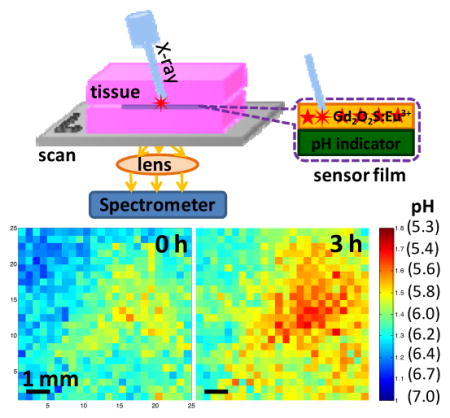
Abstract
We develop a pH sensor film that can be coated on an implant surface and imaged using a combination of X-ray excitation and visible spectroscopy to monitor bacterial infection and treatment of implanted medical devices (IMDs) through tissue. X-ray scintillators in the pH sensor film generate light when an X-ray beam irradiates them. This light first passes through a layer containing pH indicator that alters the spectrum according to pH, then passes through tissue where it is detected by a spectrometer. A reference region on the film is used to account for spectral distortion from wavelength-dependent absorption and scattering in the tissue. pH images are acquired by moving the sample relative to the X-ray beam and collecting a spectrum at each location, with a spatial resolution limited by the X-ray beam width. Using this X-ray excited luminescence chemical imaging (XELCI) to map pH through porcine tissue, we detected a pH drop during normal bacterial growth on the sensor surface, and a restoration of the pH to the bulk value during antibiotic treatment over the course of hours with millimeter resolution. Overall, XELCI provides a novel approach to noninvasively image surface pH for studying implant infections and treatments.
Keywords: X-ray excited luminescence chemical imaging, in situ pH sensor film, S. epidermidis
Graphical Abstract
A pH sensor film is coupled with an X-ray excited scintillator light source to image through thick tissue local acidosis on the film surface during bacterial growth. High spatial resolution pH imaging is achieved through tissue by irradiating the film surface with a narrowly collimated X-ray beam and collecting the luminescence spectrum point-by-point as the sample is moved relative to the beam.
1. Introduction
Although implanted medical devices (IMDs), such as fracture fixation devices, prosthetic joints, and pacemakers, have greatly improved patients’ lives, the implant surface can be colonized by bacteria resulting in the formation of antibiotic-resistant biofilms especially in vulnerable populations.[1] For example, approximately 5% of the 200,000 fracture fixation devices implanted each year in the US become infected,[2] and up to 40% of battlefield injuries with internal fixation, due to increased wound trauma and increased likelihood of debris in the wounds.[3] While infections caught early can often be treated with a combination of debridement and antibiotic therapy, after approximately three weeks, the implants usually must be removed followed by antibiotic treatment and device replacement, with associated risks from prolonged hospitalization and surgeries.[4] In addition, it is difficult to monitor bacterial infection noninvasively during treatment. Hence, there is a need for developing methods to detect bacterial infections and monitor treatment in situ. Clinic diagnostic methods such as clinical symptoms and intraoperative tissue culture have limited specificity and sensitivity (e.g. intraoperative tissue culture has a sensitivity of 65–94%.), while MRI and CT of bone resorption and sinus tracts are only useful at late infection stages.[4] None of these techniques are good for monitoring the biofilm during treatment. Genetically engineered bioluminescent and fluorescent bacteria are promising for monitoring infection in animal studies but are limited exclusively to research, and the signal intensity depends upon many factors including cell count, metabolic rate, absorption and scattering in the tissue, and collection/emission optics.[5] In addition, optical scattering severely limits the image resolution.[6]
Herein we develop a technique to non-invasively image chemical concentrations on the surface of modified IMDs in order to detect, monitor, and study bacterial infections. We selected surface pH as our target analyte because infection and inflammation cause local acidosis, especially in dormant and poorly perfused regions near the implant surface.[7] For example, Hidalgo and coworkers used confocal microscopy together with ratiometric pH nanosensors to study pH gradients within both E. coli and wastewater biofilms.[8] Although the bulk pH was 7.2, the pH on the substrate surface was as low as 5.0 and varied with lateral position and depth. This heterogeneous pH is thought to play a strong role in antibiotic resistance, in part because the heterogeneous pH affects the antibiotic chemistry and bacterial metabolism, and in part because low pH is an indication of a poorly perfused region with likely lower antibiotic penetration and dormant bacteria.[9] Although quantitative pH images of the IMDs surface have not yet been acquired in vivo, indirect evidence strongly indicates a low surface pH. X-ray and MRI images show the effects of low pH on infected bone and teeth via erosion and formation of sinus tracts. In addition, spectroscopic techniques show the acid-generated change in mineral composition such as the presence of brushite and uncarbonated apatite materials in both cavities and infected bone.[10] Low pH in vivo is attributed to a combination of inflammation, osteoclast activity and bacterial metabolization.[11] For example, Konttinen and coworkers used pH microelectrodes to measure pH adjacent to a prosthetic hip implant intraoperatively.[11d] For all measured patients, the pH in the nearby muscle, gluteus medius was 7.39 +/−0.05. For patients with aceptic loosening, the pH was generally acidic and ranged between 5–7.5. This local acidosis is consistent with studies of collegen degradation, implant pitting, and metal ion release in patients with aseptic loosening.[11a] The patient with septic loosening had particularly low pH near the implant, 5.8 in the femoral stem and 4.4 in the femoral cavity. While these studies did not measure the pH on the implant surface or generate pH images to study inhomogeneity, they do suggest significant drops in pH near an infected implant surface that should be measurable with our technique.
To study infections in situ, we designed a pH sensor film to non-invasively image through tissue the pH on the sensor surface with high spatial resolution using a novel technique we call X-ray excited luminescence chemical imaging (XELCI). The technique is related to X-ray luminescence tomography (XLT) which images scintillators embedded in tissue at high resolution using a combination of X-ray excitation and optical detection.[12] XLT constructs tomographic images of the distribution of X-ray nanoscintillators by selectively irradiating the tissue point-by-point or slice-by-slice and measuring the X-ray excited luminescence. Recently, the acquisition time for XLT has been greatly reduced via reconstructing high spatial image with mathematical methods, and the low background signal allows 3-dimensional reconstruction with as few as two angles.[13] In a numerical simulation of limited angle XLT of X-ray nanoscintillators embedded in tissue, the Xing group determined that a 6 mm diameter object with 1 μg/ mL nanoparticle concentration and 0.1 μg/mL in the surrounding tissue could be resolved in up to 4 cm tissue with a signal to noise ratio of 10 using a 1 mm X-ray beam with a dose of cGy.[14] Compared to XLT, XELCI detects luminescence from a high concentration of X-ray scintillator microparticles embedded within a film (e.g. a 100 μm thick film with 5 g/cm3 of Gd2O2S microparticles, assuming a 70% packing density) and uses the X-ray generated light in conjunction with indicator dyes to provide surface-specific images of analyte concentrations near the surface. The relatively thick and stable X-ray scintillator film provides much greater luminescence intensity than XLT for dilute nanoparticles and no background from out of focus slices. Our group has employed X-ray excited optical luminescence to spectrochemically measure pH without tissue and its high spatial resolution through tissue by irradiating two different types of radioluminescent films embedded in tissue.[15] The ability of X-ray excited optical luminescence to study silver dissolution through tissue in one dimension was also demonstrated.[16] We also showed that hollow X-ray scintillators could be used to monitor drug release in vitro and the luminescent signal could be detected at one wavelength in living mice, however, luminescent signal was detected by only irradiating a single location in living mice (not a scanned or two dimensional image) and no in vivo drug release monitoring was achieved.[17] Herein, we extended our previous work and developed an XELCI technique to non-invasively map local pH variation due to bacterial metabolic activity in two dimensions through thick tissue with high spatial resolution. This is the first time that a 2D chemical image was acquired through tissue and the first time a reference region was used to account for the tissue induced spectral distoration. In the future, XELCI will be used to noninvasively monitor bacterial infection on implant surface in animal model with our own home-built system.
2. Results and Discussion
2.1. pH sensor film principle
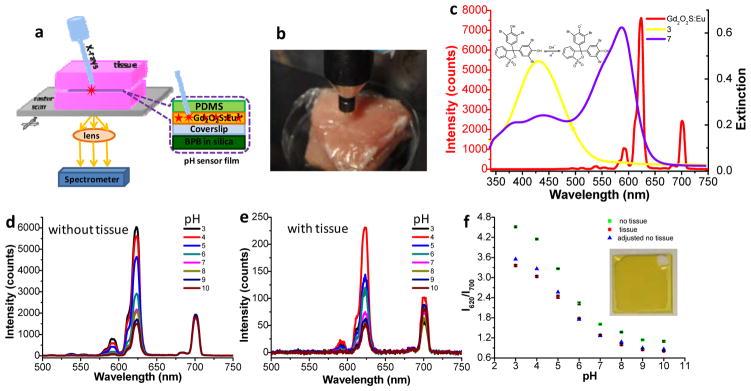
Figure 1a shows the working principle of XELCI. The scintillators (Gd2O2S: Eu phosphors sealed in a PDMS layer) are excited by a narrow X-ray beam which generates a luminescent spot at a location and time controlled by the X-ray beam. The luminescence then passes through the pH indicator layer (silica containing bromophenol blue (BPB) dye), which alters the luminescence spectrum according to pH. The light finally passes through the tissue and is detected by an external spectrometer. A two dimensional pH image is formed by scanning the sensor film in both x and y directions and determining pH from the luminescence spectrum at each position. The approach assumes that the tissue is relatively flat and homogeneous on the millimeter size scale. Figure 1b shows a photo of the X-ray source aiming at the sensor through porcine tissue. Figure 1c shows the luminescence spectrum (red line, plotted against the left y-axis). The intense peak at 620 nm is attributed to the 5D0→7F2 transition of Eu3+, while the near infrared emission at 700 nm arises from the 5D0→7F4 transition of Eu3+.[18] The BPB-doped silica film has a pH-dependent absorption spectrum, with an isobestic point indicating only two species, protonated and deprotonated (Figure S1a). Figure S1b shows a stretched calibration curve based on the absorption at the deprotonated peak, with a dynamic range between pH 4–9 indicating multiple environments within the film with different effective pKas,[19] and 2–4% variation in absorbance at the deprotonated peak from film to film. As shown in Figure 1c, the absorption spectrum of the film at pH 7 overlaps more with the red (620 nm) luminescence of the scintillators than at pH 3, while the 700 nm peak is not absorbed by BPB at any pH and serves as an intensity reference to account for changes in X-ray intensity and optical collection efficiency.
Figure 1.
XELCI setup and pH calibration curves with and without tissue. (a) Schematic drawing of the XELCI setup. (b) Photograph of the setup. (c) Luminescent spectrum of scintillators (Gd2O2S: Eu) (red line, left y-axis) and the extinction spectra of BPB-doped silica films in pH buffer 3.0 (yellow, right y-axis) and buffer 7.0 (purple, right y-axis). Inset: acidic and basic forms of BPB. (d) Luminescence spectra of pH sensor film in response to different standard buffers range from pH 3.0 to pH 10.0. (e) Luminescence spectra of pH sensor film in response to different pH buffers after passing through porcine tissue. (f) pH calibration curves without tissue (green squares), with tissue (red dots) and the adjusted no tissue calibration curve adjusted using the reference region (blue triangles). Error bars represent standard deviation of three different measurements at different positions on the same sample. Inset: photograph of a pH sensor film with uncoated reference region (white region) on top right.
2.2. pH calibration curves with and without tissue
We then studied how tissue absorption and scattering affects the luminescence spectra and pH calibration curves. Figure 1d shows the luminescence spectra of the pH sensor film in response to standard buffers ranging from pH 3.0–10.0. The same pH sensor film was then embedded between two slices of porcine muscle tissue and the experiment was repeated (Figure 1e). As is evident from Figure 1d and 1e, all the luminescence spectra are essentially background-free because tissue has a small X-ray absorption cross-section and a very low X-ray to visible conversion efficiency. The luminescence signal was attenuated 25 to 30 times after passing through tissue but still with a S/N between 10 and 28. Figure 1d and 1e also show that increasing the pH causes the luminescence intensity at 620 nm to decrease relative to that at 700 nm, as expected from the BPB absorption spectrum (see Figure 1c and Figure S1). Figure 1f shows the 620 nm to 700 nm luminescence intensity ratio as a function of pH for a sensor film alone (green squares) and for the same film placed between two slices of tissue (red dots). The ratios decrease as pH increases for both without and with tissue, indicating that the pH sensor film is indeed pH responsive. However, at each pH, placing the sensor between the tissue slices decreases the 620 nm to 700 nm intensity ratio because the tissue absorbs more light at 620 nm than at 700 nm.[20] To determine how much the tissue distorts the spectrum, a region of the film was left without any pH-dye coating to serve as an in situ spectral reference. A tissue factor of 1.27 was obtained for 700 nm/620nm light and this factor was used to correct the calibration curve without tissue (green squares). The corrected calibration curve (blue triangles in Figure 1f) overlaps well with the calibration curve acquired through tissue (red dots), indicating that the in situ reference accounted for most of the spectral distortion.
2.3. BPB-doped silica film characterization
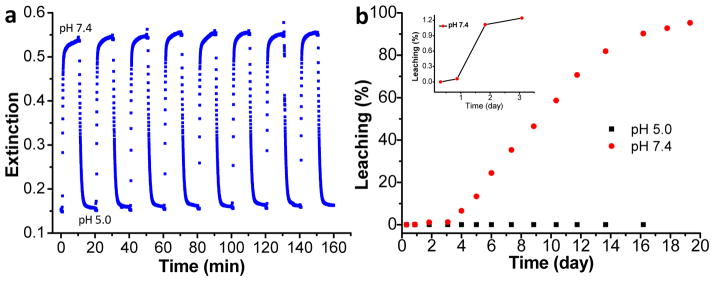
To apply these sensors to monitor pH on IMDs, the sensor layer must be reversible with a response time shorter than the pH changes due to bacterial growth. Figure 2a shows the real-time response of the pH films to alternate cycled PBS (pH 7.4) and standard buffer 5. Nearly the same response was obtained for eight repetitions with no evidence of degradation, indicating good reversibility. The response time is about 30 s for a 90% change, which is similar to the result reported from literature for dye-doped silica films.[19b] This response time is sufficiently fast to study bacterial growth mediated pH variation in real time which is expected to change over the course of hours.[19a] We also studied the sensor stability against leaching. The leaching curves for the BPB-doped silica films in standard pH 5.0 buffer and in PBS (pH 7.4) are shown in black squares and red dots in Figure 2b. In pH 5 buffer, there was no BPB leaching for more than 20 days. However, in PBS, there was no significant BPB leaching (~1% or less) for the first three days (inset figure in Figure S2b), followed by increasing leaching over the next 10 days. This leaching behavior in PBS is probably due to the dissolution of silica at slightly basic pH 7.4. Indeed, the dissolution of silica-based HPLC column in phosphate buffer pH 7.0 has been observed before.[21] The stability of the BPB-doped film is sufficient to test the principle here, although not yet for long term in vivo detection. In future, we will incorporate the pH indicators to poly (ethylene glycol)-diacrylate hydrogels[22] or titania[23] to improve the sensor stability and biocompatibility.
Figure 2.
Characterization of BPB-doped silica film. (a) Real-time reversibility of BPB-doped silica film in response to pH 7.4 and pH 5.0, alternatively, by measuring the absorbance at 595 nm.(b) Leaching study of BPB-doped silica films in pH 5.0 (black squares) and pH 7.4 (red dots). Inset: First three days of the leaching results for the BPB-doped silica film in pH 7.4.
2.4. Expected XELCI spatial resolution
Our goal is to use the pH sensor films to map pH variation caused by bacterial growth on the IMDs surface with high spatial resolution in situ. Our hypothesis, as shown in Figure 3a, is that the spatial resolution of XELCI is essentially limited by the X-ray beam size because a narrowly collimated X-ray source only generates light in the excited spot. This is unlike a scanning fluorescence excitation mode where the tissue scatters the excitation beam (typical photon mean free path is ~ 100 μm for red light) and generates fluorescence in a large region of the tissue.[20b,24] Multiple fluorescent points cannot be distinguished if they are much closer together than the tissue thickness because the excitation light spreads as it diffusively scatters through the tissue and the same scattering process also occurs for the fluorescence emission; high resolution images would require a significant fraction of either ballistic excitation or emission photons.[25] Fortunately, X-rays scatter far less by tissue than optical photons. In XELCI, the luminescence is generated only from the narrow path of the X-ray beam as X-rays have relatively small scattering coefficients in soft tissue (~0.14–0.87 cm−1, depending on the X-ray energy), thus the beam maintains focus through several centimeters of tissue.[26] For example, For 30 keV X-ray photons, the 1/e scattering depth (i.e. 1/μs) in soft tissue is 4.3 cm, which means that 87% of the incident photons travel ballistically through 6 mm of tissue.[26] The other 13% scatter according to the Klein-Nishina equation, with about 6% back scattered and 7% forward scattered. The forward scattered X-rays irradiate the film as a diffuse halo over a ~(6 mm)2 area and create a background signal. For a 1.5 mm X-ray beam, the focused spot of ballistic X-rays will be approximately two hundred fold more intense over a smaller region than the background, thus the image contrast will not be significantly affected by scattering. Although ~10% of the X-ray photons are absorbed by 6 mm of tissue (the soft tissue energy absorption coefficient for 30 keV X-ray photons is 0.1713 cm−1),[26] this absorption slightly reduces the signal intensity but does not significantly affect image resolution. Higher energy X-rays will absorb and scatter even less. As shown in Figure S2, the illumination spots of the X-ray beam with or without passing through tissue are essentially identical in size. Hence, X-rays stay focused after passing through thick tissue and the spatial resolution is primarily dictated by the X-ray beam size.
Figure 3.
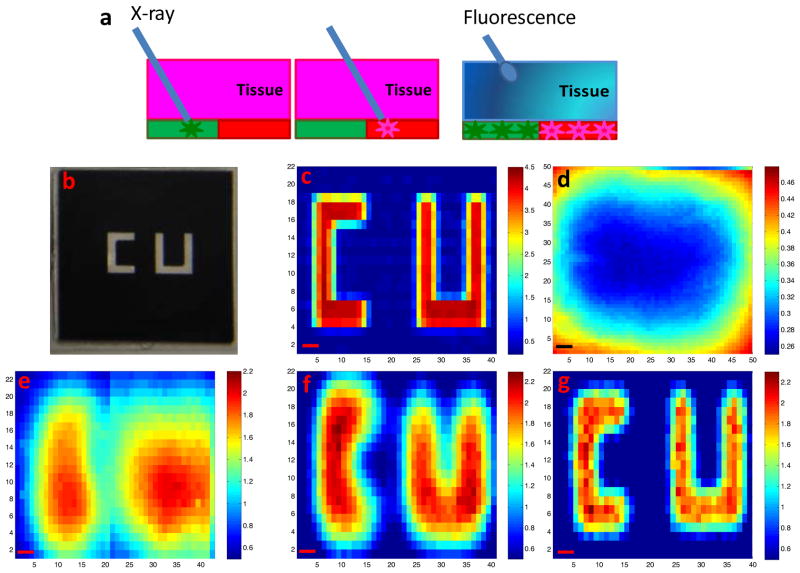
Spatial resolution of XELCI. (a) Working principle of XELCI versus fluorescence tomography. (b) Photograph of “CU” (target) on transparency. (c) Target mapping without tissue, irradiated with 1.5 mm X-ray beam. (d) Target mapped through tissue with white light as the illumination source. (e) Target mapped through tissue, irradiated with 3 mm X-ray beam. (f) Target mapped through tissue, irradiated with 1.5 mm X-ray beam; (g) Target mapped through tissue, irradiated with 1 mm X-ray beam. The color bars on the right are the ratios of the peak intensity at 620 nm over that at 700 nm. The x and y axis in c-g represent position. Step size=300 μm, scale bar= 1 mm.
2.5. “CU” target mapping
To verify that our technique can detect the luminescence signal with high spatial resolution through tissue, primarily limited by the size of the illumination source, we used XELCI to map a target through tissue. The target was a transparency with a “CU” pattern (1 mm line width) printed in black ink that absorbs more light at 620 nm than 700 nm (see Figure 3b). We placed the target over a scintillator film and mapped the ratio of 620 nm to 700 nm X-ray excited luminescence. We first mapped the XELCI ratio without tissue using a 1.5 mm X-ray beam (Figure 3c), and then with the target embedded in tissue using nominal X-ray beam widths of 3 mm (Figure 3e), 1.5 mm (Figure 3f), and 1 mm (Figure 3g); these nominal widths were determined from the beam profiles (the width at 20% of the maximum intensity) shown in Figure S3. Using a 3 mm beam, two objects are evident but their shape could not be resolved (Figure 3e). Mapping with a 1.5 mm X-ray beam could identify the two letters with a little image blurring while mapping with a 1 mm X-ray gave the best resolution. We chose 1.5 mm X-ray beam as the excitation source for the following experiments as a compromise between resolution, intensity and scanning time. To show the advantages of XELCI compared to conventional transmitted light imaging through tissue, we also acquired a ratio image of the same target but mapped through tissue using the microscope head lamp as the light source (Figure 3d). No indication of letters could be seen and only significant features come from light diffusing through from the edges of the target. The high spatial resolution of XELCI not only enables high resolution pH mapping, but also provides the ability to detect multiple analytes using different sensors at different locations and use local reference regions as intensity and spectral standards to account for spectral distortion in the tissue.
2.6. pH imaging during bacterial growth
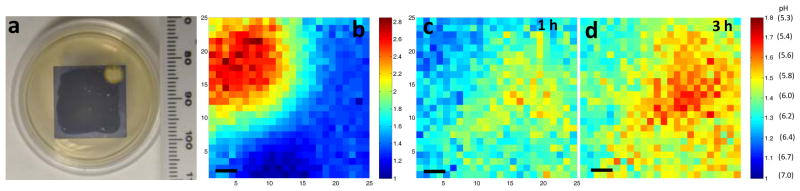
We then applied XELCI to non-invasely map through tissue the pH decrease caused by bacterial growth on the interface between our pH sensor films and TSA plates (sensor surface). Staphylococcus epidermidis (S. epid.) was chosen as the model bacterial species as it is notorious for nosocomial infections.[1] We selected two strains, ATCC 12228 and ATCC 35984, to show generality. Figure 4a is a typical photograph of a pH sensor film on the TSA plate inoculated with S. epid. 12228. The sensor film turned from purple to yellow where there was growth. The color change was attributed to bacterial metabolism which generates acidic species and lowers the local pH.[7,19a] Then, we mapped the sample through tissue with XELCI. Figure 4c is the ratio image of the bacterial region after being grown for 2 h, and Figure 4d is the ratio image of the same region 3 h later. The pH was calculated by referring to the external pH calibration curve and the nearby reference region (Figure 4b). Specifically, the effect of tissue on the 620 nm/700 nm peak ratio was first calculated by dividing the ratio of the 620 nm/700 nm of a bare luminescent film by the average of four points from the center of the reference region with a variance of 1–4% (Figure 4b). All the ratios were then adjusted by this tissue spectral distorition factor and the pH values were calculated by referring to the external pH calibration curve shown in Figure 2f (green square). For a control experiment, sterile PBS was added instead of the bacterial solution. Figure S4a is a photograph of the control plate. Figure S4b and S4c are the ratio images of the reference region and the sample region, respectively. The pH was calculated the same way as disscussed above and the pH for the sterile film was 7.0±0.2 throughout the image and the film was purple (neutral) after removal from tissue. We observed similar results using a different bacterial strain, S. epid.35984. Figure S5A is a photograph of the plate after being incubated with S. epid.35984 for 10 h and Figure S5b is the ratio image of the reference region. Figure S5c and S5d are the ratio images of the bacterial region mapped at different time points. A pH decrease over time was also seen through tissue with high spatial resolution. These results are representative of multiple experimental repetitions (at least six), where pH consistantly decreased to 5–6 within 4 h after culturing. These measurements cannot be performed with conventional approaches such as pH electrodes and indicator dye spectroscopy because conventional pH electrodes cannot acquire images of a heterogeneous surface pH through tissue (at least not without developing a pH electrode array with associated electronics, power, and telemetry), and spectroscopic techniques have poor resolution through tissue and significant autofluorescence backgrounds. The approach, however, is based upon well established and robust principles of colorimetric pH indicators,[19b] and calibrated with pH buffer standards. In addition, our approach is geared towards studying the biofilm rather than quantifying total number of colony forming units (CFU). We hope to correlate pH with the concentration of bacteria and gene expression activity using bioluminescent transfected bacterial strain in the future.
Figure 4.
pH mapping of S. epid. 12228 growth through tissue. (a) Photograph of the sample after S. epid. 12228 grew for 2 h at the interface of the pH sensor film and the TSA plate at 37 °C. (b) Ratio map of the reference region. The XELCI images appear to be mirror images of the photographs because the XELCI images were viewed from below. (c) Ratio map of the sample region after 2 h growth. (d) Ratio map of the same region 3 h later. The color bars on the right are the ratios of the peak intensity at 620 nm over intensity at 700 nm. The x and y axis in b-d represent position. Step size=300 μm, scale bar=1 mm.
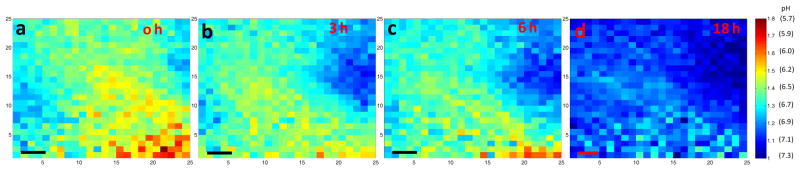
2.7. pH imaging during antibiotic treatment
We next exposed bacteria to antibiotics to watch their effect on the local pH. We chose S. epid. 12228 as a representative bacterial strain, and chose ciprofloxacin (cipro), a DNA gyrase inhibitor, as a typical antibiotic.[27] Figure S6b is a representative photograph of S. epid. 12228 grown on the sensor surface; an acidic region is visible after 2 h of incubation. Figure 5a shows the luminescence ratio map of the film through tissue after 2 h of bacterial growth. The color bar also indicates the corresponding pH calculated with the external calibration curve and the reference region (Figure S6a). Figure 5b, 5c and 5d are the ratio maps of the same region 3 h, 6 h and 18 h after cipro addition, respectively. We can see that the size of acidic region shrunk and the pH became more neutral, due to reduced metabolic activity and buffering from the agar gel. This was further verified through the photograph of the film after removal from tissue 18 h after cipro addition, Figure S6c. The yellow spot largely shrunk and the whole film returned to purple color, indicating neutral pH color. It is evident that the effect of antibiotics on local acidosis could be monitored through tissue with high temporal and spatial resolution. The effect of cipro on the growth of S. epid. 12228 was repeated multiple times (at least six), and a gradual pH restoration within 4 h after the addition was consistantly observed.
Figure 5.
pH map of S. epid. 12228 during antibiotic treatment. (a) Ratio map of the sample region where S. epid. 12228 has been grown for 2 h at 37 °C before Cipro addition. (b) Ratio map of the same region 3 h after the Cipro addition. (c) 6 h after Cipro addition. (d) 18 h after Cipro addition. The color bars on the right are the ratios of the peak intensity at 620 nm over intensity at 700 nm. The pH values are calculated by referring to the external pH calibration curve and the reference region in Figure S7a. The x and y axis represent position. Step size=300 μm; scale bar=1 mm.
3. Conclusion
In conclusion, we constructed pH sensor films using a pH indicator dye to modulate the luminescence spectrum of X-ray scintillators. Employing X-rays as the excitation source provides high tissue penetration depth with minimum beam broadening and negligible autofluorescence background. By collimating the X-ray beam and scanning the sample point-by-point, an image mapping of the “CU” letter was obtained through thick tissue with high spatial resolution. Using XELCI, we imaged through porcine tissue the pH decrease during S. epid. growth and the pH restoration after application of antibiotics. These proof-of-principle experiments demonstrate the great potential of our technique in detecting bacterial infection and evaluating treatment on IMDs in vivo in biomedical field. Future work involves developing a dedicated imaging system with higher spatial resolution, modifying dynamic compression plate surfaces for pH detection, and performing experiments on small animals.
4. Experimental Section
Materials
Bromophenol blue (BPB), tetramethoxyl orthosilicate (TMOS) and methyltrimethoxysilane (MTMOS) were all purchased from Alfa Aesar (Ward Hill, MA). Carboxymethylcellulose (CMC) and microcoverslips (#1, 18 mm x 18 mm) were purchased from Fisher Scientific (Pittsburgh, PA). Europium ions doped gadolinium oxysulfide scintillators (Gd2O2S: Eu) were purchased from Phosphor Technology Ltd. (Stevenage, UK). D (+) Glucose was purchased from Sigma-Aldrich (Saint Louis, MO). Polydimethylsiloxane (PDMS) base and its curing agent were purchased from Dow corning (Midland, MI). Hydrochloric acid (2 N) and standard pH buffers were purchased from BDH (West Chester, PA). Reagent alcohol, tryptic soy broth (TSB) and tryptic soy agar (TSA) were ordered from EMD (Gibbstown, NJ). Staphylococcus epidermidis 12228 (S. epid. 12228) and Staphylococcus epidermidis 35984 (S. epid. 35984) were purchased from ATCC (Manasssas, VA). Transparencies (3M, CG6000) were purchased from Amazon. Porcine tissue (Smithfield boneless center, thick cut chops) was purchased from a local Walmart store and was sliced to 6 mm pieces using a meat slicer (Chef’s Choice international, Graef 630). We measured seven different slices of tissue at different regions with calipers. The average thickness within the same slice was between 5.9 mm and 6.3 mm with a standard deviation ranging from 0.1 mm to 0.4 mm depending on the slice. Comparing between different slices, the average was 6.2 mm with a standard deviation of 0.1 mm. The relatively small variation in thickness within a sample is not expected to appreciably affect the luminescence spectral ratio although it may more significantly affect signal intensity.
Preparation and Characterization of BPB-doped Silica film
Organically modified silica films doped with BPB were prepared according to our previous methods with minor modifications.[19] Specifically, TMOS (320 μL) and MTMOS (138 μL) were mixed in ethanol (228 μL). Then, an appropriate amount of distilled water was added to maintain the molar ratio of silane over H2O to be 1:4, HCl (0.1M, 166 μL) was then added to initiate the polymerization. BPB (25 mg) was added subsequently. The mixture was sealed and stirred at room temperature for 24 h. Then, an aliquot of the mixture (40 μL) was spread onto a precleaned microcoverslip for 20 s and then spun cast at 2500 rpm for 15 s to form a transparent and uniform silica film. These films were dried at room temperature with a relative humidity of 40 % for 2 days. Then, the BPB-doped films were activated in 1x PBS overnight for further experiments. To measure the absorbance spectra of BPB-doped silica films in response to different pH, three films were immersed to different standard buffers ranging from 3.0 to 10.0 for 30 min. The absorbance spectra were taken with a UV-2101pc spectrometer (Shimadzu, Torrance, CA) with the respective standard buffers as references. The average absorbance at different pH was shown in Figure S1b and the error bars correspond to ± 1 standard deviation. To study the reversibility of the BPB-doped silica films, one of the BPB-doped films was fixed to a Petri-dish bottom and mounted to a Leica microscope stage (Leica DMI 5000M, Wetzlar, Germany). The absorbance variations in response to alternate pH 5.0 and 1x PBS (7.4) were recorded in real time with the microscope head lamp (tungsten filament) as the light source. The spectra were recorded with an acquisition time of 0.1 s in every 2 s for 10 min.
pH sensor film preparation
pH sensor films were prepared with the following procedures. First, Gd2O2S: Eu (1 g/mL) was prepared by dispersing an appropriate amount of scintillators in 0.5 % CMC. Then, 100 μL of the solution was spread onto the other side of the BPB-doped silica films. After being dried at room temperature overnight, PDMS with a base to curing agent ratio of 10:1 was spread onto the surface of the scintillators to fix them in position. The PDMS layer was cured at room temperature for 2 d. To create a local reference region, a small square (~2 mm) of BPB-doped silica film was removed and then PDMS was spread onto the blank region.
Measurement of X-ray beam sizes
The sizes of the X-ray beams were measured with the setup shown in Figure S4a. Specifically, a scintillator film was irradiated with an X-ray beam formed with different collimators (brass cylinder with various hole diameters) mounted on the X-ray tube, and the luminescent images were captured with a Nikon camera (D90, Japan). An image of a ruler was also taken with the same setup with room light on. The red components of the luminescent images were analyzed with Matlab to determine X-ray beam profile and the width at 20% of the maximum intensity used as a conservative measure of beam width. To study the tissue effect on beam broadening, a 6 mm thick slice of porcine tissue was placed between the X-ray source and the scintillator film target, and luminescent images were acquired with X-ray on and room light off. The images were analyzed with Matlab to calculate X-ray beam sizes and compare with/without tissue.
Acquisition of X-ray excited optical luminescence spectra
The X-ray excited optical luminescence spectrum of a film of Gd2O2S: Eu phosphors was obtained with the setup displayed in Figure 1a. The sample was irradiated with a mini X-ray tube (Amptek Inc. MA, USA), operating at a tube voltage of 40 kV and a tube current of 99 μA. The X-ray beam diameter was controlled by mounting collimators at the end of the X-ray tube window. The X-ray excited optical luminescence was collected with a 10x objective lens (NA=0.3) under dark field mode (with no excitation, dirchroic, or emission filters in the optical path). The luminescence was then dispersed by a spectrometer (DNS 300, DeltaNu, Laramie, WY) with a 150 lines/mm grating and collected with a cooled CCD camera (iDUS-DU420A-BV, Andor, South Windsor, CT), set to 100 kHz transfer rate; at this setting, each digital count corresponded to 15 photoelectrons. The spectra were recorded with Andor’s Solis software with an acquisition time of 1 s otherwise indicated.
Target mapping
The target, “CU” letters in white, with a line width of 1 mm and a space of 2 mm between lines in an 18 mm x 18 mm black background, was printed onto the rough side of a transparency twice with ink jet ink (photo black, Nano Digital), using an epson stylus photo R200 ink jet printer. The target was then attached to one side of a microcoverslip with PDMS. The other side of the microcoverslip was deposited with 100 μL of Gd2O2S: Eu (1 g/mL in 0.5 % CMC) and PDMS sequentially. The target was first mapped without tissue, irradiated with a 1.5 mm X-ray beam. Then, it was sandwiched between two pieces of porcine muscle tissue and mapped with 3 mm, 1.5 mm and 1 mm X-ray beams as excitation sources, respectively. For collecting luminescence spectra point-by-point, the target was first placed onto the microscope stage and moved with a step size of 300 μm horizontally and vertically in a raster mode. The movement of the stage was controlled with an HCI image software (Hamamatsu Corporation, PA, USA). At the same time, the luminescence spectra were collected in kinetic acquisition mode with Andor software. The synchronicity was achieved by correlating the HCI image with the Andor acquisition software. For mapping the target with white light source, no scintillators were deposited on the other side of the target and the microscope head lamp (tungsten filament) was used as the illumination source. To create a ratio-mapping image, the data acquired were analyzed with a written Matlab script. Specifically, the peak intensity at 620 nm and that at 700 nm was obtained from all the spectra and then the ratios of the peak intensity at 620 nm over that at 700 nm were calculated. The ratios were then plotted with the pcolor function built in Matlab.
Bacterial culture and pH mapping through tissue
Fresh cultures of S. epid.12228 and S. epid. 35984 were prepared according to our previous procedure.[19a] Specifically, a single S. epid.12228 or S. epid. 35984 colony from a TSA plate streaked within less than a week was inoculated to sterile TSB (5 mL). Bacteria were grown to the stationary phase under shaking at 37 °C for 16–18 h. The fresh culture was then washed with sterilized PBS twice and dispersed in sterilized PBS (5 mL). The number of bacteria was quantified by measuring the optical density at 600 nm (Smartspec 3000, Bio-Rad). The concentration of bacteria was then diluted to 107 cells/mL with sterilized PBS. This bacterial solution (2 μL) was spotted to a small TSA plate supplemented with 1 % glucose. Glucose concentrations between 0.25%–4% (w/w) are widely used for culturing biofilms in the absence of a circulatory or flow system.[28] The pH sensor film was then laid on top of the TSA plate after the bacterial spot dried. The plate grown with S. epid.12228 was left grown at 37 °C for 2 h, while the plate inoculated with S. epid. 35984 was grown for 10 h at 37 °C. A photograph of the TSA plate was taken with a Nikon camera (D90, Japan) before it was sandwiched between two pieces of tissue (6 mm) for radioluminescence acquisition. Both the region where the bacteria were spotted and the reference region were mapped sequentially with the same setup described in the target mapping section. The region with bacteria was also mapped again 3 h later. For a control study, sterilized PBS (2 μL) was added instead of bacterial solution. After incubation at 37 °C for 10 h, the region where PBS was added and the reference region were mapped through tissue using the same method. The ratio of the peak intensity at 620 nm over that at 700 nm was calculated with the same Matlab script as in the target mapping section. The corresponding pH was calculated by referring to the external pH calibration curve and the reference region to account for the tissue effect.
Antibiotic effect study: S. epid
12228 was chosen as a model to study the effect of antibiotic on the bacterial growth. A fresh culture of S. epid.12228 was prepared as described above. The culture was also washed with PBS twice and diluted to a concentration of 107 cells/mL. Then, this bacterial solution (2 μL) was spotted to a TSA plate (1% glucose). A pH sensor film was placed after the spot dried. The whole TSA plate was incubated at 37 °C for 2 h for the bacteria to grow. A photograph of the TSA plate was obtained before the bacterial spot was mapped through porcine tissue (6 mm) with the method described in the above bacterial culture and pH mapping through tissue section. The whole mapping area was 7.5 mm × 7.5 mm (a 25x25 pixel field) with an increment of 0.3 mm. Then the reference region was also mapped with the same way. Afterwards, ciprofloxacin (1 μL, 1 mg/mL) was added onto the TSA plate at a distance of about 5 mm to the bacterial spot without in contact with the pH sensor film. The same bacterial spot was mapped again in 3 h, 6 h and 18 h, respectively. A photograph of the TSA plate with the pH sensor film was obtained after the last mapping. Data analysis was done the same way as in the above section.
Supplementary Material
Acknowledgments
This research was supported by National Science Foundation (NSF) CAREER award CHE1255535 and by NIGMS of the National Institutes of Health under award number 5P20GM103444-07.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.a) Vertes A, Hitchins V, Phillips KS. Anal Chem. 2012;84:3858. doi: 10.1021/ac2029997. [DOI] [PubMed] [Google Scholar]; b) Bryers JD. Biotechnol Bioeng. 2008;100:1. doi: 10.1002/bit.21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zalavras C, Christensen T, Rigopoulos N, Holtom P, Patzakis M. Clin Orthop Relat Res. 2009;467:1715. doi: 10.1007/s11999-009-0743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mody RM, Zapor M, Hartzell JD, Robben PM, Waterman P, Wood-Morris R, Trotta R, Andersen RC, Wortmann G. J Trauma Acute Care Surg. 2009;67:758. doi: 10.1097/TA.0b013e3181af6aa6. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerli W. Best Pract Res Clin Rheumatol. 2006;20:1045. doi: 10.1016/j.berh.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Sjollema J, Sharma PK, Dijkstra RJB, van Dam GM, van der Mei HC, Engelsman AF, Busscher HJ. Biomaterials. 2010;31:1984. doi: 10.1016/j.biomaterials.2009.11.068. [DOI] [PubMed] [Google Scholar]
- 6.Xu CT, Svenmarker P, Liu H, Wu X, Messing ME, Wallenberg LR, Andersson-Engels S. ACS Nano. 2012;6:4788. doi: 10.1021/nn3015807. [DOI] [PubMed] [Google Scholar]
- 7.Stewart PS, Franklin MJ. Nat Rev Micro. 2008;6:199. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 8.Hidalgo G, Burns A, Herz E, Hay AG, Houston PL, Wiesner U, Lion LW. Appl Environ Microbiol. 2009;75:7426. doi: 10.1128/AEM.01220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart PS, William Costerton J. Lancet. 2001;358:135. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 10.Esmonde-White KA, Esmonde-White FWL, Holmes CM, Morris MD, Roessler BJ. Diabetes Care. 2013;36:3652. doi: 10.2337/dc13-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Abu-Amer Y, Darwech I, Clohisy J. Arthritis Research & Therapy. 2007;9:S6. doi: 10.1186/ar2170. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Anderson JM, Rodriguez A, Chang DT. Seminars in Immunology. 2008;20:86. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shen Y, Liu W, Wen C, Pan H, Wang T, Darvell BW, Lu WW, Huang W. J Mater Chem. 2012;22:8662. [Google Scholar]; d) Konttinen YT, Takagi M, Mandelin J, Lassus J, Salo J, Ainola M, Li TF, Virtanen I, Liljeström M, Sakai H, Kobayashi Y, Sorsa T, Lappalainen R, Demulder A, Santavirta S. J Bone Miner Res. 2001;16:1780. doi: 10.1359/jbmr.2001.16.10.1780. [DOI] [PubMed] [Google Scholar]
- 12.Pratx G, Carpenter CM, Sun C, Lei X. IEEE Trans Med Imag. 2010;29:1992. doi: 10.1109/TMI.2010.2055883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Liu X, Liao Q, Wang H. Opt Lett. 2013;38:4530. doi: 10.1364/OL.38.004530. [DOI] [PubMed] [Google Scholar]; b) Li C, Di K, Bec J, Cherry SR. Opt Lett. 2013;38:2339. doi: 10.1364/OL.38.002339. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chen D, Zhu S, Yi H, Zhang X, Chen D, Liang J, Tian J. Med Phys. 2013;40:031111. doi: 10.1118/1.4790694. [DOI] [PubMed] [Google Scholar]
- 14.Carpenter CM, Pratx G, Sun C, Xing L. Phys Med Biol. 2011;56:3487. doi: 10.1088/0031-9155/56/12/003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Patrick AL, Yang Z, VanDerveer DG, Anker JN. Anal Chem. 2011;83:5045. doi: 10.1021/ac200054v. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Longfield DE, Varahagiri VS, Nguyen KT, Patrick AL, Qian H, VanDerveer DG, Anker JN. Analyst. 2011;136:3438. doi: 10.1039/c0an00931h. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Moore T, Qi B, Colvin DC, Jelen EK, Hitchcock DA, He J, Mefford OT, Gore JC, Alexis F, Anker JN. ACS Nano. 2013;7:1178. doi: 10.1021/nn304369m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tseng T-K, Choi J, Jacobsohn L, Yukihara E, Davidson M, Holloway P. Appl Phys A. 2010;100:1137. [Google Scholar]
- 19.a) Wang F, Raval Y, Chen H, Tzeng T-RJ, DesJardins JD, Anker JN. Adv Healthcare Mater. 2014;3:197. doi: 10.1002/adhm.201300101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Makote R, Collinson MM. Anal Chim Acta. 1999;394:195. [Google Scholar]
- 20.a) Cubeddu R, Pifferi A, Taroni P, Torricelli A, Valentini G. Appl Phys Lett. 1999;74:874. doi: 10.1364/ao.38.003670. [DOI] [PubMed] [Google Scholar]; b) Farrell TJ, Patterson MS, Wilson B. Med Phys. 1992;19:879. doi: 10.1118/1.596777. [DOI] [PubMed] [Google Scholar]
- 21.Kirkland JJ, Henderson JW, DeStefano JJ, van Straten MA, Claessens HA. J Chromatogr A. 1997;762:97. doi: 10.1016/s0021-9673(96)00945-4. [DOI] [PubMed] [Google Scholar]
- 22.Lee S, Ibey BL, Coté GL, Pishko MV. Sensor Actuat B-Chem. 2008;128:388. [Google Scholar]
- 23.Ge J, Zhao L, Chen L-R, Shi Y-P. J Chromatogr Sci. 2010;48:29. doi: 10.1093/chromsci/48.1.29. [DOI] [PubMed] [Google Scholar]
- 24.a) Helmchen F, Denk W. Nat Meth. 2005;2:932. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]; b) Boas DA, Brooks DH, Miller EL, DiMarzio CA, Kilmer M, Gaudette RJ, Quan Z. IEEE Signal Proc Mag. 2001;18:57. [Google Scholar]
- 25.Ntziachristos V. Nat Meth. 2010;7:603. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 26.Seltzer SM. Radiat Res. 1993;136:147. [PubMed] [Google Scholar]
- 27.a) Drlica K, Zhao X. Microbiol Mol Biol Rev. 1997;61:377. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sanders CC. Rev Infect Dis. 1988;10:516. doi: 10.1093/clinids/10.3.516. [DOI] [PubMed] [Google Scholar]
- 28.a) StepanoviĆ S, VukoviĆ D, Hola V, Bonaventura GD, DjukiĆ S, ĆIrkoviĆ I, Ruzicka F. APMIS. 2007;115:891. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]; b) Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. J Microbiol Methods. 2000;40:175. doi: 10.1016/s0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.