Abstract
Testis of Hemidactylus flaviviridis, commonly known as Indian wall lizard, displays a lack of cellular and metabolic activity in regressed phase of testis during non-breeding season of the year. Retracted Sertoli cells (Sc), fibroid myoid cells and pre-meiotic resting spermatogonia are observed in such testis. This situation is akin to certain forms of infertility in men where hormone supplementation fails to generate sperm despite the presence of Sc and germ cells (Gc) in testis. In testis of lizard, spermatogenesis is reinitiated upon increased level of hormones during appropriate season (phase of recrudescence). Study of genes associated with generation of sperm, from regressed adult testis in lizard, may provide valuable information for understanding certain forms of male idiopathic infertility. Subtractive hybridization using testicular RNA obtained from the regressed and active phases of lizard reproductive cycle led to identify eight partial mRNA sequences that showed sequence homology with mice genes. We further evaluated the gene expression prolife by real-time PCR in three different reproductive phases of H. flaviviridis: regressed (pre-meiotic), recrudescent (meiotic) and active (post meiotic), for comparison with the corresponding testicular phases found in testis of 5 days (pre-meiotic), 20 days (meiotic) and 60 days (post-meiotic) old mouse. This is the first report where genes associated with progression of spermatogenesis during active phase, which follows a regressed state of adult testis, were identified in lizard and found to be conserved in mouse. Six important genes, Hk1, Nme5, Akap4, Arih1, Rassf7 and Tubb4b were found to be strictly associated with active spermatogenesis in both mouse and lizard. Factors interfering with the expression of any of these genes may potentially abrogate the process of spermatogenesis leading to infertility. Such information may shed light on unknown causes of idiopathic male infertility.
Introduction
Spermatogenesis is the complex biological process by which sperm are produced by successive mitosis and meiosis. This requires extensive intercellular coordination within the testis among different cell types, e.g., germ cells, Sertoli cells, Leydig cells, peritubular cells, etc. Intracellular coordination links to the timely expression of certain genes and their interactions, which collectively play a major role in the successful completion of sperm production.
Seminiferous tubules in the testis are surrounded by lymphatic endothelium [1]. The peritubular myoid cells and basal lamina provide structural support to seminiferous tubules within which Sertoli cell and germ cells resides. The Sertoli cells give structural framework for the organization of tubules and provide niche for the germ cell division and differentiation [2,3]. Rodents, mice and rats are most commonly used to study regulation of male fertility. In them, undifferentiated spermatogonial stem cells are situated at basement membrane either as single cell (As) or as pairs of cells (Apr) or as chains of aligned (Aal) cells [4]. Aal cells divide and differentiate into spermatocytes and after two rounds of meiosis develop into round spermatids. These round spermatids become elongated and eventually lose their large portion of cytoplasm to finally develop into sperm [5]. However, there is a remarkable variation in this process from species to species and from seasonal breeders to non-seasonal breeders.
In mouse (continuous breeder), various phases of testicular development appears as the animal matures from neonatal to adulthood. At around 5 days of post natal age (neonatal), the testis consist of undifferentiated spermatogonial cells and more than 50 percent of those remain in the resting phase with no cellular division and differentiation [6]. At 8–10 days of age, the beginning of meiosis (germ cells in leptotene stage) is apparent in a few tubules. At around 19 days, nearly half of all tubules exhibit the initiation of meiosis (presence of pachytene). A 60-day old mouse is considered an adult, with testicular sperms and reproductive maturity [6].
As opposed to mouse, most of the lizard species are seasonal breeders, they mate in spring, offspring hatches in summers. Seasonal reproduction is a tactic to make use of energy in an economical manner. The wall lizards have a prenuptial cycle of sperm maturation. Prenuptial reptiles generates sperm prior to or during the mating period [7]. Hemidactylus flaviviridis, commonly known as Indian wall lizard, is a seasonally breeding animal. Based on seasonal variations they have three reproductive phases within a year [8], namely- regressed (June and July), recrudescent (August and September), and active phase (October-May). Regressed Phase is the most inactive phase of spermatogenesis. There is a lack of cellular and metabolic activity in testis during this phase. Only retracted Sertoli cells and undifferentiated pre-meiotic resting spermatogonia are observed in the testis. All other advanced germ cells are absent. Peritubular myoid cells become fibroid and overall size of the testis is reduced to the smallest [9]. This is a situation akin to several forms of infertility in men [10]. In lizards, complete arrest of spermatogenesis (observed for a period of about 2 months) in regressed phase, followed by the initiation of spermatogenesis occurs through proliferation of spermatogonia in recrudescence phase. Majority of the germ cell development and maturation occurs in the active phase where sperm are released into the lumen of seminiferous tubules. In the winter months (November to January), spermatogenesis is slow and active breeding happens from February to May when sperm are continuously generated within the testis [8,9,11]. Study of genes involved in generation of sperm from regressed testis, immediately following stage of recrudescence may provide valuable information for treating infertile men having inactive spermatogenesis in spite of presence of Sc and Gc in the testis (similar to the testis of regressed phase in lizards).
We have previously reported the differential gene expression in testis of three different reproductive phases of wall lizard by microarray analysis [12]. As microarray was performed using mouse array chip, wall lizard specific gene sequences were not available for further studies. The aim of the present study was to identify the genes which are differentially expressed in the testis during active and regressed phase of reproductive cycle in wall lizards by subtractive hybridization technique. Presence of such genes, specifically expressed in testis during regressed, recrudescent and active phases of wall lizards were compared with the testis from neonatal (no spermatogenesis), pubertal (initiation of spermatogenesis) and adult phases (all stages of spermatogenesis) of mice, respectively. Such study will not only enrich our current understanding about genes involved in the onset of spermatogenesis and testicular development which are conserved across the species but will also help in addressing issues related to certain forms of infertility.
Materials and Methods
Tissue collection and RNA isolation
Laboratory mouse strain FVB/J was kept in animal house of National Institute of Immunology according to guidelines provided by The Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), government of India. Indian wall lizards Hemidactylus flaviviridis were procured from a local supplier in Delhi, India, and were kept in the animal house of Department of Zoology, University of Delhi, and maintained as described earlier by us previously [13]. Adult male wall lizards, of body weight around 8 to10 grams in the months of June (during regressed phase), September (recrudescent phase) and February (active phase) were procured and maintained (12 hr light: 12hr dark) in wooden cages with wire meshes on the side and on top, and were fed live insects as food. The animals were acclimatized to the laboratory conditions under confinement for a week prior to the commencement of experiments. All the experiments regarding animals for this study were performed according to the guidelines provided and approved by Institutional Animal Ethics Committee (IAEC), University of Delhi and National institute of Immunology under guidance of The Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), government of India. Isolation of testes for all three phases (active, recrudescent and regressed) was done according to a previously mentioned protocol by us [12]. For a single biological sample of testicular tissue from wall lizards, 10 to 15 from regressed, 4 to 5 from recrudescence and 2–3 males from active phase were sacrificed by decapitation. For the isolation of mouse testicular tissue from 5, 20 and 60 days post natal; the number of animals were sacrificed by cervical dislocation was 10–12, 4–6 and 2–3, respectively for each group. Testes were snap-frozen in liquid nitrogen and crushed to powder with sterile mortar and pestle. The powder were mixed in 1ml Tri reagents (Sigma-Aldrich,USA). This generated one biological replicate for each phase and, likewise, three such biological replicates were generated for each sample group. RNA isolation from tissue in Tri-reagent was done according to manufacturer’s recommendation (Sigma-Aldrich,USA). To analyze for any changes in testicular weight, tubule diameter and body weight during different testicular phases in lizards and mice, we have randomly taken three animals (six testes) from each phase in lizard and mice. Body and testes weight were recorded. All testes were used for preparing histological sections for measurement of tubule diameter.
Histology of testis
Testes isolated from various phases of the reproductive cycle of wall lizards and different postnatal days of mice were fixed using 10 percent neutral buffered formalin, pH 7. Paraffin blocks were prepared, 5 μm sections were made on microtome machine (2040AUTOCUT, Reichert-Jung), and hematoxylin-eosin staining was performed. After DPX mounting, the sections were observed and testicular diameters were measured under bright field upright microscope (Eclipse Ti Nikon, Japan).
Subtractive hybridization
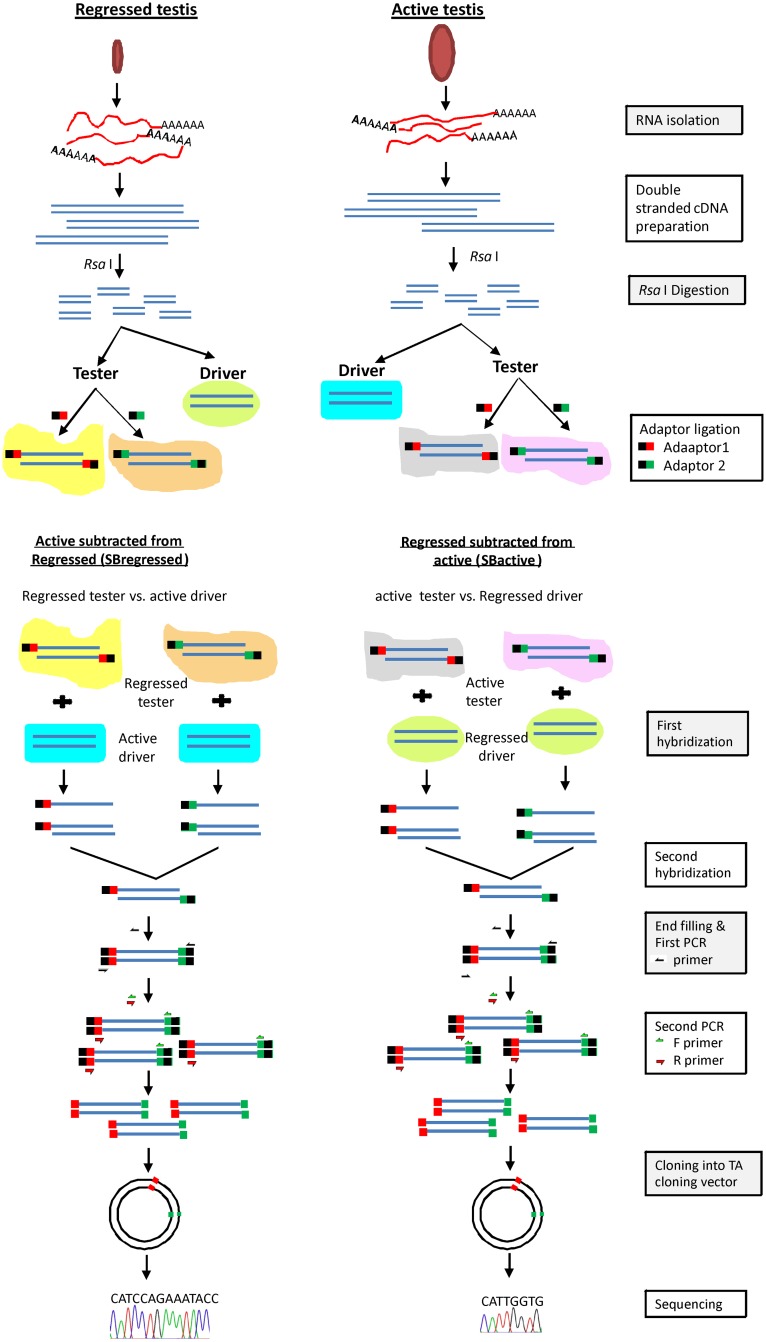
Subtractive hybridization was done according to the manufacturer’s protocol (Clontech,USA: cat.No– 637401; protocol no- PT1117-1). In brief, 2 μg of RNA isolated from testes in active and regressed phases were used to generate double-stranded cDNAs, which were digested with RsaI enzyme, to generate blunt–end partial cDNAs. The digested resultant of each phase was divided in two portions named tester and driver. Testers from the active and regressed phases were again divided in two halves; the one half was ligated with adaptor1 and to another adaptor2 (provided with the kit). The efficiency of adaptor ligation was determined as suggested in the kit protocol. Hybridization was performed between the active tester and the regressed driver, where regressed is subtracted from active (hereafter will be referred to SBactive) and the regressed tester and the active driver, where active is subtracted from regressed (hereafter will be referred to SBregressed). The hybridized samples were used for primary and secondary subtraction PCRs with primers provided with the kit. Subtraction efficiency was evaluated between the subtracted and un-subtracted samples of the active phase using primers specific to H. flaviviridis. PCR amplification was terminated in different cycles (28, 33, 38 and 42) and all PCR products were run on gel to validate the subtraction efficiency. The secondary PCR products, contained partial cDNAs, were ligated to a TA cloning vector in multiple cloning site (Promega Corp, USA), and were transformed into E. coli. From each SBactive and SBregressed sample, randomly130 positive colonies were selected to perform colony-PCR to verify the insert size in TA cloning vector. Colonies showing higher molecular mass with a single amplification product in the colony-PCRs were considered; 48 different colonies, of each SBactive and SBregressed sample were selected. pDNA from those selected colonies were isolated with GenElute plasmid miniprep kit (Sigma, USA). The region of pDNA having partial cDNAs were sequenced (Amnion Biosciences, India). A flowchart regarding the above-described subtractive hybridization process is illustrated in Fig 1.
Fig 1. Flowchart of subtractive hybridization protocol for lizard testicular samples.
Schematic representation depicting method of subtractive hybridization between RNA isolated from testes of two different reproductive phases (active and regressed phase) of wall lizard.
Sequence analysis and phylogenetic tree preparation
To find the identity of the partial cDNAs, the Basic Local Alignment Search Tool (BLAST) was used to align with nucleotide sequences deposited in the NCBI databases. Identities of partial cDNAs were assigned from the best aligned sequence having highest identity score and query coverage with least E value.
A phylogenetic tree was constructed with the partial cDNAs and their corresponding ortholog sequences in other species. MEGA5 software was used for generation of the phylogenetic tree by neighbor-joining statistical method and the bootstrap phylogenetic tree construction method, having a bootstrap replication of 1000 [14].
Reverse Transcription and Real-Time PCR
1 μg of RNA, isolated from the testes samples of lizards and mice, was treated with DNaseI according to the manufacturer’s protocol (Thermo Scientific, USA). To generate cDNAs form the treated RNAs, M-MLV reverse transcriptase (Promega Corp, USA) were used with oligo (dT)15 as primer in a final volume of 25μl. cDNA was diluted 3 times with nuclease-free water, and subsequent PCRs were carried out using 1μl of the diluted cDNA as template.
Gene-specific oligo-primers for real-time PCR were designed using software tools: PrimerBlast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/ as on 22 July, 2015) and GeneRunner (http://generunner.net/ as on 22 July, 2015). Reference sequences for gene-specific primers were acquired from NCBI database for mice and partial cDNA sequences, obtained from subtractive hybridization, for wall lizards. Primer sequences are listed in Table 1.
Table 1. List of primers for real-time PCR.
| Gene ID | Species | Primer sequences (5'-3') | Product length | NCBI Acc No | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| Hk1 | Lizard | GAATGACACGGTGGGAGTC | TGTATGCCTCCTATGTTCTC | 126 | JZ822542 |
| Mouse | CAGTGTGAAGTCGGCCTGAT | GGACCCATCATCCCCAAAGG | 141 | NM_001146100.1 | |
| Nme5 | Lizard | GGGATTATGTTCAATTAGCC | CTATTCCTCAGCTGAAAGAG | 199 | JZ822546 |
| Mouse | CAGGTTCATGTTTCCAGCCG | TCCTTGAAGTAGGGTTGGCG | 100 | NM_080637.3 | |
| Akap4 | Lizard | TGGAAAGTTGAGAGAGGAGG | GGAAGTGGAACTGTACCTAG | 105 | JZ822547 |
| Mouse | AGGACAACAAGATCAGGACCG | CAGCAGCACCCTTGGAATC | 89 | NM_001042542.2 | |
| Bco2 | Lizard | ACTATACAAAGCCCCTCATG | GCTGCAGTAGGAATAGGTC | 120 | JZ822548 |
| Mouse | ATGTTGGGACCGAAGCAGAG | GCAACGCCATTCCATCAAACC | 196 | NM_133217.3 | |
| Arih1 | Lizard | ATGTCTTGCAGCGAATCTTG | AACCAGTCCATTATCTTCG | 107 | JZ822545 |
| Mouse | CAGTGTCGTGCCACACTCAT | CGAAAGCACCTCTGTGGCAT | 120 | NM_019927.2 | |
| Rassf7 | Lizard | GGCTAGCCCTGACAAACGT | AGCCAAGTGTGGACAGTATG | 128 | JZ822549 |
| Mouse | ACAGGTCGATTTGTCCTTGT | GTATCGCCGATGGCTTTGG | 229 | NM_025886.3 | |
| Hmgb1 | Lizard | GTAGGGTATGCAGAACGAAG | GACTTGTCCTGTGTTATACC | 100 | JZ822550 |
| Mouse | GGACTCTCCTTTAACCGCCA | CCTTCGCTGGGACTAAGGTC | 148 | NM_010439.3 | |
| Tubb4b | Lizard | TACAGCTGAGGAGGAAGGAG | GATGACACCAGGAAGAAAGG | 170 | JZ822551 |
| Mouse | CACTTACCACGGAGATAGCGA | ACCTTCTGTGTAGTGCCCCTT | 229 | NM_146116.2 | |
| Gapdh | Lizard | ACACAGTCCATGCCATCACA | GACCTTGCCAACAGCCTTA | 134 | JF303078.1 |
| Ppia | Mouse | GTCGCTTTTCGCCGCTTGC | TCTGCTGTCTTTGGAACT | 127 | NM_008907.1 |
Real-time PCR was carried out with syber green using Mesa-green master mix kit (Eurogentech, Belgium) in Eppendorf Realplex mastercycler machine (Eppendorf, Germany). Reaction setup and PCR primer efficiency calculations were done by previously described procedure by us [15,16]. Primers with an efficiency of 1±0.2 were considered for further use. To evaluate the gene expression pattern in the testis of three reproductive phases of wall lizards and the three developmental phases of mice, three biological replicates were run for every phase of testicular sample, and for each biological sample, there were two technical replicates along with one control for reverse-transcriptase and one non-template control. The expression of Gapdh (Glyceraldehyde-3-Phosphate Dehydrogenase) for wall lizard and Ppia (Peptidylprolyl Isomerase A) for mice was used as reference gene for normalizing the target genes expression. Calculation for relative fold change with respect to the active phase in wall lizards and 60 days in mice was performed by 2-ΔΔct method as previously described by us [17].
A clustered image heat-map was generated with the relative fold change values, using online tool CIMminer (http://discover.nci.nih.gov/cimminer/home.do as on date 22 July, 2015) to represent the gene expression profile between wall lizards and mice contemporary testis. For the generation of heat map methods like euclidean, average linkage, and quantile were used as distance method, cluster algorithm, and binning method, respectively.
Correlation between testis weight and gene expression
Testes weights (mg) of nine testicular samples from wall lizards (three biological replicates for each of three phases) were assigned percentage values by considering the highest value amongst all of them as 100. Here, the highest value came from one biological replicate of the active phase of wall lizard. In a similar manner, mice testicular weights from nine testicular samples of mouse (three biological replicates for each three developmental phases) were assigned percentage values by considering the highest value amongst all of them as 100. A graph was generated for each gene, where y axis was assigned to the relative fold change values (obtained by 2-ΔΔct method) of testicular samples against their corresponding testis-weight in percentage on x axis. A linear regression line was drawn by connecting the expression values plotted on the graph. In the graph, independent linear regression line was generated for wall lizards and mice.
Statistical analysis
The relative fold change in gene expression data were represented as ±SEM (n = 3). Statistically significant difference among the samples were analyzed by one-way analyses of variance (ANOVA) followed by Newman-Keuls multiple comparison test. Difference in gene expression was considered significant when the p value was found to be less than 0.05. Correlations were determined between testis wt. and relative gene expression by Pearson's correlation coefficient analysis. p<0.05 was considered statistically significant correlation. All statistics and graph preparation were done by GraphPad prism software (USA) version 5.0.
Results
Comparison of seasonal testicular growth in wall lizards with the testicular development in mice
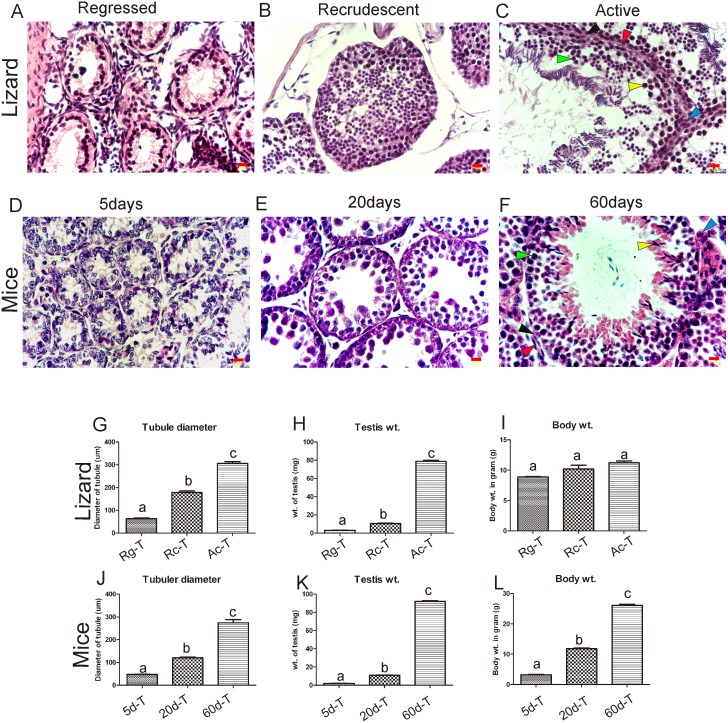
Cross section of testis from the regressed phase of a wall lizard showed small seminiferous tubules having small lumen, consisting mostly Sertoli cells and very few early germ cells (Fig 2A). The 5-day old mouse testis (Fig 2D) also had similar structure having mostly Sertoli cells and few early germ cells, except that it did not have any lumen in the seminiferous tubule. Fig 2B and 2E show cross sections of the recrudescent phase of wall lizards and 20-day old mouse testis, respectively. Testis from both species consisted of mostly pre-meiotic and meiotic stages of germ cells. Testis from active phase of wall lizards (Fig 2C) and adult 60-day old mice (Fig 2F) showed presence of all stages of germ cells along with spermatids. In the both testis, tubular diameter and testicular weight increased (Fig 2G, 2H, 2J and 2K) from regressed to active (in wall lizards) and from 5 days towards 60 days testis (in mice). The Body weight of wall lizards did not differ significantly among various reproductive phases (Fig 2I); whereas, body weight of mouse increased with age (Fig 2L).
Fig 2. Comparative analysis of testicular development in wall lizard and mice at different reproductive phase of testes.
(A), (B) and (C) represent the cross section of testis isolated from regressed, recrudescent and active phase of reproductive cycle in wall lizard, respectively. Figure (D), (E) and (F) represent testis isolated from 5 day, 20 day and 60 day old mouse respectively. In the figure (C & F), as for representation, Leydig cell (blue), peritubular cell (red), Sertoli cell (black), germ cell (green), and spermatids (yellow) are shown with respective arrowhead. Figure (G) & (J) shows comparison of tubular diameter (n = 6), (H) & (K) shows comparison of testis weight (n = 6), (I) & (L) depicts the comparative analysis body weight (n = 3) between wall lizard and mice, respectively. One way analysis of variance followed by newman-keuls multiple comparison test was performed to get P value as significance level p<0.05. In a graph, different letters above the bars denotes significant difference in values (p<0.05), Bars refers to S.D. between replicates (n = 3). Scale bar on the figure A-F indicates 0.01mm. Rg-T, Rc-T, Ac-T are testes isolated from regressed, recrudescent, and active phase of wall lizard, respectively. 5d-T, 20d-T, and 60d-Tdenotes testes isolated from 5 days, 20 days and 60 days old mouse, respectively.
Subtractive hybridization between active and regressed phase of testicular RNA from wall lizards
Subtractive hybridization was done to identify genes that are differentially expressed between active and regressed phases of the testis. To get transcripts that were highly expressed in the active phase in comparison to the regressed phase, the testicular RNA from regressed phase was subtracted from the testicular RNA of active phase (SBactive). For identification of transcripts expressed more in the regressed phase compared with the active phase, the active phase testicular RNA was subtracted from the testicular RNA of regressed phase (SBregressed).
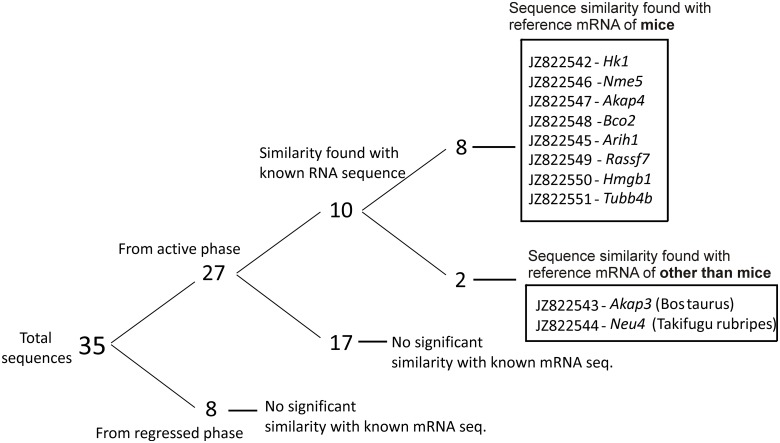
Sequencing of the fragments of transcripts, obtained from SBactive and SBregressed provided 35 unique sequences (NCBI Acc No- JZ822542 –JZ822576). Out of these, 27 sequences were from the active phase (SBactive) and 8 were from the regressed phase (SBregressed). Out of 27 sequences in the SBactive sample, 10 sequences were found to have similarity with known gene sequences from different species (Fig 3). Out of these 10, 8 partial gene sequences (Hk1, Nme5, Akap4, Tubb4b, Arih1, Rassf7, Hmgb1 and Bco2) of the wall lizards were found to have high homology with ortholog sequences in mice (Table 2). These 8 genes were from SBactive group and were used for further studies.
Fig 3. Summarized result of subtractive hybridization analysis.
Distribution of total partial sequences, obtained from subtractive hybridizations analysis between testicular RNA isolated from regressed and active phase of wall lizard.
Table 2. List of partial transcript sequences of lizard and their sequence similarity with homolog-genes of mice.
| GenBank Acc No | Length (bp) | BLAST result in NCBI database | Gene | Gene symbol | ||
|---|---|---|---|---|---|---|
| Acc No | E-value | Identity | ||||
| JZ822542 | 281 | NM_001146100.1 | 7.E-07 | 0.67 | Hexokinase 1 (Hk1) | Hk1 |
| JZ822546 | 209 | NM_080637.3 | 5E-26 | 0.75 | NME/NM23 family member 5 | Nme5 |
| JZ822547 | 665 | NM_001042542.1 | 2E-32 | 0.69 | A kinase (PRKA) anchor protein 4 | Akap4 |
| JZ822548 | 120 | NM_133217.3 | 5E-10 | 0.76 | Beta-carotene oxygenase 2 | Bco2 |
| JZ822545 | 154 | NM_019927.2 | 2E-48 | 0.88 | Ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 | Arih1 |
| JZ822549 | 486 | NM_025886.3 | 6E-36 | 0.73 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 | Rassf7 |
| JZ822550 | 420 | NM_010439.3 | 1E-24 | 0.77 | High mobility group box 1 | Hmgb1 |
| JZ822551 | 193 | NM_146116.2 | 2E-06 | 0.82 | Tubulin, beta 4B class IVB | Tubb4b |
A phylogenetic tree was constructed using the bootstarp method, with known homolog sequences found in NCBI databases (S1 Fig). The sequence of Arih1 (JZ822545) from H. flaviviridis was found to be nearest to Anolis with the highest homology, and formed an altogether different node. The sequences of Nme5 (JZ822546) and Tubb4b (JZ822551) were in greater homology with Z. albicollis. Sequences Bco2 (JZ822548), Rassf7 (JZ822549) andHmgb1 (JZ822550) were in greater homology with Xenopus. The sequence similarities of Hk1 (JZ822542) and Akap4 (JZ822547) found to be higher with Drosophila.
Comparative gene expression in the testis of wall lizards and mice by real-time PCR analysis
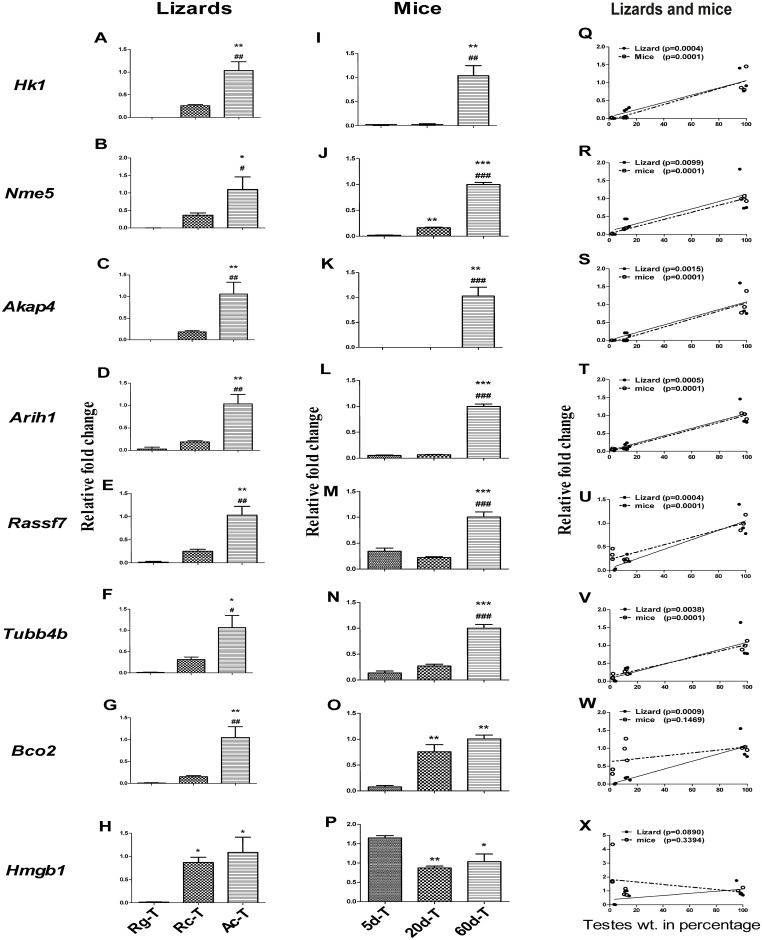
In order to analyze the mRNA expression pattern of 8 genes Hk1, Nme5, Akap4, Arih1, Rassf7, Tubb4b, Bco2and Hmgb1in different testicular phases of wall lizard and mice, real-time PCR was performed using testicular RNA. Except Hmgb1, rest other 7 genes were significantly high in the active phase of testis as compared to the regressed and recrudescent phases of wall lizards’ testis (Fig 4A–4G). Significantly higher expression of six genes Hk1, Nme5, Akap4, Arih1, Rassf7 and Tubb4b were found in the 60 days old mouse testes as compared to 5 days and 20 days old testis (Fig 4I–4N). Interestingly, the expression of Nme5 and Bco2 were significantly higher in the 20-day old testis as compared to 5-day old testis in mice (Fig 4J and 4O). In wall lizards as well as in mice, Hk1, Nme5, Akap4, Arih1, Rassf7 and Tubb4b expression were found to have significant correlation with testicular weight (Fig 4Q–4V). The increase in expression of Bco2 in 20 days old mice did not have any significant correlation with the testicular weight, although the increase in expression in the wall lizards’ testis was found to have significant positive correlation with increase in testicular weight of wall lizards (Fig 4W). The relative expression of Hmgb1 was found to be significantly higher in the recrudescence phase compared with the regressed phase of wall lizards’ testis. Comparable level of expressions was found between the recrudescence and the active phase of the wall lizards’ testis (Fig 4H). In the mice testis, the expression level of Hmgb1 was found to be similar in day 20 and day 60, however, there was significantly higher expression observed in the 5-days as compared to that at 20-day and 60-day testes (Fig 4P). Neither in wall lizards nor in mice, was the expression level of Hmgb1 found to be correlated with the increased testicular weight (Fig 4X).
Fig 4. Comparative analysis of mRNA expression of selected genes in testis of wall lizards and mice.
Shows level of mRNA expression of Hk1 (A& I), Nme5 (B& J), Akap4 (C& K), Arih1 (D &L), Rassf7 (E & M), Tubb4b (F & N), Bco2 (G & O) and Hmgb1 (H & P) in three reproductive phases of wall lizard and three developmental stages of mice testis respectively. Normalization of gene expression was done with reference gene Gapdh for wall lizard and Ppia for mouse. Correlation between relative fold change in genes expression and testis weight in wall lizard and mice for genesHk1 (Q), Nme5 (R), akap4 (S), Arih1 (T), Rassf7 (U) and Tubb4b (V). Bco2 (W), and Hmgb1 (X) were analyzed. A solid or broken line in scatter plots denotes linear regression. Open circle with dotted line represents mice and closed circle with solid line represents wall lizard relative gene expression with respect to testicular wt. in percentage. Rg-T, Rc-T, Ac-T are testes from regressed, recrudescent, and active phase of wall lizard. 5d-T, 20d-T, and 60d-T are testes from 5 days, 20 days and 60 days old mice. *p<0.05, **p<0.01 and ***p<0.001 represents the comparison of Rg-T with Rc-T and Ac-T in wall lizards and 5d-T with 20d-T and 60d-T in mice. #p<0.05, ##p<0.01 and ###p<0.001 represents the comparison of Rc-T with Ac-T in wall lizards and 20d-T with 60d-T in mice.
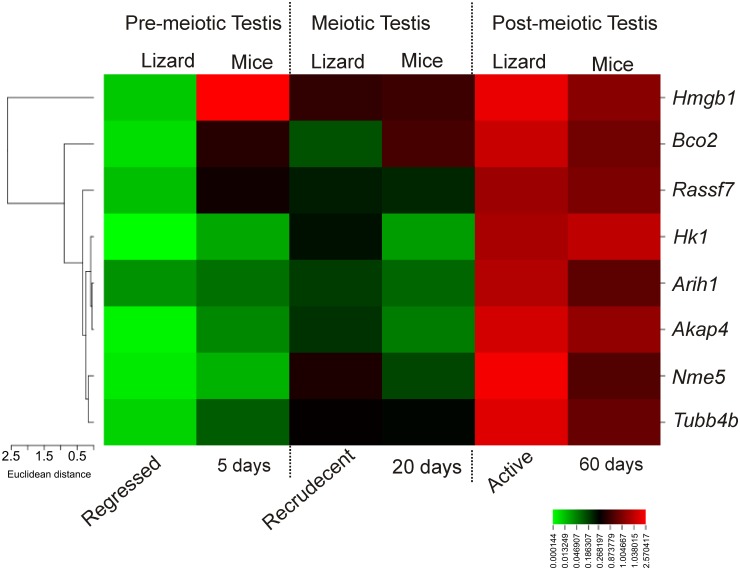
Relative fold change in gene expression values of all 8 genes were used to generate a heat map to compare the expression status between wall lizards and mice on similar testicular phases like pre-meiotic, meiotic and post-meiotic (Fig 5). The expression of Hmgb1 was found to be high in pre-meiotic testis of mice, but not in the testis of wall lizards. The expression of Hmgb1 in meiotic and post-meiotic testis in wall lizards and mice were found to be comparable. We found that Hk1, Nme5, Akap4, Arih1, Rassf7, and Tubb4b and Bco2 had a similar high expression in the testis of wall lizards during active phase and at 60days in the mice. Hk1, Arih1, Akap4 and Nme5 had comparable expressions in the regressed phase in wall lizards and 5 days old mice testis. Bco2, Rassf7 and Tubb4b had higher expression in testis of 5 days old mice compared to testis in regressed phase in the wall lizard. Rassf7, Aih1 and Tubb4b had a comparable expression in the testis of recrudescent phase of wall lizard and in 20-day old mice testis. The expression of Bco2 was found to be high in testis of 20days old mice than in the testis of recrudescent phase of the wall lizard. During recrudescent phase of the wall lizard, expression status of Hk1 and Akap4 were higher as compared to the testis of 20 days old mice. There was higher abundance of Nme5 in the testis from recrudescent phase of lizards although it was not in testis of 20 days old mice (Fig 5) which is a comparable testicular phase with recrudescent of lizards.
Fig 5. Heat map analysis of gene expression between lizards and mice.
Heat map analysis showing differential gene expression pattern in pre-meiotic, meiotic and post-meiotic testes of wall lizard and mice. The relative fold change in gene expression for genes Hk1, Nme5, Akap4, Arih1, Tubb4b, Rassf7, Bco2, and Hmgb1 were compared in corresponding testes of wall lizards and mice. Color from red to green indicates high to low expression.
Probable protein-protein interactions of differentially expression genes in testicular phases of wall lizards and mice
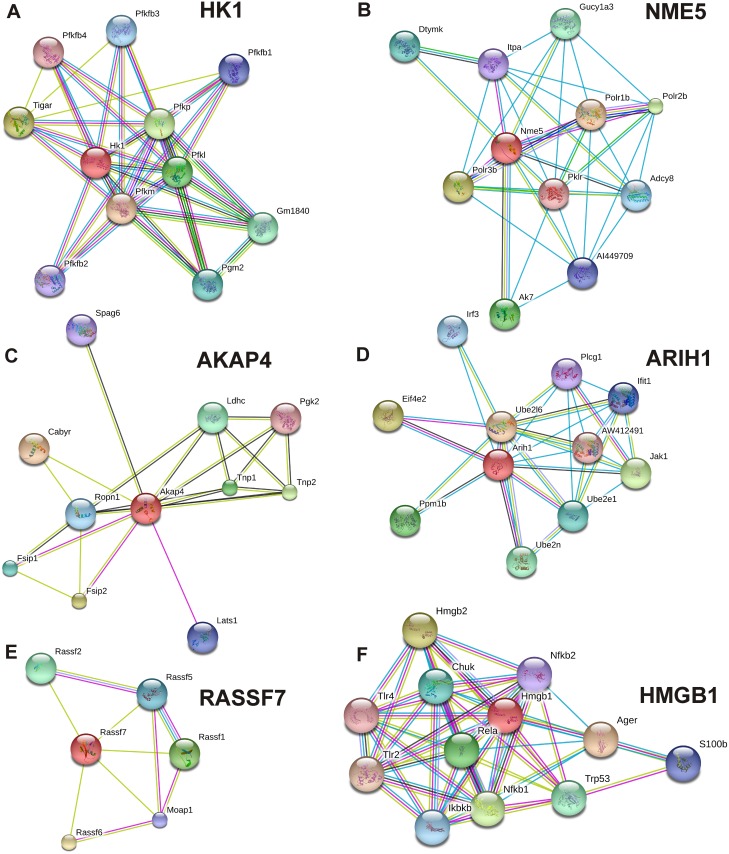
In the STRING database, there were known and predicted protein-protein interactions of Hk1, Nme5, Akap4, Arih1, Rassf7, and Hmgb1 (Fig 6). HK1 interacted with PFKM, TIGAR, PFKP, PFKL, PGM2, PFKFB3, PFKFB1, PFKFB2, and PFKFB4. NME5 interacted with POLR1B, POLR3B, POLR2B, AK7, GUCY1A3, DTYMK, ADCY8, ITPA, and PKLR. AKAP4 interacted with CABYR, FSIP2, TNP2, TNP1, LDHC, FSIP1, ROPN1, LATS1, SPAG6, and PGK2. ARIH1 interacted with UBE2L6, EIF4E2, JAK1, PPM1B, UBE2N, UBE2E1, IRF3, IFIT1, and PLCG1. RASSF7 interacted with RASSF6, RASSF1, RASSF2, RASSF5, and MOAP1. HMGB1 interacted with AGER, HMGB2, NFKB1, RELA, TRP53, CHUK, IKBKB, S100B, NFKB2, TLR4, and TLR2. The interaction score and name of interacting proteins are mentioned in S1 Table. Interactome of Bco2 and Tubb4b were not found in the STRING database.
Fig 6. Predicted interaction among proteins.
Probable interactome of different proteins, of which mRNA were differentially expressed in different testicular phase of wall lizard mice. Probable interacting partner of gene products, Hk1 (A), Nme5 (B), Akap4(C), Arih1 (D), Rassf7 (E), and Hmgb1 (F) with other proteins as per STRING database.
Discussion
In this study, we have compared the gene expression profiles in the testis of wall lizards and mice during various phases of testicular activity (pre-meiotic, meiotic and post-meiotic). Earlier we have compared the differential transcriptome of three reproductive phases of testis from wall lizard [12]. Since lizard array chip was not available, wall lizard testicular RNA was hybridized on known mouse array chip. As gene sequences of wall lizard and mouse vary widely, the expression signals due to hybridization was low in confidence. Additionally as lizards gene sequences are not available, the validation of microarray data and further expression studies with gene specific primers are not efficient. To overcome these problems, we have compared the testicular RNA expression profile between regressed and active phase of spermatogenesis in lizard by subtractive hybridization.
Thirty five unique partial RNA sequences were identified in testis of wall lizard. The 17 and 8 partial sequences from active and regressed phase, respectively, showed no significant similarity with known mRNA sequences in the database. The reason for the lack of similarity of these partial sequences with known mRNA sequences may reside in the lack of longer sequence reads (eg. JZ822572 and JZ822543), which is necessary to attain a significant match. There were other sequences, which were found to be relatively longer and showed partial similarity to short region of more than one mRNAs (eg. JZ822573, JZ822574, JZ822575); the query coverage for these sequence alignments were also very low (below 40%). Therefore, these 17 and 8 sequences could not be reliably identified as mRNA sequences of known genes. Hence, in the present study we could not consider these partial sequences for further experiments.
Ten partial RNAs among 35, showed sequence homology with mRNA sequences of orthologus genes from other species and found to be abundant in the testis during active phase compared to regressed phase of wall lizards. Out of 10 sequences, 8 showed sequence similarity with corresponding mice gene sequences. In order to evaluate testicular phase related similarity in the expression pattern between two species, we analyzed the expression profiles of these 8 genes using real-time PCR in different phases of mice testicular development and lizard reproductive cycle. 7 genes, Hk1, Nme5, Akap4, Arih1, Rassf7, Bco2 and Tubb4b were found to be uniformly associated with testis with active spermatogenesis in both the species. Expression of 6 genes Hk1, Nme5, Akap4, Arih1, Rassf7 and Tubb4b showed positive correlations with increased testicular weight in both wall lizards and mice, that represents their conserved pattern of expression between these two species. Specifically higher expression of Hmgb1 in testis of 5 days old mice suggested that the regressed testis cannot be considered as neonatal testis in terms of developmental status.
HK1 is an important enzyme for glucose metabolism. HK1 localizes on the outer membrane of mitochondria [18] and facilitates the conversion of glucose to glucose-6-phosphate. The isoform of HK1, spermatogenic cell-specific type 1 hexokinase (HK1S) was found in the germ cells and abundantly in sperm and located mainly in the principal piece of the sperm flagellum to help in sperm motility [19,20]. As glucose metabolism is very important in cellular processes, our finding of a high expression of Hk1 in the spermatogenically active testis of wall lizards and mice suggested its requirement for spermatogenic maintenance. Interaction of HK1 with glucose metabolism associated proteins (PFKM, PFKP, PFKL and PGM2) as well as fructose metabolism associated proteins (TIGER, PFKFB1, PFKFB2, PFKFB3 and PFKFB4) based on interactome database, suggested HK1 is important regulator of energy source to actively dividing spermatogonia. NME5 is a NME/NM23 family member protein, also known as Nm23-h5. This is a nucleotide diphosphate (NDP) kinase that catalyses phosphoryl exchanges between nucleoside di- and tri-phosphates [21]. The mRNA expression was found in spermatogonia and in early spermatocytes in human and mouse testis [21,22], and the proteins were located in the sperm head and flagella [23]. In the interactome data, there are a few proteins that showed interactions with NME5, like AK7 (helps in the maintenance of ciliary structure and function), DTYMK, ITPA and PKLR. In our knowledge, the expression of Nme5 has not been reported except in humans and mice [22,23]. AKAP4, which is a member of the AKAP family, helps in binding to the regulatory subunit of protein kinase A (PKA), which works as a scaffold protein. AKAP4 is found in the fibrous sheath in sperm and helps in giving mechanical support to the sperm tail [24]. Targeted disruption of this gene leads to lack of sperm motility in mice, possibly due to failure of signal transduction and glycolytic enzymes to be associated with the fibrous sheath of sperms [25]. The mRNA expression of AKAP4 also found in adult testis of Macropus eugenii, located in the cytoplasm of round and elongated spermatids, and in the sperm tail [26]. In the interactome database, it interacted with FSIP1 and FSIP2, which are fibrous sheath interacting proteins, and with germ-cell-specific proteins like SPAG6 and PGK2, and TNP1 and TNP2, which are nucleosome compaction proteins. BCO2 enzyme is a mitochondrial carotenoid-oxygenase, which catalyses asymmetric cleavage of carotenoids and protects the cells from carotenoid induced oxidative stress [27]. We found a higher expression of Bco2 at 20 days (meiotic) and 60 days (post-meiotic) in comparison to that in 5 days old mice testis. This higher expression of Bco2 in testis showing meiosis can be correlated with the requirement of retinoic acid (RA) for the meiotic initiation of germ cells. When germ cells undergo meiosis, they require retinoic acid [28] which is product of beta-carotene. Hence, there is a possibility that during meiosis, germ cells may undergo oxidative stress due to higher amount of beta-carotene; therefore, Bco2 may create a balance for maintaining the right amount of beta-carotenoid in the germ cells. ARIH1 can induce aggresome formation and helps in the ubiqutinilation of target proteins [29,30]. It is also found to be the marker for proliferation [31]. As the active testis of wall lizards and adult testis of mice are highly proliferative, a higher expression of Arih1 in those phases of testis in wall lizards and mice can be justified. In the predicted interactome database, this protein interacts with the JAK1, which is a protein-tyrosine kinase found in human spermatozoa [32] and helps in germ cell differentiation in Xenopus [33].RASSF7 is a ras association domain-containing protein 7, which negatively regulates stress-induced JNK activation [34]. It is also a regulator of microtubule dynamics in a cell and is required for mitosis [35]. The important paralog of this gene isRassf8, found to be essential for maintaining adherent junctions (AJ) in epithelial cells and has a role in epithelial cell migration [36]. We observed a higher expression Rassf7 in the testis of active phase and adult testis of wall lizards and mice, respectively. In the interactome data, it showed interaction with other ras-associated proteins. TUBB4B is a tubulin protein. We found its expression in the adult testis of mice and active testis of lizards, from which we can infer that the requirement of this component of microtubules may be higher in mature testis for providing structural support to large number of germ cells. HMGB1 is a high-mobility group binding protein. The expression of Hmgb1 was found to be comparable during recrudescence and active phase of lizards. This was also comparable in 20 and 60 days old testis of mice. However, this was highest in neonatal testis (5 days) of mice. Interactome data of HMGB1, suggested its interaction with several proteins related to PI3K-Akt signaling pathway, like NFKB1, RELA1, CHUK, and IKBKB, which are known to play important role in cellular growth and differentiation. The Hmgb1 gene was found to be one of the 159 transcripts from testicular biopsy where the expression was found to be high in idiopathic infertile patients of AZFc microdeletion [37]. In many patients the cause of infertility was due to severe hypo spermatogenesis, this can be because the testis remains in state of immaturity which fails to develop sufficient functional sperm. The higher expression of Hmgb1 in neonatal (immature) testis of mice and testis of infertile patients, led us to hypothesize that higher expression of HMGB1can keep the testis in immature state whereas moderate expression is seen during spermatogenesis.
In our knowledge this is the first report where increased expression of certain genes associated with active spermatogenesis was commonly observed in lizard and mammalian testis. The increase in six commonly identified genes during active phase was observed as compared to regressed state of the testis from lizard and neonatal testis of mice. This not only showed that these genes are conserved across the species but also indicated their strict association with spermatogenesis. Impairment in expression of any of these genes may potentially disturb the process of spermatogenesis leading to infertility. This information may be crucial in divulging unknown causes of idiopathic male infertility and hence should be studied further in detail.
Supporting Information
Phylogenetic tree for the genes, Hk1 (A), Nme5 (B), Akap4(C), Bco2 (D), Arih1 (E), Rassf7 (F), Hmgb1 (G), andTubb4b (H).
(TIF)
(DOC)
Acknowledgments
We are thankful to Dr. Indrashis Bhattacharya, of HNB Garhwal University, Uttarakhand for critical analysis of the paper and valuable suggestions.
Data Availability
All sequences obtained from subtractive hybridization are available from the NCBI database (accession numbers JZ822542 – JZ822576).
Funding Statement
SSM and UR received the funding from Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India, under the grant No-BT/PR14644/AAQ/01/448/2010. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fawcett DW, Neaves WB, Flores MN. Comparative observations on intertubular lymphatics and the organization of the interstitial tissue of the mammalian testis. Biol Reprod.1973;9(5):500–532. [DOI] [PubMed] [Google Scholar]
- 2.Russel LD, Ettlin R., Hikim AP, Clegg ED. Mammalian spermatogenesis Histological and Histopathological Evaluation of the Testis. 1st ed Cache river press, Bolesta, USA; 1990. [Google Scholar]
- 3.Clermont Y. Contractile elements in the limiting membrane of the seminiferous tubules of the rat. Exp Cell Res.1958;15(2):438–440. 10.1016/0014-4827(58)90052-1 [DOI] [PubMed] [Google Scholar]
- 4.De Rooij DG, Russell LD.All you wanted to know about spermatogonia but were afraid to ask. J Androl.2000;21(6):776–798. [PubMed] [Google Scholar]
- 5.Kerr JB, Loveland KL, O’Bryan MK, de Kretser DM. Cytology of the Testis and Intrinsic Control Mechanisms In: Neill DJ, editor. Knobil and Neill’s physiologyof reproduction. 3rd ed Elsevier, London: 2006. pp. 827–948. [Google Scholar]
- 6.Nebel BR, Amarose AP, Hacket EM. Calendar of gametogenic development in the prepuberal male mouse. Science.1961;134(3482):832–833. 10.1126/science.134.3482.832 [DOI] [PubMed] [Google Scholar]
- 7.Kumar S, Roy B, Rai U. Hormonal Regulation of Testicular Functions in Reptiles In: Norris OD and Lopez HK,editors. Hormonesand reproduction of vertebrates. 3rd Vol: Elsevier,London: 2011. pp. 63–88. [Google Scholar]
- 8.Sanyal MK, Prasad MRN. Reproductive cycle of the Indian House Lizard, Hemidactylus flaviviridis Ruppell. Copeia.1967;3:627–633. 10.2307/1442242 [DOI] [Google Scholar]
- 9.Khan UW, Rai U. Interrelationship among testicular cells in wall lizard Hemidactylus flaviviridis (Ruppell): An ultrastructural seasonal and experimental study. Indian J Exp Biol.2004;42(4):378–388. [PubMed] [Google Scholar]
- 10.Stukenborg J-B, Kjartansdóttir KR, Reda A, Colon E, Albersmeier JP, Söder O. Male germ cell development in humans. Horm Res Paediatr.2014;81(1):2–12. 10.1159/000355599 [DOI] [PubMed] [Google Scholar]
- 11.Khan UW, Rai U. Paracrine role of testicular macrophages in control of Leydig cell activities in the wall lizard, Hemidactylus flaviviridis. Gen Comp Endocrinol.2008;156(1):44–50. 10.1016/j.ygcen.2007.10.006 [DOI] [PubMed] [Google Scholar]
- 12.Gautam M, Mathur A, Khan MA, Majumdar SS, Rai U. Transcriptome Analysis of Spermatogenically Regressed, Recrudescent and Active Phase Testis of Seasonally Breeding Wall Lizards Hemidactylus flaviviridis. PLoS One; 2013;8(3):e58276 10.1371/journal.pone.0058276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan UW, Rai U. Differential effects of histamine on Leydig cell and testicular macrophage activities in wall lizards: Precise role of H1/H2 receptor subtypes. J Endocrinol.2007;194(2):441–448. 10.1677/JOE-06-0225 [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol.2011;28(10):2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharya I, Pradhan BS, Sarda K, Gautam M, Basu S, Majumdar SS. A switch in Sertoli cell responsiveness to FSH may be responsible for robust onset of germ cell differentiation during prepubartal testicular maturation in rats. Am J Physiol Endocrinol Metab.2012;303(7):E886–E898. 10.1152/ajpendo.00293.2012 [DOI] [PubMed] [Google Scholar]
- 16.Majumdar SS, Sarda K, Bhattacharya I, Plant TM.Insufficient androgen and FSH signaling may be responsible for the azoospermia of the infantile primate testes despite exposure to an adult-like hormonal milieu. Hum Reprod. 2012;27(8):2515–2525. 10.1093/humrep/des184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharya I, Basu S, Sarda K, Gautam M, Nagarajan P, Pradhan BS, et al. Low levels of Gαs and Ric8b in testicular sertoli cells may underlie restricted FSH action during infancy in primates. Endocrinology.2015;156(3):1143–1155. 10.1210/en.2014-1746 [DOI] [PubMed] [Google Scholar]
- 18.Schindler A, Foley E. Hexokinase 1 blocks apoptotic signals at the mitochondria. Cell Signal.2013;25(12):2685–2692. 10.1016/j.cellsig.2013.08.035 [DOI] [PubMed] [Google Scholar]
- 19.Nakamura N, Shibata H, O’Brien DA, Mori C, Eddy EM.Spermatogenic cell-specific type 1 hexokinase is the predominant hexokinase in sperm. Mol Reprod Dev.2008;75(4):632–640. 10.1002/mrd.20791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura N, Miranda-Vizuete A, Miki K, Mori C, Eddy EM. Cleavage of disulfide bonds in mouse spermatogenic cell-specific type 1 hexokinase isozyme is associated with increased hexokinase activity and initiation of sperm motility. Biol Reprod.2008;79(3):537–545. 10.1095/biolreprod.108.067561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang KC, Ok DW, Hong JC, Kim MO, Kim JH. Cloning, sequencing, and characterization of the murine nm23-M5 gene during mouse spermatogenesis and spermiogenesis. Biochem Biophys Res Commun.2003;306(1):198–207. 10.1016/S0006-291X(03)00916-1 [DOI] [PubMed] [Google Scholar]
- 22.Munier A, Feral C, Milon L, Pinon VPB, Gyapay G, Capeau J, et al. A new human nm23 homologue (nm23-H5) specifically expressed in testis germinal cells. FEBS Lett.1998;434(3):289–294. 10.1016/S0014-5793(98)00996-X [DOI] [PubMed] [Google Scholar]
- 23.Munier A, Serres C, Kann ML, Boissan M, Lesaffre C, Capeau J, et al. Nm23/NDP kinases in human male germ cells: role in spermiogenesis and sperm motility? Exp Cell Res. 2003;289(2):295–306. 10.1016/S0014-4827(03)00268-4 [DOI] [PubMed] [Google Scholar]
- 24.Brown PR, Miki K, Harper DB, Eddy EM.A-kinase anchoring protein 4 binding proteins in the fibrous sheath of the sperm flagellum. Biol Reprod. 2003;68(6):2241–2248. 10.1095/biolreprod.102.013466 [DOI] [PubMed] [Google Scholar]
- 25.Miki K, Willis WD, Brown PR, Goulding EH, Fulcher KD, Eddy EM.Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev Biol. 2002;248(2):331–342. 10.1006/dbio.2002.0728 [DOI] [PubMed] [Google Scholar]
- 26.Hu Y, Hu H, Park AJ, O’Brien DA, Shaw G, Renfree MB. A-kinase anchoring protein 4 has a conserved role in mammalian spermatogenesis. Reproduction. 2009;137(4):645–653. 10.1530/REP-08-0337 [DOI] [PubMed] [Google Scholar]
- 27.Amengual J, Lobo GP, Golczak M, Li HNM, Klimova T, Hoppel CL, et al. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 2011;25(3):948–959. 10.1096/fj.10-173906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogarth CA, Griswold MD. Review series The key role of vitamin A in spermatogenesis. J Clin Invest. 2010;120(4):956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parelkar SS, Cadena JG, Kim C, Wang Z, Sugal R, Bentley B, et al. The parkin-like human homolog of Drosophila ariadne-1 (HHARI) can induce aggresome formation in mammalian cells and is immunologically detectable in Lewy bodies. J Mol Neurosci. 2012;46(1):109–121. 10.1007/s12031-011-9535-1 [DOI] [PubMed] [Google Scholar]
- 30.Ardley HC, Tan NG, Rose SA, Markham AF, Robinson PA. Features of the parkin/ariadne-like ubiquitin ligase, HHARI, that regulate its interaction with the ubiquitin-conjugating enzyme, Ubch7. J Biol Chem. 2001;276(22):19640–19647. 10.1074/jbc.M011028200 [DOI] [PubMed] [Google Scholar]
- 31.Elmehdawi F, Wheway G, Szymanska K, Adams M, High AS, Johnson CA, et al. Human Homolog of Drosophila Ariadne (HHARI) is a marker of cellular proliferation associated with nuclear bodies. Exp Cell Res. 2013;319(3):161–172. 10.1016/j.yexcr.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 32.Lachance C, Leclerc P. Mediators of the Jak/STAT signaling pathway in human spermatozoa. Biol Reprod. 2011;85:1222–1231. 10.1095/biolreprod.111.092379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyakutake K, Kawasaki T, Zhang J, Kubota H, Abe S-I, Takamune K. Asymmetrical allocation of JAK1 mRNA during spermatogonial stem cell division in Xenopus laevis. Dev Growth Differ.2015;57:389–399. 10.1111/dgd.12219 [DOI] [PubMed] [Google Scholar]
- 34.Takahashi S, Ebihara A, Kajiho H, Kontani K, Nishina H, Katada T. RASSF7 negatively regulates pro-apoptotic JNK signaling by inhibiting the activity of phosphorylated-MKK7. Cell Death Differ. 2011;18(4):645–655. 10.1038/cdd.2010.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Recino A, Sherwood V, Flaxman A, Cooper WN, Latif F, Ward A, et al. Human RASSF7 regulates the microtubule cytoskeleton and is required for spindle formation, Aurora B activation and chromosomal congression during mitosis. Biochem J. 2010;430(2):207–213. 10.1042/BJ20100883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lock FE, Underhill-Day N, Dunwell T, Matallanas D, Cooper W, Hesson L, et al. The RASSF8 candidate tumor suppressor inhibits cell growth and regulates the Wnt and NF-kappaB signaling pathways. Oncogene. 2010;29(30):4307–4316. 10.1038/onc.2010.192 [DOI] [PubMed] [Google Scholar]
- 37.Gatta V, Raicu F, Ferlin A, Antonucci I, Scioletti AP, Garolla A, et al. Testis transcriptome analysis in male infertility: new insight on the pathogenesis of oligo-azoospermia in cases with and without AZFc microdeletion. BMC Genomics.2010;11:401 10.1186/1471-2164-11-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree for the genes, Hk1 (A), Nme5 (B), Akap4(C), Bco2 (D), Arih1 (E), Rassf7 (F), Hmgb1 (G), andTubb4b (H).
(TIF)
(DOC)
Data Availability Statement
All sequences obtained from subtractive hybridization are available from the NCBI database (accession numbers JZ822542 – JZ822576).