Sir,
Chikungunya virus (CHIKV) infection is an Aedes mosquito borne illness caused by an Alphavirus of the family Togaviridae1. CHIKV was first isolated from Tanzania, Africa, in 19532. Since then it has caused numerous outbreaks in continental Africa, the Indian Ocean region, and Southeast Asia. The first outbreak of CHIKV in India was reported in 1963 in Kolkata followed by epidemics in Chennai, Puducherry and Vellore in 1964; Vishakapatnam, Rajahmudry, Kakinada, Nagpur in 1965; and Barsi in 19733. Subsequently, in the absence of either active or passive surveillance it seemed that the virus had disappeared from the country until the end of 20054.
Several States in India experienced massive outbreaks of CHIK infection during 2005-2006. Andhra Pradesh, the most affected State was first to report the CHIKV epidemic in December 2005 in India5. Several districts of Karnataka State have also recorded large number of CHIKV related fever cases3,6. The outbreak continued with reports of a large number of cases from several other States (Rajasthan, Gujarat, Tamil Nadu, Orissa and Madhya Pradesh)6. A change in the CHIKV genotype, enhanced efficiency of mosquitoes to transmit the virus, an immunologically naive population, rapid means of trade and travel, global warming and lack of an efficient public health system are some of the important factors that influenced the explosive re-emergence of CHIKV7.
In Andhra Pradesh, though the outbreak was controlled by the end of 2006, sporadic cases continued to be reported from different districts in the following years8,9,10. During September-October 2013, a massive outbreak (with >1000 cases) was reported from Guntur district, with CHIK like symptoms. The present study was conducted to confirm the CHIKV infection among suspected patients using serology and molecular techniques.
A total of 1905 people from a population of 37,439 were affected with fever and poly-arthralgia in Thurakaplem, Dasaripalem, Rayapudi, Chintamotu, Thurupupslem, Donepodi, Vellataru, Kollaplem, Bhattiprolu, Addepalli, Illavarem and Pesarlanka villages of Guntur district. To investigate the causative agent, 60 serum samples (2 ml each) were selected randomly to represent all the above villages, transported to National Institute of Virology (NIV), Pune, on dry ice and screened for CHIKV using IgM enzyme linked immunosorbent assay (ELISA) (NIV kit), nested RT-PCR (reverse transcriptase-polymerase chain reaction) and real time PCR. The age of the patients from whom the samples were collected ranged from 5-65 yr. CHIKV IgM test was carried out as per manufacturer's instructions. For nested RT-PCR and real time PCR, RNA was isolated using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) as per manufacturer's instructions. Superscript II (Invitrogen, USA) was used for reverse transcription (42°C for 1 h). Nested RT-PCRs targeting the E1 and NSP 3 genes were carried out as described earlier3. Nested RT-PCR targeting the E3 gene was also done by using in-house designed primers: (F1,5’- CAG ATA CCC GTG CAC ATG AAGT-3’ and R1, 5’- TGA GCT AAG TAT GGT CTT GT-3’) that produced a 534 base pairs (bp) fragment and (F 2, 5’-CAG ACC GAT CTT CGA CAA CA-3’ and R 2, 5’-TCA TGA CGT TGT CCT CAA GC-3’) that produced a 271-bp product. Cycling conditions were one cycle at 94°C for 5 min; 35 cycles each of 94°C (1 min), 47°C (1 min), and 72°C (1 min); followed by final extension for 7 min at 72°C. The products were visualized on 1.2 per cent agarose gel11. Standard CHIKV from NIV repository (African genotype, Strain No. 061573; Andhra Pradesh 2006; Accession Number EF027134) was used as positive control and phosphate buffer saline (PBS) as negative control to compare the results.
Viral load in the serum samples was determined by real time-PCR targeting the E3 structural protein region using the standard curve method11,12,13. One step real time RT-PCR was performed in 25 μl reaction mixture containing 5μl RNA, 12.5μl TaqMan One-Step RT-PCR 2X Master Mix, 1 μl 40X (RT + RNAasin) (Applied Biosystems, USA) each 1 μl sense (μM), 1 μl anti-sense (μM) primer and 1μl TaqMan probe. Real-time one step RT-PCR was performed in a 96-well format using 7300 real time PCR system and SDS software V 1.0.2 (Applied Biosystems). The amplification programme included: reverse transcription at 48°C for 30 min, initial denaturation at 95°C for 10 min, and 50 cycles of denaturation (95°C for 15 sec) and annealing and extension (60°C for 1 min)11,12,13.
Samples were also processed for virus isolation3 but virus could not be isolated. Nucleotide sequencing was performed for the partial E1 gene of four clinical samples. The PCR products were purified by using QIAquickPCR Purification Kit (Qiagen) and sequenced using Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, USA) in an automatic sequencer (ABI PRISM 3100 Genetic Analyzer, Applied Biosystems). Multiple sequence alignment of the nucleotide sequences of the E1 gene was performed using Clustal X, version 1.8314.
Out the 60 samples (35 female and 25 males) processed, 42 (70%, 25 females and 17 males) were found to be positive for IgM antibodies. Age-wise distribution of CHIK fever cases did not vary significantly among the age groups starting from 5 to 65 in the affected areas (Fig. 1). All IgM positive samples were from 3-30 days post onset day (POD). Twelve samples were positive for nested RT-PCR (50% were IgM positive for CHIKV antibodies, POD between 2-9 days) targeting the NSP3 gene and six were positive for E3 gene region (all were negative for CHIKV antibodies, POD between 2-4 days) (Table, Fig. 2A, B). Only three samples (<3POD) which were IgM negative and nested RT-PCR positive were found to be positive by real time PCR. RNA load for positive samples was in the range of 104-105 copies/ml. A total of 12 samples were found to be negative for IgM antibodies, nested RT-PCR and real time PCR.
Fig. 1.
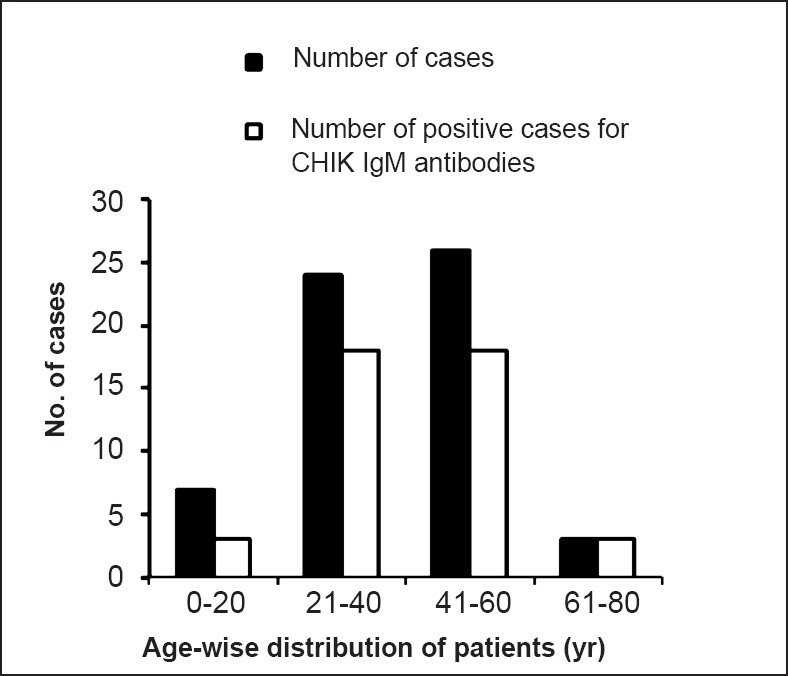
Age-wise distribution of samples from cases and IgM positive status.
Table.
Details of positive samples confirmed by nested RT- PCR and real time PCR
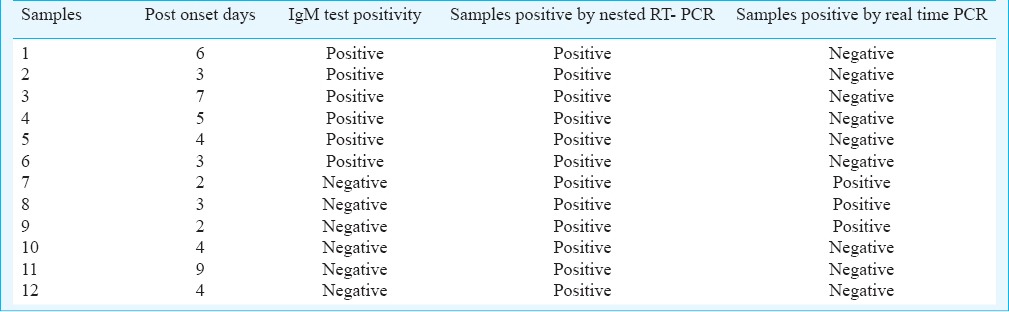
Fig. 2.
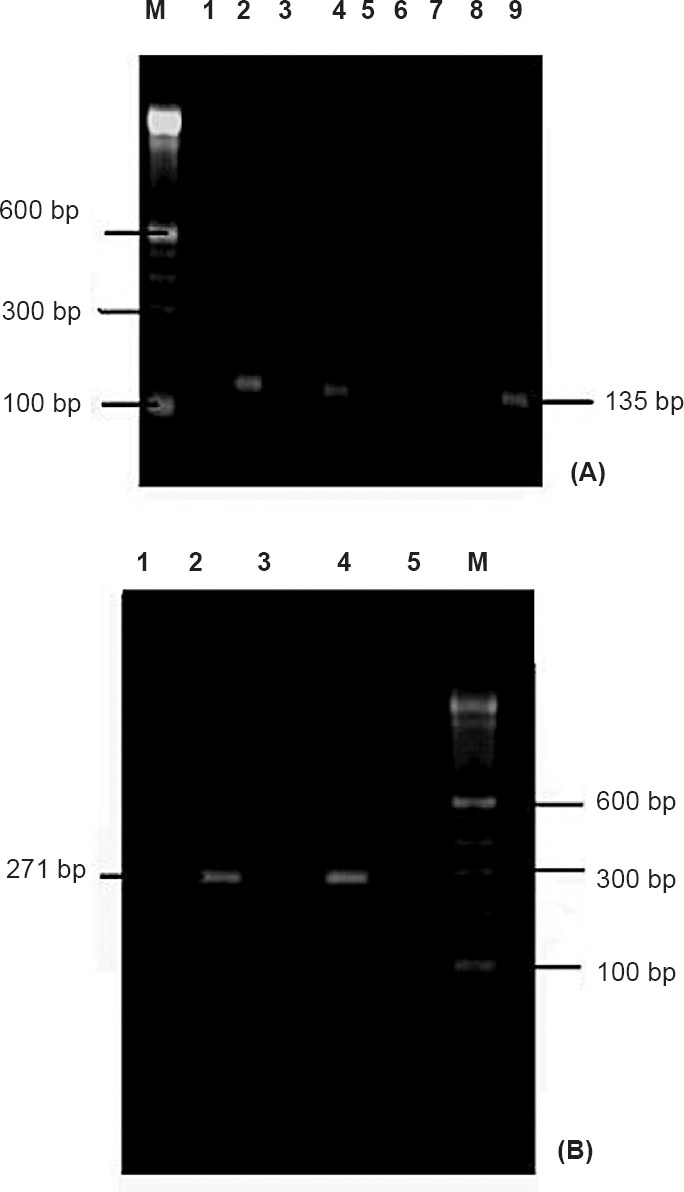
(A). Nested RT-PCR of samples targeting NSP 3 gene region; Lane M: Molecular weight marker, Lanes 1, 3, 5-8: Negative control, Lanes 2, 4: Chikungunya positive samples, Lane 9: Positive control. (B). Nested RT-PCR of samples targeting E-3 gene region; Lane M: Molecular weight marker, Lanes 1, 3, 5: Negative control, Lane 2: Chikungunya positive samples, Lane 4: Positive control.
CHIKV outbreaks of the Asian genotype have been reported from Vijaywada, Rajahmundry, Visakhapatanam, etc. in Andhra Pradesh during 1964-1965 and re-emerged as the East Central South African (ECSA) genotype in 2005-2006 in an explosive form affecting 23 districts with approximately 77,533 cases15,16. Since then, sporadic cases have been reported from different places of Andhra Pradesh3,5,8,9,16,17,18,19,20,21,22. The present investigation showed 42 IgM positive cases out of the 60 samples (70% positivity) tested though all the patients had acute CHIK like symptoms such as high fever and polyarthralgia resulting in restricted movements. The IgM positivity in the serum samples demonstrated recent CHIKV infection in the area. However, using molecular methods, only a few samples were found positive as nested RT-PCR and real time PCR tests yielded 12 and 3 positives, respectively. This may be attributed to the high PODs of the samples as rate of detection decreases with increase in PODs. This result confirms the role of CHIKV in an outbreak form during the post monsoon season in Guntur district. Post monsoon CHIKV outbreak have also been reported from other parts of the country23,24,25.
It is suggested that IgM antibodies develop after two days of the infection with CHIKV and in chronic cases virus persists upto three months26. Our results also support this as 42 samples were found to be positive for IgM antibodies. In the present study, positivity by both nested RT-PCR and real time PCR was found to be low in comparison with IgM antibody detection. Based on our earlier report11, the lower sensitivity that we observed in case of real time PCR in this study in comparison to nested RT-PCR was not expected. There might have been chances of RNA degradation as here the real time PCR was performed for those cases which were positive by nested RT- PCR.
Sequence analysis of the partial E1 gene of four samples identified the CHIKV strain as belonging to the currently circulating ECSA genotype and having an alanine residue at position 226 of the E1 protein (E1:226A). Notably none of the CHIKV isolates from Andhra Pradesh during 2005-2007 had A226V mutation that has been reported to provide a fitness advantage to Ae. albopictus mosquitoes6,27. Earlier studies based on the structural polyprotein gene have shown that the CHIKV isolates obtained from 2009-2010 outbreaks in Tamil Nadu and Andhra Pradesh formed a separate clade within the ECSA lineage with a few specific mutations including a novel E1-K211E mutation22. Several mutations in the CHIKV genome have been reported since 2005 that enable the virus to adapt to new vectors or help enhance the transmission of the virus28. The changes (M269V, D284E, and V322A) as noted in other CHIKV strains6,29,30 were also observed in our CHIKV sequences. However, no unique mutation was observed in the partial E1 gene sequenced in this study. There may be a need for full E1 gene sequencing rather than partial gene sequencing along with sequencing of the structural genes. One of the other limitations of our study was the inability to isolate the CHIKV from serum samples. Improper maintenance of cold chain during collection and transportation of the samples could be the most probable reason.
Overall, the present study provided evidence for a CHIKV outbreak in Guntur district of Andhra Pradesh. This outbreak of CHIKV in Guntur district along with the prior ones since 2005-2006 demonstrates the endemicity of the State to the CHIKV infection. Surveillance of vectors and their breeding habitats and effective control measures are needed to contain spread of the virus. Comprehensive efforts from the health authorities along with community participation in source reduction, anti-larval and mosquito management as well as diagnostic facilities for early detection are warranted to control the disease.
Acknowledgment
The authors acknowledge financial support provided by the Indian Council of Medical Research (ICMR), Ministry of Health and Family Welfare, Government of India. Authors thank Drs A.B. Sudeep and Sarah Cherian for reviewing the manuscript.
References
- 1.Sudeep AB, Parashar D. Chikungunya: an overview. J Biosci. 2008;33:443–9. doi: 10.1007/s12038-008-0063-2. [DOI] [PubMed] [Google Scholar]
- 2.Ross RW. The Newala epidemic. III. The virus: isolation, pathogenic properties and relationship to the epidemic. J Hyg (Lond) 1956;54:177–91. doi: 10.1017/s0022172400044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yergolkar PN, Tandale BV, Arankalle VA, Sathe PS, Sudeep AB, Swati SG, et al. Chikungunya outbreaks caused by African genotype, India. Emerg Infect Dis. 2006;12:1580–3. doi: 10.3201/eid1210.060529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lum FM, Teo TH, Lee WWL, Kam YW, Renia L, Ng LF. An essential role of antibodies in the control of Chikungunya virus infection. J Immunol. 2013;190:6295–302. doi: 10.4049/jimmunol.1300304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravi V. Re-emergence of Chikungunya virus in India. Indian J Med Microbiol. 2006;24:83–4. doi: 10.4103/0255-0857.25175. [DOI] [PubMed] [Google Scholar]
- 6.Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, et al. Genetic divergence of chikungunya viruses in India (1963-2006) with special reference to the 2005-2006 explosive epidemic. J Gen Virol. 2007;88:1967–76. doi: 10.1099/vir.0.82714-0. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia R, Narain JP. Re-emerging chikungunya fever: some lessons from Asia. Trop Med Int Health. 2009;14:940–6. doi: 10.1111/j.1365-3156.2009.02312.x. [DOI] [PubMed] [Google Scholar]
- 8.Naresh Kumar CVM, Sangamithra P, Rajasekhar M, Saigopal DVR. Surveillance of chikungunya virus in Andhra Pradesh, Southern India. Asian Pacific J Trop Med. 2010;38:60–5. [Google Scholar]
- 9.Sai Gopal DVR, Kumar N. Isolation and diagnosis of Chikungunya virus causing outbreaks in Andhra Pradesh, India. J Clin Sci Res. 2013;2:2–7. [Google Scholar]
- 10.Muniaraj M. Fading chikungunya fever from India: beginning of the end of another episode? Indian J Med Res. 2014;139:468–70. [PMC free article] [PubMed] [Google Scholar]
- 11.Parashar D, Paingankar MS, Sudeep AB, More A, Shinde SB, Arankalle VAA. Evaluation of in house developed laboratory diagnostic techniques for chikungunya. Curr Sci. 2014;107:2011–3. [Google Scholar]
- 12.Parashar D, Paingankar MS, Kumar S, Gokhale MD, Sudeep AB, Shinde SB, et al. Administration of E2 and NS1 siRNAs inhibit chikungunya virus replication in vitro and protects mice infected with the virus. PLoS Neg Trop Dis. 2013;7:e2405. doi: 10.1371/journal.pntd.0002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cecilia D, Kakade M, Alagarasu A, Patil J, Salunke A, Parashar D, et al. Development of multiplex real time RT PCR assay for simultaneous detection of dengue and chikungunya viruses. Arch Virol. 2014;160:323–7. doi: 10.1007/s00705-014-2217-x. [DOI] [PubMed] [Google Scholar]
- 14.Thomson JD, Gibson TJ, Plewniank F, Jeanmougin F, Higgins DG. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Vector Borne Disease Control Programme (NVBDCP), “Chikungunya cases,”. 2013. [accessed on October 14, 2014]. Available from: http://nvbdcp.gov.in/chik-cd.html .
- 16.Naresh Kumar CVM, Johnson AMA, Sai Gopal DVR. Molecular characterization of chikungunya virus from Andhra Pradesh, India & phylogenetic relationship with Central African isolates. Indian J Med Res. 2007;126:534–40. [PubMed] [Google Scholar]
- 17.Dash PK, Parida MM, Santhosh SR, Verma SK, Tripathi NK, Ambuj S, et al. East Central South African genotype as the causative agent in reemergence of Chikungunya outbreak in India. Vector Borne Zoonotic Dis. 2007;7:519–27. doi: 10.1089/vbz.2007.7272. [DOI] [PubMed] [Google Scholar]
- 18.Kumar NC, Nadimpalli M, Vardhan VR, Gopal SD. Association of ABO blood groups with Chikungunya virus. Virol J. 2010;7:140. doi: 10.1186/1743-422X-7-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murty US, Dhanwada N, Arora N. Chikungunya outbreak in Kurnool district, Andhra Pradesh. Internet J Health. 2007;6:8. [Google Scholar]
- 20.Mohan A. Chikungunya fever: clinical manifestations & management. Indian J Med Res. 2006;124:471–4. [PubMed] [Google Scholar]
- 21.Lakshmi V, Neeraja M, Subbalaxmi MV, Parida MM, Dash PK, Santhosh SR, et al. Clinical features and molecular diagnosis of chikungunya fever from South India. Clin Infect Dis. 2008;46:1436–42. doi: 10.1086/529444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sumathy K, Ella KM. Genetic diversity of Chikungunya virus, India 2006-2010: evolutionary dynamics and serotype analyses. J Med Virol. 2012;84:462–70. doi: 10.1002/jmv.23187. [DOI] [PubMed] [Google Scholar]
- 23.Santhosh SR, Dash PK, Parida MM, Khan M, Tiwari M, LakshmanaRao PV. Comparative full genome analysis revealed E1: A226V shift in 2007 Indian Chikungunya virus isolates. Virus Res. 2008;135:36–41. doi: 10.1016/j.virusres.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Dwibedi N, Mohapatra MK, Beuria MK, Kerketta AS, Sabat J, Kar SK, et al. Emergence of Chikungunya virus infection in Orissa, India. Vector Borne Zoo Dis. 2010;10:347–54. doi: 10.1089/vbz.2008.0190. [DOI] [PubMed] [Google Scholar]
- 25.Barve S, Nanda S, Javadekar TB. Chikungunya fever: The resurgence and epidemiological pattern in Western Indian. Natl J Med Res. 2013;3:159–61. [Google Scholar]
- 26.Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis. 2007;7:319–27. doi: 10.1016/S1473-3099(07)70107-X. [DOI] [PubMed] [Google Scholar]
- 27.Cherian SS, Walimbe AM, Jadhav SM, Gandhe SW, Hundekar SL, Mishra AC, et al. Evolutionary rates and timescale comparison of Chikungunya viruses inferred from the whole genome/E1 gene with special reference to the 2005-07 outbreak in the Indian subcontinent. Infec Gen Evol. 2009;9:16–23. doi: 10.1016/j.meegid.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Das B, Sahu A, Das M, Patra A, Dwibedi B, Kar SK, et al. Molecular investigations of chikungunya virus during outbreaks in Orissa, Eastern India in 2010. Infect Genet Evol. 2012;12:1094–101. doi: 10.1016/j.meegid.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Singh RK, Tiwari S, Mishra VK, Tiwari R, Dhole TN. Molecular epidemiology of Chikungunya virus: mutation in E1 gene region. J Virol Methods. 2012;185:213–20. doi: 10.1016/j.jviromet.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean Outbreak. PLoS Med. 2006;3:e263. doi: 10.1371/journal.pmed.0030263. [DOI] [PMC free article] [PubMed] [Google Scholar]


