) and thrombocytopenia (74.3%). Fifteen (7.5%) of them had mucosal bleeding manifestations, and the rest were uncomplicated dengue fever. The patients were mostly adults with a mean age of 45.75 ± 38.61 yr. Of the 52 acute serum samples tested, 15 were positive in RT-PCR. The causative virus was identified as DENV serotype 2 belonging to the Cosmopolitan genotype.
Interpretations & conclusions:
We report here the involvement of DENV serotype 2 in an outbreak in Nepal in 2013. Earlier outbreaks in the region in 2010 were attributed to serotype 1 virus. As serotype shifts are frequently associated with secondary infections and severe disease, there is a need for enhancing surveillance especially in the monsoon and post-monsoon periods to prevent large-scale, severe dengue outbreaks in the region.
Keywords: Dengue fever, dengue virus type 2, India, Nepal, phylogeny
Dengue, the vector-borne disease transmitted predominantly by Aedes aegypti and Ae. albopictus mosquitoes is a common and widespread arboviral infection in the tropical and subtropical regions causing significant morbidity and mortality1,2. The causative dengue virus (DENV) belongs to the Flaviviridae family and has a single-stranded, positive-sense RNA genome, approximately 11 kb long. Based on antigenicity, there are four serotypes of DENV referred to as DENV-1, DENV-2, DENV-3, and DENV-42.
The clinical picture of the disease ranges from a mild dengue fever (DF) to severe cases manifested with haemorrhagic complications and shock syndrome which might be fatal. A primary dengue infection usually results in DF. Secondary infection with a heterologous serotype in the presence of non-neutralizing antibodies from the primary infection may advance to severe dengue. This phenomenon of antibody dependent enhancement (ADE)3 occurs in regions where multiple serotypes co-circulate or when a new serotype is introduced into a dengue prevalent region.
The first case of dengue was reported from Nepal in 2004, followed by 32 laboratory confirmed cases in a minor outbreak in 20064. Subsequent studies have reported the circulation of all four serotypes of DENV in nine districts of the low-land Terai region5,6. A few random cases were confirmed in the following year along with the detection of Ae. aegypti larvae in the region7. Study of an epidemic in 2010 documented the circulation of dengue virus type 1 in Nepal8. The present study was aimed to investigate the nature and extent of an outbreak of febrile disease suspected as dengue in five districts of Nepal in 2013.
Material & Methods
Study design and locations: A hospital-based, prospective study was carried out from July to December 2013. Six hospitals in the outbreak regions- Narayani zone hospital and Bhawani Hospital of Birgunj, Parsa; Bharatpur Hospital in Bharatpur, Chitwan; Universal Medical College in Bhairahawa; Institute of Medicine in Kathmandu; and Janakpur Zone Hospital in Janakpur, Dhanusa, were identified as the centres for collection of biological samples. Selected samples were brought to the laboratory in the Central Department of Biotechnology, Tribhuvan University, Kathmandu in Nepal for serological and reverse transcription (RT)-PCR analysis and cDNAs were sent to the collaborator's laboratory in India for nucleotide sequencing and phylogenetic analysis.
Inclusion and exclusion criteria of patients: All the ‘dengue-suspected’ patients who visited the above hospitals were routinely screened for dengue infection using a rapid diagnostic kit (RDT) (Panbio, Australia) supplied by the Ministry of Health and Family Welfare, Government of Nepal for the detection of IgM antibodies. The WHO-2009 definition9 was used to identify patients suspected to have dengue and these patients were subjected to the rapid diagnostic test. A total of 2340 dengue-suspected patients visited these hospitals during the study period. Consent was obtained from 198 patients (Parsa n=127; Chitwan n=33; Rupandehi n=6; Janakpur n=24 and Kathmandu n=8) based on the available testing facilities in our laboratories. These patients underwent detailed clinical and laboratory investigations.
Ethical approval was taken from Nepal Health Research Council (NHRC), Kathmandu to conduct the study using human samples as well as transferring the non-infectious cDNA material to the collaborator's laboratory.
Collection of blood samples and serological assay: Blood samples (3 ml) were collected from the 198 patients and the serum was separated and stored in deep freezers at the respective hospitals. The samples were brought to our laboratory in cold-chain and stored further at -80° C. All these samples were further subjected to Dengue IgM capture ELISA (Panbio, Australia) according to the instructions given in the manufacturer's protocol. Panbio units were computed for each sample, and results were classified either as positive or negative.
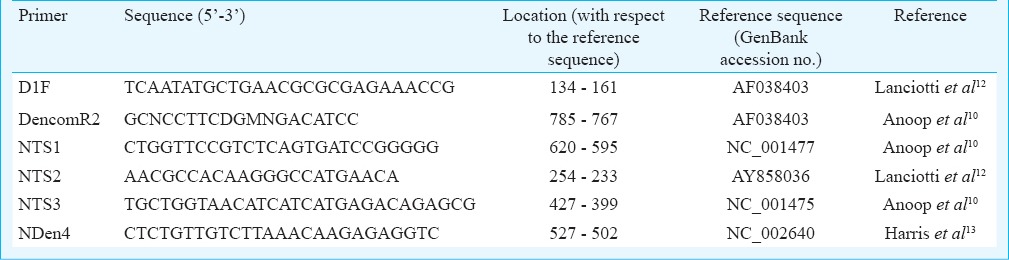
Viral RNA detection and serotype identification: Viral RNA was isolated from 52 samples using Nucleospin viral RNA isolation kit (MACHEREY-NAGEL, Germany) following the manufacturer's instructions. Dengue viral RNA detection was done by RT-PCR using the specific pair of primers D1F and Dencom R2 which amplified the region corresponding to the nucleotide positions 134-785 of the genome coding for the Core-Pre-membrane (C-PrM) region as described previously10,11 (Table I). For serotype identification, a semi-nested PCR assay was performed on the RT-PCR positive samples with the 1:10 diluted primary PCR product as the template and previously reported serotype-specific primers (D1F, NTS1, NTS2, NTS3 and NDen4; Table I)12 using GoTaq PCR Master mix (Promega, USA).
Table I.
Primers used for dengue virus confirmation and serotype-specific PCR
Nucleotide sequencing and phylogenetic analysis: For nucleotide sequencing, the C-prM region was PCR amplified using the D1F-DencomR2 primers from the first strand viral cDNA that was synthesized using AMV RT-reverse transcription system (Promega, USA). The PCR product was purified by Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare, USA). Both strands of the amplicon were directly sequenced using the specific primers D1F and DencomR2. The Big Dye Terminator kit (Applied Biosystems, USA) was used as per the manufacturer's instructions for sequencing and the reaction mix was analyzed in an ABI 3730 Genetic Analyzer automated DNA sequencer. The sequences were aligned using CLUSTAL W function of BioEdit 7.2.314 software package and the phylogenetic tree was constructed with the Maximum-likelihood (ML) method using MEGA 6 (version 6.0) software package (www.megasoftware.net/).
Results
Outbreak epidemiology and seasonality: From the disease affected areas, the number of patients with febrile illness demonstrated an unusual increase during the monsoon and post-monsoon period from July to December, 2013. The suspected dengue outbreak was found to originate in the Birgunj city of Parsa District during August which spread immediately to Bharatpur city of Chitwan District with a rapidly increasing number of cases during the first two weeks of September. The outbreak expanded further to Butwal city, the neighbouring district of Rupandehi in October. The outbreak later reached Janakpur, headquarter of Dhanusha district. Finally, the disease appeared in the highland districts of Kathmandu in mid-October. Of the total 2340 dengue-suspected cases presented in the six hospital study centres, the highest numbers were from Parsa district (1160) followed by Chitwan (n=490), Dhanusha (n=356), Rupandehi (n=178) and Kathmandu districts (n=156). The demographic distribution of the spread of the disease showed that it originated in the lower plains and later spread to the highlands of Nepal. The outbreak peaked in September and subsided around the second week of December (Fig. 1).
Fig. 1.
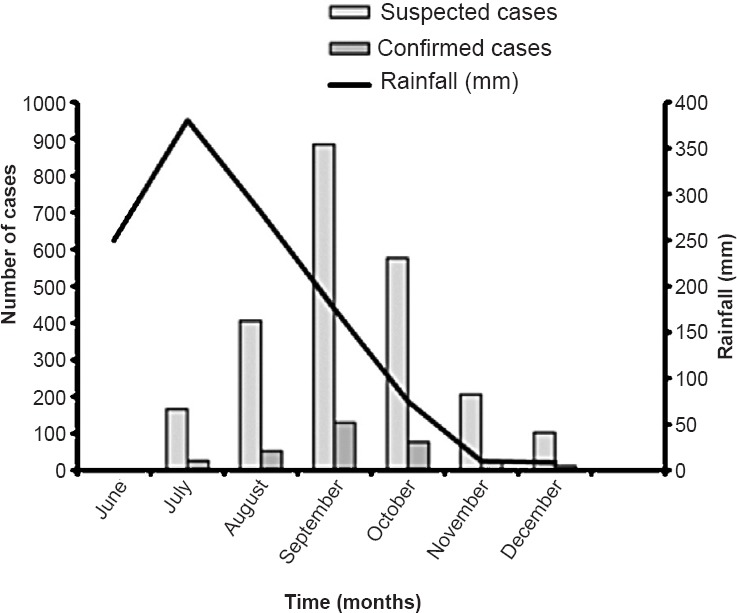
Seasonal distribution of dengue cases during the 2013 outbreak. The average rainfall figures were obtained from previous reports (http://www.weather-and-climate.com/average-monthly-Rainfall-Temperature-Sunshine, Katmandu, Nepal; accessed on April 15, 2014) and plotted in the graph.
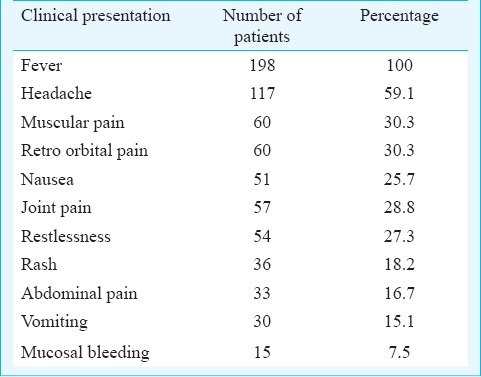
Clinical symptoms: Among the 198 patients analyzed in detail, males in the 15-50 yr age group predominated, with a child-adult ratio of 0.2:1 (33/165) and male-female ratio of 7:4. The mean age (± standard deviation) was 45.75±38.61 yr (age range 2-77 yr). Most of these patients had fever, headache, thrombocytopenia, joint pain, nausea, muscular pain and rashes (Table II) as the main symptoms. Some patients also had symptoms of restlessness, retro-orbital pain, rash, vomiting and abdominal pain; 147 (74.3%) patients had thrombocytopenia with < 100,000 platelets/ml of blood. Fifteen patients were found to have mucosal bleeding manifestation and were diagnosed to suffer from dengue with warning signs while the others were classified as dengue fever (DF) without warning signs.
Table II.
The common clinical manifestations observed in patients (n=198) during the outbreak
Anti-dengue IgM positivity: RDT positivity was 13.5 per cent among the 2340 serum samples analyzed in the hospitals. Among the 198 patients who were selected irrespective of the RDT status, 21.2 per cent were found to be positive in anti-dengue IgM capture ELISA in our laboratory.
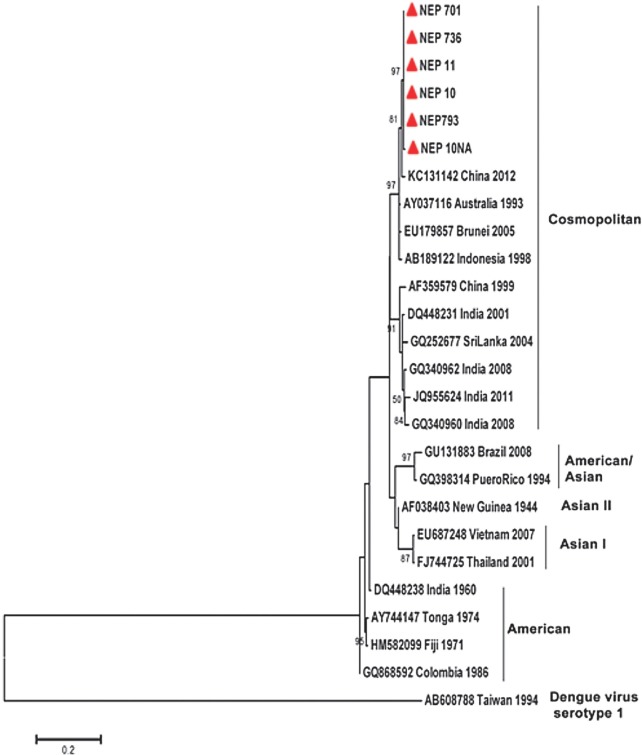
Dengue virus nucleic acid detection, serotype identification and phylogenetic analysis: Based on the clinical history and time of presentation in the hospital, only 52 of the 198 samples were in the early phase of the fever (1-4 days). These samples were used for viral RNA isolation and molecular studies. RT-PCR amplified the expected 652 bp amplicon from 15 samples (29%) indicating the presence of dengue viral RNA. Of these positive samples, 13 were from Parsa district and the remaining two were from Chitwan. A low PCR positivity could be attributed to the need for storage of samples and the possibility of freeze-thawing and RNA degradation while transportation from hospital to the laboratories. Serotype identification by semi-nested PCR showed an amplification of 119 bp confirming the virus to be of serotype 2. No case of concomitant infection with more than one serotype was observed. Phylogenetic analysis of the sequences of six of the samples (C-PrM region; 348nt, position 217-564 with respect to the reference sequence AF038403; GenBank Accession Nos. KP343870-KP343875) showed that they belonged to the Cosmopolitan genotype of DENV-2 sharing similarity to the strains isolated from Brunei and China (Fig. 2).
Fig. 2.
Phylogenetic analysis of the nucleotide sequences of the 348 bp C-PrM region of DENV-2 samples from Nepal (indicated with a red coloured triangle) and selected sequences from the GenBank. Maximum-likelihood (ML) tree was made with 1000 boot-strap replications with Kimura-2 parameter and Gamma-distributed (K2+G) settings as the substitution model. Dengue virus type 1 (AB608788) was used as the out-group.
Discussion
Over the years, the Terai belt of Nepal, located on the south of the outer foothills of Himalayan ranges, have become a dengue established region. The 2013 outbreak predominantly gripped this region, affecting the districts of Parsa, Chitwan, Rupandehi, Dhanusa and Kathmandu. The maximum number of positive dengue cases was detected in Parsa and Chitwan. Uncontrolled urbanization and the global climatic change permits the spread and breeding of Aedes mosquitoes in temperate zones in many parts of the world15,16. Establishment of these demographic and environmental factors in Kathmandu could facilitate continued persistence of the disease in the region.
The earlier occurrence of dengue cases in Nepal had been sporadic till 2010, when an epidemic occurred5. There were also reports of circulation of all the serotypes of the virus in the region in sporadic cases6. One of the major observations in the study was the involvement of DENV-2 in this outbreak. This observation also indicated a shift in serotype as against the earlier outbreak in 2010, which was caused by DENV-18. Such shifts in the circulating dengue virus serotype predispose the population to severe infections due to the phenomenon of antibody - dependent enhancement (ADE) along with the inherent virulence of the infecting strain17,18,19. A study using meta-analysis on the factors associated with severe dengue has identified primary vs. secondary infection and infection with DENV-2 as two factors that have positive correlation with dengue shock syndrome20. However, in the outbreak we studied, serious complications were minimal, which could be due to the inherent low virulence of the newer strain or the result of being primary infections. Looking into anti-dengue IgG positivity in the samples would have given a picture on the rate of past dengue infections. Along with serotype specific PCR, the sequencing of a small stretch of the genome has further confirmed the viral serotype, and phylogenetic study revealed the circulation of the Cosmopolitan genotype in the region.
In conclusion, we report here the involvement of dengue virus serotype 2 of the Cosmopolitan genotype in an outbreak in Nepal. A shift in the serotype might prove to be a major factor affecting the dengue spectrum in Nepal. The information generated will help in developing better control programmes to prevent the occurrence of severe forms of the disease in this country.
Acknowledgment
The authors acknowledge the Director, Rajiv Gandhi Center for Biotechnology (RGCB), India; Everest International Clinic, Kathmandu, for their support. This work was supported by funding from University Grants Commission (UGC), Nepal; institutional grant from Department of Biotechnology, Government of India and student fellowship from Department of Science and Technology (DST), Government of India.
Footnotes
Conflicts of Interest: None.
References
- 1.Pinheiro FP, Corber SJ. Global situation of dengue and dengue haemorrhagic fever, and its emergence in the Americas. World Health Stat Q. 1997;50:161–9. [PubMed] [Google Scholar]
- 2.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–96. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halstead SB, O’Rourke EJ. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J Exp Med. 1977;146:201–17. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takasaki T, Kotaki A, Nishimura K, Sato Y, Tokuda A, Lim CK, et al. Dengue virus type 2 isolated from an imported dengue patient in Japan: first isolation of dengue virus from Nepal. J Travel Med. 2008;15:46–9. doi: 10.1111/j.1708-8305.2007.00165.x. [DOI] [PubMed] [Google Scholar]
- 5.Pandey BD, Morita K, Khanal SR, Takasaki T, Miyazaki I, Ogawa T, et al. Dengue virus, Nepal. Emerg Infect Dis. 2008;14:514–5. doi: 10.3201/eid1403.070473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malla S, Thakur GD, Shrestha SK, Banjeree MK, Thapa LB, Gongal G, et al. Identification of all dengue serotypes in Nepal. Emerg Infect Dis. 2008;14:1669–70. doi: 10.3201/eid1410.080432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gautam I, Dhimal MN, Shrestha SR, Tamrakar AS. First record of Aedes aegypti (L.) vector of dengue virus from Kathmandu, Nepal. J Nat Hist Mus. 2009;24:154–64. [Google Scholar]
- 8.Pandey BD, Nabeshima T, Pandey K, Rajendra SP, Shah Y, Adhikari BR, et al. First isolation of dengue virus from the 2010 epidemic in Nepal. Trop Med Health. 2013;41:103–11. doi: 10.2149/tmh.2012-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nathan MB, Dayal-Drager R, Guzman M. Dengue: guidelines for diagnosis, treatment, prevention and control. Geneva: World Health Organization; 2009. Epidemiology, burden of disease and transmission; pp. 3–21. [Google Scholar]
- 10.Anoop M, Issac A, Mathew T, Philip S, Kareem NA, Unnikrishnan R, et al. Genetic characterization of dengue virus serotypes causing concurrent infection in an outbreak in Ernakulam, Kerala, South India. Indian J Exp Biol. 2010;48:849–57. [PubMed] [Google Scholar]
- 11.Anoop M, Mathew AJ, Jayakumar B, Issac A, Nair S, Abraham R, et al. Complete genome sequencing and evolutionary analysis of dengue virus serotype 1 isolates from an outbreak in Kerala, South India. Virus Genes. 2012;45:1–13. doi: 10.1007/s11262-012-0756-3. [DOI] [PubMed] [Google Scholar]
- 12.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–51. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris E, Robers TG, Smith L, Selle J, Kramer LD, Valle S, et al. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J Clin Microbiol. 1998;36:2634–9. doi: 10.1128/jcm.36.9.2634-2639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- 15.Rochlin I, Ninivaggi DV, Hutchinson ML, Farajollahi A. Climate change and range expansion of the Asian tiger mosquito (Aedes albopictus) in Northeastern USA: Implications for public health practitioners. PLoS One. 2013;8:e60874. doi: 10.1371/journal.pone.0060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roiz D, Neteler M, Castellani C, Arnoldi D, Rizzoli A. Climatic factors driving invasion of the tiger mosquito (Aedes albopictus) into new areas of Trentino, Northern Italy. PLoS One. 2011;6:e14800. doi: 10.1371/journal.pone.0014800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, et al. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230:244–51. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- 18.Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, et al. Selection-driven evolution of emergent dengue virus. Mol Biol Evol. 2003;20:1650–8. doi: 10.1093/molbev/msg182. [DOI] [PubMed] [Google Scholar]
- 19.Yamanaka A, Mulyatno KC, Susilowati H, Hendrianto E, Ginting AP, Sary DD, et al. Displacement of the predominant dengue virus from type 2 to type 1 with a subsequent genotype shift from IV to I in Surabaya, Indonesia 2008-2010. PLoS One. 2011;6:e27322. doi: 10.1371/journal.pone.0027322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huy NT, Van Giang T, Thuy DH, Kikuchi M, Hien TT, Zamora J, et al. Factors associated with dengue shock syndrome: a systematic review and meta-analysis. PLoS Negl Trop Dis. 2013;7:e2412. doi: 10.1371/journal.pntd.0002412. [DOI] [PMC free article] [PubMed] [Google Scholar]