Abstract
In tissues, macrophages are exposed to metabolic, homeostatic and immune-regulatory signals of local or systemic origin that influence their basal functions and responses to danger signals. Signal transduction pathways regulated by extracellular signals are coupled to distinct sets of broadly expressed stimulus-regulated transcription factors whose ability to elicit gene expression changes is influenced by the accessibility of their DNA binding sites in the macrophage genome. In turn, accessibility of macrophage-specific transcriptional regulatory elements (enhancers and promoters) is specified by transcription factors that determine the macrophage lineage or impose their tissue-specific properties. Here, we review recent findings that advance our understanding of mechanisms underlying priming and signal-dependent activation of macrophages, and discuss the impact of genetic variation on these processes.
Macrophages are present in virtually all tissues, where they integrate a large number of inputs to coordinate developmental, metabolic and immune functions, thus critically contributing to maintain homeostasis. The complexity of macrophage roles in tissues, their impact on homeostasis and disease, and the possibility to exploit their functional plasticity for therapeutic purposes has increased the general interest towards these cells and prompted a large number of mechanistic studies.
Macrophage activation and conditioning by a broad panel of stimuli
Many functional and nearly all molecular studies of macrophages by necessity have until now mainly focused on primary macrophages and macrophage cell lines exposed in vitro to single, strongly polarizing ligands, with lipopolysaccharide (LPS), interferon gamma (IFNγ) and interleukin 4 (IL-4) providing the most intensively studied paradigms. In vivo, however, macrophages are - often simultaneously - exposed to a broad range of stimuli whose integration eventually determines a continuum of distinct transcriptional and functional outputs1–3.
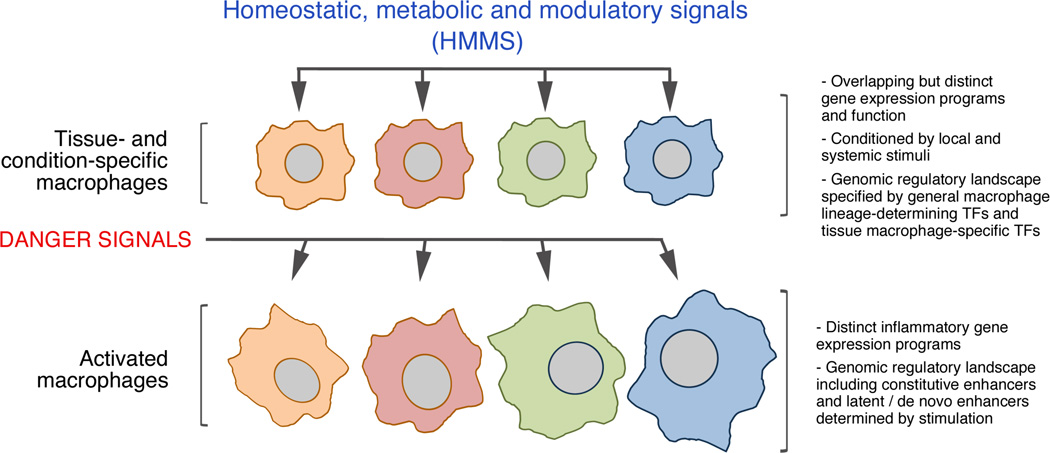
For the sake of clarity, we can categorize stimuli acting on macrophages and influencing their biology into two main classes, danger signals on the one hand and homeostatic, metabolic and modulatory signals (HMMS) on the other (Figure 1).
Figure 1.
The interplay between homeostatic tissue signals and danger signals in the control of macrophage function. Tissue macrophages are exposed to micro-environmental signals that impact their gene expression programs and function and also affect the quality of their response to danger signals, resulting in distinct inflammatory gene expression programs in different tissues.
Danger signals include virtually all microbial components that don't have a counterpart in the animal kingdom (Pathogen Associated Molecular Patters, such as LPS)4, 5 or that reach intracellular sites where they are not normally present (such as viral DNA in the cytoplasm of infected cells)6, 7, but also endogenous molecules whose presence at high levels in the extracellular milieu sampled by macrophages denotes a local loss of cellular or tissue integrity. The cellular site of initial detection of a specific danger signal varies, which in the specific case of microbial signals reflects both the distinct route of entry of the pathogen and, correspondingly, the different subcellular localization of cognate Pattern Recognition Receptors8. While the trans-membrane Toll-like receptors (TLR) can be associated with either the cell surface (e.g. TLR4, sensing LPS) or the endosomes (e.g. TLR3, sensing double stranded RNA after virus uptake into phagosomes), a panel of sensors including the dsRNA-specific RIG-I helicase and the DNA-specific cyclic GMP-AMP synthase (cGAS) constantly monitor the anomalous presence of these nucleic acids in the cytoplasm6, 7, 9.
The endogenous danger signals are collectively indicated as alarmins10 and include the chromatin protein HMGB1 as well as extracellular ATP, which are both actively released during sterile injury (such as trauma) and determine responses analogous to those triggered by their microbial counterparts. Macrophage exposure to either endogenous or microbial danger signals should be considered as an exceptional event triggering an emergency response that aims at eliminating the invaders, repairing the tissue and restoring homeostasis.
A conceptually much different situation is the stimulation of tissues macrophages by a broad range of HMMS. In fact, macrophage exposure to these stimuli impacts on their ability to respond to danger signals (Figure 1). One paradigmatic example is provided by the response to apoptotic bodies: while their ingestion by monocytes recruited to an inflamed environment results in additional inflammation, their ingestion by tissue macrophages is usually immunologically silent11. The spectrum of HMMS is broad and includes: i) immune-modulatory cytokines metabolites and nutrients; iii) apoptotic bodies; iv) mechanical signals.
The detailed analysis of these stimuli is beyond the scope of this review and below we refer the reader to several excellent review articles. We only briefly discuss selected instances that are more relevant to our main focus. As regards immune-modulatory cytokines, some have a prominent and well-characterized role in macrophage biology. IL-4 and the closely related IL-13 induce alternative macrophage activation in the context of the response to helminths and in allergic diseases12 but also modulate macrophage function in tissues, notably the adipose tissue, where they control normal homeostasis and the thermogenic response to cold stress13. Most importantly, pre-exposure to IL-4 alters the ability of macrophages to respond to pathogen infection14, providing a paradigmatic example of how tissue conditioning may regulate inflammatory responses generated by macrophage recognition of danger signals. Among metabolic stimuli and nutrients that control macrophage biology, heme released upon erythrocyte disposal triggers the formation of highly specialized red pulp macrophages via induction of the transcription factor SPI-C15, while Retinoic Acid promotes the generation of peritoneal macrophages via induction of the transcription factor GATA6, and fatty acids contribute to macrophage activation in obesity, thus subverting their conditioning by locally produced IL-416–18. Other notable examples of the impact of a locally produced metabolite are provided by lactate generated by aerobic glycolysis in tumors -which induces macrophage expression of some genes critical for tumor growth19- and by succinate produced upon macrophage activation by LPS, which stabilizes the Hypoxia Inducible Factor 1α (HIF1α) thus enhancing IL-1b production20. Apoptotic bodies normally generated during developmental and tissue remodeling processes are recognized by dedicated receptors expressed by macrophages recruited in response to eat-me signals and, as discussed above, have a differential potential to activate macrophages depending on their pre-existing state11, 21. Finally, mechanical stimulation in tissues also affects macrophage function, with elongation stress promoting an M2 like gene expression program and reduced secretion of inflammatory cytokines22.
Relaying signals to the nucleus by stimulus-regulated transcription factors
Specific coupling of such individual signals to distinct transcriptional outputs is enabled by two distinct groups of mechanisms: first, the selective activation of a limited number of signaling pathways and downstream transcription factors by each receptor; and second, the pre-existing repertoire of accessible genomic regulatory sequences available for such transcription factors to land and subsequently regulate gene expression (discussed in the next chapters).
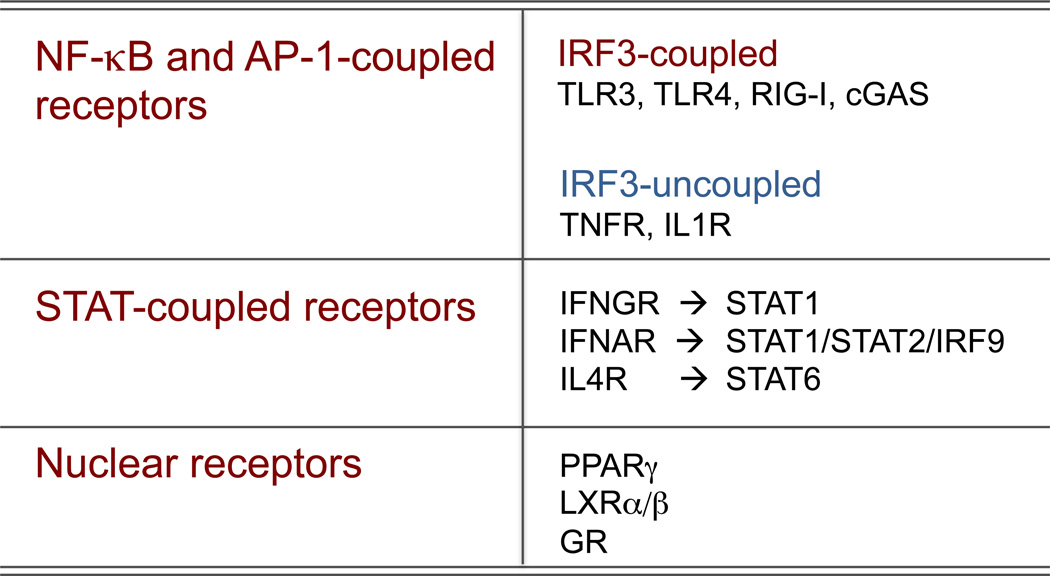
Based on their coupling to pathways and transcription factors, three broad groups of receptors particularly relevant for macrophage activation and priming can be identified (Figure 2): i) receptors coupled to the NF-κB and AP-1 family of transcription factors, which control a large number of canonical inflammatory genes; ii) receptors coupled to STAT family transcription factors; iii) nuclear receptors.
Figure 2.
Three main groups of receptors relevant for macrophage activation were schematically classified based on the main transcription factors coupled to them. The coupling to the IRF3 transcription factor is critical for the activation of the Ifnb1 gene and the ensuing interferon response, which accounts for most of the secondary gene expression in macrophages activated by LPS.
Receptors that activate NF-κB (via the IκB-kinase complex, IKK)23 and AP-1 (via Jun kinases, JNKs, which represent a distinct branch of MAP kinases)24, include all TLRs, many members of the Tumor Necrosis Factor receptor superfamily (TNFRSF) such as TNF receptors I and II (TNFRSF1A and TNFRSF1B) and CD40, as well as the type I Interleukin 1 receptor (IL1R1 or CD121a). These receptors act through distinct signal transducers and adapters, which contributes to explain the quantitative and kinetic differences in NF-κB and AP-1 activation. More importantly, they can be subdivided based on their ability (TLR3, TLR4, RIG-I, cGAS) or inability (TNFRs and IL1R) to trigger the phosphorylation, dimerization and nuclear entry of the transcription factor Interferon Regulatory Factor 3 (IRF3), which is essential for the activation of the IFNγ1 gene and the ensuing IFN-dependent deployment of an anti-microbial response25.
STAT-coupled receptors26 include the IFNγ receptor (IFNGR, which activates mainly STAT1 homodimers), the IFNα/γ receptor (IFNAR1, which activates STAT1:STAT2:IRF9 trimers with a distinct DNA-binding specificity compared to that of STAT1:STAT1)27 and the IL-4 receptor (IL4R, which is coupled to STAT6 homodimers). The sets of genes activated by these receptor-transcription factor pairs reflect the distinct biological functions of the activating cytokines. While IFNγ and IFNα/γ are mainly dedicated to the activation of genes involved in the containment of intracellular pathogens, IL-4 triggers an alternative activation program that is relevant for immunity against helminths and allergic reactions but also, as discussed above, for metabolic homeostasis.
In addition to signals that result from activation of cell surface receptors, macrophage phenotypes can be strongly influenced by intra- or extra-cellular signals that regulate members of the nuclear receptor superfamily. Several members of this family, including the glucocorticoid receptor, PPARγ, and liver X receptors (LXRs), function as counter-regulatory factors that suppress transcriptional activities of NF-κB and other pro-inflammatory transcription factors through direct and indirect mechanisms28–31. Recent studies further indicate important roles of nuclear receptors in specifying tissue-specific programs of macrophage gene expression. For example, local production of retinoic acid has been suggested to activate retinoic acid receptors in peritoneal macrophages, which in turn induces expression of GATA6 and other transcription factors that collaborate with PU.1 to drive a peritoneal macrophage-specific program of gene expression16, 17, 32. Conversely, induction of PPARγ expression by GM-CSF is critical for the proper development of alveolar macrophages and expression of genes required for their specific functions in clearance of pulmonary surfactant33.
Decoding signals at the genome level. General principles about the cis-regulatory information of mammalian genomes
Once activated, stimulus-regulated transcription factors control gene expression by binding to promoter and enhancer elements of target genes. As discussed above, most stimulus-regulated transcription factors are broadly expressed, yet their activation in different cell types results in cell-specific responses. For example, LPS activates NF-κB in both macrophages and B cells. While some NF-κB target genes are induced to a similar extent in both cell types, many others are preferentially activated in one cell type or the other34. These cell-specific responses at the level of gene expression are linked to different functional consequences; for example B cells35, but not macrophages, proliferate in response to LPS signaling. In contrast, the transcriptional response in macrophages is much more strongly coupled to the induction of genes that promote tissue inflammation36. An important question is to understand how specific signals, such as LPS, direct distinct patterns of gene expression required for the specialized functions of different cell types.
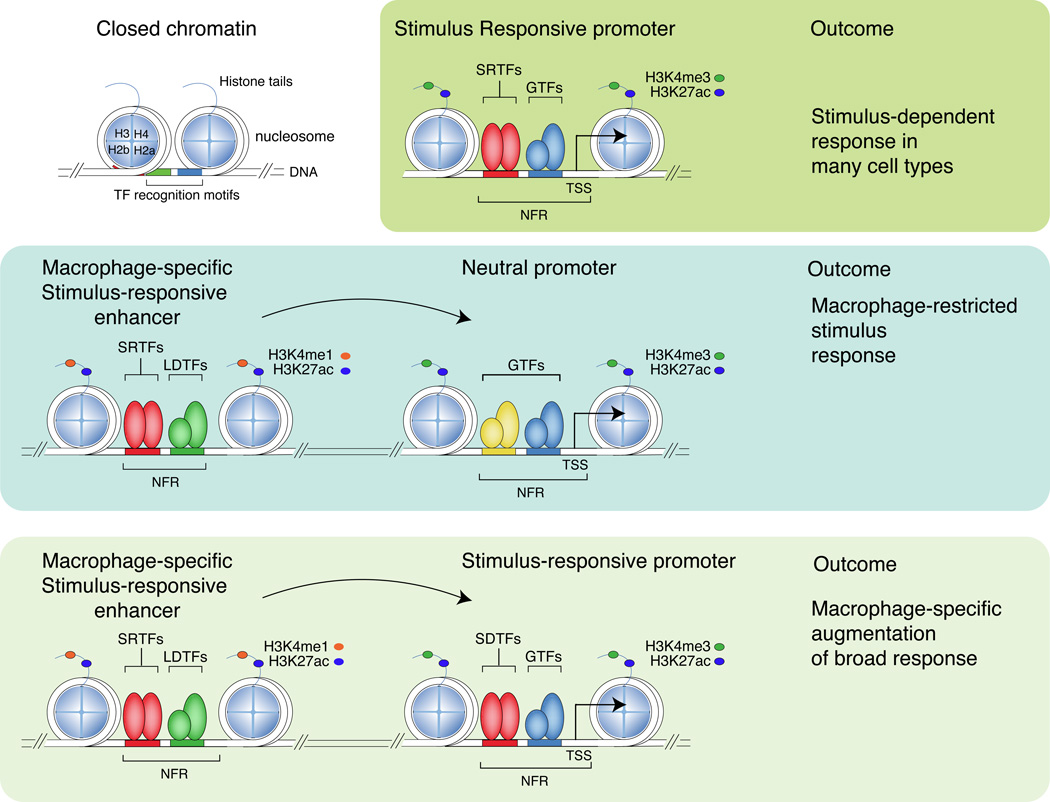
Signal-dependent gene expression is achieved at the level of individual genes through the actions of stimulus-regulated transcription factors (SRTFs) on promoters and enhancers, heretofore collectively indicated as cis-regulatory elements (Figure 3). These genomic elements must be recognized and interpreted within the context of chromatin, in which the fundamental repeating subunit is the nucleosome. Each nucleosome consists of an octamer of two copies of histones H2a, H2b, H3 and H4 that is encircled by approximately two turns of DNA (146 bp). Any given DNA fragment has a different affinity for core histones and thus a different ability to efficiently assemble nucleosomes37, 38. Affinity for histones is influenced by the DNA sequence and in general increases with G+C content39 up to a specific point beyond which the extremely high G+C content disfavors nucleosome assembly. In fact, the very high local concentration of G+C in CpG islands, which account for about 70% of mammalian gene promoters, interferes with nucleosome assembly in vivo and in vitro and thus accounts for their nucleosome depletion40–42. The regulatory relevance of the association of cis-regulatory sequences with nucleosomes is indicated by the observation that transcription factor binding sites are usually embedded in sequences with a high affinity towards nucleosomes43, implying that transcription factors must compete with nucleosomes to gain access to regulatory DNA sequences (Figure 3).
Figure 3.
Stimulus-regulated transcription factors (SRTFs) act at promoters and enhancers to direct broad or cell restricted transcriptional responses. Promoters and enhancers are primed by lineage-determining transcription factors (LDTFs) that collaborate with each other and other transcription factors to displace nucleosomes. Promoters, which are distinguished by high levels of H3K4me3 in comparison to H3K4me1, are generally primed in many cell types by broadly expressed transcription factors. Enhancers, which are distinguished by high levels of H3K4me1 in comparison to H3K4me3, are more likely to be primed by cell-type specific combinations of lineage determining transcription factors. PU.1, C/EBPs, AP-1 factors and IRFs are important macrophage lineage determining factors that drive the selection of a large fraction of macrophage-specific enhancers. Stimulus-regulated transcription factors primarily bind to DNA and activate gene expression at primed promoters and enhancers that contain their DNA recognition motifs and result in in recruitment of co-activator complexes that deposit H3K27ac. Binding of a stimulus regulated transcription factor to a promoter that is primed in many cell types is likely to result in a broad signal-dependent response. Binding of a stimulus regulated transcription factor to a cell-specific enhancer is likely to result in a cell restricted response or cell-specific potentiation of a broad response. H3K27Ac: histone H3 acetylated at lysine 27. H3K4me1: histone H3 monomethylation at lysine 4. NFR; nucleosome free region. TSS; transcriptional start site. SRTF; stimulus responsive transcription factor. GTF; general transcription factor (e.g., SP-1). LDTF; lineage determining TF (e.g. PU.1), SRTF; Stimulus responsive transcription factor, e.g., NFκB. H2a, H2b, H3, H4; histones H2a, 2b, 3 and 4, respectively.
Each histone within the octamer has the potential to be altered by a large number of post-translational modifications that primarily occur at residues within their protruding amino termini, known as histone tails44. Specific modifications are associated with different functional states. Although the precise functional role of different chromatin modifications and their readers in this specific and other responses is only partially understood45, evidence for a direct role of chromatin in regulating macrophage responses has been reported46, 47. Promoters are characterized by high levels of trimethylation of histone H3 lysine 4 (H3K4me3) in comparison to mono- or di-methylation of this residue, whereas enhancers exhibit high levels of monomethylation of H3 lysine 4 (H3K4me1) in comparison to tri-methylation. Histone acetylation of lysine residues, including H3 lysines 9 and 27 (H3K9ac and H3K27ac), and H4 lysines 5, 8 and 12, is associated with active promoters and enhancers. Conversely, alternative histone modifications are associated with transcriptional repression, including tri-methylation of H3 lysine 27 (H3K27me3) and H4 lysine 20. By using antibodies that recognize specific histone modifications to immunoprecipitate fragmented chromatin and performing massively parallel sequencing of the enriched DNA (ChIP-Seq), it is possible to derive genome-wide maps of these histone modifications48. This allows a global inference of enhancers and promoters and their relative activity states. For example, a genomic region marked by high levels of H3K4me1 in comparison to H3K4me3 and also marked by H3K27ac would be scored as a putative active enhancer (Figure 3, right).
By using ChIP-Seq to map promoter and enhancer regions in diverse cell types and tissues, it has been estimated that the mammalian genome contains hundreds of thousands of enhancer-like elements, greatly exceeding the number of protein coding genes and their respective promoters49–51. Each cell type selects from this large repertoire of potential regulatory elements on the order of 20 – 40 thousand enhancer-like regions49–51 -namely, regions that based on their histone modification profile and sequence composition are likely enhancers, although this is difficult to experimentally validate on such a large scale. A large fraction of such enhancers are active in a cell-specific manner. Because stimulus-regulated transcription factors (STRFs) operate at both enhancers and promoters, these observations suggest that their actions at cell-specific enhancers are important determinants of cell-specific transcriptional responses to a given signal.
A requirement for understanding stimulus-dependent responses is thus to understand the mechanisms by which specific promoter and enhancer elements are selected from the genome. Cis-regulatory elements consist of combinations of binding sites for sequence-specific transcription factors clustered within a short distance (usually less than 150–200 base pairs)52. However, the structural organization of the nucleosome results in a physical barrier to the binding of transcription factors to the side of DNA helix facing the nucleosome core. Of the hundreds of sequence specific transcription factors that are expressed within a cell, only a small fraction are able to recognize their respective recognition sequences in the context of a chromatin environment53. Such transcription factors are often referred to as pioneer factors and largely coincide with lineage-determining transcription factors (LDTFs)(Figure 3), namely transcription factors determining cell fate choices. Since cis-regulatory sequences have a high affinity for nucleosomal histones42, 43 and are thus partially occluded (Figure 3), effective binding to DNA occurs when there is a concordance of a combination of closely spaced sequence motifs for factors that are expressed in that cell type that have pioneering activity. In general, promoters contain combinations of DNA binding sites that are recognized by relatively broadly expressed transcription factors, such as SP-1, YY-1, NFY and Gabpa54. Consistent with this, the H3K4me3 mark associated with promoters can be detected for one half to two thirds of all protein-encoding genes within a particular cell type. In resting macrophages, H3K4me3 not only marks promoters of genes that are constitutively expressed, but is also present at most of the TLR4-responsive promoters prior to exposure to LPS55. Furthermore, many of the most stimulus-responsive promoters are occupied by RNA polymerase II that has initiated transcription of a short mRNA but is paused 30–60 nucleotides from the transcriptional start site56. In general, the presence of a very high G+C content or a CpG island in a gene promoter strongly correlates with its constitutive association with RNA polymerase II and H3K4me340, 57, which may relate both to the nucleosome depletion associated with these sequence features40–42 and with the high G+C content of binding sites for broadly expressed transcription factors such as SP-158. Instead, the presence at promoters of an intermediate G+C content that favors nucleosome assembly imposes the requirement for chromatin remodeling enzymes in gene activation40, 58. In turn, this results in a tighter regulation of gene expression and a higher dynamic range of LPS-induced expression compared to genes containing a CpG island58. Moreover, changes in chromatin states at promoters may underlie complex functional interactions between macrophage-activating stimuli. While IFNγ is unable to activate many LPS-inducible genes, it can induce histone acetylation and chromatin remodeling at their promoters, thus priming them for enhanced activation in response to LPS59.
Collectively, these features indicate that promoter regions of a large fraction of signal responsive genes are poised for rapid responses.
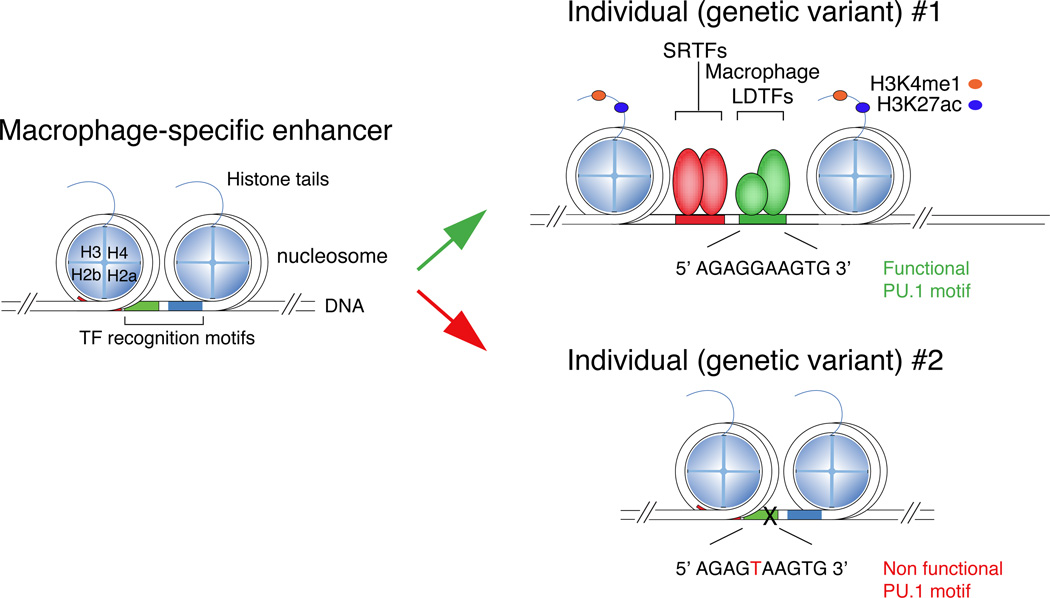
In contrast to promoters, the enhancers within a particular cell type are more highly enriched for recognition motifs for lineage-determining/pioneer factors that are required for the development of that cell type51 (Figure 3). In macrophages, enhancer-like regions are highly enriched for motifs for PU.1, C/EBPs, AP-1, IRFs and RUNX transcription factors, all of which are required for macrophage development and function60–62. Furthermore, these factors reside at a large fraction of the H3K4me1-marked regions in these cells. For example, ChIP-Seq experiments for PU.1 and C/EBPα indicate that they are present, alone or together, at nearly two thirds of the H3K4me1-marked regions60 (this overlap being influenced by experimental and analytical issues such as the efficiency of the ChIP, the sequencing depth and the statistical thresholds used to call ChIP peaks). At enhancer-like regions occupied by both PU.1 and C/EBP or by PU.1 and IRF8, binding is frequently mutually dependent60, 63. Using natural genetic variation as a ‘mutagenesis’ approach, mutations in C/EBP motifs were not only found to abolish binding of C/EBP factors, but also abolished the binding of PU.1 to nearby PU.1 recognition motifs that were not directly affected by mutations64. Reciprocally, mutations in PU.1 motifs affected both PU.1 binding and the binding of C/EBP factors to nearby intact C/EBP recognition motifs (Figure 4). These results indicate that while each factor is ‘pioneering’, it is not alone able to compete effectively with histones without collaborative interactions with the other. In line with this notion, among the hundreds of thousands of PU.1 binding sites that are present in the mouse genome (which include bona fide sites and many random occurrences of nucleotide combinations resembling PU.1 sites), those that are bound in vivo (as determined by ChIP-seq in different PU.1-expressing cell types) are located nearby binding sites for other lineage-determining transcription factors42. By extension, PU.1 and C/EBP factors are predicted to require collaborative interactions with other partners to bind to locations where one is present without the other. Consistent with this, a systematic analysis of motif mutations that result in altered binding of PU.1 in peritoneal macrophages and microglia provided evidence for many additional transcription factors beyond PU.1 that function as collaborative partners for PU.1, including AP-1 factors, IRFs, KLF family members and GATA factors32. These findings suggest that PU.1, C/EBP and a handful of other macrophage lineage-determining transcription factors collaborate with each other and most likely with dozens of other transcription factors to set up the majority of the macrophage-specific enhancer landscape.
Figure 4.
Effect of natural genetic variation on enhancer selection and function. Two allelic forms of the genomic region shown at left are distinguished by a single nucleotide polymorphism (SNP) in the DNA recognition motif for PU.1. Genetic variant 1 (at top) preserves the PU.1 recognition motif, enabling PU.1 to bind and collaborate with other LDTFs and provide access to SRTFs. Genetic variant 2 disrupts the PU.1 motif, preventing PU.1 from binding and resulting in corresponding loss of binding of collaborative LDTFs and SRTFs. Because this genomic region only achieves features of active enhancers in cells that express the correct combinations of LDTFs and active SRTFs, effects of this SNP on gene expression are specific for those cell types, e.g., macrophages.
Actions of stimulus-regulated transcription factors at pre-existing and latent enhancers
An important observation to emerge from the study of signal-dependent gene expression in macrophages is that many enhancer-like regions exist in a ‘primed’ (H3K4me1-positive/H3K27ac negative) and transition to an ‘active’ state (H3K4me1-positive/H3K27ac-positive) in a signal-dependent manner61, 64, 65. Furthermore, ChIP-Seq studies of several signal responsive transcription factors, including p65 (NF-κB), STAT factors, and nuclear receptors, indicate that the great majority (80–90%) of signal dependent DNA binding events occur at accessible genomic regions, many of which exhibit features of primed or active enhancers61, 64–66 (Figure 3). Because these open regions of chromatin are established by macrophage lineage-determining factors (LDTFs) in a cell-specific manner, the DNA binding patterns of the stimulus-regulated transcription factors (SRTFs) to these sites exhibit corresponding cell-specific binding patterns. DNA binding to enhancer-like regions in the vicinity of positively regulated genes is generally associated with a gain in H3K27ac, consistent with local recruitment of histone acetyltransferases that promote transition of primed enhancers to an active state61, 64, 65. These observations support the concept that one mechanism by which broadly expressed stimulus-regulated transcription factors exert cell specific functions is by being directed to pre-established enhancers that are associated with the appropriate set of target genes for that factor in that cell type.
While the majority of the binding of stimulus-regulated transcription factors occurs at pre-existing enhancer-like regions, some stimuli promote the binding of a small percentage of such factors (e.g., 10–20%) to closed regions of the genome and lead to the acquisition of histone modifications associated with enhancers65, 67. These regions, referred to as ‘latent’ or ‘de novo’ enhancers are of great interest because they provide the opportunity to visualize time-dependent intermediates in the process of enhancer selection and to determine functional consequences of histone modifications following cessation of the inducing stimulus. Binding of stimulus regulated transcription factors to latent/de novo enhancers co-occurred with and was dependent on macrophage lineage determining factors65, 67, indicating that they functioned as obligatory collaborative partners at these sites. However, not all stimulus-regulated transcription factors are similarly able to bind and modify regions not pre-marked by PU.1 and C/EBP TFs. In response to LPS, AP-1 and IRF8 may have a dominant role in opening previously inaccessible genomic regions. Indeed, a large fraction of LPS-inducible IRF8 binding events occurred at regions that were devoid of PU.1 binding and more than 50% of them at over 5 kb of distance from the nearest PU.1 bound site63. Importantly, many of these latent / de novo enhancers retained their histone modification signatures following withdrawal of the initiating stimulus and were associated with a faster, stronger and more diversified response to re-stimulation67. These observations suggest that the writing of the histone modification signature provides an epigenomic memory of the prior stimulus that facilitates subsequent responses.
Intriguingly, enhancers have been found to be sites of RNA-Pol II-dependent transcription, resulting in the production of capped but unstable and short enhancer RNAs (eRNAs)68–71. Emerging evidence suggests that at least some of these eRNAs contribute to enhancer activity72. Several mechanisms have been proposed, including contributions of eRNAs to enhancer/promoter looping73, recruitment of mediator (a multi-subunit complex tightly associated with RNA Pol II and required for its recruitment and transcriptional activation at most genes)74, and displacement of NELF (a factor that negatively regulates the early phases of transcription elongation)75. Not all eRNAs exhibit activity, however, and the diversity of proposed mechanisms implies that substantial additional work will be required to determine the spectrum and general importance eRNA-dependent mechanisms in enhancer function76. Enhancer transcription itself, independent of the eRNA product, has also been shown to contribute to the writing of the H3K4me1/2 enhancer signature at de novo enhancers65. At these locations, transcription factor binding was tightly coupled to histone acetylation and eRNA production and preceded histone H3K4me1 and H3K4me2 deposition. H3K4 methylation was blocked by inhibitors of RNA Pol II elongation and knockdown of MLL3/4, two paralogous and partially redundant methyltransferases previously implicated in the deposition of H3K4me1 at enhancers77, 78. These results suggest that the writing of H3K4me1/2 at de novo enhancers results from the recruitment of MLL proteins, possibly mediated by their binding to the C-terminal domain of RNA Pol II during elongation from enhancer transcriptional start sites.
Communication between enhancers and promoters is considered to be through looping, in which regions of DNA that are far apart in one dimensional space are brought into close proximity in 3 dimensions79. The definitive test of an enhancer’s function is to delete the corresponding genomic region and assess consequences with respect to gene expression. This has been performed thus far for only a small number of putative enhancer elements. However, physical interactions can be evaluated in a gene-specific or genome-wide manner through chromatin conformation capture methods that fix intra and inter chromosomal interactions by formaldehyde crosslinking79. These assays indicate that the genome is organized into compartments of different scales. At the largest scale, megabase regions of active and repressed chromatin are sequestered into separate compartments. Within these compartments, the genome is subdivided into self-associating regulatory blocks or ‘topological domains’ (TADs) - with an average size of 0.8–1 Mb in the mouse - that are generally similar across cell types. TADs can be further subdivided into regulatory blocks of genes and their associated enhancers. The boundaries of TADs and the subdomains within them are frequently found associated with the DNA sequence-specific binding protein CTCF, which is thought to play a role in constraining actions of enhancers to their specific target genes. These studies suggest that the organization of TADs and subTADs determines the range over which a stimulus responsive transcription factor could operate following binding to a particular enhancer.
Enhancer landscapes in tissue resident macrophages
Nearly all studies of macrophage activation have been performed in vitro, in which single or simple combinations of signaling molecules are added in the context of a controlled tissue culture environment. These experimental systems have been and will continue to be very powerful for elucidation of molecular mechanisms underlying specific programs of macrophage activation. For example, it was recently demonstrated that it is possible to perform a genome-wide CRISPR screen in bone marrow-derived dendritic cells to identify genes essential for LPS activation of TNFα expression80. This screen identified many new and unexpected genes required for TLR-dependent regulation of TNFα, indicating that there is much to learn even for intensively studied signaling pathways in a well-defined model system. However, recent genome-wide studies also indicate that different populations of tissue macrophages can exhibit highly divergent patterns of gene expression that are linked to their tissue-specific homeostatic functions81. Furthermore, these subset-specific patterns of gene expression are strongly influenced by the local tissue environment32, 82. For example, transplantation of peritoneal macrophages to the lung results in a substantial transition of gene expression from that observed in the peritoneal cavity to a pattern more like that expressed in alveolar macrophages82. Consistent with this, the enhancer landscapes associated with different tissue-resident macrophages are also shaped by the tissue environment32, 82. Approximately 15–20% of the enhancer-like regions found in large peritoneal macrophages are relatively specific for that subset in comparison to microglia, and vice versa.
Based on the in vitro observation that the majority of binding of signal-dependent transcription factors occurs at pre-established regulatory elements, an implication of the existence of subset-specific enhancers is that the responses of different tissue macrophage populations to the same signal may be qualitatively and/or quantitatively different as well. Similar responses to the same signal would be expected for genes that exhibit the same response element organization, but genes exhibiting gain or loss of signal-dependent enhancers in one macrophage subset as compared to another would be expected to respond differently. Also by extension from in vitro studies revealing latent/de novo enhancers, tissue-specific signals acting on stimulus-regulated transcription factors would be expected to drive gene expression through both pre-existing enhancers and the selection of new enhancers. There is emerging evidence that this is the case. For example, the retinoic acid receptor gene, which is auto-regulated by retinoic acid, exhibits evidence of enhancer priming in many macrophage subsets, but is particularly active in peritoneal macrophages that are exposed to relatively high concentrations of endogenous retinoic acid17, 32. This results in a peritoneal macrophage-specific expression of retinoic acid receptor target genes, which include transcription factors such as GATA6 that can collaborate with PU.1 to initiate selection of new enhancers that are specific to the peritoneal macrophage subset.
These observations raise the interesting question of the extent to which there is a ‘core’ or 'reference' macrophage epigenome -namely, a shared and subset-independent regulatory landscape- and whether or to what extent it is possible to predict responses of one macrophage subset based on the responses in another. This could have practical utility in that macrophages contribute to a broad range of diseases, but are difficult to access in most tissues. An important question is whether responses of monocytes, or monocyte-derived macrophages, could be used to predict pathological responses of macrophages in disease settings such as atherosclerosis, Alzheimer’s disease and cancer. Alternatively, knowledge of enhancer landscapes of different macrophage subsets and the corresponding transcription factors required for their selection could be useful in programming macrophages in vitro to assume more in vivo-like properties.
Regulatory variation: impact on regulation, health and disease
An important outcome of genome wide association studies (GWAS) that link common single nucleotide polymorphisms (SNPs) to phenotypes of interest is that regardless of the phenotype, ~90% of the variants identified are in non-coding regions of the genome83. These findings are consistent with an important role of such variants in affecting the function of transcriptional regulatory elements. However, identification of a SNP does not indicate mechanism of action, the cell type in which it acts or even the gene that is affected84. A further approach is to relate genetic variants to gene expression, thereby defining expression quantitative trait loci (eQTLs). For example, correlation of SNPs with gene expression in monocytes and T cells of a multi-ethnic cohort of individuals identified hundreds of eQTLs, a significant fraction of which had an association that was restricted to either T cells or monocyte gene expression85. This is consistent with the causal variant (the reference SNP itself or a linked variant) acting at a T cell or monocyte-specific regulatory element. Interestingly, an over-representation of monoctyte-specific eQTLs was observed among Alzheimer’s and Parkinson’s disease variants85, suggesting that studies of monocytes may have value for predicting functions of tissue macrophages and their impact on disease. Going forward, an important goal will be to directly examine the consequences of genetic variation on the selection and function of transcriptional control elements, taking advantage of the ability to measure transcription factor binding, enhancer and promoter activity states, and gene expression on a genome-wide level. An important example is provided by a recent study of the effect of natural genetic variation on the binding and function of PPARγ, a master regulator of adipogenesis and the molecular target of anti-diabetic drugs86. SNPs that alter PPARγ binding were found to alter adipose gene expression and modulate human metabolic disease risk. Such studies applied to macrophages are thus likely to provide substantial new insights into mechanisms that regulate gene expression and how non-coding genetic variation affects phenotypic diversity and disease.
Conclusions and future perspectives
Progresses in genomics have greatly increased our understanding of the transcriptional bases of macrophage-specific responses to environmental perturbations. Current models mechanistically explain how transcription factors controlling macrophage development set the stage for the activity of stimulus-dependent transcription factors, and how in turn activation can change the genomic cis-regulatory landscape. The same models provide an interpretative framework for the impact of genetic variability on macrophage-specific gene expression programs. However, most of the data converging into such models, albeit comprehensive, are correlative: a systematic and quantitative analysis of the role of individual cis-regulatory elements in the regulation of specific genes and in macrophage activation in response to different stimuli is the next frontier in the field.
Footnotes
Competing interests:
The authors declare no competing interests.
References
- 1.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nature reviews. Immunology. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nature reviews. Immunology. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 3.Murray PJ, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor symposia on quantitative biology. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annual review of immunology. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 6.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hornung V, Hartmann R, Ablasser A, Hopfner K-PP. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nature reviews. Immunology. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Cai X, Chiu Y-HH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Molecular cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 10.Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. Journal of leukocyte biology. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 11.Hochreiter-Hufford A, Ravichandran KS. Clearing the Dead: Apoptotic Cell Sensing, Recognition, Engulfment, and Digestion. Cold Spring Harbor Perspectives in Biology. 2013 doi: 10.1101/cshperspect.a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Dyken SJ, Locksley RM. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annual review of immunology. 2013;31:317–343. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu Y, et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell. 2014;157:1292–1308. doi: 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varin A, Mukhopadhyay S, Herbein G, Gordon S. Alternative activation of macrophages by IL-4 impairs phagocytosis of pathogens but potentiates microbial-induced signalling and cytokine secretion. Blood. 2010;115:353–362. doi: 10.1182/blood-2009-08-236711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haldar M, et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 2014;156:1223–1234. doi: 10.1016/j.cell.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosas M, et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chawla A, Nguyen KD, Goh YP. Macrophage-mediated inflammation in metabolic disease. Nature reviews. Immunology. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colegio OR, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor PR, et al. Macrophage receptors and immune recognition. Annual review of immunology. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 22.McWhorter FY, Wang T, Nguyen P, Chung T, Liu WF. Modulation of macrophage phenotype by cell shape. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17253–17258. doi: 10.1073/pnas.1308887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Science's STKE : signal transduction knowledge environment. 2006;2006:re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 24.Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nature reviews. Immunology. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 25.Doyle S, et al. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 26.O'Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nature reviews. Immunology. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- 29.Ito A, et al. LXRs link metabolism to inflammation through Abca1-dependent regulation of membrane composition and TLR signaling. Elife. 2015;4 doi: 10.7554/eLife.08009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P, et al. NCoR repression of LXRs restricts macrophage biosynthesis of insulin-sensitizing omega 3 fatty acids. Cell. 2013;155:200–214. doi: 10.1016/j.cell.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlenhaut NH, et al. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol Cell. 2013;49:158–171. doi: 10.1016/j.molcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider C, et al. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- 34.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li SK, Abbas AK, Solomon LA, Groux GM, DeKoter RP. Nfkb1 activation by the E26 transformation-specific transcription factors PU.1 and Spi-B promotes Toll-like receptor-mediated splenic B cell proliferation. Mol Cell Biol. 2015;35:1619–1632. doi: 10.1128/MCB.00117-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 37.Jiang C, Pugh B. Nucleosome positioning and gene regulation: advances through genomics. Nature reviews. Genetics. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struhl K, Segal E. Determinants of nucleosome positioning. Nature structural & molecular biology. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fenouil R, et al. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome research. 2012;22:2399–2408. doi: 10.1101/gr.138776.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barozzi I, et al. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Molecular cell. 2014;54:844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lidor Nili E, et al. p53 binds preferentially to genomic regions with high DNA-encoded nucleosome occupancy. Genome research. 2010;20:1361–1368. doi: 10.1101/gr.103945.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. J Mol Biol. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annual review of immunology. 2014;32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 46.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan CH, et al. BET bromodomain inhibition suppresses transcriptional responses to cytokine-Jak-STAT signaling in a gene-specific manner in human monocytes. European journal of immunology. 2015;45:287–297. doi: 10.1002/eji.201444862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roadmap Epigenomics C, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Escoubet-Lozach L, et al. Mechanisms establishing TLR4-responsive activation states of inflammatory response genes. PLoS Genet. 2011;7:e1002401. doi: 10.1371/journal.pgen.1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogatsky I, Adelman K. Preparing the first responders: building the inflammatory transcriptome from the ground up. Mol Cell. 2014;54:245–254. doi: 10.1016/j.molcel.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhatt DM, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiao Y, et al. Synergistic activation of inflammatory cytokine genes by interferon-gamma-induced chromatin remodeling and toll-like receptor signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 62.Lichtinger M, et al. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. The EMBO journal. 2012;31:4318–4333. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mancino A, et al. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev. 2015;29:394–408. doi: 10.1101/gad.257592.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heinz S, et al. Effect of natural genetic variation on enhancer selection and function. Nature. 2013 doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaikkonen MU, et al. Remodeling of the Enhancer Landscape during Macrophage Activation Is Coupled to Enhancer Transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barish GD, et al. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes & development. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 68.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koch F, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nature structural & molecular biology. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 71.Arner E, et al. Gene regulation. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schaukowitch K, et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annual review of genetics. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 77.Hu D, et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33:4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee JE, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol. 2013;25:387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parnas O, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015 doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30:1095–1096. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Soccio RE, et al. Genetic Variation Determines PPARgamma Function and Anti-diabetic Drug Response In Vivo. Cell. 2015;162:33–44. doi: 10.1016/j.cell.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]