Abstract
Identifying key molecules that launch regeneration has been a long sought goal. Multiple regenerative animals show an initial wound-associated proliferative response that transits into sustained proliferation if a significant portion of the body part has been removed 1-3. In the axolotl, appendage amputation initiates a round of wound-associated cell cycle induction followed by continued proliferation that is dependent on nerve-derived signals 4,5. A wound-associated molecule that triggers the initial proliferative response to launch regeneration has remained obscure. Using an expression cloning strategy followed by in vivo gain- and loss-of-function assays, we identified axolotl MARCKS like Protein (MLP) as an extracellularly released factor that induces the initial cell cycle response during axolotl appendage regeneration.
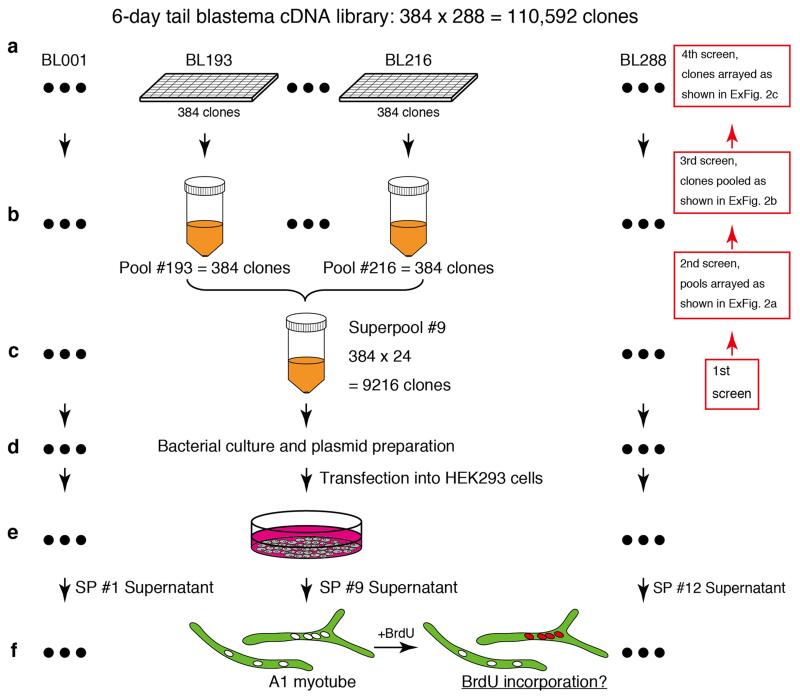
To identify a regeneration-initiating molecule in the salamander, Ambystoma mexicanum (axolotl), we aimed to functionally screen6,7 axolotl cDNAs using an in vitro salamander myotube cell cycle re-entry assay (Notophthalmus viridescens--newt) 8 with the aim of performing in vivo analysis in the axolotl that is convenient for molecular analysis 9-11. To establish if axolotl blastema tissue expresses a myotube cell cycle entry inducing factor, we injected Xenopus oocytes with mRNAs from tail blastema, limb blastema or mature limb and assayed the extracellular media on myotubes (Fig. 1a). Tail or limb blastema mRNAs scored positively, comparable to serum, whereas the mature tissue mRNAs showed little inducing activity. We next screened an arrayed 6-day tail blastema cDNA eukaryotic expression vector library for the activity12. Transfection of DNA representing the entire library as a single pool into HEK293 cells (Fig 1b, sample WL) yielded cell media that stimulated myotube cell cycle entry (Fig. 1b). This library was fractionated into 12 superpools, which yielded four positive superpools (superpool #6, 9, 10 and 12, Fig. 1b, ExFig. 1a-f). Sib-selection of superpool #9 through three subfractionation steps resulted in identification of a single clone responsible for the activity (ExFig. 2a-c).
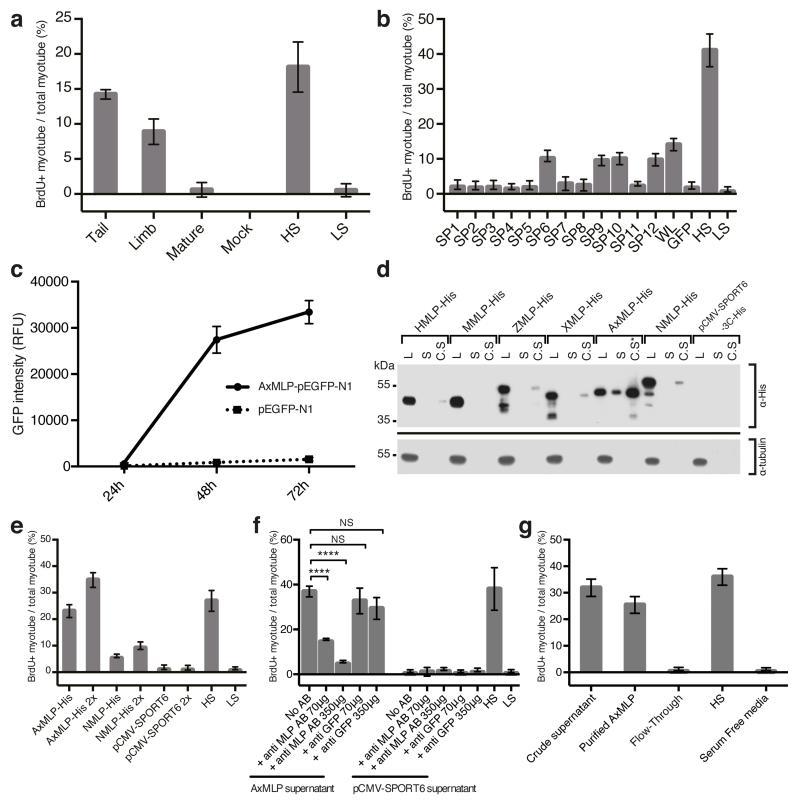
Figure 1. Extracellular AxMLP identified by expression cloning is necessary and sufficient for cell cycle re-entry in vitro.
a, Supernatants from Xenopus oocytes injected with total mRNA from tail and limb blastema induced robust BrdU incorporation in cultured new myotubes (“Myotube assay”). (n=6: 2 biological, 3 technical replicates each; mean±s.d.). b, A screen for the cell cycle inducing clone. Culture media from HEK293 cells transfected with 6-day tail blastema cDNA library pools and assayed on myotubes for cell cycle induction (see ExFig. 1 for scheme) identified four positive superpools (n=12: 4 biological, 3 technical replicates each; mean±s.d.). Superpool 9 was sib-selected to a single clone, see ExFig. 2. c, AxMLP-GFP fusion protein detection in culture media of transfected HEK293 cells by fluorescence luminometry. (n=3: biological replicates; mean±s.d.). d, MLP orthologs show differing levels of extracellular protein. AxMLP was readily detectable by Western Blotting in cell culture supernatant (S). Human, Zebrafish, Xenopus and Newt MLPs (HMLP, ZMLP, XMLP and NMLP) were only detectable in 40-fold concentrated supernatants (C.S). C.S.*: 5-fold concentrated supernatant. Loading control: anti-tubulin. e. AxMLP and NMLP supernatants both induce myotube cell cycle with response corresponding to protein levels in supernatant (n=6: 2 biological, 3 technical replicates each; mean±s.d.). f. Induction of myotube cell cycle re-entry by AxMLP is specifically blocked by addition of polyclonal anti-AxMLP antibodies to culture supernatant. (n=6: 2 biological, 3 technical replicates each; mean±s.d.). g, Purified AxMLP induces myotube cell cycle re-entry (n=6: 2 biological, 3 technical replicates each; mean±s.d.). SP, superpool; WL, whole library; HS, high serum; LS, low serum. L, cell lysate; S, supernatant. C.S, concentrated supernatant. NS, not significant; ****P<0.0001 with Student’s t-test.
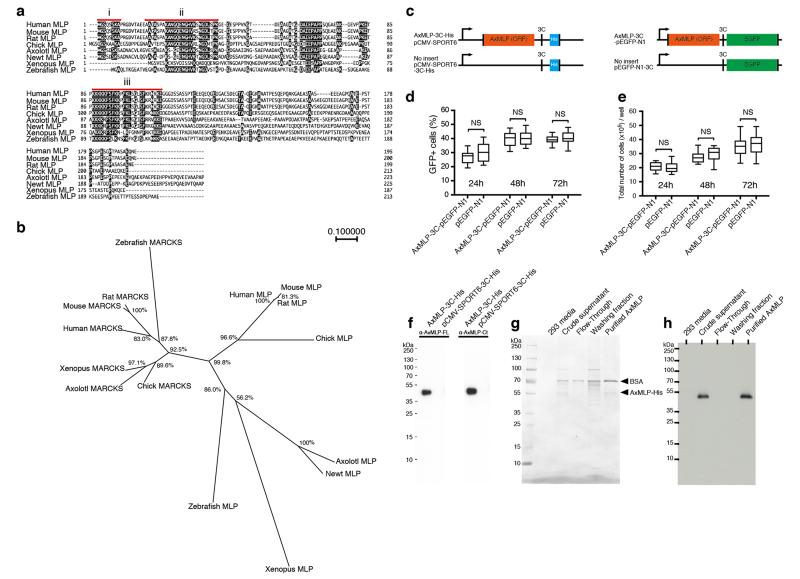
The positive clone encoded a 224 amino acid protein containing three conserved domains (myristoylated N-terminus, MARCKS homology domain and effector domain) similar to MARCKS-like Protein (MLP) (ExFig. 3a) showing 74.1%, 68.0% and 80% amino acid sequence identity to human MLP, including the glycine G2 in the myristoylated domain and two serines in the effector domain (S94 and S105) important for plasma membrane binding (for review see 13). The C-terminal region showed low (14.4%) sequence conservation. Phylogenetic analysis showed that the axolotl sequence clustered with other vertebrate MLPs (ExFig. 3b).
Previous work in other species had indicated that MLP is an intracellular substrate for protein kinase C associated with the plasma membrane, phagocytic vesicles, and actin while phosphorylation by PKC induced dissociation to the cytoplasm (for review see 14). We asked if AxMLP acted on myotubes as a secreted factor or whether it induced expression of a secreted factor in the HEK293 cells. To determine if AxMLP was extracellularly released we transfected an AxMLP-C-terminal EGFP fusion construct or the pEGFP-N1 control construct into HEK293 cells (ExFig. 3c). Increasing levels of GFP fluorescence intensity were observed in AxMLP transfected but not EGFP-N1 transfected culture media (Fig. 1c). The percentage of GFP+ cells and the cell number in AxMLP- and GFP-transfected samples remained equivalent over time (ExFig. 3d,e). Timelapse imaging further showed that AxMLP-transfected cells grew similarly to the control cells (ExMovie 1,2) indicating that extracellular AxMLP did not derive from dying cells. When comparing expression of AxMLP to zebrafish, Xenopus, mouse, newt and human MLPs in HEK293 cells, we found extracellular MLP from all species but the AxMLP yielded a high proportion of extracellular protein compared to other species (Fig. 1d). Bioassay of the axolotl versus newt MLP media induced a myotube response corresponding to the amount of protein seen by Western blot (Fig. 1e).
To establish necessity, we exposed AxMLP-containing culture media to a polyclonal antibody against AxMLP which inhibited the myotube response indicating that extracellular AxMLP is required for the activity (Fig. 1f, ExFig. 3f). To test sufficiency, we purified AxMLP-His which displayed a characteristic high gel mobility (ExFig. 3g,h) (for review see 13). Exposure of myotubes to purified AxMLP in serum free conditions yielded a robust myotube response with an approximate half-maximal response at 50.5 ng/μl (Fig. 1g, ExFig. 2d-f). We conclude that extracellular AxMLP is sufficient to induce myotube cell cycle re-entry.
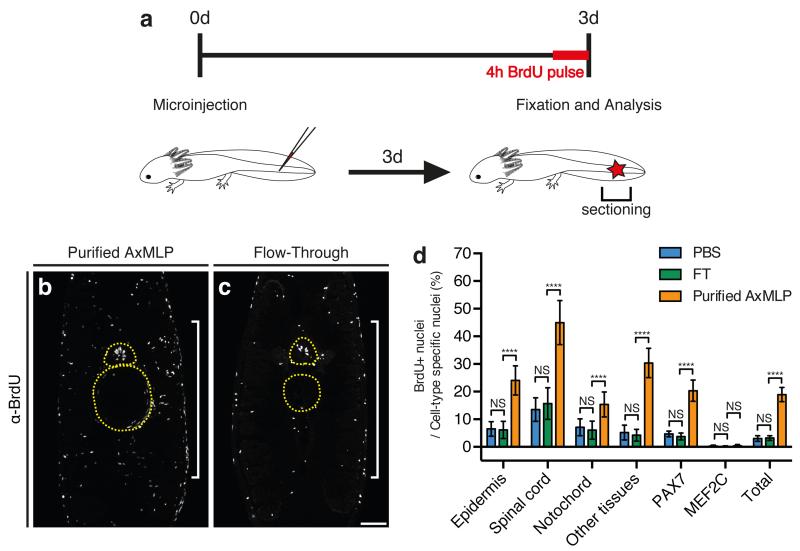
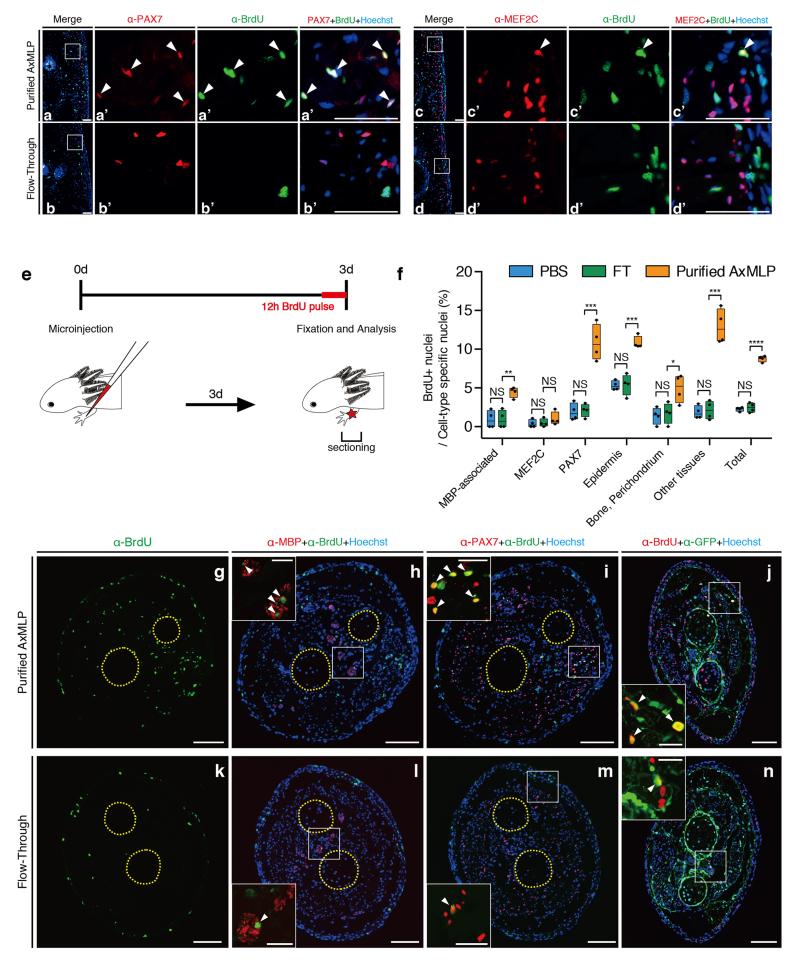
To determine the in vivo function of extracellular AxMLP we first queried if purified AxMLP protein injected into uninjured axolotl tail (Fig. 2) and limb (ExFig. 4) tissue was sufficient to induce cell cycle re-entry. We injected 270 ng of AxMLP followed by injection of BrdU at 3 days post-amputation (dpa) (Fig. 2a, ExFig. 4e). AxMLP-injected tails contained significantly more BrdU-positive cells (18.9±2.59%) than control tails injected with media depleted of AxMLP (Flow-Through --FT, 3.20±0.863%; PBS, 3.04±1.00%) (Fig. 2b-d). AxMLP injection caused increased BrdU-uptake in all counted cell types in limbs and tails except for MEF2C+ (Myocyte Enhancer Factor) muscle nuclei (Fig. 2b-d, ExFig. 4a-d,f-n). Interestingly it was recently found that muscle fibers can dedifferentiate during newt limb regeneration, but not in axolotl 15. The responsiveness of axolotl PAX7+ satellite cells but not MEF2C+ muscle nuclei to AxMLP corresponds with PAX7+ satellite cells being the main contributors to muscle regeneration in axolotl15.
Figure 2. AxMLP is sufficient to induce cell cycle entry in vivo.
a, Schematic illustration of in vivo protein injection experiment. b,c,Transverse sections of tails injected with purified AxMLP (b) or Flow-through (fraction depleted of AxMLP) (c) immunostained for BrdU. d, Quantification of BrdU+ cells in injected tails. Quantification of BrdU+/PAX7+ cells and BrdU+/MEF2C+ shows that AxMLP induces cell cycle entry in PAX7+ cells (d). NS, not significant; ****P<0.0001 with Student’s t-test, (n=15: 5 biological, 3 technical replicates each; mean±s.d.). Bar in c, 200 μm. White brackets in b, c, indicate injection site. Yellow circles in b, c, indicate spinal cord (top) and notochord (bottom), respectively.
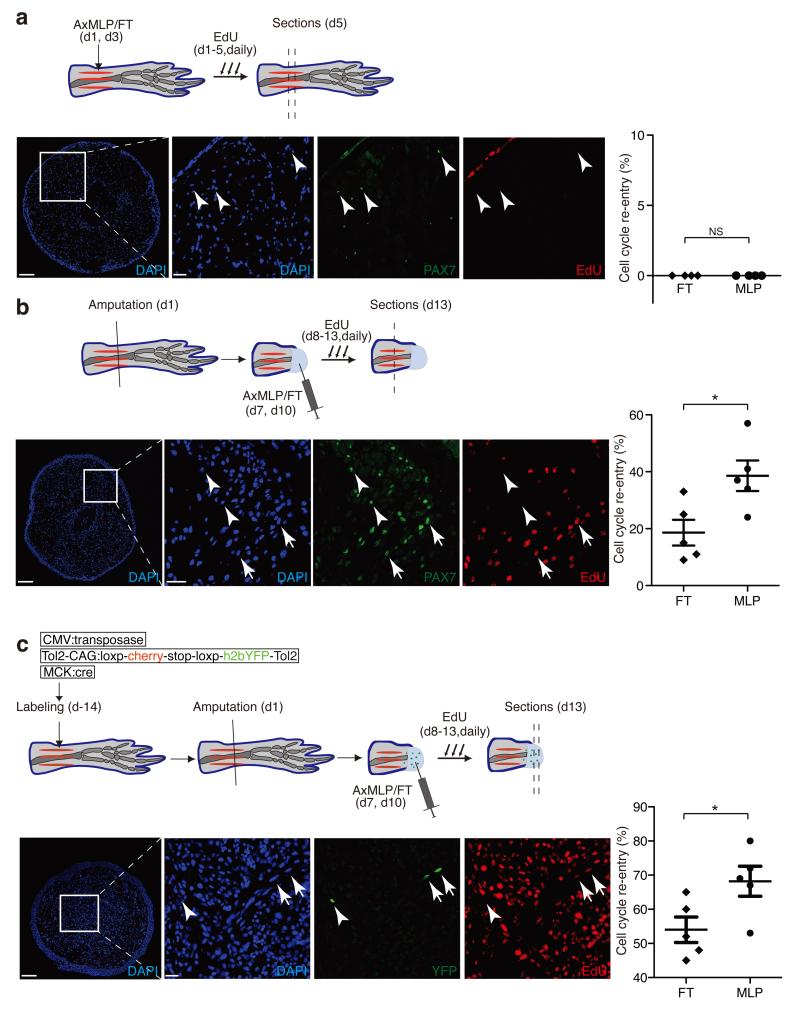
Since we had used the newt myogenic cell line for the original screen we asked whether AxMLP could promote in vivo cell cycle entry during muscle dedifferentiation in the newt. AxMLP protein was injected either into uninjured newt limbs or after limb amputation during the muscle dedifferentiation phase (Fig. 3). Injection of AxMLP into uninjured newt tissue was not sufficient to induce a cell cycle response (Fig. 3a). Injection during regeneration, however, resulted in an increased EdU uptake in PAX7+ satellite cells as well as dedifferentiating myofibre-derived cells (Fig. 3b, c) 15. These data indicate that AxMLP can also promote cell cycle entry of at least two cell types in newt, including dedifferentiating muscle. The requirement for an additional injury signal to induce cell cycle entry in newt correlates with a higher propensity of axolotl stem cells to cycle in homeostasis compared to their newt counterparts16,17.
Figure 3. AxMLP induces cell cycle re-entry of muscle-derived cells in newt limb.
a-c top panels, Schematic representation of the experimental paradigm testing the effect of AxMLP on PAX7+ satellite cell proliferation in uninjured limb (a), on PAX7+ satellite cell activation in injured limb (b) or myofiber dedifferentiation after injury (c). a-lower panels: Transverse section of uninjured limb injected with purified AxMLP shows no induction of EdU incorporation. Graph showing no difference in either purified AxMLP or Flow-through (FT) injected uninjured limbs (right side; n=4: biological replicates). b-lower panels: Transverse section of the regenerating limb injected with AxMLP shows increased EdU incorporation of PAX7+ cells. Graph showing more EdU+/PAX7+ satellite cells in AxMLP-injected limbs (right side; n=5: biological replicates; mean±sem). c-lower panels: Transverse section from myofiber-labeled, regenerating limb injected with AxMLP. Graph showing more EdU+/YFP+ myofiber progeny in AxMLP injected blastemas (right side; n=5: biological replicates; mean±sem). NS, not significant; *P<0.05 with Student’s t-test. Bars in lower magnification images, 200 μm; in higher magnification images, 20 μm. Arrowheads indicate marker+/EdU− cells. Arrows indicate marker+/EdU+ cells.
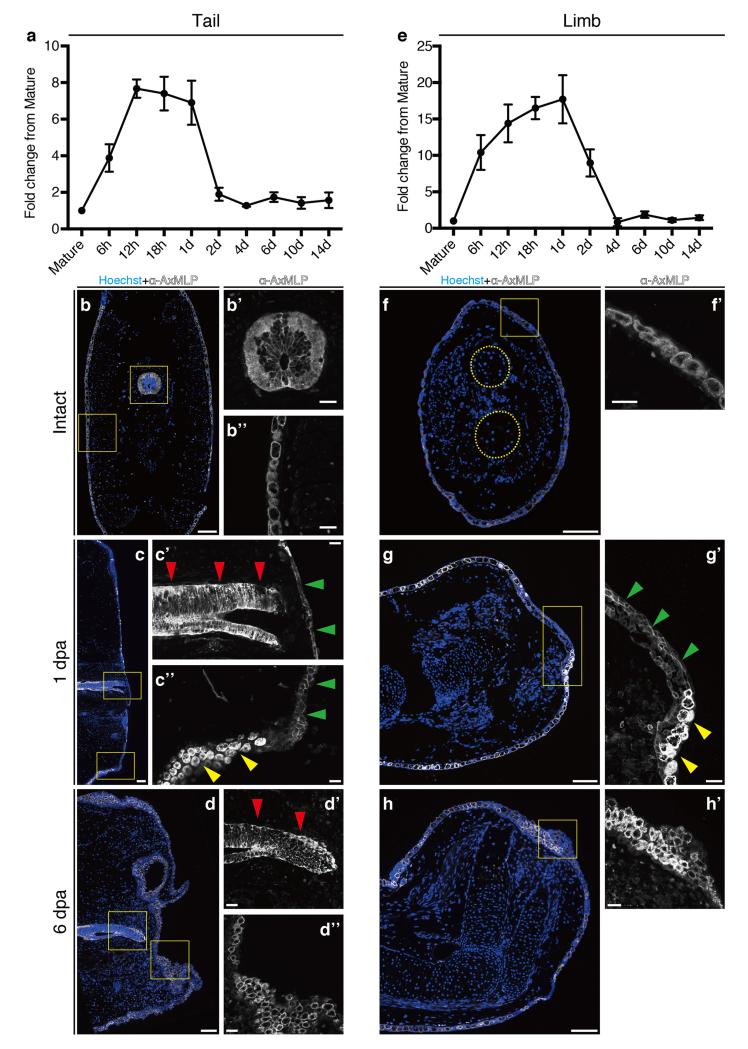
We next asked if AxMLP is important for cell proliferation during axolotl regeneration. Microarray and quantitative reverse transcription PCR (qRT-PCR) detected expression in mature limb and tail tissue followed by up-regulation with a peak of expression at 12 to 24 hours post amputation (hpa), returning to basal levels at 2 dpa in the tail and 4 dpa in the limb (ExFig. 5a,e). These observations are consistent with a role for AxMLP in early events of regeneration. Protein localization via immunofluorescence showed that in uninjured tissue, AxMLP was observed cytoplasmically localized in epidermis and in tail spinal cord cells, including radial glia and axonal tracts (ExFig. 5b-b”,f,f’). At 1 and 6 dpa, expression was maintained in the epidermis and spinal cord (ExFig. 5c,d-d”,g,h,h’). However the protein in the regenerating wound epidermis at 1 dpa was plasma membrane-associated (ExFig. 5c’,c”,g’, green arrowheads). Such localization changes have previously been described for MARCKS proteins and is dependent on phosphorylation state 18. In summary, AxMLP protein is cytoplasmically localized in mature tissue. Upon amputation, mRNA levels rise by at least eight-fold. Concomitantly the AxMLP protein in the wound epidermis shows juxtamembrane localization, consistent with its N-terminal myristoylation sequence and suggestive of extracellular release. These data suggest that both the level and intracellular localization are critical in AxMLP’s role as a non-autonomous inducer of initial cell cycles. A detailed understanding of the different cytoplasmic pools and their relationship to the extracellular form will require further study.
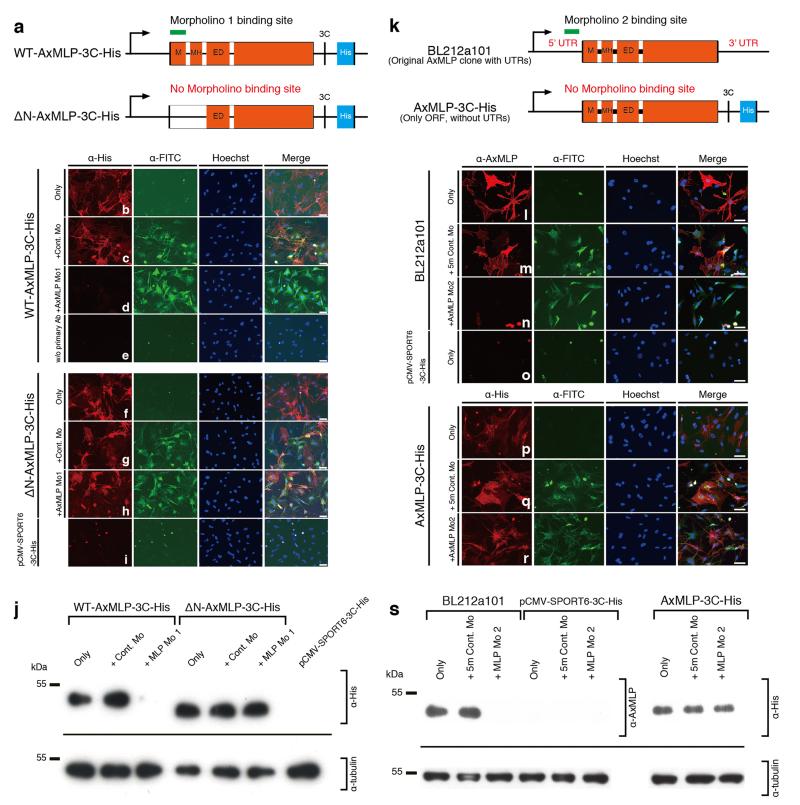
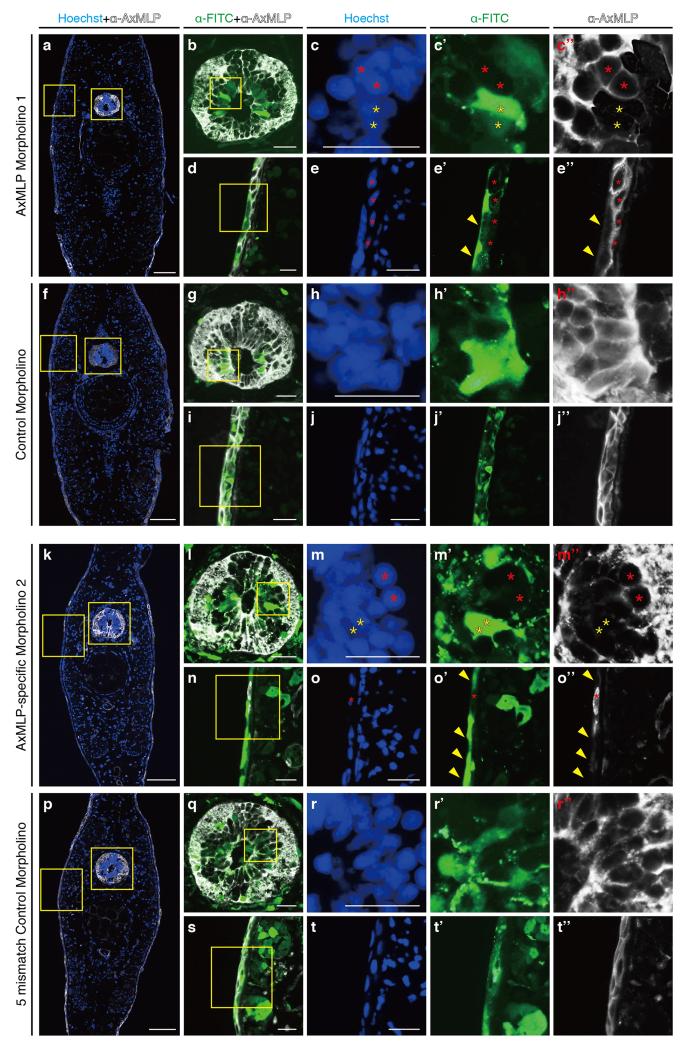
To test the function of endogenous AxMLP during regeneration we implemented two different FITC-conjugated morpholinos directed against 5′ sequences of the AxMLP mRNA. We validated the effectiveness of the morpholinos in vitro by co-electroporation with plasmid encoding full length-AxMLP or ΔNterminal-AxMLP-His lacking the target sequence into cultured newt cells (ExFig. 6a,k). Immunostainings and Western Blots showed that the morpholinos strongly reduced AxMLP expression but did not affect the expression of ΔN-AxMLP (ExFig. 6b-j,l-s). No effects were observed with two different negative control morpholinos including a five-mismatch morpholino (ExFig. 6b-j,l-s).
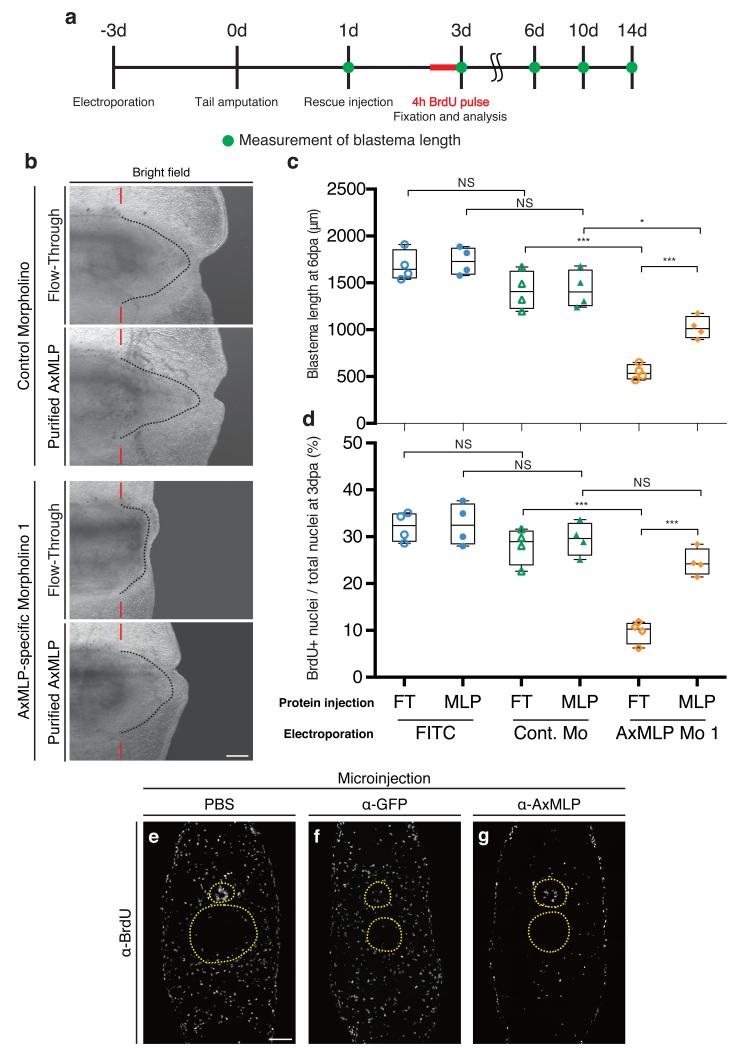
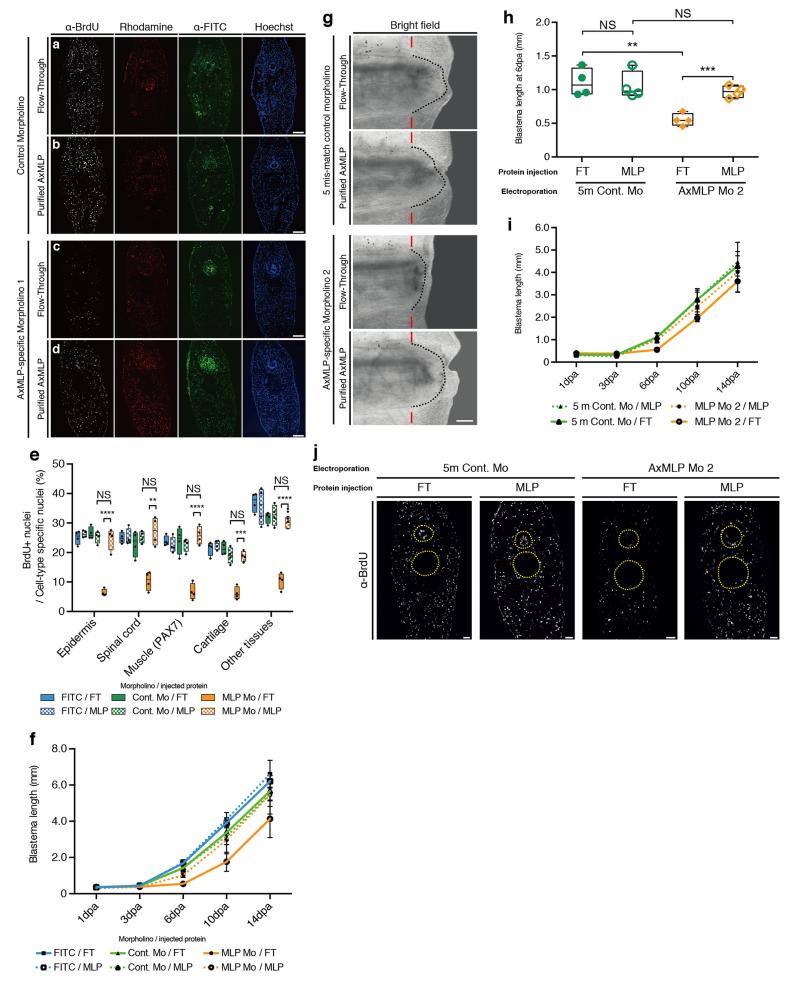
To knockdown AxMLP in vivo, we electroporated AxMLP or control morpholinos into the tail epidermis and spinal cord 3-days prior to amputation. Reduction of protein levels in electroporated cells was confirmed by immunostaining (ExFig. 7). To test whether exogenously provided AxMLP protein would rescue the knockdown phenotype, the electroporated tails were injected at 1 dpa with purified AxMLP or inactive FT. Blastema length was measured at 1, 3, 6, 10 and 14 dpa, and BrdU incorporation was assayed at 3 dpa (Fig. 4a, ExFig. 8). Incorporation of BrdU in the AxMLP morpholino-electroporated samples was significantly reduced (Fig. 4d, ExFig. 8a-e). Correspondingly at 6 dpa, the blastema length of the AxMLP morpholino/FT-injected tails was 59% smaller than that of the control morpholino/FT-injected ones (specific/FT, 546.7±80.1μm; control/FT, 1318±206μm; Fig. 4b,c). In contrast, AxMLP morpholino-electroporated tails injected with purified AxMLP protein showed 50% rescue in blastema length and 85% rescue of BrdU incorporation (Fig. 4c,d). The partial rescue in blastema length is likely due to the limited amounts of AxMLP provided by a single injection. The specificity of the phenotype was confirmed by implementing the second morpholino and the control five-mismatch morpholino (ExFig. 8g-j). These results show that knockdown of AxMLP via morpholino results in reduced cell proliferation that can be rescued by provision into the muscle/blastema tissue of exogenous AxMLP protein.
Figure 4. AxMLP is necessary for cell proliferation during early tail regeneration.
a, Schematic diagram of the morpholino electroporation experiment. b, Brightfield images of the morpholino electroporated/protein injected tails at 6 dpa showing inhibition of regeneration by anti-AxMLP morpholino and rescue by protein injection. c, Blastema length at 6 dpa. The sample order on the X-axis is the same as in (d) (n=4: biological replicates; center values as median; points represent each sample). d, Quantification of BrdU+ cells in tail blastema sections at 3 dpa (n=4: biological replicates; center values as median; points represent each sample). e-g, Injection of anti-AxMLP antibody inhibits proliferation after tail amputation. Transverse sections of 3 day regenerating tails that had been injected with PBS (e), anti-GFP antibody (f) or anti-AxMLP antibody (g) (For details see ExFig. 9). Sections immunostained for BrdU. *P<0.05, ***P<0.0005 with Student’s t-test. Bars in b, 500 μm; in e, 200 μm. Red bars in b, indicating amputation planes. Dashed lines in b, delineate shape of the blastema. Yellow circles in e-g, delineate the spinal cord (top) and notochord/cartilage (bottom), respectively.
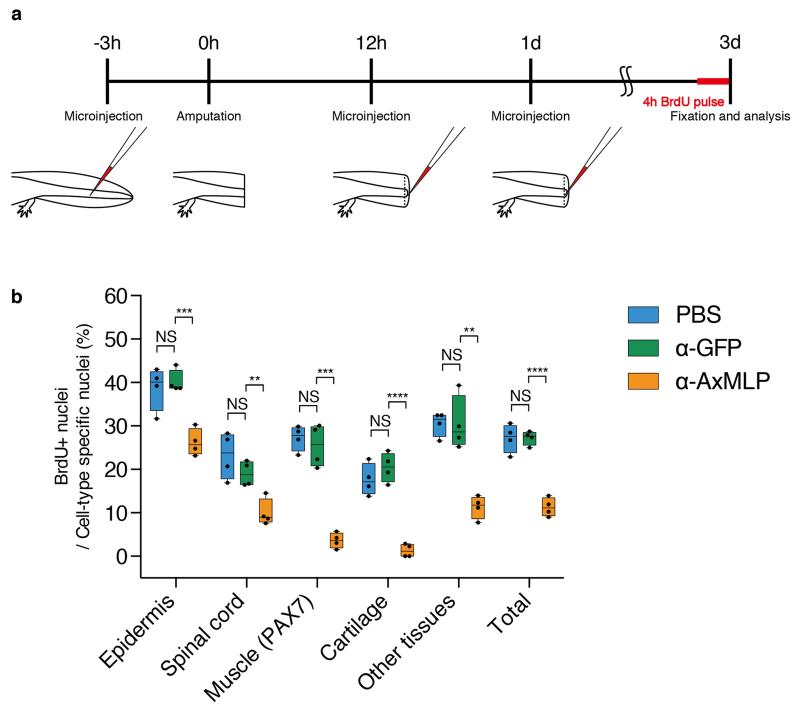
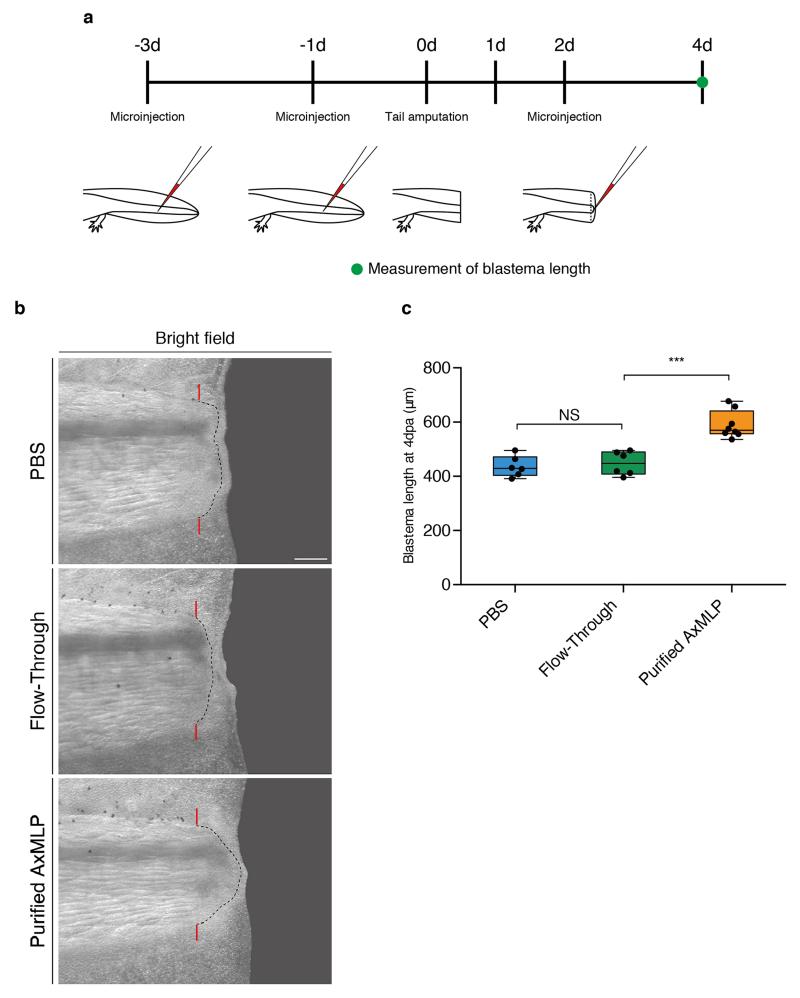
To corroborate the morpholino experiments, we injected the anti-AxMLP blocking antibody into the tail before and during regeneration (ExFig. 9a) which strongly reduced BrdU incorporation in multiple tissues (Fig. 4e-g, ExFig. 9b). These results show that in vivo knockdown of AxMLP activity by two methods resulted in reduced cell proliferation during early regeneration. To determine if an excess of AxMLP could accelerate regeneration, we performed multiple injections of AxMLP protein before and during early phases of regeneration (ExFig. 10a). The oversupply of protein resulted in a larger blastema at 4 dpa (ExFig. 10b,c).
This work represents the first identification of a molecule, AxMLP, by functional expression cloning and in vivo testing in appendage regeneration and therefore sets an experimental paradigm for future studies. Previous work indicated that spinal cord neural stem cells accelerate their cell cycle kinetics resulting in increased mitoses between 3-4 dpa19. Our work indicates that AxMLP is a major factor responsible for the induction of these cell cycle kinetics. How AxMLP is delivered extracellularly, its mode of intracellular signaling and whether orthologs beyond salamanders are associated with regenerative events will be important topics of future investigation.
Materials and methods
Animals
All animal experiments were performed in accordance with the European Community and local ethics committee guidelines. Xenopus laevis were purchased (Nasco) and maintained in our animal facility. Ambystoma mexicanum (axolotls) were bred and maintained in our facility, where they were kept at 18 °C in Dresden tap water and fed daily with artemia or fish pellets. 5 to 6 cm (snout to tail tip) axolotls were used for all the experiments. Animals were anesthetized for all the surgical process as previously described20. Labelling of connective tissue was achieved by transplanting lateral plate mesoderm from GFP transgenic embryos to normal host embryos as previously described20.
Protein expression by Xenopus Oocytes
Total RNA was isolated from 1-day, 3-day and 5-day limb and tail blastemas with TRIzol reagent (Invitrogen) according to the manufacturer’s manual. Total RNA from mature (not regenerating) limb tissues was isolated using the same procedure as blastema samples. Blastema RNAs from the different time points were equivalently pooled as limb-blastema total RNA or tail-blastema total RNA, respectively. mRNA was purified from limb-blastema or tail-blastema total RNA with the Poly (A) Quick mRNA isolation kit (Stratagene). Xenopus oocyte preparation and microinjection were essentially as previously described6,7,21. Briefly, mature oocytes were defolliculated with collagenase (Sigma). Purified mRNA (5.0 ng) was injected into the selected healthy oocytes after the defolliculation. 8 injected oocytes were cultured together in one well of 96-well plate (Nunc) for 48 hours and the supernatants were harvested for myotube assay (see below).
Functional expression cloning
110,592 clones from a 6-day tail blastema library were arrayed into 288 × 384-well plates22. To prepare the “pools”, all the saturated bacterial cultures on one 384-well plate were pooled in one conical tube, and 288 pools were prepared from the library in total. To prepare the “superools”, 24 pools were combined together in one conical tube, and 12 superpools were prepared from the pools in total (ExFig. 1a-c). To obtain superpool plasmid, 500 μl of superpool bacteria was cultured in 50 ml LB medium (ExFig. 1d). To avoid losing low frequency clones in the superpools, the optical density (O.D.) of each culture was controlled and the cultures were harvested around O.D. 0.6. Superpool plasmids were purified with QIAGEN Plasmid Midi Kit (QIAGEN) according to the manufacturer’s manual. To reconstitute the whole library, 5 μg of each superpool plasmids were pooled in one tube prior to transfection. HEK293FT cells (Invitrogen) were maintained with the standard protocol from Invitrogen. To obtain the superpool supernatants, 8.0×105 of HEK293 cells were plated on one well of 6-well plate (Nunc) and 1 μg of each individual superpool plasmid was transfected into HEK293 cells with Fugene 6 (Roche, ExFig. 1e) according to the manufacturer’s manual. For the first 24 hours, the transfected HEK293 cells were kept in the 10% fetal calf serum (FCS) medium. Then the cells were rinsed with FreeStyle 293 expression medum (Gibco) that is a serum free media and cultured in the medium at 72 hours after transfection. Individually harvested supernatants were concentrated approximately ten-fold with a Vivaspin 10,000 MWCO (Sartorius). These concentrated supernatants were tested on A1 myotubes (ExFig. 1f). It should be noted that given the injury-specific extracellular activity of AxMLP, we infer that the Xenopus oocyte and HEK293 cell systems are likely to be in “wound epithelium like” signalling states that permit atleast some extracellular release of AxMLP, and that the 6-day regenerating tail blastema cDNA had a sufficient number of AxMLP clones for detection in the expression cloning system. We only detected 1 AxMLP clone among 100,000 clones, and this may reflect the levels of mRNA present at later regeneration timepoints.
Maintenance of A1 myoblasts, myotube differentiation from A1 myoblasts, myotube purification and subsequent myotube assay were performed essentially described previously8,23,24. Briefly, concentrated supernatants were individually added into myotube culture medium in a 96-well plate and incubated for 5 days, and BrdU (Sigma, final 10 μg/ml) was added to the culture media for 18 hours prior to the fixation with 1.5% PFA (Sigma)/PBS. Fixed myotubes were stained with anti-MHC and anti-BrdU antibodies (mouse monoclonal, 4a1025 and Bu20a) conjugated to FITC or rhodamine with DyLight Antibody Labeling Kit (Thermo Scientific) according to the manufacturer’s manual. For the quantification of BrdU incorporation activity, the total number of myotubes and BrdU+ myotubes were counted by hand under the microscope (Zeiss Axioplan 2). This biological evaluation of the BrdU-incorporation activity on myotubes is called the “Myotube assay”. The high serum (15% FCS) condition was used as a positive control for myotube assays and low serum (0.5% FCS) or serum free condition was used as a negative control.
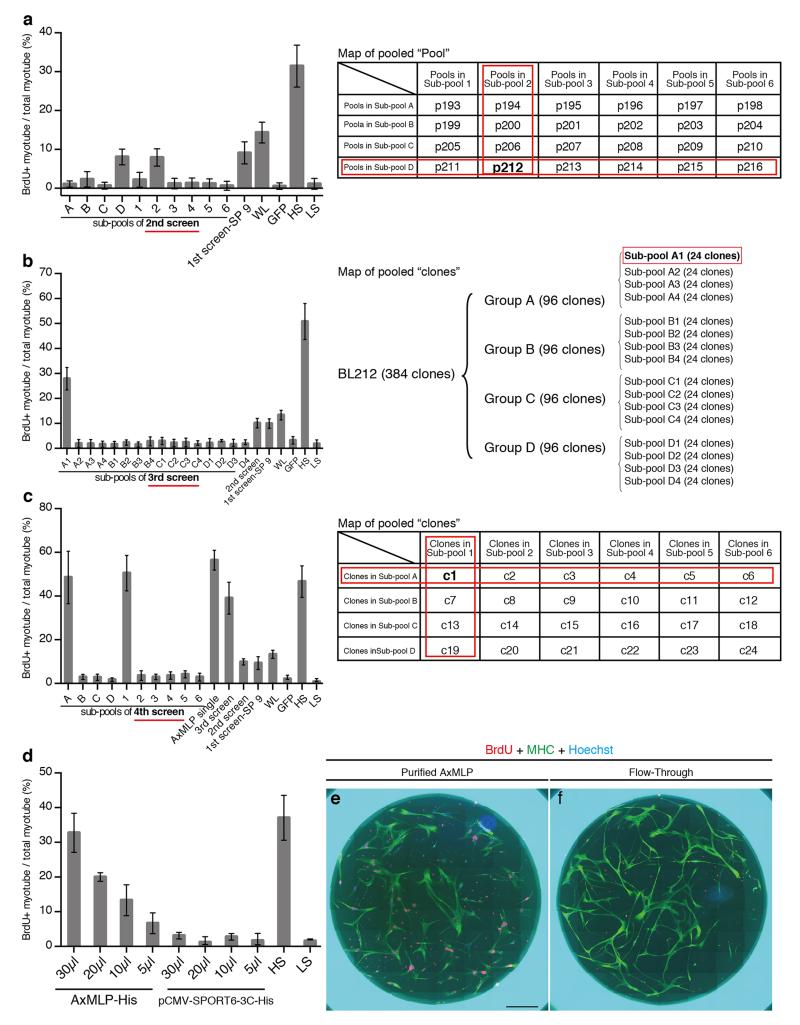
For the second screen from superpool #9, 24 pools in superpool #9 were divided into smaller sub-pools, i.e., sub-pool A contained Pool #193 to #198 (6 pools) and sub-pool #1 contained Pool #193 to #211 (4 pools) (ExFig. 2a). These sub-pools were cultured to O.D. 0.6 before plasmid preparation. For the third screen from Pool #212, 384 single clones were arrayed by 96-pin plastic replicators (Genetix) on 96-well plates (SARSTEDT) filling 150 μl LB medium/well (ExFig. 2b; Group A-D). Individual clones on the 96-well plates were statically cultured until they were saturated and 24 clones were pooled together (ExFig. 2b; sub-pool A1-D4). Plasmids from each pool were purified with QIAprep spin miniprep kit (QIAGEN). For the fourth screen from A1, 24 clones were individually cultured in LB medium and the plasmids were purified with QIAprep spin miniprep kit (QIAGEN). To construct sub-pools, 1 μg of the plasmid from each single clone was pooled according to the diagram in ExFig. 2c. The process from the transfection into HEK293 cells to myotube assay in the second to the fourth screen was the same as the first screen. To validate transfection efficiency during whole expression cloning, 50 pg of SEAP (secreted alkaline phosphatase)-pCMV-SPORT6 plasmid was co-transfected with the samples as a spike and a portion of the supernatants was assayed by Great EscAPe SEAP Chemiluminescence Kit 2.0 (Clontech). The luminescence of the supernatants was measured by GENiosPro Microplate Reader (TECAN). We confirmed there was no significant difference of transfection efficiency during the expression cloning (data not shown).
Plasmid construction
Human and mouse MLP cDNA clones were purchased from OriGene Technologies (clone ID: human, SC112373; mouse, MC208965). Zebrafish and Xenopus MLP cDNA clones were purchased from Source BioScience (clone ID: zebrafish, 6795591; Xenopus, 8330180). All oligo sequences and the restriction enzyme sites using for cloning are showed in Supplementary Table 2. Since the backbone vector of the cDNA library is pCMV-SPORT6, we sub-cloned following genes into pCMV-SPORT6 vector (Invitrogen) or pCMV-SPORT6-3C-His vector. PCR amplified fragments with the oligos #1 and #2 from pSEAP2-Basic (Clontech) were sub-cloned into pCMV-SPORT6. The oligos #3 and #4 were attached to pCMV-SPORT6 to generate a backbone vector, pCMV-SPORT6-3C-His (ExFig. 3c, left bottom). AxMLP ORF (open reading frame) was amplified by PCR with the oligos #5 and #6 from the original AxMLP clone (BL212a101) and sub-cloned in the pCMV-SPORT6-3C-His vector (ExFig. 3c, left top). N-terminal deletion AxMLP was amplified by PCR with the oligos #7 and #8 from the original AxMLP clone (BL212a101) and sub-cloned in the pCMV-SPORT6-3C-His vector (ExFig. 6a, bottom). Human, mouse, zebrafish and Xonopus MLP ORFs were amplified from purchased cDNA clones with specific primers (for human, oligo #9, #10; mouse, oligo #9, #11; zebrafish, oligo #12, #13; Xenopus, oligo #14, #15, respectively), and were sub-cloned in the pCMV-SPORT6-3C-His vector. The oligos #16 and #17 were attached to the pEGFP-N1 (Clontech) to generate a backbone vector, pEGFP-N1-3C (ExFig. 3c, right bottom). AxMLP ORF was amplified by PCR with the oligos #18 and #19 from the original AxMLP clone (BL212a101) and sub-cloned into the pEGFP-N1-3C (ExFig. 3c, right top). Newt MLP ORF was amplified by PCR from newt limb blastema cDNA with the oligos #28 and #29 and the PCR fragments were sub-cloned in the pCMV-SPORT6-3C-His vector. These constructs were confirmed by sequencing.
Assessment of AxMLP extracellular secretion
For measuring the GFP intensity of supernatants, 8.0×105 of HEK293 cells were plated on 6-well plates and 1 μg of AxMLP-3C-pEGFP-N1 plasmid or 1 μg of empty-pEGFP-N1 plasmid were transfected into HEK293 cells. The supernatants were harvested at 24 hours post transfection (hpt), 48 hpt and 72 hpt and concentrated with Vivaspin 10,000 MWCO (Sartorius) individually. The fluorescence intensity was measured using a GENiosPro Microplate Reader (TECAN). To determine the percentage of GFP+ cells in the culture, the transfected cells were detached with Trypsin-EDTA (Gibco, final 0.05%)/PBS from the well, then spread on improved Neubauer chamber. The number of GFP+ cells and total cells in the grids were counted by hand and the percentage was calculated. Time-lapse imaging was performed under Axiovert 200M (Zeiss) with humidity, temperature and CO2 control chamber. Images were taken every 30 minute from 5 to 72 hours post transfection.
Antibody blocking
For the antibody-based blocking assay in vitro, 1 μg of AxMLP-3C-His-pCMV-SPORT6 plasmid or empty-pCMV-SPORT6-3C-His plasmid was transfected into HEK293 cells with Fugene 6 (Roche). The supernatants were harvested at 72 hpt and concentrated. 10 μg of AxMLP-3C-His protein (22.7 kDa) were treated with 70 μg or 350 μg of anti-AxMLP polyclonal antibody (see below, antibody part) or anti-GFP polyclonal antibody (MPI-CBG antibody facility) as a negative control, respectively at room temperature for 2 hours. These antibody-treated supernatants were used for myotube assay.
For in vivo antibody blocking assay, anti-full-length AxMLP polyclonal antibody (see below, antibody part), anti-GFP polyclonal antibody (MPI-CBG antibody facility) or PBS as a negative control were injected into mature (not regenerating) tail as the first injection (3 hours prior to amputation) and injected into the blastema as the second injection (12 hours post amputation) and as the third injection (1 day post amputation) (ExFig. 9a). These samples were co-injected with tetramethylrhodamine dextran MW, 70,000 (Molecular Probes, final 2.5 mg/ml) as a tracer. The injection efficiency was confirmed based on the intensity of the rhodamine under the fluorescence dissecting microscope (SZX 16, OLYMPUS). No animals were excluded from the analysis. In each injection 500 ng, then, in total 1.5 μg antibody or equivalent volume of PBS were injected. Injected animals were kept in clean tap water for 3 days at room temperature. The animals were injected intraperitoneally with 30 μl of 2.5 mg/ml BrdU (Sigma) 4 hours prior to collecting the tails. The injected blastemas were fixed, embedded, cryosectioned and immunostained as described below (see immunohistochemistry part). For the imaging, the tiled images of the entire cross-section of the tails taken on a Zeiss Observer.Z1 (Zeiss) were then stitched by Axiovision software or Zen 2 (Zeiss). For the quantification at least a total 1000 cells per one animal were counted from 4 different animals in each condition (PBS, anti-GFP antibody or anti-AxMLP injection, respectively), and the marker positive nuclei (BrdU+, PAX7+, MEF2+ or Hoechst+) on the sections were counted by hand. The cells in spinal cord, epidermis and cartilage/notochord were separately counted based on morphology. “The other tissues” containing mainly mesenchymal cells and endothelial cells were calculated by the subtraction from the total number to the number of all the other specific cell types.
AxMLP purification
For his-tagged AxMLP purification, AxMLP-3C-His-pCMV-SPORT6 plasmid was transfected into HEK293 cells and the supernatant was harvested at 72 hpt. His tagged protein in the supernatant was purified in native conditions on a 1 ml HisTrap HP column (GE Healthcare) using FreeStyle 293 expression medium including 500 mM imidazole step elution. The eluate (purified AxMLP) and depleted media (flow-through) were concentrated with Vivaspin 10,000 MWCO (sartorius) 40 fold and the final concentration of purified AxMLP was 1.31 μg/μl. Both concentrated eluate (purified AxMLP) and flow-through fractions were dialyzed with Spectra/Por Dialysis Membrane MWCO 6-8000 (Spectrum Laboratories) in AMEM (MEM medium (Gibco) diluted 25% with distilled water) for biological assays. The fractions from the purification were tested by Silver Staining and Western Blotting (ExFig. 3g,h). The washing fraction was concentrated about 10 fold to load the same volume as other fractions on 4-20% gradient SDS-PAGE gels (anamed Elektrophorese). Western Blotting and Silver Staining were performed with a standard protocol. Briefly, the fractions were treated with 2xSample Buffer including DTT (Sigma, final 0.2 M) and boiled at 95°C for 10 minutes. The proteins were blotted on PROTRAN nitrocellulose transfer membrane (Whatman) by TE 77 Semi-Dry Transfer Unit (Amersham). The membrane was blocked with 5% skim milk. Primary antibodies used: mouse anti-His (QIAGEN, 1/5000), mouse anti-α-tubulin (MPI-CBG antibody facility, DM1A 1/5000), rabbit anti-AxMLP-full length (1/2500), rabbit anti-AxMLP-C terminus (1/2500). Secondary antibodies used: goat anti mouse-HRP (Jackson ImmunoResearch Laboratories, 1/5000), goat anti rabbit-HRP (Jackson ImmunoResearch Laboratories, 1/5000). Cell lysates for Western Blotting were obtained by directly adding 2xSample Buffer on top of the cultured cells and were boiled at 95°C for 10 minutes.
Antibodies and Immunohistochemistry
For the preparation of anti-full-length AxMLP polyclonal antibody, a GST-fusion protein with full-length amino acids of AxMLP was expressed in bacteria and purified by standard methods on GS-trap, glutathione sepharose (GE Healthcare). Purified GST-fusion protein as an antigen was used to immunize rabbits (Charles River). Anti-serum was affinity purified using maltose binding protein fused with full-length AxMLP conjugated to NHS-Sepharose resin (GE Healthcare). To raise C-terminal AxMLP polyclonal antibody, KLH (Keyhole Limpet Hemocyanin) tagged peptides, PPVEPQVEEVAAPAP was used to immunize rabbits and the affinity purified polyclonal antibody was provided (Eurogentec). Both anti-full-length and anti-C-terminal AxMLP polyclonal antibodies were tested on the cell lysate from AxMLP transfected HEK293 cells (ExFig. 3f).
Limb blastema and tail blastema preparations for sectioning were produced essentially as previously described25. Briefly, limb blastemas amputated at the wrist level were collected from the level of the shoulder and tail blastemas amputated at 12th myotome from the cloaca were collected at 10th myotome of the regenerating tail. These limb and tail blastemas were immunostained as previously described15,25,26. Briefly, the samples were fixed with MEMFA fixative at 4 °C over night, and were rinsed with PBS several times. The buffer was replaced from PBS to 10%, 20% and 30% sucrose (Sigma)/PBS, then the samples were embedded in tissue-tek, O.C.T. Compound (Sakura) for cryosection and the tissues were sectioned at 10 μm thickness with Microm HM 560 cryostat (Thermo). Primary antibodies used: mouse anti-BrdU (MPI-CBG antibody facility, Bu20a 1/400), rabbit anti-BrdU (antibodies-online, 1/600), mouse anti-PAX7 (MPI-CBG antibody facility, PAX7 1/450), rabbit anti-MEF2 (Santa Cruz, 1/200), rabbit anti-AxMLP-C terminus (1/200), rabbit anti-GFP (Rockland, 1/400), rabbit anti-FITC (Invitrogen, 1/400). mous anti-FITC (Jackson ImmunoResearch Laboratories, 1/400), rat anti-MBP (GeneTex, 1/200). Following appropriate fluorophore-conjugated secondary antibodies were used (all in 1/200 dilution): donkey anti-mouse Alexa Fluor (AF) 647 (Molecular Probes), goat anti-mouse AF 647 (Jackson ImmunoResearch Laboratories), donkey anti-mouse AF 488 (Molecular Probes), goat anti-rabbit AF 647 (Jackson ImmunoResearch Laboratories), donkey anti-rabbit AF 488 (Molecular Probes), donkey anti-rat AF 488 (Molecular Probes). The cell nuclei were stained with Hoechst 33342 (Sigma, final 0.5 μg/ml). Imaging for the stained sections was performed with Zeiss Observer.Z1 (Zeiss) controlled by Axiovision software or Zen2 (Zeiss).
Quantitative Reverse Transcription PCR (qRT-PCR)
Total RNA preparation, reverse transcription and qRT-PCR were essentially described in the previous work2. Briefly, 3 biological replicas were prepared for each time point and they were technically independent in all the processes (tissue collection, RNA preparation, cDNA synthesis and qRT-PCR). 8~10 limb or tail blastemas from one time point were collected in one tube and homogenized by POLYTRON PT1600E (KINEMATICA). Total RNA was purified with RNeasy Mini or Midi Kit (QIAGEN) according to the manufacturer’s manual. Complementary DNA was synthesized from 300 ng of total RNA using SuperScript III First-Strand Synthesis System (Invitrogen) and qRT-PCR was performed with Power SYBR Green Master Mix (Invitrogen) in total volume of 12 μl with the final primer concentration of 300 nM on the LightCycler 480 (Roche). To obtain the values of fold change for each time point, the relative concentration of the PCR products was calculated by standard curve method. To obtain the standard curves of the limb time course or the tail time course respectively , the dilution series (1/4, 1/16, 1/64, 1/256 and 1/1024) were made from the mixture of cDNAs that were equivalently collected from the cDNA samples in all the different time points. These dilution series were used as the template for the PCR and the relative concentrations were calculated by LightCycler 480 Software (Roche) based on the standard curves. The concentration of AxMLP was normalized with that of Rpl4 (large ribosomal protein 4). Primers used for PCR were showed in Supplementary Table 2 (AxMLP, #20, #21; Rpl4, #22, #23). The raw values of qPCR data were shown in Supplementary Table 1.
Protein injection into axolotl tail and limb
The dialyzed protein samples: purified AxMLP, depleted media (flow-through) (see AxMLP purification part) or PBS as a negative control were injected into mature (not regenerating) tails with a pressure injector, PV830 Pneumatic Picopump (World Precision Instruments). These protein samples were co-injected with tetramethylrhodamine dextran MW, 70,000 (Molecular Probes, final 2.5 mg/ml) as a tracer. A glass capillary (Harvard Apparatus) for the injection was pulled with P-97 Micropipette Puller (Sutter Instrument) and sharpened manually (external tip diameter: 30 μm). The injection efficiency was confirmed based on the intensity of the rhodamine under the fluorescence dissecting microscope (SZX 16, OLYMPUS). No animals were excluded from the analysis. In total, 270 ng of purified AxMLP or equivalent volume of controls were injected into one side of the tail. Injected animals were kept in clean tap water for 3 days at room temperature. The animals were injected intraperitoneally with 30 μl of 2.5 mg/ml BrdU (Sigma) 4 hours prior to collecting the tails (Fig. 2a). The injected part of the tails was identified by rhodamine positive myotomes and these tails were fixed, embedded, cryosectioned and immunostained as described above (see immunohistochemistry part). For the quantification, the tile images of whole corss-section of the tails from Zeiss Observer.Z1 (Zeiss) were stitched by Axiovision software (Zeiss). Three sections from 5 different animals in each condition (PBS, flow-through or purified AxMLP injection, respectively) were taken, and the marker positive nuclei (BrdU+, PAX7+, MEF2+ or Hoechst+) on the sections were counted by hand. The cells in spinal cord, epidermis and notochord were separately counted based on morphology. “The other tissues” containing mainly mesenchymal cells and endothelial cells were calculated by the subtraction from the total number to the number of all the other specific cell types.
For the protein injection into the limb, the procedure was essentially described as the tail injection above. Purified AxMLP protein was injected into the mature (not regenerating) right lower limbs at the center between the elbow and the wrist. The control samples (flow-through fraction or PBS) were injected into the left limbs of the same animal that were injected with purified AxMLP on their right limbs. In total 2.0 μg purified AxMLP or equivalent volume of controls were injected into the limbs. The animals were injected intraperitoneally with 30 μl of 2.5 mg/ml BrdU (Sigma) 12 hours prior to collecting the limbs (ExFig. 4e). For the quantification, at least a total 1000 cells per one animal were counted from 4 different animals in each condition (PBS, flow-through or purified AxMLP injection, respectively), and the marker positive nuclei (BrdU+, PAX7+, MEF2+, MBP + or Hoechst+) on the sections were counted by hand. The cells in epidermis and bone/perichondrium were separately counted with their morphology. “The other tissues” containing mainly mesenchymal cells and endothelial cells were calculated by the subtraction from the total number to the number of all the other specific cell types.
For the acceleration experiment, purified AxMLP, flow-through or PBS as a negative control were injected into mature (not regenerating) tail as the first (3 days prior to amputation) injection and as the second (1 day prior to amputation) injection and injected into the blastema as the third (2 day post amputation) injection (ExFig. 10a). These samples were co-injected with tetramethylrhodamine dextran MW, 70,000 (Molecular Probes, final 2.5 mg/ml) as a tracer. The injection efficiency was confirmed based on the intensity of the rhodamine under the fluorescence dissecting microscope (SZX 16, OLYMPUS). No animals were excluded from the analysis. The samples were injected into both side of the tail and in each injection, 600 ng, then, in total 1.8 μg protein or equivalent volume of controls were injected. Injected animals were kept in clean tap water for 4 days at room temperature. The length of the blastema was measured from the amputation plane to the tip at the spinal cord level on 4 days post amputation based on the stereoscope images (SZX 16, OLYMPUS).
Morpholino electroporation
In vitro assay
A1 myoblasts were transfected with original clone: BL212a101, AxMLP-3C-His, ΔN-AxMLP-3C-His or empty pCMV-SPORT6-3C-His plasmids and co-transfected with AxMLP-specific morpholinos (Gene Tools, Supplementary Table 2: #24, #26) or control morpholinos (Gene Tools, Supplementary Table 2: #25, #27) using Microporator (Digital Bio) according to the manufacturer’s manual with some modifications. All morpholinos were modified with C-terminus FITC. A1 myoblasts were re-suspended in 1xSteinberg solution at a density of 5.0 × 106 cells/ml followed by incubation of 10 μl cell suspension with 0.5 μg of plasmid and 1 μl of the morpholino (final 100 μM in the incubation). Electroporation was performed at 1000 volts, 35 mS pulse length and 3 pulses and the electroporated cells were spread in 10% FCS AMEM media24 on 24-well plate (Nunc), immediately after the electroporation. The culture medium was replaced by new media at 24 hours post electroporation and the cells were kept in culture at 72 hours post electroporation. The electroporated cells were fixed with 1.5% PFA/PBS, and the cell lysates were prepared for Western Bloting. Primary antibodies used for immunostaining: mouse anti-His (QIAGEN, 1/200), mouse anti-FITC (Jackson ImmunoResearch Laboratories, 1/400), rabbit anti-FITC (Invitrogen, 1/400), rabbit anti-AxMLP-full length (1/1000). Secondary antibodies used for immunostaining (all in 1/250 dilution): goat anti-mouse Cy3 (Jackson ImmunoResearch Laboratories), goat anti-mouse AF488 (Jackson ImmunoResearch Laboratories), donkey anti-rabbit AF 488 (Molecular Probes), goat anti-rabbit Cy3 (Jackson ImmunoResearch Laboratories). Images of the stained cells were taken with Zeiss Observer.Z1 (Zeiss) controlled by Axiovision software (Zeiss).
In vivo assay with rescue protein injection
The electroporation to the spinal cord was performed as previously described with some modification27. To deliver morpholino into the spinal cord and the both sides of the tail epidermis, the tail required electroporation twice with NEPA 21 electroporator (Nepa Gene). The first electroporation was for the spinal cord and one side (left) of the epidermis, and the second electroporation was for the other side (right) of the epidermis. 1.5 μl of morpholino (1.0 mM) was loaded at a small piece of tissue paper on the left side of the epidermis. Approximately, 3 μl of morpholino (1.0 mM) was injected into the spinal cord and immediately electroporated (first electroporation). Sequentially, 1.5 μl of morpholino (1.0 mM) was loaded at a small piece of tissue paper on the right side of the epidermis and electroporated (second electroporation). The first electroporation conditions: Poring pulse, 70 volts, 5.0 mS pulse length and 1 pulse; Transfer pulse, 55 volts, 55 mS pulse length, 5 pulses and 15% decay. The second electroporation conditions: Poring pulse, 70 volts, 10 mS pulse length and 1 pulse; Transfer pulse, 30 volts, 30 mS pulse length, 7 pulses and 5% decay. FITC dextran MW, 70,000 (Molecular Probes, final 5 mg/ml) was used as a negative control, since morpholinos were labeled with FITC. The electroporation efficiency in the spinal cord and epidermis was examined based on the intensity of the FITC under the fluorescence dissecting microscope (SZX 16, OLYMPUS). The animals with low FITC intensity were excluded from the next step of the experiments. 3 days post electroporation, the tails were amputated at the level of maximumal morpholino electroporated part. 1 day post amputation, a total 360 ng (180 ng for the spinal cord and 180 ng for blastema) of purified AxMLP or equivalent volume of control flow-through fraction was injected into the spinal cord and the blastema to rescue morpholino effect. The length of the blastema was measured from the amputation plane to the tip at the spinal cord level on 1, 3, 6, 10 and 14 days post amputation based on the stereoscope images (SZX 16, OLYMPUS). To detect BrdU incorporation, the animals were injected intraperitoneally with 30 μl of 2.5 mg/ml BrdU (Sigma) 4 hours prior to collecting the tails at 3 day post amputation. Fixation, embedding, cryosection, staining and imaging were described above (see Protein injection part). For the quantification, 3 cross-sections of the blastema (200 – 300 μm posterior to the amputation plane) from 4 different animals in each condition (FITC/flow-through, FITC/purified AxMLP, Control Morphlino/flow-through, Control morpholino/purified AxMLP, AxMLP-specific morpholino/flow-through, AxMLP-specific morpholino/purified AxMLP, respectively) were taken, and the marker positive nuclei (BrdU+, PAX7+, MEF2+ or Hoechst+) on the sections were counted by hand.
Newt experiments
Animals
Red-spotted newts, Notophthalmus viridescens, were supplied by Charles D. Sullivan Co. (Nashville, TN, USA). Animals were anesthetized in 0.1% ethyl 3-aminobenzoate methanesulfonate (Sigma) for 15 min. Forelimbs were amputated above the elbow, and the bone and soft tissue were trimmed to produce a flat amputation surface. Animals were left to recover overnight in an aqueous solution of 0.5% sulfamerazine (Sigma). At specified time-points, the uninjured or regenerating limbs were collected. All surgical procedures were performed according to the European Community and local ethics committee guidelines.
Protein injection and cell cycle assays in newt limbs
The general condition in the newt experiments: 2 μl of 5 mg/ml purified AxMLP protein or equivalent volume of flow-through (AxMLP depleted fraction) was injected into the newt limbs. For EdU-labelling, animals were injected intraperitoneally with 50-100 μl of 1 mg/ml EdU. To investigate the effect of AxMLP on intact newt limbs, purified AxMLP or flow-through was injected into the uninjured limb twice at day-1 and day-3. EdU was administered daily from day-1 to day-5 (Fig. 3a, upper panel). To investigate the effect of AxMLP on regenerating limbs purified AxMLP or flow–through was injected into the regenerating limbs at 7 and 10 dpa (Fig. 3b, upper panel). EdU was administered daily from 8 to 13 dpa. For labeling myofiber progeny, a H2B-YFP reporter construct was introduced into myofibers prior to amputation as previously described15 (Fig. 3c, upper panel). Cell-cycle re-entry was quantified by EdU incorporation in the YFP+ myofiber progeny at 13 dpa.
Immunohistochemistry
Frozen sections (5-10 μm) were thawed at room temperature and fixed in 4% formaldehyde for 5 min. Sections were blocked with 5% donkey serum and 0.1% Triton-X for 30 min at room temperature. Sections were incubated with anti-GFP (Abcam 6673), anti-PAX7 (DSHB) or anti-MHC (DSHB) overnight at 4°C and with secondary antibodies for 1 hour at room temperature. Antibodies were diluted in blocking buffer and sections were mounted in mounting medium (DakoCytomation) containing 5 μg/ml DAPI (Sigma). EdU detection was performed as previously described15. An LSM 700 Meta laser microscope with LSM 6.0 Image Browser software (Carl Zeiss) was used for confocal analyses. One in every 8 sections was selected and labelled. For PAX7+ satellite cell counting, 3 sections were randomly selected and counted. For blastema YFP+ cell counting, all the sections in the region from regenerate tip to the bone were counted.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software). Student’s t-test, parametric, two-tail testing was applied to populations to determine the p values indicated in the figures. The significance was considered with p values from < 0.05.
Extended Data
Extended Figure 1. Schematic illustration of the expression cloning approach.
a, 110,592 clones from a 6-day tail blastema library were arrayed on 288 × 384-well plates. b, One 384-well plate was pooled into one conical tube and called a “pool”. In total, 288 pools were prepared from the library. c, 24 pools were combined in one conical tube and called “superpool” (SP) containing 9216 clones. In total 12 superpools were prepared. d, Bacteria of each superpool was cultured and plasmid was prepared. e, The superpool plasmids were transfected into HEK293 cells. f, Individual supernatants were tested on A1 myotubes for cell cycle re-entry activity (myotube assay) (See Fig. 1b). Positive superpools were successively subfractionated and the assay process was repeated back from the positive superpool (first screen) to come to a single clone (fourth screen) (right side, a-c) (See ExFig. 2a-c).
Extended Figure 2. Expression cloning of AxMLP as a myotube cell cycle inducer.
a, The results of the second round screen of superpool 9 (see Fig. 1b and ExFig 1) and its sub-pooling diagram (right side). Sub-pool D and sub-pool 2 showed higher BrdU incorporation activity than the others, identifying pool #212 as positive (n=12: 4 biological, 3 technical replicates each; mean±s.d.). b, The result of the third round screen of pool #212 from superpool 9 and its sub-pooling diagram (right side). Sub-pool A1 showed activity (n=6: 2 biological, 3 technical replicates each; mean±s.d.). c, Fourth round screen of SP9 identified a single active clone (c1), AxMLP. (n=12: 4 biological, 3 technical replicates each; mean±s.d.). The pooling diagram is shown on the right side. d, AxMLP supernatant induces an S-phase response in a dose dependent manner in the newt myotube assay. Different amounts of AxMLP-containing supernatant (30 μl, 20 μl, 10 μl and 5.0 μl respectively) were provided to the myotube cell culture medium. The myotube BrdU incorporation correlated with the amount of supernatant provided, whereas pCMV-SPORT6 supernatant did not provoke cell cycle entry at any dose (n=6: 2 biological, 3 technical replicates each; mean±s.d.). e,f, Newt myotubes treated with purified AxMLP (e) or Flow-through (f) were immunostained for BrdU and MHC. More BrdU incorporated nuclei (red) in myotubes (green) were observed in culture supplied with purified AxMLP compared to Flow-Through-treated cultures. Bar in e, 1 mm
Extended Figure 3. AxMLP is classified as a member of the MARCKS family and characterization of its extracellular release in HEK293 cells.
a, Amino-acid sequence alignment of AxMLP with sequences from other vertebrates, human, mouse, rat, chick, newt, Xenopus and zebrafish. AxMLP contains three conserved domains, i) Myristoylated N-terminus domain, ii) MARCKS homology domain, iii) Effector domain. b, A phylogenetic tree of vertebrate MARCKS family proteins. The tree was constructed by the neighbor joining method with the ClustalW program. The percentage beside the nodes shows that a node was supported in 1000 bootstrap pseudo replications. The scale bar indicates evolutionary distance. c, Schematic illustration of His tagged AxMLP (left) and EGFP fused AxMLP (right). 3C protease PreScission site was inserted between AxMLP and the tag for both constructs. d, e, AxMLP does not induce significant cell death. The percentage of GFP-expressing HEK293 cells (d) and absolute number of the cells (e) (d,e: n=16: 4 biological, 4 technical replicates each; mean±s.d.; center values as median; whiskers as maximum and minimum, respectively) at the indicated time points of culture. There was no significant difference with Student’s t-test between AxMLP transfected cells and the control in any time points. f, Characterization of anti-AxMLP antibodies by Western Blot. Cell lysates from HEK293 cells transfected with the indicated plasmids were tested for the full-length AxMLP polyclonal antibody (left) and C-terminal AxMLP polyclonal antibody (right). g, Silver Staining of the fractions from AxMLP-His purification. BSA was added to purified fraction as a carrier protein. h, AxMLP-His purification analyzed by anti-His-tag Western Blotting.
Extended Figure 4. AxMLP is sufficient to induce cell cycle entry in axolotl tail and limb.
a-d, Sections from AxMLP-injected tails immunostained for BrdU / PAX7 (a,b) and BrdU / MEF2C (c,d) (refers to data in Fig. 2). e, Schematic illustration of the protein injection into axolotl limb. f, Quantification of BrdU+ cells in the limbs injected with PBS, Flow-through or purified AxMLP (n=4: biological replicates; center values as median; points represent each sample). Transverse sections from purified AxMLP injected (g-j) or Flow-through injected limbs (k-n). Sections were immunostained for BrdU (g,k), BrdU/MBP-Myelin Basic Protein (h,l), BrdU/PAX7 (i,m) and BrdU/GFP (j,n). GFP+ cells represent connective tissues in LPM (Lateral Plate Mesoderm)-GFP transplanted axolotls. All molecular markers used except MBP had nuclear expression, and therefore allowed one-to-one colocalization of nuclear BrdU with nuclear staining of the marker. Therefore, we refer to the MBP data as “MBP-associated.” White boxes, highlight the magnified images. Yellow circles, indicating two bones in the lower limb. NS, not significant; *P<0.05, **P<0.005, ***P<0.0005, ****P<0.00005 with Student’s t-test. Bars in a-d: 100 μm, Bars in g-n: lower magnisfication images, 200μm; in higher magnification images, 50μm. White arrowheads, indicating marker+/BrdU+ cells.
Extended Figure 5. Upregulation of AxMLP transcript during early regeneration and alteration of AxMLP protein localization in wound epidermis cells.
Measurement of AxMLP expression by qPCR at the indicated time points during tail (a) and limb (e) regeneration (n=3: biological replicates; mean±s.d.). To obtain the values of fold-change for each time point, the relative concentration of the PCR products were calculated by the standard curve method. The concentration of AxMLP was normalized with that of Rpl4 (large ribosomal protein 4). b-h, Immunostaining with anti-AxMLP antibody (white) on tails (b-d) and limbs (f-h), of intact (b,f: transverse sections), 1day post amputation (dpa) (c: sagittal; g: horizontal) and 6 dpa (d: sagittal; h:horizontal) samples. By 6 dpa, the epithelial organization and AxMLP expression appeared to be returning to a less tightly adherent, less membrane associated appearance (d-d”,h,h’). Bars in b, c, d, f, g, h, 200 μm; in b’ b” c’ c” d’ d” f’ g’ h’, 50 μm. Red arrowheads in c’d’, indicating spinal cord; Green arrowheads in c’, c”, g’ indicating wound epidermis; Yellow arrowheads in c”, g’ indicating normal epidermis. Yellow circles in f, indicating two bones in the lower limb.
Extended Figure 6. AxMLP morpholinos specifically and efficiently reduce AxMLP translation in cultured cells.
a, Schematic illustration of wild type (WT) AxMLP (top) and N-terminal deletion AxMLP (bottom) constructs used to characterize AxMLP Morpholino 1. The N-terminally deleted AxMLP lacks the morpholino binding site. Both constructs have a His-tag on their C-terminus (a). M, Myristoylated N-terminus domain; MH, MARCKS homology domain; ED, Effector domain. b-i, Electroporated A1 myoblasts were stained with indicated markers. b-e, WT-AxMLP plasmid was co-electroporated with the control morpholino (c) or the AxMLP-specific morpholino (d), whereas WT-AxMLP plasmid only (b) or WT-AxMLP only without any primary antibody staining was used as negative controls (e). f-h, ΔN-AxMLP plasmid was co-electroporated with the control morpholino (g) or the AxMLP-specific morpholino (h), whereas ΔN-AxMLP plasmid only (f) or pCMV-SPORT6-3C-His (empty vector) plasmid only served as negative controls (i). j, Western Blotting for the cell lysates from the experiment above. AxMLP morpholino specifically reduced AxMLP protein expression. k, Schematic illustration of the constructs used to characterize AxMLP Morpholino 2. The original AxMLP expression clone from the cDNA library (BL212a101: top) was used as it included the 5’UTR target site for AxMLP Morpholino 2. The sub-cloned AxMLP-His construct lacks the binding site for AxMLP Morpholino 2 and was used as the control construct. l-r, Electroporated A1 myoblasts were stained with indicated markers. l-n, BL212a101 plasmid was co-electroporated with the five-mismatch control morpholino (m) or the AxMLP-specific morpholino 2 (n), or BL212a101 plasmid only (l) or pCMV-SPORT6-3C-His (empty vector) plasmid only served as negative controls (o). AxMLP was detected using an anti-AxMLP antibody (Red), and morpholinos detected via FITC conjugation (Green). p-r, AxMLP-3C-His plasmid was co-electroporated with the five-mismatch control morpholino (q) or the AxMLP-specific morpholino 2 (r) or AxMLP-3C-His plasmid only (p). AxMLP was detected using an anti-His-tag antibody (Red), and Morpholinos detected via FITC conjugation (Green). s, Western Blotting for the cell lysates from the experiment above. AxMLP morpholino 2 specifically reduced AxMLP protein expression. Bars, 100 μm.
Extended Figure 7. AxMLP morpholinos knockdown endogenous AxMLP in vivo.
The morpholinos shown in a-j” were used in Fig. 4 and ExFig. 6a-j,8a-f and the morpholinos shown in k-t” were used in ExFig. 6k-s,8g-j. Transverse sections from AxMLP-specific morpholino 1 (a-e) or control morpholino (f-j) electroporated tail. b, the spinal cord (SC) boxed in a. c-c”, the higher magnification images of the SC boxed in b. AxMLP expression was detected in morpholino negative cells (red asterisks), whereas it was reduced in morpholino positive cells (yellow asterisks). d, the epidermis boxed in a. e-e”, AxMLP expression was unaffected in morpholino negative cells (red asterisks), whereas it was reduced in morpholino positive cells (yellow allowheads). In the control morpholino electroporated tail (f), there was no morpholino specific knockdown phenotype in either SC (g-h”) or epidermis (i-j”). The same experiments were performed with AxMLP specific morpholino 2 (k-o) and the corresponding five-mismatch control morpholino (p-t). The data sets were the same as a-j above. Bars, in a,f,k,p 200μm; in b-e, g,-j, l-o, q-t 50μm.
Extended Figure 8. AxMLP is necessary for initial cell proliferation during tail regeneration.
a-d, Representative transverse sections of the morpholino electroporated/protein injected blastemas that were used for quantification of BrdU incorporation in Fig. 4d. Rhodamine was co-injected with the protein samples. e, Quantification of BrdU+ cells in blastema sections of morpholino electroporated/protein injected tails at 3 dpa (n=4: biological replicates; center values as median; points represent each sample). f, The length of the blastema during tail regeneration. The data at 6 dpa was plotted in Fig. 4c. By 14 days the difference in total regenerate length among the samples was not statistically significant. g-j, The same experimental scheme (shown in Fig. 4a) as was used for AxMLP morpholino 1 was implemented for a second specific morpholino (AxMLP-specific morpholino 2). g, Brightfield images of the morpholino 2 electroporated/protein injected tails at 6 dpa. h, Blastema length at 6 dpa (n=4: biological replicates; center values as median; points represent each sample). i, The length of the blastema during tail regeneration. The data at 6 dpa was plotted in h. j, Transverse sections immunostained for BrdU from morpholino electroporated/protein injected tails at 3 dpa. AxMLP specific morpholino 2 combined with flow-through (FT) injection shows reduction of BrdU incorporation, whereas AxMLP protein injection rescues the phenotype. The corresponding five-mismatch control morpholino does not affect BrdU incorporation. NS, not significant, **P<0.005, ***P<0.0005, ****P<0.00005 with Student’s t-test. Bars in a-d,j, 200 μm; in g, 500 μm. Red bars in g, indicating amputation planes. Dashed lines in g, delineate the shape of the mesenchymal blastema. Yellow circles in j, indicating spinal cord (top) and notochord/cartilage (bottom), respectively.
Extended Figure 9. Anti-AxMLP antibody significantly blocks BrdU incorporation during tail regeneration.
a, Schematic illustration of antibody injection into axolotl tail. b, Quantification of BrdU+ cells in blastema sections of antibody injected tails at 3 dpa (n=4: biological replicates; center values as median; points represent each sample). NS, not significant; **P<0.005, ***P<0.0005, ****P<0.00005 with Student’s t-test.
Extended Figure 10. Exogenous AxMLP accelerates normal tail regeneration.
a, Schematic illustration of the protein injection into axolotl tail and blastema. b, Bright field images of the protein injected tails at 4 dpa. c, Blastema length at 4 dpa. (n=6: PBS, FT; n=8: AxMLP, biological replicates; center values as median; points represent each sample). The blastema from purified AxMLP injected tails significantly increased the regenerate length. NS, not significant; ***P<0.0005 with Student’s t-test. Bar in b, 500 μm. Red bars in b, indicating amputation planes. Dashed lines in b, delineate the shape of the mesenchymal blastema.
Supplementary Material
Extended Movie 1. AxMLP transfected HEK293 cells proliferate and did not show additional cell death.
Time-lapse movies of HEK293 cells transfected with AxMLP-3C-His plasmid. Between 5 and 72 hours after transfection. The culture medium was changed at 24 hours post transfection from 10% serum medium to the serum free, FreeStyle 293 medium.
Extended Movie 2. pCMV-SPORT6-3C-His transfected HEK293 cell proliferation during culture.
Time-lapse movies of pCMV-SPORT6-3C-His as a negative control to compare to AxMLP transfected HEK293 cells (ExMovie 1). Between 5 and 72 hours after transfection. The culture medium was changed at 24 hours post transfection from 10% serum medium to the serum free, FreeStyle 293 medium.
Supplementary Table 1. Raw values of qPCR.
Raw values of qPCR in tail regeneration time course (a, ExFig. 5a) and limb (b, ExFig. 5e).
Supplementary Table 2. The oligos used in this manuscript.
See “Plasmid construction” section in the Materials and Methods.
Acknowledgements
We thank Akira Tazaki , Yuka Taniguchi, Ines Wagner, Aida Rodrigo-Albors, Dunja Knapp, Prayag Murawala, Barbara Borgonovo and David Drechsel for technical advice and important discussions; We thank Maritta Schuez , Anja Telzerow and Yuka Taniguchi for assistance; Beate Gruhl, Anja Wagner, Sabine Mögel for animal care and Josh Currie, Tatiana Sandoval-Guzman and Eugeniu Nacu for comments on the manuscript. EMT was supported by a BMBF Biofutures grant, DFG grant TA274/5-1, ERC Advanced Grant, Institutional funding from MPI-CBG, and the DFG CRTD and AS by the Swedish Research Council and Cancerfonden. NCBI GenBank accession number for AxMLP is KT367888.
Footnotes
Full Materials and Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knapp D, et al. Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration-specific gene program. PLoS One. 2013;8:e61352. doi: 10.1371/journal.pone.0061352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tassava RA, Mescher AL. The roles of injury, nerves, and the wound epidermis during the initiation of amphibian limb regeneration. Differentiation. 1975;4:23–24. doi: 10.1111/j.1432-0436.1975.tb01439.x. [DOI] [PubMed] [Google Scholar]
- 4.Endo T, Bryant SV, Gardiner DM. A stepwise model system for limb regeneration. Dev Biol. 2004;270:135–145. doi: 10.1016/j.ydbio.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Holtzer S. The inductive activiy of the spinal cord in urodele tail regeneration. Journal of Morphology. 1956;99:1–39. [Google Scholar]
- 6.Julius D, MacDermott AB, Axel R, Jessell TM. Molecular characterization of a functional cDNA encoding the serotonin 1c receptor. Science. 1988;241:558–564. doi: 10.1126/science.3399891. [DOI] [PubMed] [Google Scholar]
- 7.Yang YC, et al. Human IL-3 (multi-CSF): identification by expression cloning of a novel hematopoietic growth factor related to murine IL-3. Cell. 1986;47:3–10. doi: 10.1016/0092-8674(86)90360-0. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka EM, Gann AA, Gates PB, Brockes JP. Newt myotubes reenter the cell cycle by phosphorylation of the retinoblastoma protein. J Cell Biol. 1997;136:155–165. doi: 10.1083/jcb.136.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khattak S, et al. Foamy virus for efficient gene transfer in regeneration studies. BMC Dev Biol. 2013;13:17. doi: 10.1186/1471-213X-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khattak S, et al. Germline Transgenic Methods for Tracking Cells and Testing Gene Function during Regeneration in the Axolotl. Stem cell reports. 2013;1:90–103. doi: 10.1016/j.stemcr.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnapp E, Tanaka EM. Quantitative evaluation of morpholino-mediated protein knockdown of GFP, MSX1, and PAX7 during tail regeneration in Ambystoma mexicanum. Dev Dyn. 2005;232:162–170. doi: 10.1002/dvdy.20203. [DOI] [PubMed] [Google Scholar]
- 12.Gruber CE. Production of cDNA libraries by electroporation. Methods Mol Biol. 1995;47:67–79. doi: 10.1385/0-89603-310-4:67. [DOI] [PubMed] [Google Scholar]
- 13.Sundaram M, Cook HW, Byers DM. The MARCKS family of phospholipid binding proteins: regulation of phospholipase D and other cellular components. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2004;82:191–200. doi: 10.1139/o03-087. [DOI] [PubMed] [Google Scholar]
- 14.Aderem A. The MARCKS brothers: a family of protein kinase C substrates. Cell. 1992;71:713–716. doi: 10.1016/0092-8674(92)90546-o. [DOI] [PubMed] [Google Scholar]
- 15.Sandoval-Guzman T, et al. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell. 2014;14:174–187. doi: 10.1016/j.stem.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Berg DA, et al. Efficient regeneration by activation of neurogenesis in homeostatically quiescent regions of the adult vertebrate brain. Development. 2010;137:4127–4134. doi: 10.1242/dev.055541. doi:dev.055541 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Maden M, Manwell LA, Ormerod BK. Proliferation zones in the axolotl brain and regeneration of the telencephalon. Neural Dev. 2013;8:1. doi: 10.1186/1749-8104-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seykora JT, Myat MM, Allen LA, Ravetch JV, Aderem A. Molecular determinants of the myristoyl-electrostatic switch of MARCKS. J Biol Chem. 1996;271:18797–18802. doi: 10.1074/jbc.271.31.18797. [DOI] [PubMed] [Google Scholar]
- 19.Rodrigo Albors A, et al. Planar cell polarity-mediated induction of neural stem cell expansion during axolotl spinal cord regeneration. Elife. 2015;4 doi: 10.7554/eLife.10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- 20.Kragl M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 21.Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis : a laboratory manual. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 22.Habermann B, et al. An Ambystoma mexicanum EST sequencing project : analysis of 17,352 expressed sequence tags from embryonic and regenerating blastema cDNA libraries. Genome Biol. 2004;5:R67. doi: 10.1186/gb-2004-5-9-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferretti P, Brockes JP. Culture of newt cells from different tissues and their expression of a regeneration-associated antigen. J. Exp. Zool. 1988;247:77–91. doi: 10.1002/jez.1402470111. [DOI] [PubMed] [Google Scholar]
- 24.Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roensch K, Tazaki A, Chara O, Tanaka EM. Progressive specification rather than intercalation of segments during limb regeneration. Science. 2013;342:1375–9. doi: 10.1126/science.1241796. [DOI] [PubMed] [Google Scholar]
- 26.Zarzosa A, et al. Axolotls with an under- or oversupply of neural crest can regulate the sizes of their dorsal root ganglia to normal levels. Dev. Biol. 2014;394:65–82. doi: 10.1016/j.ydbio.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigo-Albors A, Tanaka EM. High-efficiency electroporation of the spinal cord in larval axolotl. In: Kumar A, Simon A, editors. Salamanders in Regeneration Research: Methods and Protocols. Methods in Molecular Biology. Springer; USA: 2015. pp. 115–126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Movie 1. AxMLP transfected HEK293 cells proliferate and did not show additional cell death.
Time-lapse movies of HEK293 cells transfected with AxMLP-3C-His plasmid. Between 5 and 72 hours after transfection. The culture medium was changed at 24 hours post transfection from 10% serum medium to the serum free, FreeStyle 293 medium.
Extended Movie 2. pCMV-SPORT6-3C-His transfected HEK293 cell proliferation during culture.
Time-lapse movies of pCMV-SPORT6-3C-His as a negative control to compare to AxMLP transfected HEK293 cells (ExMovie 1). Between 5 and 72 hours after transfection. The culture medium was changed at 24 hours post transfection from 10% serum medium to the serum free, FreeStyle 293 medium.
Supplementary Table 1. Raw values of qPCR.
Raw values of qPCR in tail regeneration time course (a, ExFig. 5a) and limb (b, ExFig. 5e).
Supplementary Table 2. The oligos used in this manuscript.
See “Plasmid construction” section in the Materials and Methods.