Abstract
Objective
MicroRNA-150 (miR-150) was revealed to be an attractive prognostic biomarker in recent studies. However, the prognostic significance of miR-150 expression in cancer remains inconclusive. The aim of this study was to summarize the global predicting role of miR-150 in survival in patients with various carcinomas.
Methods
Eligible studies were identified through multiple search strategies. Data were extracted from the studies by investigating the relationship between miR-150 expression and survival in patients with cancer. A meta-analysis of the hazard ratio (HR) was then performed to evaluate the prognostic role of miR-150 in different tumors. Pooled HRs of miR-150 for overall survival and progression-free survival were calculated to measure the effect of miR-150 expression on prognosis.
Results
This meta-analysis included nine published studies concerning various carcinomas. Our results indicate that an elevated miR-150 expression is associated with an enhanced overall survival in the digestive tract cancer subgroup (HR =0.57, 95% confidence interval [CI]: 0.37–0.90) and a poor progression-free survival in various cancers (HR =3.08, 95% CI: 2.00–4.75).
Conclusion
miR-150 may have the potential to become a new useful prognostic factor to monitor cancer prognosis and progression. However, given the current insufficient relevant data, further clinical studies are needed.
Keywords: microRNA-150, carcinomas, prognosis, meta-analysis
Introduction
Cancer is a major public health challenge worldwide. Although the overall cancer mortality decreased by 20% between 1991 and 2010, cancer remains one of the most common causes of death worldwide.1 MicroRNAs (miRNAs) are a family of small, endogenous noncoding RNAs with a length of 19–22 nucleotides. These molecules can negatively regulate gene expression at the posttranscriptional level by binding to the 3′-untranslated region (3′-UTR) of target messenger RNA, resulting in messenger RNA degradation and/or translational repression.2,3 miRNAs participate in crucial biological processes, such as differentiation, proliferation, and apoptosis.4 Furthermore, the aberrant expression of miRNAs has been observed in various diseases, including human carcinomas.5 Therefore, miRNAs may be diagnostic or prognostic biomarkers of carcinomas.
In recent years, microRNA (miR-150) has been investigated in many clinical studies and was found to have a potential prognostic value. miR-150, localized on chromosome 19q13, has been indicated as a hematopoietic-specific miRNA in malignant lymphoma. In chronic lymphocytic leukemia, it has been observed to be significantly downregulated in cases with unfavorable clinicobiological markers and a worse prognosis.6 Abnormal expression of miR-150 has also been found in various other solid tumor tissues, such as lung cancer, breast cancer, esophagus cancer, colorectal cancer, and pancreatic cancer.7–11 miR-150 has been demonstrated to be significantly downregulated in esophageal squamous cell carcinoma and primary colorectal cancer, contributing to a poor prognosis.9,10 By contrast, in breast cancer cell lines, miR-150 is highly expressed, promotes growth and clonogenicity, and reduces apoptosis.12 Additional studies have demonstrated elevated miR-150 expression in other carcinomas, such as prostate cancer and thyroid cancer.13,14
Although various studies have reported contradictory results, miR-150 is undeniably an attractive mediator of specific target genes that play an important role in survival and progression. However, the expression of miR-150 in cancers and the prognostic significance remain unclear. Therefore, we conducted this systematic review using a meta-analysis to clarify the accurate relationship between miR-150 expression and the prognosis of patients with carcinoma.
In this study, we conducted a meta-analysis of miR-150 for overall survival (OS) and progression-free survival (PFS) to predict the clinical results of patients with cancer in multiple human malignant neoplasms.
Materials and methods
Search strategy
We carefully searched the online PubMed and EMBASE databases until August 31, 2015, to identify relevant studies. For literature retrieval, combinations of the keywords, such as microRNA-150, miRNA-150, miR-150, cancer, carcinoma, neoplasm, and tumor, were utilized. A manual review of the references of relevant publications was also carefully performed to obtain additional information. All of the searches were conducted independently by two reviewers, and the differences were checked and resolved by discussion.
Inclusion and exclusion criteria
The eligible criteria of studies were as follows: 1) the study subjects were patients with any type of cancer, 2) miR-150 expression was measured in tumor tissue or plasma, 3) the relationship between miR-150 expression and clinical outcomes was reported, and 4) the full-text article was available in English. Studies were excluded based on the following criteria: 1) reviews, letters, and laboratory reports; 2) overlapping or duplicate data; 3) studies of nondichotomous miR-150 expression levels; and 4) an absence of key information regarding survival outcome, such as hazard ratios (HRs) and 95% confidence intervals (CIs), or the lack of mean to calculate such parameters.
Data extraction
Eligible articles were reviewed independently by two investigators (Wei Wang and Yali Zhang) under the guidelines of a critical review checklist from the Dutch Cochrane Centre proposal for the Meta-Analysis of Observational Studies in Epidemiology.15 The following items were extracted: 1) publication details, such as the first author’s name and the publication year; 2) the characteristics of the studied population, including ethnicity, type of disease, detected sample, design of the study, detection method, cutoff definition, and follow-up time; and 3) HR of miR-150 expression for OS and PFS along with the 95% CI and P-value. If HRs were not reported in the article, we used Engauge Digitizer Version 4.1 (free software downloaded from http://sourceforge.net)16 to read the Kaplan–Meier survival curves to obtain the HRs and their 95% CIs. If there were insufficient data, controversies, or any other uncertainties in an article that might be related to our meta-analysis, we asked the corresponding authors for additional information.
Statistical analysis
All of the HRs and their corresponding 95% CIs were used to calculate the pooled HR. The overall HR was >1, and the 95% CI did not overlap in the forest plot, indicating a poor prognosis in patients with high expression of miR-150. The heterogeneity among the studies was measured using Cochran’s Q test and Higgins’s I2 statistic. Heterogeneity was defined as P<0.05 or I2>50%. If significant heterogeneity was observed (P<0.05 or I2>50%), a random effects model was applied; otherwise, the fixed effects model was used.17 Publication bias was evaluated using the funnel plot with the Egger’s bias indicator test.18 All P-values were two sided, and a P-value <0.05 was considered to be statistically significant. All of the statistical analyses were conducted using Stata12 (StataCorp LP, College Station, TX, USA), Review Manager V.5.3 (Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration), and Microsoft Excel (V.2007; Microsoft Corporation, Redmond, WA, USA).
Results
Summary of enrolled studies
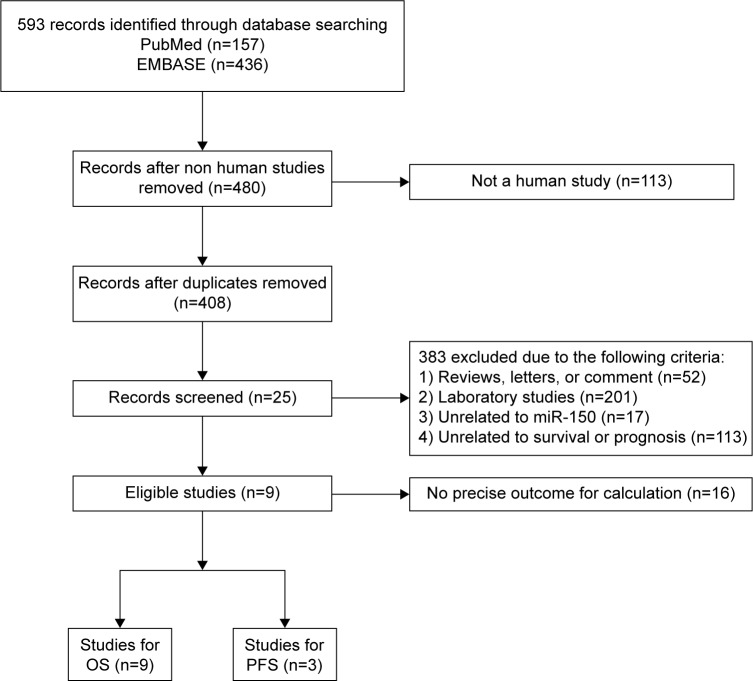
A total of 593 studies were collected from a primary literature survey using PubMed and EMBASE. One hundred and thirteen studies were excluded because they did not use human subjects. Seventy-two studies were removed because of duplication. After manually screening the titles and abstracts, 383 studies were excluded because they were review articles, letters, laboratory studies, and unrelated to miR-150 or unrelated to survival or prognosis. Upon further full-text review of 25 studies, 15 were eliminated due to inadequate data for meta-analysis and one study was eliminated because the method used for detecting miR-150 was immunohistochemical staining, which was inadaptable to detect miRNA.13 Finally, nine studies were considered eligible for this meta-analysis (Figure 1).6–11,14,19,20
Figure 1.
Flow diagram of study selection process.
Abbreviations: PFS, progression-free survival; OS, overall survival.
The main features of the nine enrolled studies are systematically summarized in Tables 1 and 2. All of the studies reported the patient’s OS, and two studies focused on PFS as well as OS. Four studies referred to a different subtype and provided survival data for both outcomes, OS and PFS; these four studies were listed twice. The included studies were published between 2011 and 2015. There were 1,999 participants from the People’s Republic of China, Japan, Switzerland, Korea, USA, and Belgium. The patients had various carcinomas, including breast cancer, melanoma, esophageal cancer, thyroid cancer, colorectal cancer, small-cell lung cancer, pancreatic cancer, and chronic lymphocytic leukemia. Cancerous tissues were usually examined to determine the miR-150 expression level, while serum samples were tested in three studies and cellular circulating miR-150 from purified CD19+ was tested in one study. Quantitative real-time polymerase chain reaction was widely used in nine studies to assess miRNA-150 expression. Five studies directly reported HRs and 95% CIs. We calculated these necessary statistical variables by survival curves in four studies.
Table 1.
Main features of the enrolled studies
| References | Country | Tumor type | Study design | Number of studies | Age (median) | Dominant ethnicity | Sample | Survival | MicroRNA type |
|---|---|---|---|---|---|---|---|---|---|
| Yokobori et al9 | Japan | Esophageal cancer | R | 108 | 64.9 | Asian | Tissue | OS | miR-23b |
| Dettmer et al14 | Switzerland | Thyroid cancer | R | 26 | NR | Caucasian | Tissue | OS | |
| Ma et al10 | People’s Republic of China | Colorectal cancer | R | 239 | NR | Asian | Tissue | OS | |
| Mraz et al6 | USA | CLL | R | 154 | 56.5 | Caucasian | Tissue | OS | |
| Ciruelos et al8 | USA | Breast cancer | R | 172 | 53.7 | Caucasian | Tissue | OS/PFS | miR-30e, miR-30a, miR-21, miR-210, miR-93, miR-99b, and miR-572 |
| Lee et al11 | Korea | Pancreatic cancer | R | 221 | 60.2 | Asian | Tissue | OS | miR-99a, miR-194, miR-375, miR-342-3p, and miR-487b |
| Lee et al11 | Korea | Pancreatic cancer | R | 189 | NR | Asian | Tissue | OS | miR-99a, miR-194, miR-375, miR-342-3p, and miR-487b |
| Stamatopoulos et al19 | Belgium | CLL | R | 273 | 63 | Caucasian | Cell | OS | |
| Stamatopoulos et al19 | Belgium | CLL | R | 252 | 69 | Caucasian | Serum | OS | |
| Bi et al7 | People’s Republic of China | SCLC | R | 40 | 50 | Asian | Tissue | OS/PFS | |
| Bi et al7 | People’s Republic of China | SCLC | R | 42 | 60.1 | Asian | Tissue | OS/PFS | |
| Fleming et al20 | People’s Republic of China | Melanoma | R | 201 | 61 | Asian | Serum | OS | miR-30d, miR-15b, and miR-425 |
| Fleming et al20 | People’s Republic of China | Melanoma | R | 82 | 59.9 | Asian | Serum | OS | miR-30d, miR-15b, and miR-425 |
Note: Study design is described as P or R.
Abbreviations: CLL, chronic lymphocytic leukemia; NR, not reference; OS, overall survival; P, prospective; PFS, progression-free survival; R, retrospective; SCLC, small-cell lung cancer.
Table 2.
Difference in the overall survival and progression-free survival between high- and low-expression cases of microRNA-150 in the enrolled studies
| References | Method | Cutoff value | Follow-up time (months) | miR-150 expression (number of cases)
|
OS
|
PFS
|
|||
|---|---|---|---|---|---|---|---|---|---|
| High | Low | HR (95% CI) | P-value | HR (95% CI) | P-value | ||||
| Yokobori et al9 | qRT-PCR | Mean expression level | 1–128 | 30 | 78 | 0.77 (0.31, 1.92) | 0.5734 | – | – |
| Dettmer et al14 | qRT-PCR | Fold change =0.2 | 5.1–203.6 | 11 | 15 | 1.57 (0.30, 8.14) | 0.5932 | – | – |
| Ma et al10 | qRT-PCR | The median value | NR | NR | NR | 0.52 (0.31, 0.85) | 0.0132 | – | – |
| Mraz et al6 | qRT-PCR | The median value | NR | NR | NR | 0.36 (0.17, 0.77) | 0.0076 | – | – |
| Ciruelos et al8 | qRT-PCR | NR | NR | NR | NR | 4.51 (2.36, 8.61) | 0.0000 | 3.75 (2.27, 6.19) | 0.0000 |
| Lee et al11 | qRT-PCR | The median value | NR | 111 | 110 | 1.69 (1.25, 2.27) | 0.0007 | – | – |
| Lee et al11 | qRT-PCR | The median value | NR | 93 | 96 | 1.74 (1.26, 2.41) | 0.0008 | – | – |
| Stamatopoulos et al19 | qRT-PCR | The median value | 7–380 | 180 | 93 | 0.53 (0.31, 0.93) | 0.0203 | – | – |
| Stamatopoulos et al19 | qRT-PCR | The median value | 7–380 | 133 | 119 | 2.06 (1.02, 4.15) | 0.0439 | – | – |
| Bi et al7 | qRT-PCR | NR | 4.4–104.2 | 20 | 20 | 3.05 (0.57, 16.39) | 0.1926 | 1.6 (0.52, 4.97) | 0.4124 |
| Bi et al7 | qRT-PCR | NR | 9–81 | 21 | 21 | 2.12 (0.54, 8.22) | 0.2816 | 2.02 (0.56, 7.30) | 0.2828 |
| Fleming et al20 | qRT-PCR | Risk score cutoff: 0.2 | 17.5–128.9 | 106 | 95 | 2.21 (0.68, 7.19) | 0.1873 | – | – |
| Fleming et al20 | qRT-PCR | Risk score cutoff: 0.2 | 0.8–95.0 | 41 | 41 | 2.34 (0.43, 12.78) | 0.3253 | – | – |
Note: HR, high vs low.
Abbreviations: CI, confidence interval; HR, hazard ratio; miR-150, microRNA-150; NR, no reference; OS, overall survival; PFS, progression-free survival; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
OS is associated with miR-150 expression
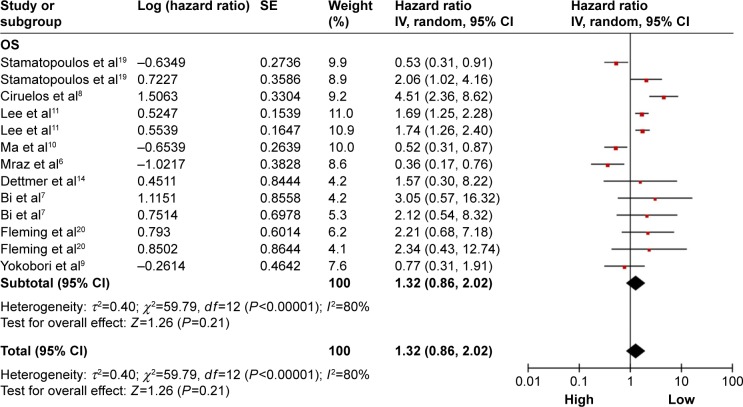
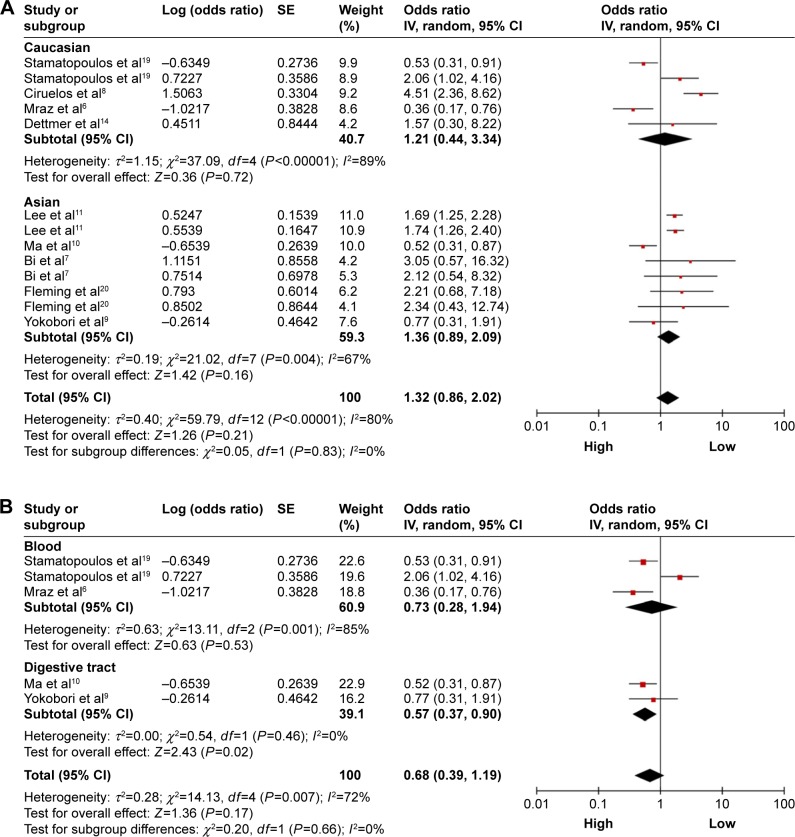
A total of nine studies were used for OS analysis, and significant heterogeneity between studies was found (P<0.00001, I2=80%). Therefore, a random effects model was applied. Our results failed to demonstrate any significant association between miR-150 expression and OS (HR =1.32, 95% CI: 0.86–2.02; Figure 2). To obtain further insight, we performed subgroup analysis with ethnicity, disease system, and detected sample type to evaluate the miR-150 prognostic value in cancer (Table 3). However, only a pooled analysis of two studies in the digestive tract cancer subgroup indicated that an increased miRNA-150 expression was significantly correlated with an enhanced OS (pooled HR =0.57, 95% CI: 0.37–0.90) because of the low heterogeneity (P=0.46, I2=0%). No other subgroups exhibited any significant results as shown by subgroup analyses (Figure 3).
Figure 2.
Forest plots of merged analyses of OS in association with microRNA-150 expression.
Notes: Squares and horizontal lines represent study-specific HRs and 95% CIs, respectively. The areas of the squares correspond to weights, and the diamonds represent the overall HRs and 95% CIs.
Abbreviations: CIs, confidence intervals; df, degrees of freedom; HRs, hazard ratios; OS, overall survival; SE, standard error; IV, inverse variance.
Table 3.
Pooled HRs, 95% CIs, and P-values for OS stratified by ethnicity, disease system, and detected sample for overall and subgroup analyses
| Subgroup | Number of studies | OS
|
|
|---|---|---|---|
| pHR (95% CI) | P-values | ||
| Total | 9 | ||
| Ethnic subtotal | |||
| Caucasian | 4 | 1.21 (0.44, 3.34) | 0.72 |
| Asian | 5 | 1.36 (0.89, 2.09) | 0.16 |
| Disease system subtotal | |||
| Blood | 2 | 0.73 (0.28, 1.94) | 0.53 |
| Digestive tract | 2 | 0.57 (0.37, 0.90) | 0.02 |
| Detected sample subtotal | |||
| Tissue | 6 | 1.27 (0.81, 2.01) | 0.3 |
| Serum | 3 | 1.30 (0.45, 3.72) | 0.63 |
Abbreviations: CIs, confidence intervals; HRs, hazard ratios; OS, overall survival; pHR, pooled hazard ratio.
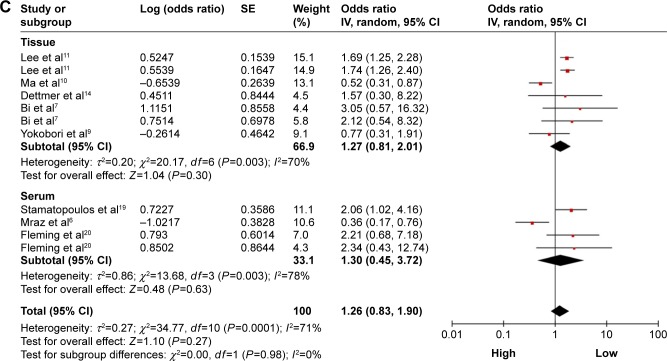
Figure 3.
Forest plots of merged analyses of OS in different subgroups in association with microRNA-150 expression.
Notes: (A) Forest plots for the subgroup analysis of OS in different ethnicity. Squares and horizontal lines represent study-specific HRs and 95% CIs, respectively. The areas of the squares correspond to weights, and the diamonds represent the overall HRs and 95% CIs. (B) Forest plots for the subgroup analysis of OS in different diseases systems. (C) Forest plots for the subgroup analysis of OS in different detected samples.
Abbreviations: CIs, confidence intervals; df, degrees of freedom; HRs, hazard ratios; OS, overall survival; SE, standard error; IV, inverse variance.
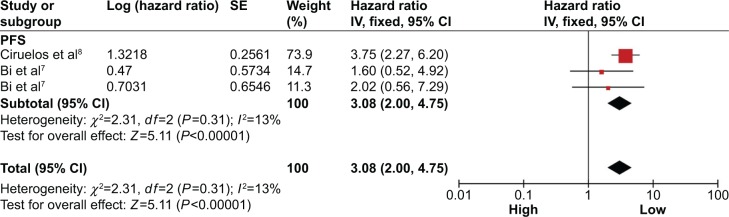
Progression associated with miRNA-150 expression
A total of two studies included in the PFS analysis revealed that high miR-150 expression predicted a worse PFS with a combined HR of 3.08 (95% CI: 2.00–4.75) via a fixed effects model (P=0.31, I2=13%; Figure 4).
Figure 4.
Forest plots of merged analyses of PFS in association with microRNA-150 expression.
Notes: Squares and horizontal lines represent study-specific HRs and 95% CIs, respectively. The areas of the squares correspond to weights, and the diamonds represent the overall HRs and 95% CIs.
Abbreviations: CIs, confidence intervals; df, degrees of freedom; HRs, hazard ratios; PFS, progression-free survival; SE, standard error; IV, inverse variance.
Publication bias
Publication bias was evaluated by Begg’s funnel plot and Egger’s test in Figure 5. In the pooled analyses of OS, Egger’s test P-value was 0.897, as shown by symmetric funnel plots. Therefore, no evidence of publication bias was noted. For the pooled analyses of PFS, due to the small number of included documents, publication bias has a little importance.
Figure 5.
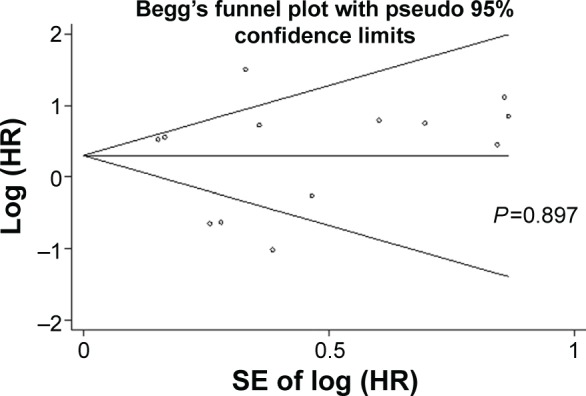
Begg’s funnel plots of the publication bias for overall merged analysis of OS.
Note: Each point represents a separate study.
Abbreviations: HR, hazard ratio; OS, overall survival; SE, standard error.
Discussion
In recent years, miRNAs have been considered as potential biomarkers of cancer prediction, diagnosis, and prognosis. They are more stably expressed than miRNAs and can be easily assessed by quantitative real-time polymerase chain reaction (qRT-PCR) in the tissues or serum of patients with cancer.21 Of the available miRNAs, miR-150 has been particularly notable, attracting the increasing cancer research.
Many studies have indicated that the aberrant expression of miR-150 is closely associated with tumorigenesis, cancer development, malignant behavior, and a curative effect by influencing oncogenes and/or tumor suppressor genes.22–24 In hematological malignancies, miR-150 dysregulation has been demonstrated to be involved in tumorigenesis.23 Gu et al demonstrated that miR-150 promotes the proliferation and migration of lung cancer through specifically targeting the 3′-UTR of p53, SRCIN1, and BAK1.25
Yokobori et al demonstrated that, in esophageal squamous cell carcinoma (ESCC), through targeted degradation of ZEB1, miR-150 induces mesenchymal -epithelial transition (MET)-like changes, and evidently inhibits tumorigenicity and tumor proliferation.9 In pancreatic cancer cells, Srivastava et al revealed that the overexpression of miR-150 inhibits growth, clonogenicity, and invasion, as well as enhances intercellular adhesion by downregulating MUC4.26
In chronic myeloid leukemia, miR-150 has been demonstrated to be involved in the mechanism of apoptosis induced by cisplatin in the human chronic myeloid leukemia cell line K562.27 Currently, many clinical studies have shown a significant correlation between the expression level of miR-150 and its potential application as an index of prognosis. However, the results are inconsistent and even contradictory. Therefore, it is necessary to conduct stratified pooled analyses to identify the prognostic value of miR-150 in survival and progression.
In this meta-analysis, the expression of miR-150 was not significantly correlated with OS in cancer (HR =1.32, 95% CI: 0.86–2.02). However, in our subgroup analysis, we only found that the high expression of miR-150 was associated with an enhanced OS in digestive tract cancer (HR =0.57, 95% CI: 0.37–0.90). Yokobori et al evaluated miR-150 expression in 108 curative resected ESCC samples to reveal the low expression of miR-150 in ESCC contributed to malignant potential, such as tumor depth, lymph node metastasis, lymphatic invasion, venous invasion, clinical staging, and poor prognosis.9 Ma et al assessed the expression of miR-150 and survival in 239 patients with colorectal cancers, demonstrating that tumors with the low expression of miR-150 were associated with a poor survival outcome.10 For studies evaluating PFS, the expression of miR-150 was significantly associated with a worse PFS in cancer (HR =3.08, 95% CI: 2.00–4.75). Empirically, HR >2 is considered as strongly predictive.28
The overexpression of miR-150 has been shown in many studies to be associated with poor outcomes in several cancer types, and our results from PFS support these conclusions. However, the reason that miR-150 is associated with the progress of cancers remains poorly understood.
Recently, several studies have reported the underlying mechanisms that may play key roles, especially in invasion and metastasis. Wang et al identified that miR-150 targets the 3′-UTR of p53, and p53 protein promotes the expression of miRNAs, including miR-34a, miR-184, miR-181a, and miR-148, which affect the cell cycle progression of non-small-cell lung cancer.29 Cao et al demonstrated that the repression of SRCIN1 by miR-150 consequently triggers the activation of the Src/focal adhesion kinase and Src/Ras/extracellular signal-regulated kinase pathways, which eventually promote the proliferation and migration of lung cancer cells. This promotion by miR-150 could be reversed by over-expressing SRCIN1.30 The results suggest that miR-150 plays a key role in cancer progression with different mechanisms in various cancer subtypes.
However, our conclusions should be considered with caution for the following five reasons. First, there are only nine eligible articles included in our analyses, leading to the relative insufficiency of studies in subgroup analyses. There are only two studies carried out for PFS. Moreover, only articles in English were included in our meta-analysis. Second, some HRs were calculated according to the data extracted from the survival curve; thus, several small errors may have been introduced. Third, the studies did not propose a clear miR-150 cutoff value. Although most of them defined the median as the cutoff of elevated miR-150 expression, accurate values could vary in the different study populations. The lack of abundant miR-150 expression data in the global population makes it difficult to set a standard cutoff. Fourth, the current statistical analysis could not render miR-150 independently predictive, due to methodological limits. Fifth, the lack of individual HR data for other markers makes it difficult to exclude the influence of confounding factors in meta-analysis. Therefore, relevant studies are required to address the aforementioned problems before miRNA-150 may be used as a prognostic biomarker in clinical applications.
Conclusion
In summary, this meta-analysis demonstrated that miR-150 expression was not significantly associated with multiple cancers for OS, but the high expression of miR-150 was significantly associated with worse progression. However, the current data are insufficient. These findings must be confirmed in further prospective clinical studies.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Lee S, Vasudevan S. Post-transcriptional stimulation of gene expression by microRNAs. Adv Exp Med Biol. 2013;768:97–126. doi: 10.1007/978-1-4614-5107-5_7. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Guz M, Rivero-Muller A, Okon E, et al. MicroRNAs-role in lung cancer. Dis Markers. 2014;2014:218169. doi: 10.1155/2014/218169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mraz M, Chen L, Rassenti LZ, et al. miR-150 influences B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1. Blood. 2014;124(1):84–95. doi: 10.1182/blood-2013-09-527234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi N, Cao J, Song Y, et al. A microRNA signature predicts survival in early stage small-cell lung cancer treated with surgery and adjuvant chemotherapy. PLoS One. 2014;9(3):e91388. doi: 10.1371/journal.pone.0091388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciruelos E, De Velasco GA, Gamez A, et al. Clinical significance of microRNA expression as aprognostic factor in early N+ breast cancer (BC) Cancer Res. 2012;23(1):96. [Google Scholar]
- 9.Yokobori T, Suzuki S, Tanaka N, et al. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104(1):48–54. doi: 10.1111/cas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Zhang P, Wang F, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61(10):1447–1453. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- 11.Lee KH, Lee JK, Choi DW, et al. Postoperative prognosis prediction of pancreatic cancer with seven microRNAs. Pancreas. 2015;44(5):764–768. doi: 10.1097/MPA.0000000000000346. [DOI] [PubMed] [Google Scholar]
- 12.Huang S, Chen Y, Wu W, et al. miR-150 promotes human breast cancer growth and malignant behavior by targeting the pro-apoptotic purinergic P2X7 receptor. PLoS One. 2013;8(12):e80707. doi: 10.1371/journal.pone.0080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dezhong L, Xiaoyi Z, Xianlian L, et al. miR-150 is a factor of survival in prostate cancer patients. J BUON. 2015;20(1):173–179. [PubMed] [Google Scholar]
- 14.Dettmer MS, Perren A, Moch H, Komminoth P, Nikiforov YE, Nikiforova MN. MicroRNA profile of poorly differentiated thyroid carcinomas: new diagnostic and prognostic insights. J Mol Endocrinol. 2014;52(2):181–189. doi: 10.1530/JME-13-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis of observational studies in epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, Fang Z, Xu J, Zhu W, Li Y, Yu Y. Prognostic value and clinicopathology significance of microRNA-200c expression in cancer: a meta-analysis. PLoS One. 2015;10(6):e0128642. doi: 10.1371/journal.pone.0128642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 18.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamatopoulos B, Van Damme M, Crompot E, et al. Opposite prognostic significance of cellular and serum circulating microRNA-150 in patients with chronic lymphocytic leukemia. Mol Med. 2015;21:123–133. doi: 10.2119/molmed.2014.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleming NH, Zhong J, da Silva, et al. Serum-based miRNAs in the prediction and detection of recurrence in melanoma patients. Cancer. 2015;121(1):51–59. doi: 10.1002/cncr.28981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10(3):297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 22.Jiang X, Huang H, Li Z, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22(4):524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe A, Tagawa H, Yamashita J, et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25(8):1324–1334. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- 24.Zhao JJ, Lin J, Lwin T, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115(13):2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu XY, Wang J, Luo YZ, et al. Down-regulation of miR-150 induces cell proliferation inhibition and apoptosis in non-small-cell lung cancer by targeting BAK1 in vitro. Tumour Biol. 2014;35(6):5287–5293. doi: 10.1007/s13277-014-1688-4. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava SK, Bhardwaj A, Singh S, et al. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 2011;32(12):1832–1839. doi: 10.1093/carcin/bgr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie SY, Li YJ, Wang PY, Jiao F, Zhang S, Zhang WJ. miRNA-regulated expression of oncogenes and tumor suppressor genes in the cisplatin-inhibited growth of K562 cells. Oncol Rep. 2010;23(6):1693–1700. doi: 10.3892/or_00000813. [DOI] [PubMed] [Google Scholar]
- 28.Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia. 2001;6(4):375–392. doi: 10.1023/a:1014778713034. [DOI] [PubMed] [Google Scholar]
- 29.Wang DT, Ma ZL, Li YL, et al. miR-150, p53 protein and relevant miRNAs consist of a regulatory network in NSCLC tumorigenesis. Oncol Rep. 2013;30(1):492–498. doi: 10.3892/or.2013.2453. [DOI] [PubMed] [Google Scholar]
- 30.Cao M, Hou D, Liang H, et al. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur J Cancer. 2014;50(5):1013–1024. doi: 10.1016/j.ejca.2013.12.024. [DOI] [PubMed] [Google Scholar]