Abstract
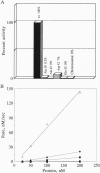
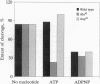
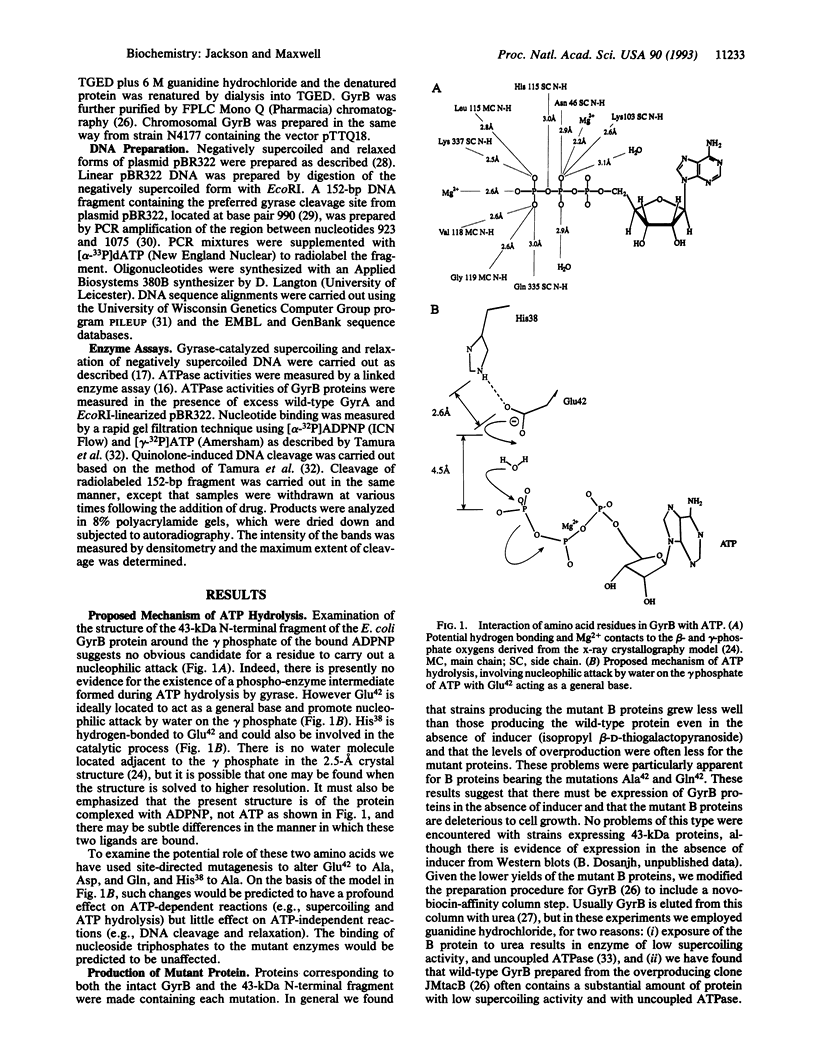
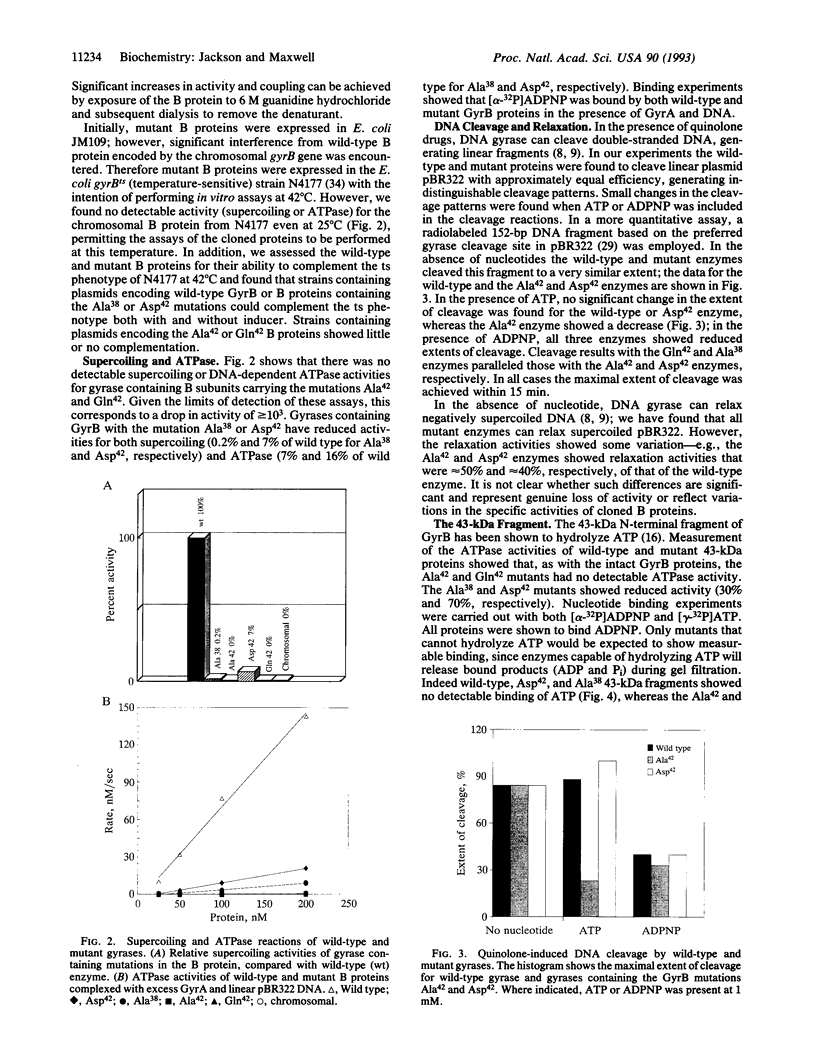
We propose a mechanism for the hydrolysis of ATP by the DNA gyrase B protein in which Glu42 acts as a general base and His38 has a role in aligning and polarizing the glutamate residue. We have tested this mechanism by site-directed mutagenesis, converting Glu42 to Ala, Asp, and Gln, and His38 to Ala. In the presence of wild-type A protein, B proteins bearing the mutations Ala42 and Gln42 show no detectable supercoiling or ATPase activities, while Asp42 and Ala38 proteins have reduced activities. In the DNA cleavage and relaxation reactions of gyrase, which do not require ATP hydrolysis, wild-type and mutant proteins have similar activities. When the 43-kDa N-terminal fragment of the gyrase B protein (which hydrolyzes ATP) contained the mutations Ala42 or Gln42, ATP was bound but not hydrolyzed, supporting the idea that Glu42 is involved in hydrolysis but not nucleotide binding.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi T., Mizuuchi M., Robinson E. A., Appella E., O'Dea M. H., Gellert M., Mizuuchi K. DNA sequence of the E. coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 1987 Jan 26;15(2):771–784. doi: 10.1093/nar/15.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali J. A., Jackson A. P., Howells A. J., Maxwell A. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry. 1993 Mar 16;32(10):2717–2724. doi: 10.1021/bi00061a033. [DOI] [PubMed] [Google Scholar]
- Bates A. D., Maxwell A. DNA gyrase can supercoil DNA circles as small as 174 base pairs. EMBO J. 1989 Jun;8(6):1861–1866. doi: 10.1002/j.1460-2075.1989.tb03582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. O., Peebles C. L., Cozzarelli N. R. A topoisomerase from Escherichia coli related to DNA gyrase. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6110–6114. doi: 10.1073/pnas.76.12.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras A., Maxwell A. gyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol Microbiol. 1992 Jun;6(12):1617–1624. doi: 10.1111/j.1365-2958.1992.tb00886.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher L. M., Mizuuchi K., O'Dea M. H., Ohmori H., Gellert M. Site-specific interaction of DNA gyrase with DNA. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4165–4169. doi: 10.1073/pnas.78.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Fisher L. M., O'Dea M. H. DNA gyrase: purification and catalytic properties of a fragment of gyrase B protein. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6289–6293. doi: 10.1073/pnas.76.12.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett P., Grimshaw A. J., Wigley D. B., Maxwell A. Cloning of the DNA gyrase genes under tac promoter control: overproduction of the gyrase A and B proteins. Gene. 1990 Sep 1;93(1):139–142. doi: 10.1016/0378-1119(90)90148-k. [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989 Apr 15;77(1):51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Jackson A. P., Maxwell A., Wigley D. B. Preliminary crystallographic analysis of the ATP-hydrolysing domain of the Escherichia coli DNA gyrase B protein. J Mol Biol. 1991 Jan 5;217(1):15–17. doi: 10.1016/0022-2836(91)90606-7. [DOI] [PubMed] [Google Scholar]
- Lockshon D., Morris D. R. Sites of reaction of Escherichia coli DNA gyrase on pBR322 in vivo as revealed by oxolinic acid-induced plasmid linearization. J Mol Biol. 1985 Jan 5;181(1):63–74. doi: 10.1016/0022-2836(85)90324-9. [DOI] [PubMed] [Google Scholar]
- Maxwell A., Gellert M. Mechanistic aspects of DNA topoisomerases. Adv Protein Chem. 1986;38:69–107. doi: 10.1016/s0065-3233(08)60526-4. [DOI] [PubMed] [Google Scholar]
- Maxwell A., Gellert M. The DNA dependence of the ATPase activity of DNA gyrase. J Biol Chem. 1984 Dec 10;259(23):14472–14480. [PubMed] [Google Scholar]
- Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983 Aug;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., O'Dea M. H., Gellert M. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5960–5963. doi: 10.1073/pnas.75.12.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Higgins N. P., Cozzarelli N. R. Interaction between DNA gyrase and its cleavage site on DNA. J Biol Chem. 1980 Mar 10;255(5):2211–2219. [PubMed] [Google Scholar]
- Reece R. J., Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26(3-4):335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- Reece R. J., Maxwell A. Probing the limits of the DNA breakage-reunion domain of the Escherichia coli DNA gyrase A protein. J Biol Chem. 1991 Feb 25;266(6):3540–3546. [PubMed] [Google Scholar]
- Reece R. J., Maxwell A. The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 1991 Apr 11;19(7):1399–1405. doi: 10.1093/nar/19.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., Maxwell A. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J Biol Chem. 1989 Nov 25;264(33):19648–19653. [PubMed] [Google Scholar]
- Staudenbauer W. L., Orr E. DNA gyrase: affinity chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res. 1981 Aug 11;9(15):3589–3603. doi: 10.1093/nar/9.15.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Cozzarelli N. R. The intrinsic ATPase of DNA gyrase. J Biol Chem. 1980 Jul 10;255(13):6299–6306. [PubMed] [Google Scholar]
- Sugino A., Higgins N. P., Brown P. O., Peebles C. L., Cozzarelli N. R. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4838–4842. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura J. K., Bates A. D., Gellert M. Slow interaction of 5'-adenylyl-beta,gamma-imidodiphosphate with Escherichia coli DNA gyrase. Evidence for cooperativity in nucleotide binding. J Biol Chem. 1992 May 5;267(13):9214–9222. [PubMed] [Google Scholar]
- Tse Y. C., Kirkegaard K., Wang J. C. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J Biol Chem. 1980 Jun 25;255(12):5560–5565. [PubMed] [Google Scholar]