Abstract
Background
The long noncoding RNA (lncRNA) colorectal neoplasia differentially expressed – h (CRNDE-h) plays important roles in the early stages of human development and cancer progression. We investigated the expression and clinical significance of lncRNA CRNDE-h in colorectal cancer (CRC).
Methods
The expression level of lncRNA CRNDE-h was analyzed in 142 CRC tissues and 142 paired adjacent nontumorous tissues, along with 21 inflammatory bowel diseases, 69 hyperplastic polyp, and 73 colorectal adenoma samples, using quantitative real-time polymerase chain reaction. The association between lncRNA CRNDE-h, and Iroquois homeobox protein 5 (IRX5) mRNA was examined in the same 142 CRC tissues.
Results
We found that lncRNA CRNDE-h level was elevated in the CRC and adenoma groups compared with the other groups (all at P<0.001). In CRC, upregulation of lncRNA CRNDE-h was significantly correlated with large tumor size, positive regional lymph node metastasis, and distant metastasis (all at P<0.05). Area under the curve for lncRNA CRNDE-h showed diagnostic capability for distinguishing CRC from other groups. Patients with CRC with high lncRNA CRNDE-h expression level had poorer overall survival than those with low lncRNA CRNDE-h expression (log-rank test, P<0.001). Further, multivariable Cox regression analysis suggested that increased expression of lncRNA CRNDE-h was an independent prognostic indicator for CRC (hazard ratio [HR]=2.173; 95% confidence interval [CI], 1.282–3.684, P=0.004). Furthermore, lncRNA CRNDE-h expression was positively correlated with IRX5 mRNA in CRC tissues.
Conclusions
Our data offers convincing evidence for the first time that lncRNA CRNDE-h is associated with adverse clinical characteristics and poor prognosis, which suggests that it might play an important role in CRC development and progression and might have clinical potential as a useful prognostic predictor.
Keywords: colorectal cancer, long noncoding RNA, CRNDE-h, diagnosis, prognosis, IRX5
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors and the third leading cause of cancer deaths worldwide.1 Although current detection methods, such as fecal occult blood testing and fiber-optic colonoscopy, have significantly improved outcomes for patients with CRC, more than half of those at an advanced stage of disease die of carcinoma recurrence after undergoing radical resection.2 Therefore, to provide better treatment strategies, there is an urgent need to further understand the precise molecular mechanism of CRC and to identify new prognostic biomarkers and effective individualized therapeutic targets.
Genome sequencing projects have revealed that protein-coding RNAs account for only 2% of the total genome, while more than 90% of the genome is transcribed as non-coding RNAs (ncRNAs), among which are long noncoding RNAs (lncRNAs), which are more than 200 nucleotides (nt) in length with no protein-coding capacity.3 Expression of lncRNAs, which was previously believed to represent random transcriptional noise, has been observed to vary by disease, tissue, and developmental stage, indicating specific functions for lncRNAs in human development and disease, including cancers.4–7
The lncRNA colorectal neoplasia differentially expressed – h (CRNDE-h) (colorectal neoplasia differentially expressed transcript variant 1), 1,059 nt in length, is encoded by an uncharacterized gene locus (Chr16:hCG_1815491). Expression level of lncRNA CRNDE-h is highest in the early stages of human development and progressively decreases, with detected expression ranking as follows: embryoid body > fetus > juvenile > adult (UniGene ESTdatabase) (www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.237396). In the group of acute myeloid leukemias (AMLs), lncRNA CRNDE-h expression is particularly increased in FAB Type M2 AML and in promyelocytic Type M3 AML.8 A similar mode of expression occurs in prostate cancer, in which lncRNA CRNDE-h expression level is significantly upregulated and associated with poor prognosis and distant metastases.9–11 However, some tumors, such as oncocytomas and clear cell kidney carcinomas, show significantly downregulated expression of lncRNA CRNDE-h.12 These results suggest that lncRNA CRNDE-h might play a major role in cancer progression, and exert distinct effects on different types of cancers. Nevertheless, the exact expression pattern of lncRNA CRNDE-h and its clinical and prognostic significance in CRC remains unclear.
A survey within the GENCODE project showed that the expression of almost 3% of lncRNAs shows high positive correlation with their neighboring mRNA.13 Iroquois homeobox protein 5 (IRX5) mRNA, which is transcribed from chromosome 16 on the strand opposite to the adjacent CRNDE gene, is a member of the Iroquois homeobox gene family. In a previous study, the association between CRNDE and IRX5 expression was analyzed for different tissues,14 such as bronchial epithelium, ovary epithelium, and prostate basal epithelium, but there was little information on the correlation between lncRNA CRNDE-h and IRX5 mRNA expression in CRC. Therefore, further investigation is needed to estimate the relationship between lncRNA CRNDE-h and IRX5 mRNA in CRC.
To date, there has been no investigation of the correlation between lncRNA CRNDE-h expression and the clinicopathological characteristics of CRC. In the current study, the expression levels of lncRNA CRNDE-h in clinically resected human colorectal disease tissues were examined by quantitative real-time polymerase chain reaction (qRT-PCR), and their clinicopathological significance and potential prognostic value for CRC were further assessed. We then evaluated the potential relationship between lncRNA CRNDE-h expression and IRX5 mRNA amplification by qRT-PCR. These data demonstrated that expression of lncRNA CRNDE-h was increased in CRC and closely correlated with tumor progression, recurrence, and survival, suggesting that lncRNA CRNDE-h may represent a novel indicator of poor prognosis in CRC and may be a potential therapeutic target for diagnosis and gene therapy.
Materials and methods
Patients and tissue samples
In total, 305 subjects who had undergone radical resection in the Department of General Surgery, Qilu Hospital of Shandong University, between July 2007 and October 2009 were enrolled in this study. Patients with CRC for whom medical records were incomplete, who had received prior radiation or chemotherapy, were lost to follow-up, or withdrew consent, were excluded from this study. Medical history, such as age, sex, tumor location, tumor size, differentiation, lymphovascular invasion, T stage, regional lymph nodes metastasis, and distant metastasis were recorded. The tissues involved were 142 CRC tissues and 142 corresponding adjacent nontumorous tissues (at least 5 cm from the margin of the tumor), plus 21 inflammatory bowel disease (IBD), 69 hyperplastic polyp, and 73 adenoma tissues. All of the tissues were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction. The postoperative pathological staging was determined for each individual patient according to the Union for International Cancer Control/American Joint Committee on Cancer TNM staging system for CRC (seventh edition), and followed up at every third month until the patient’s death or until March 2015, whichever occurred first. Detailed information on the clinical features of all patients in this study is shown in Table 1. Follow-up studies included physical examination, laboratory analysis, and computed tomography if necessary. The Human Research Ethics Committee of Qilu Hospital of Shandong University approved all aspects of this study and oral informed consent was obtained from each patient. All specimens were handled and made anonymous according to ethical and legal standards.
Table 1.
Clinicopathological characteristics and lncRNA CRNDE-h expression in patients with CRC (n=142)
| Variables | No of cases | Percent % |
|---|---|---|
| Age (years) | ||
| <60 | 80 | 56.3 |
| ≥60 | 62 | 43.7 |
| Sex | ||
| Male | 87 | 61.3 |
| Female | 55 | 38.7 |
| Tumor location | ||
| Rectum | 93 | 65.5 |
| Colon | 49 | 34.5 |
| Tumor differentiation | ||
| Well | 31 | 21.8 |
| Moderate | 83 | 58.5 |
| Poor | 28 | 19.7 |
| Lymphovascular invasion | ||
| No | 91 | 64.1 |
| Yes | 51 | 35.9 |
| T stage | ||
| T1–T2 | 30 | 21.1 |
| T3–T4 | 112 | 78.9 |
| Regional lymph node metastasis | ||
| Negative | 76 | 53.5 |
| Positive | 66 | 46.5 |
| Distant metastasis | ||
| No | 112 | 78.9 |
| Yes | 30 | 21.1 |
| Tumor size | ||
| <5 cm | 87 | 61.3 |
| ≥5 cm | 55 | 38.7 |
| Expression of lncRNA CRNDE-h | ||
| Low | 71 | 50.0 |
| High | 71 | 50.0 |
Abbreviations: lncRNA, long noncoding RNA; CRNDE-h, colorectal neoplasia differentially expressed – h; CRC, colorectal cancer.
Cell culture
Human CRC cell line (HT-29) was purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). Cells were cultured in RPMI-1640 (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in a 5% CO2 incubator.
RNA extraction and reverse transcription
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen) in accordance with the manufacturer’s instructions. Preparation and handling of RNA took place under RNase-free conditions. The RNA concentration was determined by using spectrophotometry to measure absorbance at 260 nm, and the isolated RNA was stored at −80°C until use. cDNA was synthesized from 1,000 ng of RNA using Primerscript™ RT Reagent Kit (TaKaRa, Dalian, People’s Republic of China) in a 20 μL reaction volume.
qRT-PCR
qRT-PCR analysis was performed using Power SYBR Green (TaKaRa) on a CFX96 Real-Time PCR Detection System, which was also used for data collection. Results were normalized to an endogenous control (GAPDH). The thermal cycling program used for quantification was as follows: an initial denaturation step at 95°C for 15 seconds, followed by 40 cycles of denaturation at 95°C for 5 seconds and annealing and extension at 58°C for 34 seconds. After amplification, the melting curve of the product was obtained by a temperature gradient at 75°C under continuous fluorescence monitoring. The qRT-PCR measurement was performed in triplicate to remove any outliers. The specificity of the amplified product (CRNDE-h) was confirmed by sequencing (Figure S1). The relative expression of CRNDE-h in HT-29 cells was used as a calibrator in each run; no-template controls were included as negative control. The relative expression of CRNDE-h was calculated and normalized relative to GAPDH, using the comparative threshold cycle (Ct) method. The qRT-PCR primers for lncRNA CRNDE-h, and mRNA GAPDH were as follows:
CRNDE-h: F: 5′-CGCGCCCGCGCGGCGGAGGA-3′, R: 5′-TATGAATTGCAGACTTTGCAGA-3′;
IRX5: F: 5′-ACCCAGCGTACCGGAAGAA-3′, R: 5′-CGGCGTCCACGTCATTTTAT-3′;
GAPDH: F: 5′-GTCAACGGATTTGGTCTGTATT-3′, R: 5′-AGTCTTCTGGGTGGCAGTGAT-3′.
Statistical analysis
All statistical data were analyzed by Statistical Program for Social Sciences (SPSS) software (version 17.0; SPSS, Chicago, IL, USA) and GraphPad Prism (version 5.0; GraphPad Software, LaJolla, CA, USA). We used the Kolmogorov–Smirnov test to determine the distribution of each group. Data were expressed as median (interquartile range). The differences between groups were estimated using Mann–Whitney U-test or Kruskal–Wallis test, as appropriate. The diagnostic performance of lncRNA CRNDE-h was determined by the receiver operating characteristic curve using MedCalc software (version 9.3.9.0; MedCalc, Mariakerke, Belgium). The Youden index (sensitivity + specificity −1) was used to determine the optimal cut-off value of lncRNA CRNDE-h expression level. Overall survival (OS) curves were plotted according to the Kaplan–Meier method, with the log-rank test applied for comparison. The Cox proportional-hazards regression model was employed to evaluate the independent prognostic factors. Spearman correlation analysis was used to examine the association between lncRNA CRNDE-h and IRX5 mRNA. Statistical significance was defined as a two-sided P-value of less than 0.05.
Results
lncRNA CRNDE-h was greatly upregulated in primary CRC samples
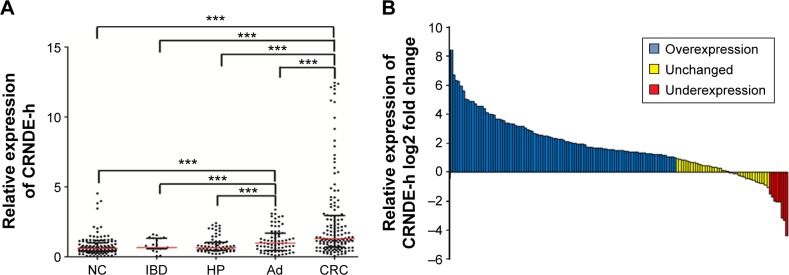
The Kruskal–Wallis test indicated that there was a large variability in lncRNA CRNDE-h among CRC, adjacent noncancerous tissue, IBD, hyperplastic polyp, and adenoma samples. As shown in Figure 1A, the expression levels of lncRNA CRNDE-h were significantly higher in CRC (1.640; interquartile range 0.883–3.737) compared with adjacent noncancerous tissues (0.769; 0.525–1.264), hyperplastic polyp (0.826; 0.574–1.311), IBD (0.846; 0.734–1.682), and adenoma (1.249; 0.579–2.142) (all at P<0.001), and were also significantly higher in the adenoma group compared with the adjacent noncancerous tissue, hyperplastic polyp, and IBD groups (all at P<0.001). Moreover, no significant differences were found between the adjacent noncancerous tissues, hyperplastic polyp, and IBD groups (all at P>0.05). Remarkably, we found that lncRNA CRNDE-h was strongly expressed in CRC tissues compared with paired adjacent noncancerous tissues (2−ΔΔCt method), and the majority of results displayed greater than two-fold increases (CRC/adjacent noncancerous tissues). In several CRC specimens, more than 50-fold increases of lncRNA CRNDE-h expression were found (Figure 1B).
Figure 1.
Quantitative analysis of lncRNA CRNDE-h using qRT-PCR in 305 colorectal disease tissues.
Notes: (A) The expression level of lncRNA CRNDE-h was significantly higher in the CRC and Ad group compared with the NC, IBD, and HP group, and its expression was normalized by GAPDH in each sample (***P<0.001). Red line represents the median value and the black line means the 25% and 75% interquartile range. (B) IncRNA CRNDE-h relative expression was expressed as log2 fold change (CRC/NC), and the log2 fold changes were defined as follows: >1, overexpression; <−1, underexpression; the remaining were defined as unchanged.
Abbreviations: CRC, colorectal cancer; Ad, colorectal adenomas; NC, adjacent noncancerous tissues; IBD, inflammatory bowel disease; HP, hyperplastic polyp; lncRNA, long noncoding RNA; CRNDE-h, colorectal neoplasia differentially expressed – h; qRT-PCR, Quantitative real-time polymerase chain reaction.
Correlation of lncRNA CRNDE-h expression with the clinicopathologic characteristics of human CRC tissues
In further evaluations of the correlation between lncRNA CRNDE-h expression and clinicopathologic characteristics, the data showed that the expression level of CRNDE-h was significantly correlated with tumor size (P=0.017), regional lymph nodes metastasis (P=0.006), and distant metastasis (P=0.003) (Table 2). However, no significant associations were found between CRNDE-h overexpression and age, sex, tumor location, tumor differentiation, lymphovascular invasion, or T stage.
Table 2.
Correlations of clinicopathological parameters and expression level of lncRNA CRNDE-h in patients with CRC (n=142)
| Variables | No of cases | IncRNA CRNDE-h expression | P-value |
|---|---|---|---|
| Age (years) | |||
| <60 | 80 | 1.466 (0.844–3.182) | 0.177 |
| ≥60 | 62 | 2.043 (0.996–4.424) | |
| Sex | |||
| Male | 87 | 1.686 (1.136–3.898) | 0.247 |
| Female | 55 | 1.427 (0.659–3.167) | |
| Tumor location | |||
| Rectum | 93 | 1.628 (0.858–3.733) | 0.613 |
| Colon | 49 | 1.651 (1.251–3.315) | |
| Tumor differentiation | |||
| Poor | 31 | 2.875 (1.016–5.229) | 0.218 |
| Moderate | 83 | 1.427 (0.855–3.133) | |
| Well | 28 | 1.694 (1.125–3.554) | |
| Lymphovascular invasion | |||
| No | 91 | 1.682 (1.121–3.674) | 0.413 |
| Yes | 51 | 1.398 (0.744–3.220) | |
| T stage | |||
| T1–T2 | 30 | 1.321 (0.574–2.414) | 0.067 |
| T3–T4 | 112 | 1.687 (1.053–3.899) | |
| Regional lymph node metastasis | |||
| Negative | 76 | 1.353 (0.885–2.277) | 0.006* |
| Positive | 66 | 2.385 (1.138–4.808) | |
| Distant metastasis | |||
| No | 112 | 1.408 (0.774–2.850) | 0.003* |
| Yes | 30 | 2.972 (1.603–5.315) | |
| Tumor size | |||
| <5 cm | 87 | 1.419 (0.724–2.468) | 0.017* |
| ≥5 cm | 55 | 2.384 (1.243–4.418) |
Notes:
P-values are statistically significant. Data presented as median (25%–75% interquartile range).
Abbreviations: lncRNA, long noncoding RNA; CRNDE-h, colorectal neoplasia differentially expressed – h; CRC, colorectal cancer.
Diagnostic value of lncRNA CRNDE-h for CRC
Receiver operating characteristic curve analyses indicated that lncRNA CRNDE-h has some capability for distinguishing CRC tissues from normal and benign tissues, with an area under the curve value of 0.757 (95% confidence interval [CI]=0.715–0.796) (Figure 2). At a cut-off value of 1.256, the optimal sensitivity and specificity were 70.4% and 70.8%, respectively, in identifying patients with CRC.
Figure 2.
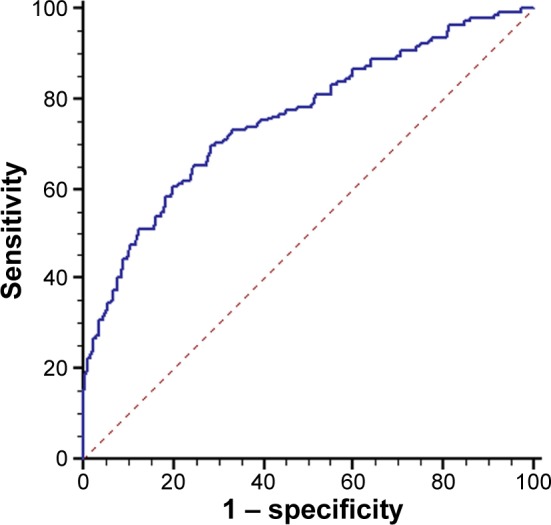
ROC curve analysis for the detection of CRC using lncRNA CRNDE-h.
Notes: ROC analysis showed the diagnosis capability for distinguishing CRC tissues from normal and benign disease groups.
Abbreviations: ROC, receiver operating characteristic; lncRNA, long noncoding RNA; CRNDE-h, colorectal neoplasia differentially expressed – h; CRC, colorectal cancer.
Prognostic implications of lncRNA CRNDE-h expression in patients with CRC
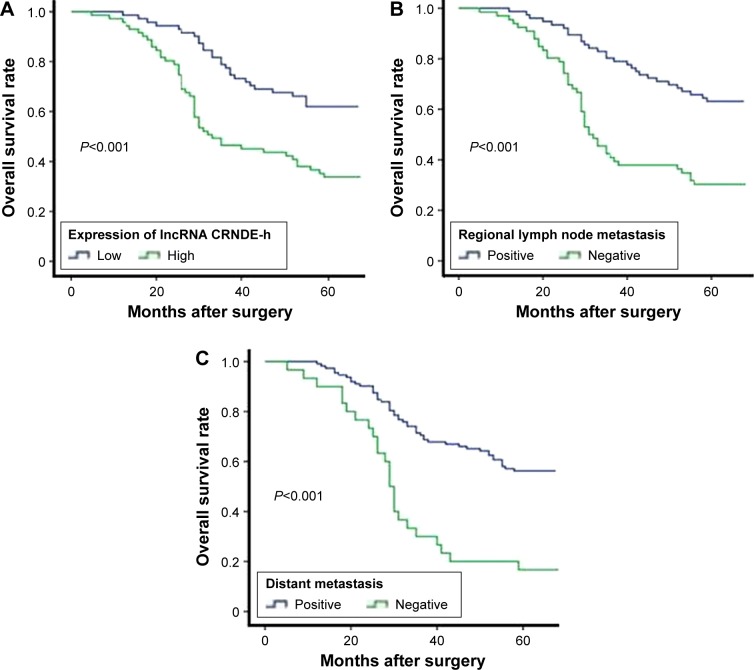
In total, 142 patients with CRC were followed-up, with a median follow-up duration of 55 months (range 5–65 months). OS was calculated from the time of diagnosis to the date of death or to study completion, and the cumulative 5-year OS rate was 47.2%. The median value (1.640) of lncRNA CRNDE-h expression level was used to categorize patients with CRC into a high-level group (n=71) and a low-level group (n=71). Kaplan–Meier analysis showed that the 5-year cumulative probability of OS for patients with high lncRNA CRNDE-h expression (29.6%) was significantly lower than that for patients with low lncRNA CRNDE-h expression (64.8%) (log-rank test, P<0.001) (Figure 3A). Moreover, Kaplan–Meier analysis and log-rank tests showed that OS was also associated with regional lymph node metastasis (log-rank test, P<0.001) (Figure 3B) and distant metastasis (log-rank test, P<0.001) (Figure 3C).
Figure 3.
Association between lncRNA CRNDE-h expression and overall survival in colorectal cancer patients.
Notes: Kaplan–Meier curves for overall survival in the 142 colorectal cancer patients according to (A) lncRNA CRNDE-h overexpression, (B) regional lymph nodes metastasis, (C) and distant metastasis, P<0.001.
Abbreviations: lncRNA, long noncoding RNA; CRNDE-h, colorectal neoplasia differentially expressed – h.
We further estimated the clinical significance of various prognostic factors that might influence survival in the subjects. Univariate analysis was performed for OS in 142 patients with CRC. As shown in Table 3, overexpression of lncRNA CRNDE-h (hazard ratio [HR]=2.906; 95% CI, 1.794–4.709; P<0.001), T stage (HR=2.086; 95% CI, 1.071–4.062, P=0.031), distant metastasis (HR=3.102; 95% CI, 1.905–5.053, P<0.001), and regional lymph node metastasis (HR=2.683; 95% CI, 1.680–4.284, P<0.001) were all statistically significant risk factors affecting the OS of patients. The other clinicopathological features (age, sex, tumor location, lymphovascular invasion, tumor differentiation, and tumor size) were not statistically significant prognostic factors (P>0.05). More importantly, to evaluate the robustness of the prognostic value of lncRNA CRNDE-h expression, we then put the four significant prognostic variables into the Cox proportional-hazards regression model for multivariate analysis, and found that only overexpression of lncRNA CRNDE-h (HR=2.173; 95% CI, 1.282–3.684; P=0.004), distant metastasis (HR=2.599; 95% CI, 1.496–4.516; P=0.001), and regional lymph node metastasis (HR=3.012; 95% CI, 1.746–5.197; P<0.001) maintained their significance as independent prognostic factors for OS rate (Table 3).
Table 3.
Univariate and multivariate Cox proportional hazards regression model analysis of overall survival in patients with CRC (n=142)
| Variables | Univariate analysis
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Age | 1.042 | 0.659–1.648 | 0.860 | |||
| Sex | 0.927 | 0.578–1.486 | 0.753 | |||
| Tumor location | 0.820 | 0.502–1.340 | 0.429 | |||
| Tumor differentiation | 1.130 | 0.786–1.625 | 0.509 | |||
| Lymphovascular invasion | 0.807 | 0.499–1.305 | 0.383 | |||
| T stage | 2.086 | 1.071–4.062 | 0.031* | 1.365 | 0.624–2.986 | 0.436 |
| Regional lymph node metastasis | 2.683 | 1.680–4.284 | <0.001* | 3.012 | 1.746–5.197 | <0.001* |
| Distant metastasis | 3.102 | 1.905–5.053 | <0.001* | 2.599 | 1.496–4.516 | 0.001* |
| Tumor size | 1.529 | 0.970–2.411 | 0.067 | |||
| High CRNDE-h expression | 2.906 | 1.794–4.709 | <0.001* | 2.173 | 1.282–3.684 | 0.004* |
Note:
P-values are statistically significant.
Abbreviations: CI, Confidence interval; HR, hazard ratio; lncRNA, long noncoding RNA; CRNDE-h, colorectal neoplasia differentially expressed – h; CRC, colorectal cancer.
Association between lncRNA CRNDE-h and IRX5 mRNA in CRC tissues
We further explored the potential underlying mechanisms involved in lncRNA CRNDE-h overexpression-induced tumorigenesis by examining expression of IRX5 mRNA in the same CRC tissues. Spearman’s rank correlation analysis showed that there was a statistically positive correlation between lncRNA CRNDE-h amplification and IRX5 mRNA expression in CRC tissue samples (r=0.692, P<0.001) (Figure 4).
Figure 4.
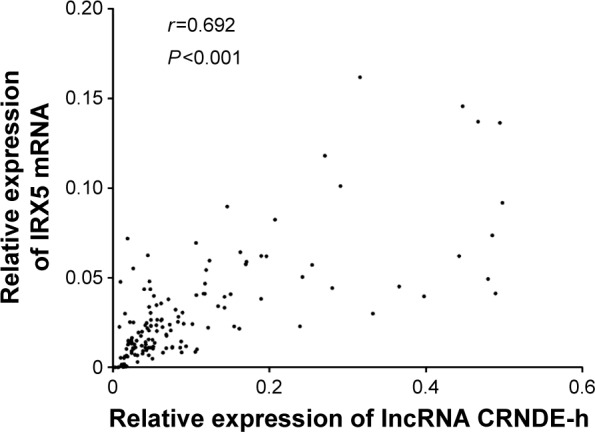
Spearman’s correlation analysis between lncRNA CRNDE-h and IRX5 mRNA.
Notes: Positive correlation of lncRNA CRNDE-h and IRX5 mRNA expression level in 142 colorectal cancer tissues was analyzed using Spearman’s correlation analysis. (r=0.692, P<0.001).
Abbreviations: lncRNA, long noncoding RNA; CRNDE-h, colorectal neoplasia differentially expressed – h; mRNA, messenger RNA; IRX5, Iroquois homeobox protein 5.
Discussion
lncRNAs function in different ways, including regulation of gene transcription in basal transcription machinery, post-transcriptional regulation of RNA splicing, and epigenetic regulation. Previous studies have indicated that about 18% of the noncoding genes that produce lncRNAs are associated with cancer, compared with only 9% of all human coding genes.15 Emerging evidence has indicated that modification in the expression of lncRNAs is associated with tumor formation.16 During the past few years, the importance of lncRNA in CRC tumorigenesis and progression has been gradually recognized. Some lncRNAs, such as CCAT1, MALAT1, PVT-1, and HOTAIR, were found to be significantly elevated in CRC, and could promote the proliferation and metastasis of CRC.17–20 However, Evf-2 and GAS5 lncRNAs were downregulated in colon cancer, which could inhibit cell-cycle progression and promote apoptosis of colon cancer cells.21,22 Nonetheless, little is currently known about the clinical value of lncRNA CRNDE-h in CRC.
CRNDE-h, an lncRNA, was originally isolated from a screen for potential oncogenic genes expressed at high levels during CRC formation and development. The locus of its encoding gene, CRNDE, spans six exons, E1 to E6, located at nt 54,952,774 to 54,963,101 on Chr16. Interestingly, at least ten alternative RNA transcripts are derived from the CRNDE locus. However, the transcript isoforms in CRC cell lines and tissues are CRNDE-a, -b, -c, -d, -e, -f, -g, -h, and -j.23 Two major CRNDE lncRNAs, CRNDE transcript variant 2 (also known as CRNDE-g) [RefSeq: NR_034106.1] and CRNDE transcript variant 1 (also known as CRNDE-h) [RefSeq: NR_034105.2], have also been identified in humans. These result from the presence of alternative 5′-splice donor sites in exon 1. In addition, CRNDE-g is only 924 nt in length, missing 135 nt at its 5′ end. Emerging evidence suggests that lncRNAs, which show high evolutionary conservation, play critical roles in a diverse array of cellular processes such as cell-cycle control,24 epigenetic regulation,25 self-renewal of stem cells,5 embryonic development, and multiple human diseases, including cancer.26 CRNDE-h includes five exons (E1A, E2, E4, E5, and E6) and two genomic vertebrate conserved (gVC) regions, one in intron 1 (gVC1) and the other in intron 4 (gVC4). In particular, gVC4 behaves like both a transcribed ultra-conserved region and an additional or alternative exon.14 An lncRNA with high evolutionary conservation, CRNDE-h has been reported to be involved in carcinogenesis. Graham et al23 showed that the phenotypic effects associated with knock-in of small open reading frames into CRC cell lines is unaffected by deletion of their start codons, which indicates that any effects are mediated by the transcripts rather than by any polypeptides that they may encode. Khalil et al27 found that there was an overlap in the lists of genes affected by short interfering RNA-mediated knockdown of CRNDE-h and polycomb repressive complex 2, potentially implicating CRNDE in the epigenetic remodeling of chromatin, and particularly in the downregulation of gene expression via targeted histone methylation or demethylation by PRC2 or corepressor for element-1-silencing transcription factor complexes, respectively. Meanwhile, downregulation of lncRNA CRNDE-h in glioma cell lines was shown to inhibit cell self-renewal, restrain cell proliferation, stop the cell cycle, and promote cell apoptosis.28 Collectively, these reports suggest that lncRNA CRNDE-h plays an important role in cancer tumorigenesis. The present study indicated that lncRNA CRNDE-h is significantly upregulated in colorectal adenomas and cancer compared with adjacent noncancerous tissues, IBD, and hyperplastic polyps. Moreover, the majority of CRC tissues displayed greater than two-fold increases of lncRNA CRNDE-h expression compared with paired adjacent noncancerous tissues. We also found that lncRNA CRNDE-h concentrations were significantly associated with factors indicating poor clinical outcome, such as large tumor size, positive distant metastasis, and positive lymph node metastasis. These data indicate that lncRNA CRNDE-h may have a role not only in the promotion of growth of the primary tumor, but also in the development of lymph node and distant metastases in CRC. Taken together, these results suggest that the dysregulated expression of lncRNA CRNDE-h might be an early event, and that a higher level of lncRNA CRNDE-h expression might be related to advanced CRC. Nonetheless, the precise mechanisms of lncRNA CRNDE-h, which induce adverse clinical characteristics in CRC, need to be further investigated.
So far, our report is among the few to testify to the significance of lncRNA CRNDE-h in identification of CRC, with an area under the curve value of 0.757 derived from comparisons between CRC tissues and normal or benign disease groups (sensitivity 70.4%; specificity 70.8%). Thus, we suggest that detection of lncRNA CRNDE-h could be an effective complement to current CRC diagnostic strategy. Moreover, accurate prediction of prognosis is essential for selection of the correct therapy for patients with CRC. Thus, we explored the prognostic value of lncRNA CRNDE-h, using Kaplan–Meier survival analysis. Patients with CRC who had high CRNDE-h levels had poor OS compared with those with low levels. Taking this a step further, Cox proportional hazards regression model analyses showed that lncRNA CRNDE-h was an independent factor influencing OS. Thus, hypothetically, lncRNA CRNDE-h expression levels may serve to identify patients at higher or lower risk in a similar way to classic prognostic factors such as lymph node metastasis or distant metastasis.
lncRNAs can act to regulate expression of their genetically neighboring protein-coding genes in cis-effection, or of distant protein-coding genes in trans-effection.29,30 IRX5 mRNA, which is encoded by CRNDE neighboring protein-coding genes IRX5, has been found in multiple cancers and contributes to development of many tumors by acting as an important transcript factor regulating key regulator genes that control cell apoptosis.31,32 The activity of IRX5 could reduce the sensitivity to the cytostatic effect of transforming growth factor-β (TGF-β), conferring a growth advantage to tumor cells prior to the acquisition of mutations in TGF-β/SMADS pathway components.33,34 In our study, lncRNA CRNDE-h expression levels plotted against IRX5 mRNA expression levels for each tissue indicated that the two RNAs had strongly correlated expression, supporting the observation that the expression of both genes is under a level of coordinate control. Based on these data, we speculate that lncRNA CRNDE-h might partly function as a tumor oncogene through contact with IRX5 mRNA in CRC. However, a limitation of the current study is that we did not clearly demonstrate this molecular function of lncRNA CRNDE-h in tumorigenesis; further investigation is needed.
Although the findings of our current study are promising, there were some limitations. First, because patients with premalignant adenomas were not followed-up, it was unclear whether those with higher CRNDE-h levels had an increased risk of developing CRC. Second, because we assessed clinical materials from Chinese patients only, lncRNA CRNDE-h expression levels as a diagnostic and prognostic biomarker must be verified in different ethnic populations before clinical application. Thus, further multicenter studies that include higher numbers of patients collected from several hospitals and diverse ethnic populations are required to validate whether CRNDE-h can be incorporated into routine clinical practice.
In conclusion, this study used an qRT-PCR assay to detect the lncRNA CRNDE-h, and found that this lncRNA might have clinical potential not only as a promising predictor of aggressive phenotype but also as an independent prognostic predictor to identify individuals with particularly low rates of survival following surgery. Further, detection of circulating lncRNA CRNDE-h in plasma by qRT-PCR could provide a noninvasive approach for analyses of early diagnosis and prognosis of CRC.
Supplementary material
Sequence data of CRNDE-h amplified production.
Notes: (A) The sequence data of CRNDE-h amplified production – Forward. (B) The sequence data of CRNDE-h amplified production – Reverse. (C) Comparison between amplified production sequences and CRNDE-h sequence in NCBI database.
Abbreviations: lncRNA, long noncoding RNA; CRNDE-h, colorectal neoplasia differentially expressed – h; Sbjct, sequence from NCBI database; NCBI, National Center for Biotechnology Information.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant numbers 81301506 and 81472025), Science and Technology Development Program of Shandong Province (2014GSF118016), Research Fund for the Doctoral Program of Higher Education of China (20130131120067 and 20120131110055), Shandong Key Research and Development Program (2015GSF118075), and Taishan Scholar Foundation.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Edwards BK, Noone AM, Mariotto AB, et al. Annual Report to the Nation on the status of cancer, 1975–2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer. 2014;120(9):1290–1314. doi: 10.1002/cncr.28509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nature Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Le Dieu R, Taussig DC, Ramsay AG, et al. Peripheral blood T cells in acute myeloid leukemia (AML) patients at diagnosis have abnormal phenotype and genotype and form defective immune synapses with AML blasts. Blood. 2009;114(18):3909–3916. doi: 10.1182/blood-2009-02-206946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lapointe J, Li C, Higgins JP, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101(3):811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pressinotti NC, Klocker H, Schafer G, et al. Differential expression of apoptotic genes PDIA3 and MAP3K5 distinguishes between low- and high-risk prostate cancer. Mole Cancer. 2009;8:130. doi: 10.1186/1476-4598-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cifola I, Spinelli R, Beltrame L, et al. Genome-wide screening of copy number alterations and LOH events in renal cell carcinomas and integration with gene expression profile. Mol Cancer. 2008;7:6. doi: 10.1186/1476-4598-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis BC, Molloy PL, Graham LD. CRNDE: A Long Non-Coding RNA Involved in CanceR, Neurobiology, and DEvelopment. Front Genet. 2012;3:270. doi: 10.3389/fgene.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khachane AN, Harrison PM. Mining mammalian transcript data for functional long non-coding RNAs. PloS One. 2010;5(4):e10316. doi: 10.1371/journal.pone.0010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliva J, Bardag-Gorce F, French BA, Li J, French SW. The regulation of non-coding RNA expression in the liver of mice fed DDC. Exp Mol Pathol. 2009;87(1):12–19. doi: 10.1016/j.yexmp.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, et al. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130(7):1598–1606. doi: 10.1002/ijc.26170. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi Y, Sawada G, Kurashige J, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110(1):164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue Y, Gu D, Ma G, et al. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2015;30(2):303–310. doi: 10.1093/mutage/geu076. [DOI] [PubMed] [Google Scholar]
- 20.Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 21.Berghoff EG, Clark MF, Chen S, Cajigas I, Leib DE, Kohtz JD. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development. 2013;140(21):4407–4416. doi: 10.1242/dev.099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):253. doi: 10.1007/s12032-014-0253-8. [DOI] [PubMed] [Google Scholar]
- 23.Graham LD, Pedersen SK, Brown GS, et al. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2(8):829–840. doi: 10.1177/1947601911431081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17(5):556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367(2):122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 29.Grote P, Wittler L, Hendrix D, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senner CE, Nesterova TB, Norton S, et al. Disruption of a conserved region of Xist exon 1 impairs Xist RNA localisation and X-linked gene silencing during random and imprinted X chromosome inactivation. Development. 2011;138(8):1541–1550. doi: 10.1242/dev.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martorell O, Barriga FM, Merlos-Suarez A, et al. Iro/IRX transcription factors negatively regulate Dpp/TGF-beta pathway activity during intestinal tumorigenesis. EMBO Rep. 2014;15(11):1210–1218. doi: 10.15252/embr.201438622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myrthue A, Rademacher BL, Pittsenbarger J, et al. The iroquois homeobox gene 5 is regulated by 1,25-dihydroxyvitamin D3 in human prostate cancer and regulates apoptosis and the cell cycle in LNCaP prostate cancer cells. Clin Cancer Res. 2008;14(11):3562–3570. doi: 10.1158/1078-0432.CCR-07-4649. [DOI] [PubMed] [Google Scholar]
- 33.Loomans HA, Andl CD. Intertwining of activin A and TGFbeta signaling: dual roles in cancer progression and cancer cell invasion. Cancers. 2014;7(1):70–91. doi: 10.3390/cancers7010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boye A, Kan H, Wu C, et al. MAPK inhibitors differently modulate TGF-beta/Smad signaling in HepG2 cells. Tumour Biol. 2015;36(5):3643–3651. doi: 10.1007/s13277-014-3002-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence data of CRNDE-h amplified production.
Notes: (A) The sequence data of CRNDE-h amplified production – Forward. (B) The sequence data of CRNDE-h amplified production – Reverse. (C) Comparison between amplified production sequences and CRNDE-h sequence in NCBI database.
Abbreviations: lncRNA, long noncoding RNA; CRNDE-h, colorectal neoplasia differentially expressed – h; Sbjct, sequence from NCBI database; NCBI, National Center for Biotechnology Information.