Abstract
Background & objectives:
Although having immense clinical relevance, yet only a few studies have been targeted to understand the chikungunya virus (CHIKV) susceptibility and growth in Aedes aegypti populations from India. This study was undertaken to investigate CHIKV susceptibility and growth kinetics in Ae. aegypti along with genetic heterogeneity of Ae. aegypti populations.
Methods:
Dose dependent CHIKV susceptibility and growth kinetic studies for three CHIKV strains reported from India were carried out in Ae. aegypti mosquito populations. The phenotypic variation and genetic heterogeneity in five Ae. aegypti populations were investigated using multivariate morphometrics and allozyme variation studies.
Results:
The dissemination and growth kinetics studies of the three CHIKV strains showed no selective advantage for a particular strain of CHIKV in Ae. aegypti. At 100 per cent infection rate, five geographic Ae. aegypti populations showed differences in dissemination to three CHIKV strains. Morphometric studies revealed phenotypic variation in all the studied populations. The allelic frequencies, F statistics, and Nei's genetic identity values showed that genetic differences between the populations were small, but significant.
Interpretation & conclusions:
The results obtained in this study suggest that genetic background of the vector strongly influences the CHIKV susceptibility in Ae. aegypti.
Keywords: Chikungunya virus, F statistics, genetic heterogeneity, morphometry analysis
Several environmental, physiological and genetic factors are known to govern the vector competence of mosquitoes1,2. Susceptibility to infection, permissiveness for pathogen development, duration of incubation period and transmission efficiency contribute to vector competence. To establish the successful infection in vector, virus has to overcome numerous barriers to infection and dissemination within the mosquito, many of which are under genetic control1,2. All these factors are known to influence the association between the vector, the pathogen transmitted by the vector and the vertebrate host into which the pathogen is transmitted1.
Chikungunya virus (CHIKV) is endemic in Africa and Southeast Asia and is transmitted by Aedes mosquitoes through an urban or sylvatic transmission cycle3. Three distinct CHIKV phylogenetic groups viz. one containing all the isolates from West Africa, one containing the isolates from Asia, and one corresponding to Eastern, Central and Southern African (ECSA) isolates, have been reported4,5,6,7. A variant of CHIKV harbouring a substitution in the residue of the E1 glycoprotein (E1-226V) has been demonstrated to be efficiently transmitted by the Aedes albopictus8,9.
The susceptibility of Aedes mosquitoes to CHIKV infection varies widely among individual mosquitoes and between the mosquito populations10,11,12. The genetic heterogeneity in the mosquito populations and in the virus strains might be crucial for differential susceptibility of vectors. Only a few in depth studies on CHIKV susceptibility and growth kinetics in Ae. aegypti and the genetic heterogeneity in Ae. aegypti mosquito populations have been reported in literature9,11. We, therefore, investigated the susceptibility, dissemination and growth kinetics of three CHIKV strains in Ae. aegypti and genetic variability in five different Ae. aegypti populations.
Material & Methods
The experiments were done in a biosafety level-2 animal facility at the National Institute of Virology, Pune, India. The study protocol was approved by the Institutional Animal Ethics Committee (IAEC) and Institutional Biosafety Committee (IBSC).
Mosquito collection and study sites: The sites (Alappuzha, Gorakhpur, Jalgaon, Tirupati and Surat) were visited during outbreaks and Ae. aegypti survey was conducted in and around the patients’ houses. Adults and larvae of Ae. aegypti were collected from Alappuzha (Kerala State, India, Collection Date: October 2010), Gorakhpur (Uttar Pradesh State, India, Collection Date: October 2010), Jalgaon (Maharashtra State, India, Collection Date: January 2010), Surat (Gujarat State, India, Collection Date: December 2009) and Tirupati (Andhra Pradesh State, India, Collection Date: March 2010). All available indoor water storage containers present in patients’ house and nearby houses were examined for presence of Ae. aegypti larvae. Mosquito larvae were examined in all available indoor water storage containers present in the houses by netting (four times per container). The containers were classified into high prolific breeding (more than 50 larvae in 4 collection attempts) and low prolific breeding sites (less than 10 larvae in 4 collection attempts). Among the larvae positive containers examined, sample collections were done from six-eight most prolific breeding container sites, harbouring large number of Ae. aegypti larvae. To establish the mosquito colony and morphometric analysis, collection sites from study areas were selected in such a way that the distance between the two sites was more than 500 m. Based on data obtained from Health Departments of these areas, the collection sites were broadly classified into three categories viz. (i) Frequent cases (Alappuzha, Jalgaon, Tirupati), (ii) High number of cases (Surat), and (iii) Rare cases (Gorakhpur). Mosquitoes were identified and colonies were established. Field collected larvae were used for the morphometric analysis. Colonies of these populations were maintained in the standard laboratory conditions at 28±1°C, 70±5 per cent relative humidity (RH) and light: dark (LD) 12:12 h cycle. Mosquitoes of Filial generation (FG) 2 to FG6 were used for CHIKV susceptibility (FG2-FG5) and allozyme studies (FG2 and FG3).
Chikungunya virus strains: CHIKV strains were passaged twice in Vero E-06 (VE-06) cells. VE-06 cells were used for the propagation of CHIKV stock. VE-06 cells were infected with CHIKV strains; CHIKV (A226) (MOI 5, African genotype, Strain No. 061573; Andhra Pradesh 2006; Accession Number EF027134), CHIKV (Asian) [MOI 5, Asian genotype, Strain No. 634029, Calcutta (now Kolkata) 1963; Accession Number EF027140] and CHIKV (A226) (MOI 5, African genotype with A226V mutation in E1 protein, Strain No. 74831, Kerala 2007; Accession Number FJ000069) and CHIKV amplified for four days. Virus titre was determined using real time PCR13 (CHIKV A226 9.16 ×108 RNA copies/ml; CHIKV A226V 1.25×109 RNA copies/ml; CHIKV Asian 9.65×108 RNA copies/ml).
Oral infection of mosquitoes: Infection assays were performed with 4-6 days old female mosquitoes. Mosquitoes were allowed to feed for one h through a goat intestine membrane covering the base of a glass feeder containing the blood-virus mixture maintained at 37°C.
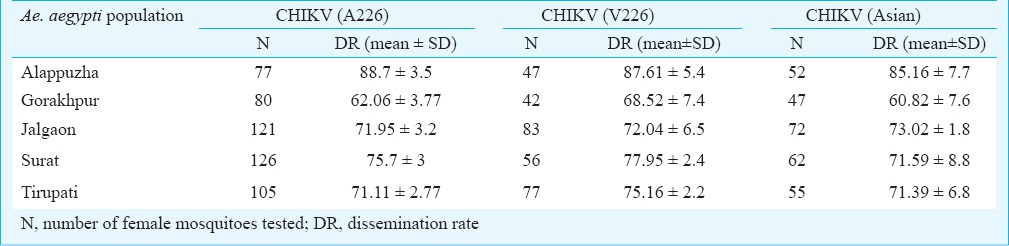
CHIKV susceptibility and dissemination: The mosquitoes (Ae. aegypti) were exposed to the 10-fold dilution of viral stocks. The CHIKV suspension was diluted in goat blood to obtain the infectious blood meal with different CHIKV titres (5.08×105 RNA copies/ml, 5.08×106 RNA copies/ml, 5.08×107 RNA copies/ml and 5.08×108 RNA copies/ml). The presence of CHIKV antigen in head squashes, salivary gland, fat bodies of individual mosquito was evaluated by indirect immuno fluorescence assay (IFA)13 after seven days post-infection (p.i.). For virus susceptibility and dissemination experiments, two independent experiments were performed. (The number of female mosquitoes tested for different CHIKV strains and titres is given in parenthesis of Fig. 1). The batches of each mosquito population (Jalgaon, Surat, Alappuzha, Tirupati and Gorakhpur) were exposed to each CHIKV strain (CHIKV A226, CHIKV V226 and CHIKV Asian, viral titres of 5.08×107 RNA copies/ml) and processed similarly for determination of dissemination rates. The number of female mosquitoes tested for each Ae. aegypti population and for each CHIKV strain is given in Table I.
Fig. 1.
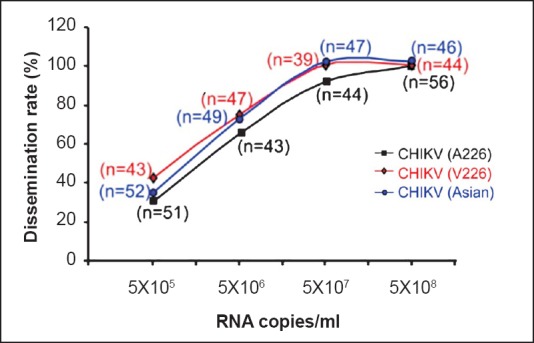
Dissemination rates of Aedes aegypti infected with CHIKV (A226), (V226) and (Asian) strains at different viral titres. The data of two independent experiments were pooled and the number of females tested is given in the parenthesis (n).
Table I.
Dissemination rates of different Aedes aegypti populations for three different CHIKV trains
Quantification of CHIKV in whole mosquito: Ae. aegypti mosquitoes were orally fed with CHIKV (A226) (5.08×107 RNA copies/ml), CHIKV (V226) (5.68×107 RNA copies/ml) and CHIKV (Asian) (5.70×107 RNA copies/ml), and ten mosquitoes were sampled everyday starting from day 1 p.i. to day 7 p.i. The number of CHIKV RNA copies in individual mosquitoes was estimated using qPCR13. Ten mosquitoes were sacrificed every day till day 7 p.i. Individual mosquitoes were homogenized in T10E5 (10 mM Tris-Cl pH8, 5 mM EDTA pH8) and supernatants were collected by centrifugation at 10000g, 4°C for five min. RNA from mosquitoes was extracted using QIAmp viral RNA minikit (QIAGEN, USA) following the manufacturer's instructions. One step qPCR was performed according to procedures described earlier13.
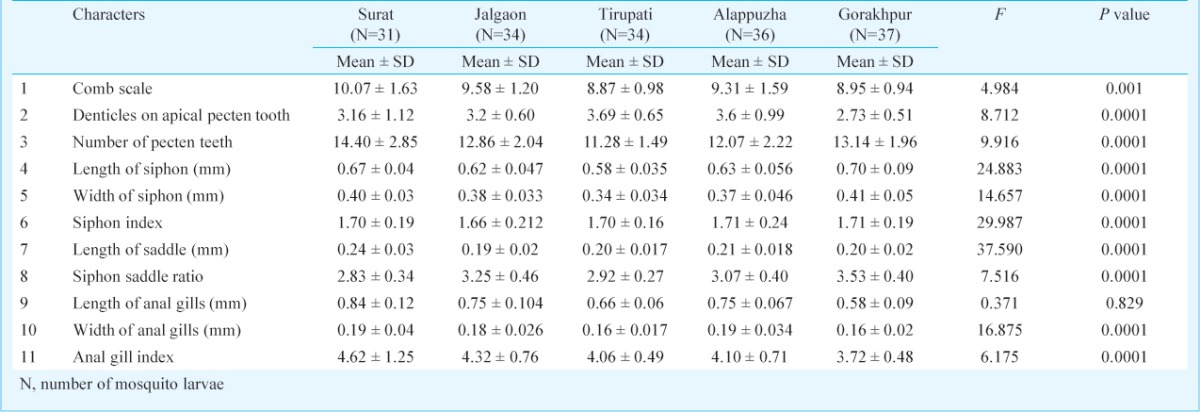
Morphological analysis: Field collected Ae. aegypti larvae were mounted in Hoyer's solution (gum arabic 15 g, chloral hydrate 75 g, distilled water 25 ml, Glycerine 5 ml). Eight morphological characters and three ratios (Siphon index, anal gill index and siphon saddle ratio) were scored for morphological analysis of field collected fourth instar larvae (Table II). The characters of fourth instar larvae were measured using micrometric oculars with the least count 0.01 mm (Leica, Germany and Olympus, Japan).
Table II.
Results of multivariate analysis of variance (MANOVA) for comparing means of morphological characters in four populations of Aedes aegypti
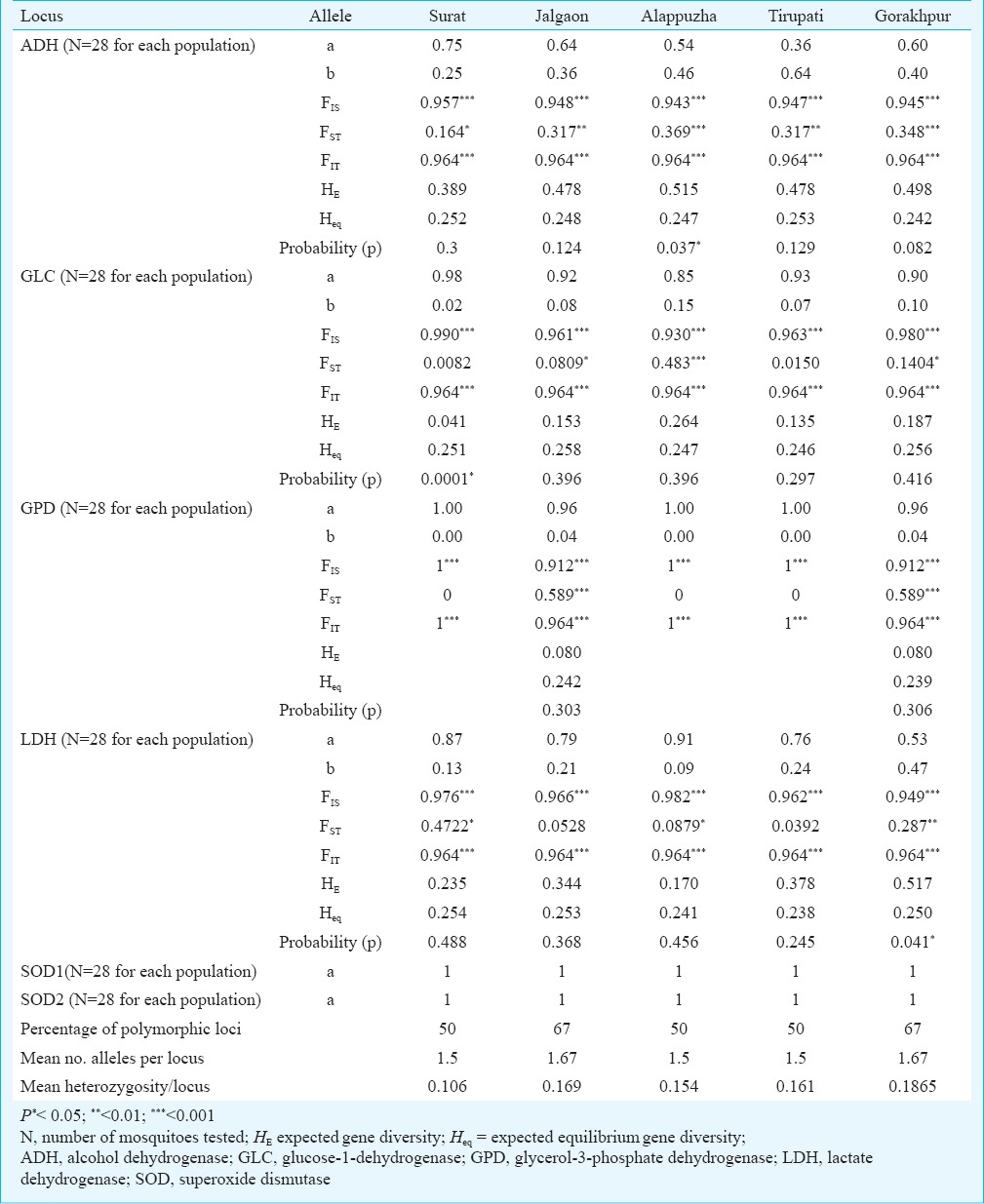
Electrophoresis and detection of enzyme activity: Adult mosquitoes (2-3 days old, randomly collected male/female mosquitoes) from each population were used for genetic analysis (n=28 for each locus; 140 individuals from each population). Electrophoresis was performed according to procedures described earlier14. Staining was carried out for five different allozymes viz., alcohol dehydrogenase (ADH), glucose 1-dehydrogenase (GLC), glycerol 3-phosphate dehydrogenase (NAD+) (GPD), lactate dehydrogenase (LDH), and superoxide dismutase (SOD)15 (Table III).
Table III.
Allelic frequencies and F statistics for the five populations of Ae. aegypti populations from India
Analysis of data
Determination of midgut infection and dissemination rates - Midgut infection rate (MIR) was determined as the number of midguts containing CHIKV antigen divided by the number of midguts examined. The dissemination rate (DR) was determined as the number of mosquitoes with detectable CHIKV antigen in non-midgut tissues (e.g. head squash tissues, salivary glands, fat body, etc.), divided by the number of mosquitoes with detectable virus antigen in the midgut. For each CHIKV strain, disseminated infection rates were compared using a χ2 test, the Fisher's exact test being used in the case of small sample sizes.
Analysis of CHIKV growth and dissemination in Ae. aegypti mosquitoes - For comparison between virus genotype and CHIKV titres at each post infection day in Ae. aegypti mosquitoes, we performed two-way factorial analysis of covariance (ANCOVA) with interactions. The normality of dissemination data was checked using Jarque-Bera test. The homogeneity of comparison groups was tested using Leven's test of homogeneity of variance. The dissemination rates of three CHIKV strains in five populations of Ae. aegypti were compared using a two-way analysis of variance (ANOVA) with Games-Howell post hoc test.
Analysis of morphological data - Multivariate ANOVA (MANOVA) was performed to evaluate the morphological differences in five populations of Ae. aegypti. Pair-wise comparison of each character was carried out using a t test, with the Bonferroni adjustment to the probabilities (as 44 comparisons were made, 0.05/44 = 0.001136 was used as cut-off value). Morphological data of five populations were analyzed using discriminant function analysis (DFA) to access the morphological differences among the populations. Multivariate statistical analyses were conducted in SPSS version 16. (SPSS Inc, Chicago, USA).
Analysis of genetic data - The observed allelic frequencies for each population were used to estimate the mean number of alleles per locus (A), effective number of alleles per locus (A0), percentage of polymorphic loci (P), and mean heterozygosity per locus (H) with respect to the Hardy-Weinberg expectation16. Agreement with Hardy-Weinberg proportions was tested using both F-statistics17 and a χ2 test for goodness of fit with Levene's correction for small samples18. To determine whether Inbreeding coefficient of an individual (I) relative to the sub (S) population (FIS) and Inbreeding coefficient of an individual (I) relative to the total (T) population (FIT) estimations for each locus were significantly different from zero, chi-square statistics [χ2 = F(2N) (k–1)] were obtained, with k(k–1)/2 degrees of freedom, where N is the sample size and k is the number of alleles. To determine the significance of the Inbreeding coefficient of subpopulation (S) relative to the total (T) population FST statistic per locus, the chi-square statistic was used: χ2 = (2N)Fst (k–1), with (k–1) (s–1) degrees of freedom, where s is the number of populations. As the χ2 test is likely to be unreliable when expected values are low19, the χ2 test was repeated with the genotypes pooled into the three classes (i) homozygotes for the most common allele; (ii) heterozygotes for the most common allele; and (iii) other genotypes. Departures from Hardy-Weinberg were only considered significant if both χ2 tests were significant. The gene frequency data were analysed using BOTTLENECK software20 to assess the evidence of recent bottlenecks. Deviations from expected heterozygosity were computed for each locus for each population. The infinite allele model was used since it is the most appropriate for allozyme data20. To determine the significance of deviations, a two-tailed Wilcoxon signed-rank test was conducted. This method tests whether the expected gene diversity (HE) is higher than the expected equilibrium gene diversity (Heq) calculated from the observed number of alleles for each locus in each population under the assumption of mutation-drift equilibrium and the infinite allele model. Population genetic structure was analyzed based on allozyme data by means of analysis of molecular variance (AMOVA) using Genalex21.
Cluster analysis: Morphological data of the five Ae. aegypti populations were used to construct the similarity and distance matrices with the Bray-Curtis index PHYLIP ver 3.68. Pair-wise genetic similarity among the populations was calculated according to the Nei's genetic similarity index and used in constructing the similarity and distance matrices. To compare the populations based on morphological and allozyme variation, the Neighbor Joining cluster analysis was performed using PHYLIP software22. Neighbor program in Phylip ver. 3.68 software was used to construct the Neighbor joining trees.
Results
Susceptibility of Ae. aegypti to three different CHIKV strains: Immuno-fluorescence assay of infected mosquitoes showed a linear progression between dissemination rates and CHIKV titres. The plateau corresponding to 100 per cent of disseminated infection rate was reached for blood-meal titres higher than 5.68×107 RNA copies/ml for CHIKV (V226) and CHIKV (Asian) while blood meal with 5.08×107 RNA copies/ml of CHIKV (A226) was sufficient to infect 92 per cent of female Ae. aegypti (Fig. 1). The effect of the infection dose on dissemination rate for each of the CHIKV strain at different virus doses (CHIKV 226A/CHIKV 226V, CHIKV 226A/CHIKV Asian and CHIKV 226V/CHIKV Asian at 5×105 RNA copies/ml, 5.08×106 RNA copies/ml, 5.08×107 RNA copies/ml and 5.08×108 RNA copies/ml) was evaluated by contingency analysis and found to be similar dissemination for each CHIKV strain at each CHIKV titre. Similar dissemination rate was observed when compared in different organs such as salivary gland, head squash, fat bodies and legs. However, such selective advantage for particular CHIKV strain was not observed in Ae. aegypti populations.
Quantification of CHIKV in whole mosquito females: Immediately after blood meals, Ae. aegypti mosquitoes had an average of 2.29×104 (±63), 3.23×104 (±51) and 5.01×104 (±31) viral RNA copies per mosquito when they ingested the blood-meal containing CHIKV A226, V226 and Asian strains, respectively. Virus RNA replication rapidly increased during the first three days. Following the blood-meal, viral load increased to reach a maximum of 4.67×107 (±151) at day 6 p.i. for CHIKV (A226), 7.24×107 (±2690) at day 5 p.i. for CHIKV (V226) and 3.16×107 (±53) viral RNA copies at day 2 p.i. and 7 p.i. for CHIKV (Asian) (Fig. 2). Using two-way factorial ANCOVA no significant difference was found in titres of the three strains of CHIKV. This indicated that there was no measurable difference in CHIKV titres for any specific CHIKV strain at different time points of CHIKV infection in Ae. aegypti mosquitoes, whereas titres of three CHIKV strains were significantly changed in due course of CHIKV infection (P= 0.002) (Fig. 2).
Fig. 2.
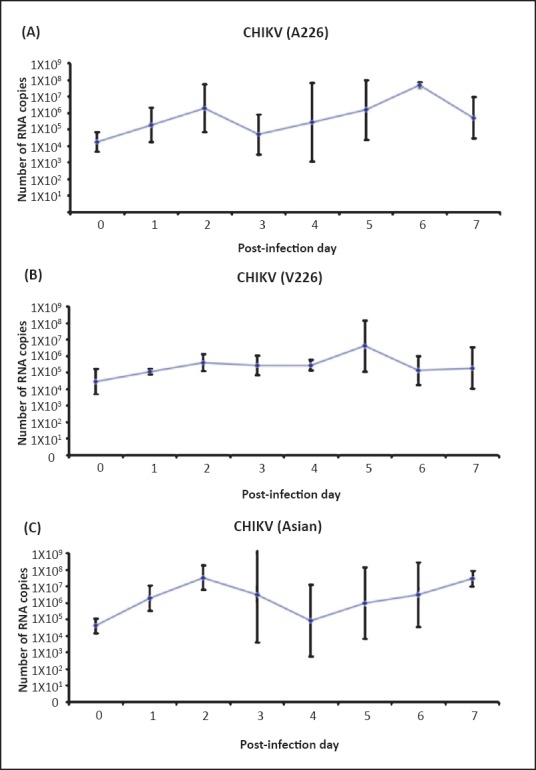
Growth kinetics of different CHIKV strains in Aedes aegypti (A) CHIKV (A226), (B) CHIKV (V226), and (C) CHIKV (Asian). Ten mosquitoes were sampled at each time point, and CHIKV genome copies were measured by q-RT-PCR. Values are expressed as mean RNA copies and error bars indicate the standard deviation.
Dissemination of CHIKV in different geographic strains of Ae. aegypti: Dissemination and growth kinetic studies suggested that 3×107 virus particles were enough to cause 100 per cent dissemination for all three CHIKV strains in laboratory reared Alappuzha populations of Ae. aegypti. We checked whether a similar dose was enough to cause 100 per cent infection in five different geographical populations of Ae. aegypti. The dose of about 5 ×107 RNA copies/ ml of all three CHIKV strains was enough to cause 100 per cent infection in midgut of all Ae. aegypti populations. Ae. aegypti from Alappuzha showed the highest dissemination rate for CHIKV strains followed by Surat, Jalgaon, Tirupati, and Gorakhpur (Table I). The null hypothesis that the dissemination data were normally distributed was accepted (Jarque Bera = 2.234 P=0.03272). Leven's test of homogeneity of variance suggested that variances were not equal across the groups (Levene statistics=1.563 P= 0.148), therefore, we used two way-ANOVA with Games-Howell post-hoc test. Two way-ANOVA showed significant differences in dissemination rate among the populations (F = 276.782, P=0.001) and CHIKV strains (F=35.235, P=0.001). There was significant difference in dissemination in population × CHIKV strain (F=9.839, P =0.001).
Phenotypic variation among the populations: A MANOVA analysis for morphometric measurements, which included geographic location as factors for all the variables analyzed, showed significant differences between the geographic populations (Pillai's Trace F=8.591, P<0.001; Wilks’Lambda F=10.147, P<0.001). Among the geographic populations, 10 variables revealed statistical differences (Table II). In discriminant function analysis, first factor (F1) explained 57.58 per cent of the total variability while the second factor (F2) explained 31.58 per cent of total variability and together the first two factors explained 89.15 per cent of the total variability. DFA results suggested that at least one population was significantly different from others (Pillai's trace = 1.485, F = 1.394, P<0.0001). DFA cluster revealed three clusters, rare cases area (Gorakhpur), high number cases area (Surat) and frequent cases areas (Alappuzha, Jalgaon and Tirupati) (Fig. 3A). Anal gill index, siphon index and denticles on apical pectin tooth were important variables which discriminated among the clusters (Fig. 3B).
Fig. 3.
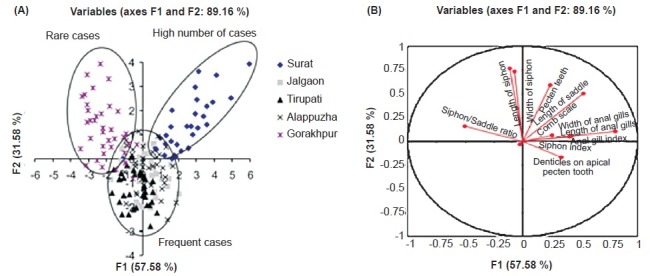
Discriminant factor analysis (DFA) of morphological data. (A) Clusters of different populations. Analysis revealed three clusters, rare cases area (Gorakhpur), high number cases area (Surat) and frequent cases areas (Alappuzha, Jalgaon and Tirupati) (B) Variables which discriminated between the clusters. Anal gill index, siphon index and denticles on apical pectin tooth characters discriminate among the clusters. Ellipses of probabilities are shown as circles.
Allozyme variations among the populations: Allozyme electrophoresis resulted in clear and consistent staining for five enzymes encoded by putative six loci: ADH, GPD, GLC, LDH, SOD1 and SOD2. A total of 10 alleles were detected from the five populations of Ae. aegypti (Table III). The allele frequency analysis revealed that all populations were monomorphic for SOD1 and SOD2 and Alappuzha, Surat and Tirupati populations were monomorphic at loci GPD. All five populations were dimorphic for ADH, GLC and LDH.
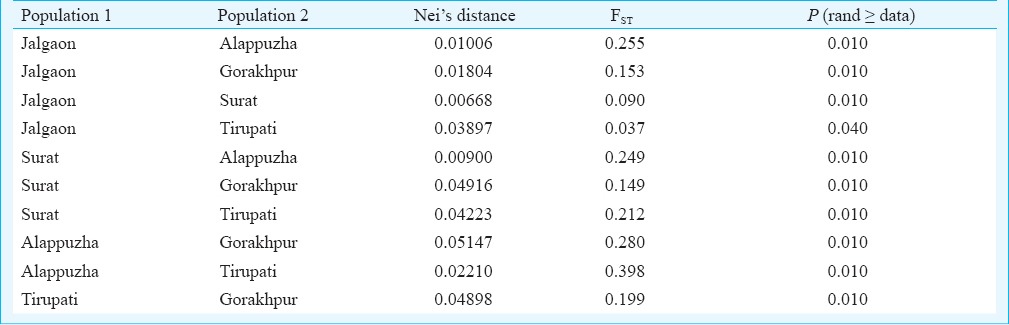
Genetic variation within the population: Based on four polymorphic loci genetic variations among the five populations of Ae. aegypti ware calculated. Mean number of alleles per locus were 1.5, 1.67, 1.5, 1.5, and 1.67 for the Surat, Jalgaon, Tirupati, Alappuzha and Gorakhpur populations, respectively. Percentages of polymorphic loci were 50, 67, 50, 50, and 67 per cent, respectively, and the mean heterozygosity per locus were 0.106, 0.169, 0.154, 0.161 and 0.1865, respectively. F statistics for six loci (Table III) described a high degree of geographic uniformity and suggested random mating between individuals within the populations. FST, FIT, and FIS values were significantly different from zero. Fst values ranged between 0 and 0.589, suggesting significant genetic differentiation within these populations. Fis values ranged from 0-1 with most of the values closer to 1 (Table III). AMOVA for the studied populations revealed that most of the genetic variation (87%) was within populations (Table IV). To estimate genetic similarity among the studied populations, Nei's genetic distances (Table V) were calculated.
Table IV.
Analysis of molecular variance (AMOVA) among the Ae. aegypti populations in India
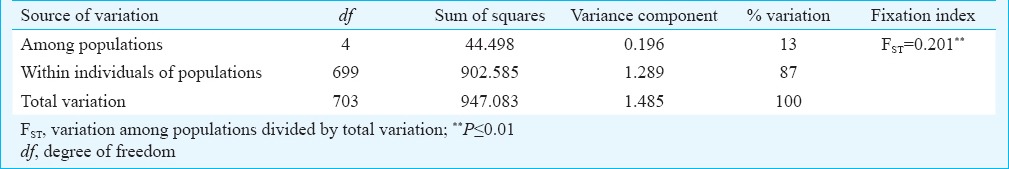
Table V.
Pairwise Nei's genetic distance and fixation index FST of Ae. aegypti populations
Bottleneck effect: Table III shows the significant test results for a recent bottleneck by polymorphic loci in each population. For the Surat population GLC locus and for Alappuzha population ADH locus showed significant difference between expected heterozygosity (HE), which was found to be higher than the expected heterozygosity at mutation equilibrium drift (Heq) (Table III). Gorakhpur population ADH and LDH loci showed significant difference between expected heterozygosity (HE) and expected heterozygosity at mutation equilibrium drift (Heq) (Table III).
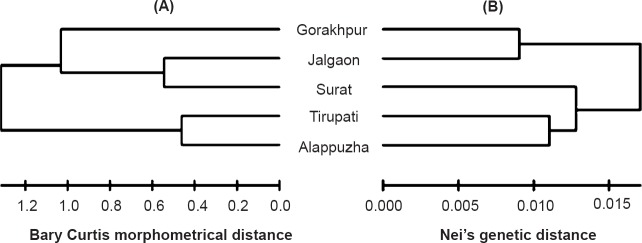
Cluster analysis of morphological and allozyme data: The dendrogram obtained by the Neighbor joining clustering method revealed the morphological similarities among the five different populations of Ae. aegypti (Fig. 4A). All five populations of Ae. aegypti formed a single cluster, where Alappuzha and Tirupati populations showed high similarity followed by the Surat and Jalgaon populations. The genetic similarities of the five populations depicted high similarity between Jalgaon and Gorakhpur followed by Alappuzha, Surat and Tirupati populations (Fig. 4B).
Fig. 4.
Neighbour joining tree showing (A) morphological relationship among five populations of Ae. aegypti (B) genetic relationships among five populations of Ae. aegypti based on Nei's pair-wise genetic distance.
Discussion
Our experiments demonstrated that all the CHIKV strains used in the study were able to infect and replicate in Ae. aegypti, but strain variations were apparent. At higher titre of CHIKV, midgut infection barriers are not able to restrict the CHIKV infection. The leaky midgut phenomenon and infection threshold might be playing important role in CHIKV infection. The head squashes, salivary gland and fat bodies of Ae. aegypti were found positive for CHIKV at day 7 p.i. and was in accordance with the short extrinsic incubation period of CHIKV in Aedes mosquitoes9. CHIKV (V226) mutation gives a selective advantage in Ae. albopictus8,9. However, such selective advantage for a particular CHIKV strain was not observed in Ae. aegypti populations.
The vector competence of CHIKV has been studied worldwide23,24,25. Tsetsarkin et al26 demonstrated that the E1-226V mutation was able to increase the vector competence for CHIKV in Ae. albopictus but not in Ae. aegypti strains. This was further confirmed in other studies11,27. Under laboratory conditions, Girod et al28 showed that Ae. aegypti populations were more competent than Ae. albopictus and Ae. aegypti from Libreville (Gabon)8 or Yaounde´ (Cameroun)29. The results obtained in the present study were similar with Girod et al28. Aedes aegypti populations from India showed high vector susceptibility for CHIKV. It has been well documented that difference in vector competence may, at least in part, be due to the presence of specific midgut epithelial receptors30. Most of the well characterized arbovirus receptors are house keeping molecules and are also present ubiquitously in the midgut brush border membrane of mosquitoes30. Therefore, we hypothesized that at 100 per cent infection rate mosquito will show similar growth kinetics. In our study, the midgut appeared infected during the entire incubation time. However, the titre of CHIKV varied among individual mosquitoes belonging to the same population. Aedes aegypti mosquito populations in the present study exhibited two distinct profiles of infection for CHIKV strains: (i) females strongly susceptible (106-107 RNA copies/female mosquito), and (ii) weakly susceptible (103-104 RNA copies/female mosquito). The results of dissemination studies showed that the susceptibility to infection and dissemination of CHIKV varied within Ae. aegypti populations and the CHIKV strain. It has been well documented that vector competence may differ among mosquito populations, which have different genetic backgrounds1,2. It has also been demonstrated that variation in vector competence for dengue viruses depends on virus replication and it does not depend on mosquito migut binding affinity31. These observations suggest that the genetic variation in populations might play an important role in determining the susceptibility of Ae. aegypti to CHIKV.
A distinct intraspecific variation has been observed worldwide in Ae. agypti populations32,33. However, very few studies have been carried out to understand the Ae. aegypti population structure in India. Using multivariate analysis of morphological characters, a significant morphological differentiation was observed in the five populations of Ae. aegypti. Discriminant function analysis of the data suggested that siphon, saddle, and anal gills related variables were the most important distinguishing characters. The highest genetic variability and diversity were found in the Gorakhpur population, whereas the Surat population showed the lowest genetic variability. The mean heterozygosity per allele of Ae. aegypti populations was similar to the average values found in other diptera14,34. In concordance with high levels of heterozygosity, the BOTTLENECK test results indicated an excess of heterozygosity relative to allele numbers at several of the gene loci studied20. This indicates that founder effects (bottlenecks) may have played a role in the history of the species. A high genetic variation was observed within populations and low variation among populations of Ae. aegypti indicating a high within-population differentiation. Similar results were obtained for Ae. aegypti populations in various parts of the world35,36,37. Low levels of genetic structuring and gene flow were observed in the studied populations. These results were consistent with those obtained in Mexico36, Rio de Janeiro37 and in Southeastern and Southern Brazil38.
In current study five populations of Ae. aegypti were studied, however, the studies on more Ae. aegypti populations from different geographical locations will give better insights in CHIKV disease dynamics. Microsatellites analysis would be more useful in resolving the population structure of Ae. aegypti.
In conclusion, the dissemination rates and growth kinetics of the three CHIKV strains showed no selective advantage for a particular strain of CHIKV in Ae. aegypti. Results from this study illustrate the complexity of population variation in the mosquito Ae. aegypti. The allelic frequencies, F statistics, and Nei's genetic identity values showed that genetic differences among the populations were small, but significant. The detected morphological and phenotypic variations may be related to differential environmental conditions such as temperature, food availability, and water quality. The genetic variability in these populations might be responsible for the differential vector susceptibility in these Ae. aegypti populations.
Acknowledgment
The study was supported by the Indian Council of Medical Research, New Delhi.
References
- 1.Beerntsen BT, James AA, Christensen BM. Genetics of mosquito vector competence. Microbiol Mol Biol Rev. 2000;64:115–37. doi: 10.1128/mmbr.64.1.115-137.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kramer LD, Ebel GD. Dynamics of flavivirus infection in mosquitoes. Adv Virus Res. 2003;60:187–232. doi: 10.1016/s0065-3527(03)60006-0. [DOI] [PubMed] [Google Scholar]
- 3.Gubler DJ. Human arbovirus infections worldwide. Ann NY Acad Sci. 2001;951:13–24. doi: 10.1111/j.1749-6632.2001.tb02681.x. [DOI] [PubMed] [Google Scholar]
- 4.Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006;3:e263. doi: 10.1371/journal.pmed.0030263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, et al. Genetic divergence of Chikungunya viruses in India (1963-2006) with special reference to the 2005-2006 explosive epidemic. J Gen Virol. 2007;88:1967–76. doi: 10.1099/vir.0.82714-0. [DOI] [PubMed] [Google Scholar]
- 6.Santhosh SR, Dash PK, Parida MM, Khan M, Tiwari M, Lakshmana Rao PV. Comparative full genome analysis revealed E1: A226V shift in 2007 Indian chikungunya virus isolates. Virus Res. 2008;135:36–41. doi: 10.1016/j.virusres.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Gurav YK, Gopalkrishna V, Shah PS, Patil DR, Mishra M, Paingankar MS, Sathe PS, Mishra AC, et al. An outbreak of chikungunya in Jamshedpur, Jharkhand in 2011. Indian J Med Res. 2012;136:886–9. [PMC free article] [PubMed] [Google Scholar]
- 8.Vazeille M, Moutailler S, Coudrier D, Rousseaux C, Khun H, Huerre M, et al. Two chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One. 2007;2:e1168. doi: 10.1371/journal.pone.0001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubrulle M, Mousson L, Moutailler S, Vazeille M, Failloux AB. Chikungunya virus and Aedes mosquitoes: saliva is infectious as soon as two days after oral infection. PLoS One. 2009;4:e5895. doi: 10.1371/journal.pone.0005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tesh RB, Gubler DJ, Rosen L. Variation among geographic strains of Aedes albopictus in susceptibility to infection with chikungunya virus. Am J Trop Med Hyg. 1976;25:326–35. doi: 10.4269/ajtmh.1976.25.326. [DOI] [PubMed] [Google Scholar]
- 11.Reiskind MH, Pesko K, Westbrook CJ, Mores CN. Susceptibility of Florida mosquitoes to infection with chikungunya virus. Am J Trop Med Hyg. 2008;78:422–5. [PMC free article] [PubMed] [Google Scholar]
- 12.Martin E, Moutailler S, Madec Y, Failloux AB. Differential responses of the mosquito Aedes albopictus from the Indian Ocean region to two chikungunya isolates. BMC Ecol. 2010;10:8. doi: 10.1186/1472-6785-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parashar D, Paingankar MS, Kumar S, Gokhale MD, Sudeep AB, Shinde SB, et al. Administration of E2 and NS1 siRNAs inhibit chikungunya virus replication in vitro and protects mice infected with the virus. PLoS Negl Trop Dis. 2013;7:e2405. doi: 10.1371/journal.pntd.0002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanojia PC, Paingankar MS, Patil A, Gokhale MD, Deobagkar DN. Morphometric and allozyme variation in Culex tritaeniorhynchus mosquito populations from India. J Insect Sci. 2010;10:138. doi: 10.1673/031.010.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasteur N, Pasteur G, Bonhomme F, Catalan J, Britton-Davidian J. Chichester: Ellis Horwood; 1988. Practical isoenzyme genetics. [Google Scholar]
- 16.Hedrick PW. 2nd ed. Sudbury, MA: Jones and Bartlett Publishers; 2000. Genetics of populations. [Google Scholar]
- 17.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–70. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 18.Levene H. On a matching problem arising in genetics. T Ann Math Stat. 1949;20:91–4. [Google Scholar]
- 19.Sokal RR, Rohlf F. 3rd ed. San Fransisco: WH Freeman; 1995. Biometry: the principal and practice of statistics in biological research. [Google Scholar]
- 20.Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered. 1999;90:502–3. [Google Scholar]
- 21.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research - an update. Bioinformatics. 2012;28:2537–9. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felsenstein J. PHYLIP Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–6. [Google Scholar]
- 23.Mangiafico JA. Chikungunya virus infection and rransmission in five species of mosquito. Am J Trop Med Hyg. 1971;20:642–5. doi: 10.4269/ajtmh.1971.20.642. [DOI] [PubMed] [Google Scholar]
- 24.Banerjee K, Mourya DT, Malunjkar AS. Susceptibility & transmissibility of different geographical strains of Aedes aegypti mosquitoes to chikungunya virus. Indian J Med Res. 1988;87:134–8. [PubMed] [Google Scholar]
- 25.Turell MJ, Beaman JR, Tammariello RF. Susceptibility of selected strains of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) to Chikungunya virus. J Med Entomol. 1992;29:49–53. doi: 10.1093/jmedent/29.1.49. [DOI] [PubMed] [Google Scholar]
- 26.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pesko K, Westbrook CJ, Mores CN, Lounibos LP, Reiskind MH. Effects of infectious virus dose and bloodmeal delivery method on susceptibility of Aedes aegypti and Aedes albopictus to chikungunya virus. J Med Entomol. 2009;46:395–9. doi: 10.1603/033.046.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girod R, Gaborit P, Marrama L, Etienne M, Ramdini C, Rakotoarivony I, et al. High susceptibility to chikungunya virus of Aedes aegypti from the French West Indies and French Guiana. Trop Med Int Health. 2011;16:134–9. doi: 10.1111/j.1365-3156.2010.02613.x. [DOI] [PubMed] [Google Scholar]
- 29.Paupy C, Ollomo B, Kamgang B, Moutailler S, Rousset D, Demanou M, et al. Comparative role of Aedes albopictus and Aedes aegypti in the emergence of dengue and chikungunya in Central Africa. Vector Borne Zoonotic Dis. 2010;10:259–66. doi: 10.1089/vbz.2009.0005. [DOI] [PubMed] [Google Scholar]
- 30.Paingankar MS, Gokhale MD, Deobagkar DN. Dengue-2-virus-interacting polypeptides involved in mosquito cell infection. Arch Virol. 2010;155:1453–61. doi: 10.1007/s00705-010-0728-7. [DOI] [PubMed] [Google Scholar]
- 31.Cox J, Brown HE, Rico-Hesse R. Variation in vector competence for dengue viruses does not depend on mosquito midgut binding affinity. PLoS Negl Trop Dis. 2011;5:e1172. doi: 10.1371/journal.pntd.0001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabachnick WJ, Powell JR. A world-wide survey of genetic variation in the yellow fever mosquito, Aedes aegypti. Genet Res. 1979;34:215–29. doi: 10.1017/s0016672300019467. [DOI] [PubMed] [Google Scholar]
- 33.Failloux AB, Vazeille M, Rodhain F. Geographic genetic variation in populations of the dengue virus vector Aedes aegypti. J Mol Evol. 2002;55:653–63. doi: 10.1007/s00239-002-2360-y. [DOI] [PubMed] [Google Scholar]
- 34.Graur D. Gene diversity in Hymenoptera. Evolution. 1985;39:190–9. doi: 10.1111/j.1558-5646.1985.tb04091.x. [DOI] [PubMed] [Google Scholar]
- 35.Herrera F, Urdaneta L, Rivero J, Zoghbi N, Ruiz J, Carrasquel G, et al. Population genetic structure of the dengue mosquito Aedes aegypti in Venezuela. Mem Inst Oswaldo Cruz. 2006;101:625–33. doi: 10.1590/s0074-02762006000600008. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Franco F, Munoz Mde L, Lozano-Fuentes S, Fernandez-Salas I, Garcia-Rejon J, Beaty BJ, et al. Large genetic distances among Aedes aegypti populations along the south Pacific coast of Mexico. Am J Trop Med Hyg. 2002;66:594–8. doi: 10.4269/ajtmh.2002.66.594. [DOI] [PubMed] [Google Scholar]
- 37.de Costa-Ribeiro MC, Lourenço-de-Oliveira R, Failloux AB. Geographic and temporal genetic patterns of Aedes aegypti populations in Rio de Janeiro, Brazil. Trop Med Int Health. 2006;11:1276–85. doi: 10.1111/j.1365-3156.2006.01667.x. [DOI] [PubMed] [Google Scholar]
- 38.da Costa-Ribeiro MC, Lourenço-de-Oliveira R, Failloux AB. Low gene flow of Aedes aegypti between dengue-endemic and dengue-free areas in southeastern and southern Brazil. Am J Trop Med Hyg. 2007;77:303–9. [PubMed] [Google Scholar]