Abstract
Background
Cardiovascular disease, a progressive manifestation of α-L-iduronidase deficiency or mucopolysaccharidosis type I, continues in patients both untreated and treated with hematopoietic stem cell transplantation or intravenous enzyme replacement. Few studies have examined the effects of α-L-iduronidase deficiency and subsequent glycosaminoglycan storage upon arterial gene expression to understand the pathogenesis of cardiovascular disease.
Methods
Gene expression in carotid artery, ascending, and descending aortas from four non-tolerized, non-enzyme treated 19 month-old mucopolysaccharidosis type I dogs was compared with expression in corresponding vascular segments from three normal, age-matched dogs. Data were analyzed using R and whole genome network correlation analysis, a bias-free method of categorizing expression level and significance into discrete modules. Genes were further categorized based on module-trait relationships. Expression of clusterin, a protein implicated in other etiologies of cardiovascular disease, was assessed in canine and murine mucopolysaccharidosis type I aortas via Western blot and in situ immunohistochemistry.
Results
Gene families with more than two-fold, significant increased expression involved lysosomal function, proteasome function, and immune regulation. Significantly downregulated genes were related to cellular adhesion, cytoskeletal elements, and calcium regulation. Clusterin gene overexpression (9-fold) and protein overexpression (1.3 to 1.62-fold) was confirmed and located specifically in arterial plaques of mucopolysaccharidosis-affected dogs and mice.
Conclusions
Overexpression of lysosomal and proteasomal-related genes are expected responses to cellular stress induced by lysosomal storage in mucopolysaccharidosis type I. Upregulation of immunity-related genes implicates the potential involvement of glycosaminoglycan-induced inflammation in the pathogenesis of mucopolysaccharidosis-related arterial disease, for which clusterin represents a potential biomarker.
Introduction
Mucopolysaccharidosis type I (MPS I), caused by a deficiency of the lysosomal enzyme α-L-iduronidase (IDUA), results in shortened lifespan, multisystemic somatic involvement, and variable neurocognitive degeneration because of accumulation of heparan sulfate (HS) and dermatan sulfate (DS) glycosaminoglycan (GAG) substrates in body tissues such as brain, soft tissues, chondrocytes, liver, and spleen [1]. Cardiovascular disease is a cardinal manifestation of MPS I, characterized by progressive thickening and compromised function of the heart valves, left ventricular hypertrophy, and diffuse coronary artery stenosis [2–6].
The advent of treatments to replace the missing IDUA enzyme, whether with intravenous enzyme replacement therapy (ERT) or via hematopoietic stem cell transplant (HSCT), has enabled MPS I patients to survive into adulthood [7, 8]. Although ERT and HSCT are able to mitigate many symptoms of MPS I, clinical experience has demonstrated that these treatments attenuate, but do not cure, the disease. Certain tissues remain resistant to treatment and continue to manifest GAG storage. Consequently, as MPS I patients survive into adulthood they face a different set of potentially life-threatening disease complications such as those involving the cardiovascular system [9, 10]. Cardiac sudden death, left-sided valvular insufficiency, ventricular dysfunction, and coronary intimal proliferation with stenosis all have been reported in stably treated patients [11–14]. Accumulation of GAG within cardiovascular structures in the face of ongoing treatment is the likely origin of these symptoms, as well as childhood-onset carotid intima-media thickening and abnormally reduced elasticity [15–19].
The pathogenesis and etiologies of treatment resistance of cardiovascular disease in MPS I are not well characterized, but studies in the murine and canine models of the disease indicate that accumulation of undegraded HS and DS GAG in the heart, valves, and vasculature alone do not adequately describe the pathophysiology. Both untreated and treated MPS I canines develop similar cardiovascular findings to human MPS I, with cardiac hypertrophy, nodular valve thickening, and vascular smooth muscle proliferation of the aorta with luminal stenosis [20–22]. Detailed histopathology of canine aortic lesions demonstrates vascular smooth muscle proliferation, activated CD68+ macrophages, and fragmented elastin fibrils in addition to GAG storage [23]. The murine MPS I model also manifests with cardiac enlargement, valvular thickening and dysfunction, and dilatation of the aorta with vascular wall thickening with elastin fibril degradation [24, 25].
Gene expression studies are a useful method to identify potential mechanisms of disease progression, but have not been comprehensively assessed for cardiovascular disease in any model of MPS I. The primary focus of expression studies for the mucopolysaccharidoses has been neurodegeneration in the Sanfilippo syndromes (MPS III) and Sly syndrome (MPS VII) [26–30]. Assessment of aortic mRNA expression for dogs with MPS I and VII, and mice with MPS VII has centered on quantification of cytokine, complement, and other inflammation-related genes [31–34]. Herein, we report alterations in arterial gene and protein expression in the canine MPS I model system, the identification of a potential marker for MPS I cardiovascular disease, and data supporting the hypothesis that a GAG-induced inflammatory process is responsible for the pathogenesis of MPS I cardiovascular disease.
Materials and Methods
Test animals and husbandry
This study was reviewed and approved by the Institutional Animal Care and Use Committees of both Iowa State University (IACUC #12-04-5791-K) and the Los Angeles Biomedical Research Institute at Harbor-UCLA (LA BioMed IACUC #20013–01). Studies were conducted in compliance with both UDSA and NIH guidelines for the use of dogs in research. Four MPS I (IDUA-/-) canines were produced from artificial insemination breedings of parental stock, diagnosed via α-L-iduronidase enzyme assay and PCR, and maintained at Iowa State University until 1 year of age, after which they were transported to LA BioMed, an AAALAC accredited facility overseen by a licensed veterinarian. Animals had ad libitum access to food (standard Teklad canine lab chow) and water, housing with enrichment, and a 12 hour light/dark cycle. Housing consisted of expanded mesh flooring, and involved housing with compatible conspecifics as required for social species unless medically or behaviorally contraindicated. Animal care was provided by the Laboratory Animal Resources veterinary staff of the respective institutions. The dog colony has a null mutation in intron 1 of the canine α-L-iduronidase gene that results in abnormal mRNA splicing, introduces a premature termination codon, and completely eliminates IDUA protein expression [35]. The four dogs were neither tolerized nor treated with IV rhIDUA, and had received six monthly intra-articular rhIDUA injections as previously reported [36].
Mouse-related protocols were approved by the University of California San Diego IACUC Animal Subjects Committee (IACUC #S99127). The MPS I (Idua-/-) mouse colony was housed, maintained, and veterinarian-supervised at The University of California San Diego, an AAALAC accredited facility. The colony, which is on the C57BL/6 background, was originally obtained from the Jackson Laboratories (B6.129-Iduatm1Clk/J) and was bred locally for 9 generations. Idua genotyping was performed as per S1 Appendix. Mice utilized for this study were fed standard chow and were neither tolerized nor treated with any form of rhIDUA.
Tissue collection and euthanasia
At 18 months of age, dogs were euthanized with a 1 cc / 10 lb dose of euthasol and 1 cm rings from the ascending aorta, descending aorta proximal to the renal arteries, and common carotid arteries were immediately obtained post-mortem. Arterial rings were processed as follows: 1) one section from each site was embedded in a cryomold with OCT (Tissue-Tek CRYO-OCT Compound, ThermoFisher Scientific, Waltham, MA), frozen and stored at -80°C, 2) one section placed in TRIzol solution (Life Technologies, Grand Island, NY) for subsequent RNA extraction, 3) and one section frozen and stored at -80°C for subsequent Western blotting.
At 9–12 months of age, female MPS I (mean age, 9.4 months; n = 8) and age-matched wild-type (mean age, 10.2 months n = 3) mice were euthanized with a combination of Isoflurane inhalation (Abbott, North Chicago, IL), cervical dislocation, and exsanguination. Heart and aorta collection was performed as per Daugherty and Whitman, and summarized as follows: after exsanguination, the right atrium was incised and the animal was perfused with 20 mL cold phosphate buffered saline (PBS) through the left ventricle [37]. The abdominal viscera were removed and the heart and aorta were carefully dissected en bloc to the iliac bifurcation. The heart was cut from the ascending aorta approximately 5 mm from the point of emergence from the left ventricle, embedded in a cryomold with OCT, and stored frozen at -80°C.
RNA isolation, amplification, and labeling
RNA isolation from canine aortic specimens was performed using the TRIzol RNA Purification System (Life Technologies, Grand Island, NY). RNA concentration and quality, as well as dye incorporation efficiency, was checked with an ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE, USA) and the Agilent Bioanalyzer 2100. A total of 100 ng RNA was used as initial starting template, which was labeled with Cy-3 or Cy-5 cytidine 5’-triphosphate using the low input fluorescent linear amplification kit (Agilent, Santa Clara, CA).
Microarray hybridizations and data analysis
Comparisons of canine arterial gene expression between control and untreated MPS animals were conducted with a canine-specific microarray covering 43,803 probes (Agilent G2519F 4x44k, Santa Clara, CA), for a total of four comparison groups: MPS ascending aorta vs. control ascending aorta, MPS descending aorta vs. control descending aorta, MPS carotid artery vs. control carotid artery, and finally pooled MPS artery (ascending aorta, descending aorta, carotid artery) vs. pooled control artery. Each comparison used four pairs of MPS vs corresponding age- and gender- matched animals to produce four biologic replicates.
Bioinformatic analysis
The data was analyzed by the WGCNA package in R 3.0.2, a package we have used previously to analyze large datasets and identify novel genomic targets [38–41]. Co-expression networks were formed on genes with similar behavior. Trait information (e.g. ascending aorta, carotid artery, and descending aorta) and Pearson correlation data were obtained.
Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 was used to functionally annotate the genes that clustered together [42,43]. DAVID’s functional annotation tool allows for pathway analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG), gene ontology annotation for biological processes, molecular function, and cellular components, assessment of transcription factor binding sites, and identification of tissues matching the gene clusters. All relationships determined had corresponding statistics within DAVID. The functional annotations presented here had a p-value ≤ 0.05 with Bonferroni correction.
Network analysis was performed using two methods. The first was WGCNA to identify networks without any prior knowledge. This enabled discovery of novel nodes and edges that could be visualized using Cytoscape version 3.1.1 [44]. The second was with GeneMania (a plugin available in Cytoscape) to examine connections based on prior evidence including co-expression, shared protein domains, pathway, physical interactions, genetic interactions, and co-localization [39, 45].
Western blotting
Flash frozen sections of harvested ascending and descending aorta that had been harvested from homozygous MPS I and heterozygous carrier dogs and stored at -80°C were thawed on ice. Sections of approximately 30 to 50 mg of each tissue sample were cut and mixed in a 1:3 weight to volume ratio with lysis buffer containing 4% SDS, EDTA, a protease inhibitor cocktail (P8340, Sigma-Aldrich, St. Louis, MO) and phosphatase inhibitor cocktails (P0044 and P5726, Sigma-Aldrich, St. Louis, MO). Standard Dounce homogenization was performed while each tissue sample was kept on ice in a 4°C room. After homogenization, each sample was sonicated in a 4°C water bath for 1 hour and then centrifuged at 10,000 rpm for 10 minutes. The supernatant was collected and centrifuged again at 10,000 rpm for 10 minutes. This supernatant was collected and total protein content of each sample was determined with a standard colorimetric assay (Bio-Rad, Philadelphia, PA) converting absorbance measured as optical density units at 595 nm (Shimadzu BioSpec 1601 Double Beam UV-Visible Spectrophotometer, Shimadzu Scientific Instruments, Columbia, MD) to protein concentration using bovine serum albumin as a standard.
Samples were diluted to a final concentration of 4 μg/μL by addition of the appropriate volume of a standard Laemmli loading buffer. These were then heated to 95°C for 20 minutes prior to loading onto a gel (Novex Tris-Glycine 4–20% Mini Protein Gel, Life Technologies, Grand Island, NY). Each well was loaded with 20 μg of total protein and a protein ladder covering the size range from 10 to 180 kDa (PageRuler Prestained Protein Ladder, Life Technologies, Grand Island, NY) was loaded in one well of each gel for reference. Separation was performed via 4–20% Tris-Glycine SDS-PAGE followed by transfer to a nitrocellulose membrane (BioTrace NT nitrocellulose transfer membrane, Pall Corp., Port Washington, NY).
Standard immunoblotting was then performed with mouse monoclonal anti-human clusterin alpha chain (1:1,000 or 1 ng/μL; EMD Millipore, Billerica, MA) diluted in 2.5% nonfat milk and incubated overnight at 4°C. After washing, the membrane was incubated for one hour at room temperature with goat anti-mouse IgG HRP conjugated secondary antibody (1:50,000; Southern Biotechnology, Birmingham, AL, USA) diluted in 2.5% nonfat milk, and then detected with Immobilon Chemiluminescent HRP Substrate (EMD Millipore, Billerica, MA). Membranes were then stripped, washed and re-probed with mouse monoclonal anti-human smooth muscle alpha-actin (1:10,000; Dako North America, Inc., Carpinteria, CA) diluted in 2.5% nonfat milk. After washing, the membrane was incubated for one hour at room temperature with goat anti-mouse IgG HRP conjugated secondary antibody (1:100,000; Southern Biotechnology, Birmingham, AL) diluted in 2.5% nonfat milk, and then detected with Immobilon Chemiluminescent HRP Substrate (EMD Millipore, Billerica, MA).
Western blot band intensities were quantified using ImageJ. After conversion of Western blot images to black and white and subtraction of background, the signal intensities of clusterin bands were subsequently measured and normalized to signal intensities of corresponding α-actin bands. The ratio of mean normalized MPS I clusterin intensity to mean heterozygote clusterin intensity was calculated to determine fold-change expression in ascending and descending aorta homogenates.
Canine aorta immunohistochemistry
Transverse cuts through each OCT-embedded tissue sample were taken with a cryostat at -20°C to produce 12 micron-thick cross-sectional slices that were mounted on glass slides. Slide mounted tissue sections were covered with neutral PBS at room temperature for twenty minutes, rinsed, and then fixed with 4% paraformaldehyde in PBS for 20 minutes. Slides were washed in PBS and then each tissue section was blocked with hydrogen peroxide for 20 minutes. After washing in PBS each tissue section was then blocked with 5% normal goat serum in PBS for one hour at room temperature. Tissue sections were then incubated overnight at 4°C with mouse monoclonal anti-human clusterin alpha chain antibody (1:2,500 or 0.4 ng/μl; EMD Millipore, Billerica, MA) diluted in 3% BSA in neutral PBS, mouse monoclonal anti-human smooth muscle alpha actin antibody (1:2,500; Dako North America, Inc., Carpinteria, CA) diluted in 3% BSA in neutral PBS, or 3% BSA in neutral PBS alone. Detection of specific staining was performed with an anti-mouse HRP-conjugated secondary antibody and diaminobenzidine (DAB) substrate-chromogen (EXPOSE Mouse and Rabbit HRP/DAB Detection Kit, Abcam, Cambridge, MA) according to manufacturer’s protocol. Tissue sections were counterstained with hematoxylin prior to visualization. Cover slips were applied to each slide and standard bright field optical microscopy was performed. In the transverse aorta sections positive (DAB) signal appears brown while the counterstain appears blue.
Murine aorta preparation and immunohistochemistry
6 micron-thick cross-sections of heart and aorta were obtained from OCT-embedded samples with a cryostat at -20°C, mounted on glass slides, dried at room temperature for 30 minutes and then fixed in fresh 10% neutral buffered formalin (Fisher #SF94-4, Waltham, MA) for 15 minutes. Slides were washed 3 times in PBS with Tween-20® (PBST). Endogenous peroxidases were blocked with a 0.03% solution of hydrogen peroxide in PBST, then washed another 3 times in PBST. Slides were overlaid with 1% BSA in PBST (Sigma #A4503, St Louis, MO) for 10 minutes, endogenous biotin blocked with Vector Labs #SP-2001 kit, and washed another 3 times in PBST. Slides were then overlaid with 1% BSA in PBST and polyclonal goat anti-mouse clusterin alpha chain (1:100; Santa Cruz #SC-6420, Dallas, TX), incubated at room temperature for 1 hour, and washed 3 times in PBST. Secondary staining was accomplished with biotinylated donkey anti-goat antibody (1:500; Jackson ImmunoResearch Laboratories #805-065-180, West Grove, PA) incubated 30 minutes at room temperature, then washed 3 times in PBST. Slides were then incubated with streptavidin-HRP (1:500; Jackson ImmunoResearch Laboratories #016-030-084, West Grove, PA) for 30 minutes, rinsed, then developed with 3-amino-9-ethylcarbazole chromogen (Vector Labs #SK-4200, Burlingame, CA) for 10 minutes. Slides were counterstained in Mayer’s Hematoxylin (Sigma #MHS16, St Louis, MO) for 1 minute, washed in PBST, air dried, and cover slips applied with aqueous mounting media (Vector Labs #H-5501, Burlingame, CA).
Results
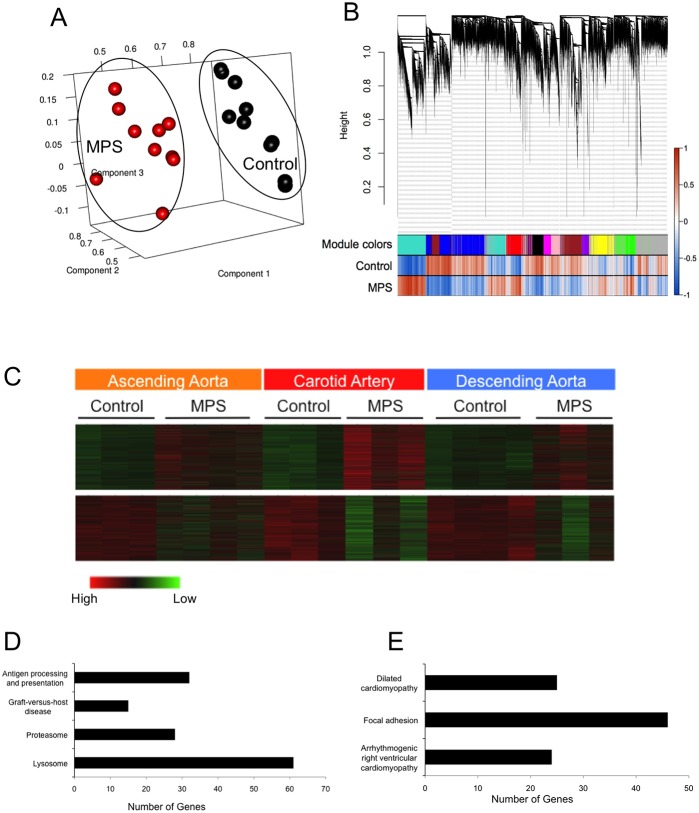
Microarray was performed on IDUA-/- and control (IDUA+/-) canine arterial samples obtained from common carotid artery, ascending aorta, and descending aorta. Data was initially analyzed using principal component analysis (PCA). Two clusters were visualized, clearly differentiating the IDUA-/- arterial expression patterns from the control patterns (Fig 1A). Dendrograms and trait heatmaps were also made using the adjacency matrix technique (S1 Fig). Using both of these dimension reduction methods, it can easily be determined if the samples cluster together or if there are outliers. Again, two main clusters were observed, one for IDUA-/- and the other for control. In addition, expression patterns from different animals at the same arterial sites demonstrated clustering according to IDUA genotype. In other words, carotid artery expression in the MPS I animals clustered together and was distinct from both MPS I ascending aorta expression and descending aorta expression patterns.
Fig 1. Microarray Results From IDUA-/- Canine Arterial Samples.
A. Principal component analysis of IDUA-/- canine arteries compared to control canine arteries clearly shows the formation of two sample clusters separating IDUA-/- and control expression. B. WGCNA dendrogram on the microarray samples showing different modules related to the Pearson correlation of the IDUA-/- and control samples. C. Heatmaps of the two major modules (turquoise and blue) formed by WGCNA showing genes upregulated (top panel) or downregulated (bottom panel) by IDUA deficiency. D. Genes significantly upregulated in IDUA-/- canine arteries include those related to immunity / inflammation (KEGG pathways: antigen processing and presentation, graft-versus-host disease), proteasomal function, and lysosomal function. E. Genes significantly downregulated in IDUA-/- canine arteries are primarily related to cellular adhesion and the cytoskeleton (placed into the dilated cardiomyopathy, focal adhesion, arrhythmogenic right ventricular cardiomyopathy KEGG pathways).
Data was subsequently analyzed using whole genome correlation network analysis (WGCNA) to identify specific modules that related back to the MPS I trait. Use of this method loosens the stringency in the analysis, since groups of genes are examined together allows a significance matrix to be made based on the networks formed [46]. Genes were further categorized based on fold-change and additional statistical tests within each module. The lists of probes/genes 2-fold or more upregulated or downregulated compared to control across modules can be found in Tables 1 and 2, respectively; full listings are included in S1 and S2 Tables. Subsequently, a dendrogram was made to represent the modules formed with the Pearson correlations for each trait shown (Fig 1B). There were ten total modules that formed (S2 Fig). Specific attention was paid to the turquoise and blue modules because those were the most statistically significant modules representing genes consistently upregulated in all three sampled vascular segments of the IDUA-/- dogs (upper panel, Fig 1C), or genes consistently downregulated in all three sampled vascular segments of the IDUA-/- dogs (lower panel, Fig 1C). Further downstream analysis with a volcano plot to identify the top targets within the turquoise and blue modules is seen in S3 Fig.
Table 1. Upregulated Genes in IDUA-/- Canine Arteries.
The 50 upregulated genes with significant (p-value ≤ 0.05) and greater than 2-fold level of upregulation in IDUA-/- canine arteries, compared to controls. Note that some genes are represented more than once due to existence of multiple mRNA probes per gene. For the full list of significantly upregulated genes, please refer to S1 Table.
| Fold Change | Gene Symbol | Systematic Name | Description |
|---|---|---|---|
| 27.126 | GPNMB | XM_858105 | PREDICTED: Canis familiaris similar to glycoprotein (transmembrane) nmb isoform b precursor, transcript variant 3 (LOC482355), mRNA [XM_858105] |
| 20.245 | GPNMB | XM_858105 | PREDICTED: Canis familiaris similar to glycoprotein (transmembrane) nmb isoform b precursor, transcript variant 3 (LOC482355), mRNA [XM_858105] |
| 12.432 | ENSCAFT00000039657 | ENSCAFT00000039657 | Osteopontin (Fragment) |
| 11.926 | CSTB | XM_535601 | PREDICTED: Canis familiaris similar to Cystatin B |
| 11.566 | ENSCAFT00000039661 | ENSCAFT00000039661 | Osteopontin (Fragment) |
| 10.506 | DLA-DRA1 | ENSCAFT00000001254 | MHC class II DR alpha chain |
| 10.220 | CLU | NM_001003370 | Canis lupus familiaris clusterin (CLU) |
| 9.595 | CLU | NM_001003370 | Canis lupus familiaris clusterin (CLU) |
| 9.464 | CTSK | ENSCAFT00000035613 | Cathepsin K precursor |
| 8.985 | TC71707 | TC71707 | Q49RE5_CANFA (Q49RE5) Cytochrome b, partial (35%) |
| 7.766 | CSTB | XM_535601 | PREDICTED: Canis familiaris similar to Cystatin B |
| 7.587 | LOC479746 | XM_536874 | PREDICTED: Canis familiaris similar to Ferritin light chain 2 |
| 7.022 | TC57368 | TC57368 | HUMLPLSTN2 L-plastin {Homo sapiens} |
| 6.975 | GPNMB | XM_858105 | PREDICTED: Canis familiaris similar to glycoprotein (transmembrane) nmb isoform b precursor, transcript variant 3 (LOC482355), mRNA [XM_858105] |
| 6.968 | CLU | NM_001003370 | Canis lupus familiaris clusterin (CLU) |
| 6.947 | CD86 | NM_001003146 | Canis lupus familiaris CD86 molecule (CD86) |
| 6.769 | DR104862 | DR104862 | Canine cardiovascular system biased cDNA |
| 6.738 | FTL | NM_001024636 | Canis lupus familiaris ferritin, light polypeptide |
| 6.718 | CTSS | NM_001002938 | Canis lupus familiaris cathepsin S (CTSS) |
| 6.704 | CLU | NM_001003370 | Canis lupus familiaris clusterin (CLU) |
| 6.677 | APOE | XM_847504 | PREDICTED: Canis familiaris apolipoprotein E, transcript variant 2 (APOE) |
| 6.666 | CTSK | NM_001033996 | Canis lupus familiaris cathepsin K (CTSK) |
| 6.655 | CTSK | NM_001033996 | Canis lupus familiaris cathepsin K (CTSK) |
| 6.605 | LGMN | XM_537355 | PREDICTED: Canis familiaris similar to Legumain precursor |
| 6.453 | CTSK | NM_001033996 | Canis lupus familiaris cathepsin K (CTSK) |
| 6.423 | DLA-DRB1 | NM_001014768 | Canis lupus familiaris MHC class II DLA DRB1 beta chain (DLA-DRB1) |
| 6.193 | FTL | NM_001024636 | Canis lupus familiaris ferritin, light polypeptide (FTL) |
| 6.178 | CTSK | NM_001033996 | Canis lupus familiaris cathepsin K (CTSK) |
| 6.055 | TYRP1 | XM_859450 | PREDICTED: Canis familiaris tyrosinase-related protein 1 (TYRP1) |
| 6.026 | CTSK | NM_001033996 | Canis lupus familiaris cathepsin K (CTSK) |
| 6.013 | BPI | XM_534417 | PREDICTED: Canis familiaris similar to Bactericidal permeability-increasing protein precursor (BPI) |
| 6.006 | CTSB | XM_543203 | PREDICTED: Canis familiaris similar to cathepsin B preproprotein |
| 5.833 | TC50899 | TC50899 | Q9N2I5_FELCA (Q9N2I5) CD16 |
| 5.828 | TC62836 | TC62836 | FCU92795 fusin {Felis catus} |
| 5.727 | LGMN | XM_537355 | PREDICTED: Canis familiaris similar to Legumain precursor |
| 5.680 | DLA-DQB1 | NM_001014381 | Canis lupus familiaris MHC class II, DQ beta 1 (DLA-DQB1) |
| 5.660 | CSGALNACT1 | XM_539946 | PREDICTED: Canis familiaris similar to chondroitin beta1,4 N-acetylgalactosaminyltransferase |
| 5.651 | MT2A | NM_001003149 | Canis lupus familiaris metallothionein 2A (MT2A) |
| 5.493 | TC51307 | TC51307 | Unknown |
| 5.331 | TGM2 | XM_542991 | PREDICTED: Canis familiaris similar to Protein-glutamine gamma-glutamyltransferase |
| 5.306 | SMOC2 | XM_858080 | PREDICTED: Canis familiaris similar to secreted modular calcium-binding protein 2 |
| 5.190 | ASAH1 | XM_540012 | PREDICTED: Canis familiaris similar to N-acylsphingosine amidohydrolase (acid ceramidase) |
| 5.152 | APOE | ENSCAFT00000007432 | Apolipoprotein E (Apo-E) |
| 5.104 | TC54889 | TC54889 | Unknown |
| 5.069 | DLA-DRA1 | NM_001011723 | Canis lupus familiaris MHC class II DR alpha chain (DLA-DRA1) |
| 4.984 | RRAGD | XM_532231 | PREDICTED: Canis familiaris similar to Ras-related GTP binding D |
| 4.921 | E02824 | E02824 | Canis familiaris cDNA clone LIB30321_005_G04 |
| 4.912 | RRAGD | XM_532231 | PREDICTED: Canis familiaris similar to Ras-related GTP binding D |
| 4.902 | TYROBP | XM_533687 | PREDICTED: Canis familiaris similar to TYRO protein tyrosine kinase binding protein isoform 2 precursor |
| 4.882 | FTH1 | ENSCAFT00000025200 | Ferritin heavy chain (EC 1.16.3.1) (Ferritin H subunit) |
Table 2. Down-regulated Genes in IDUA-/- Canine Arteries.
The 50 down-regulated genes with significant (p-value ≤ 0.05) and greater than 2-fold level of down-regulation in IDUA-/- canine arteries, compared to controls. Again, some genes are represented more than once due to existence of multiple mRNA probes per gene. For the full list of significantly down-regulated genes, please refer to S2 Table.
| Fold Change | Gene Symbol | Systematic Name | Description |
|---|---|---|---|
| 4.948 | TC67209 | TC67209 | HUMRPS17A S17 ribosomal protein {Homo sapiens} |
| 3.970 | LRRK2 | XM_543734 | PREDICTED: Canis familiaris similar to leucine-rich repeat kinase 2 |
| 3.666 | CNN1 | XM_862565 | PREDICTED: Canis familiaris similar to calponin 1, basic, smooth muscle, transcript variant 4 |
| 3.392 | CO611926 | CO611926 | DG9-109j22 DG9-ovary Canis familiaris cDNA 3' |
| 3.309 | SMTN | XM_860837 | PREDICTED: Canis familiaris similar to smoothelin isoform c |
| 3.218 | NRGN | XM_536537 | PREDICTED: Canis familiaris similar to neurogranin (LOC479401) |
| 3.212 | CSRP1 | XM_843516 | PREDICTED: Canis familiaris similar to Cysteine and glycine-rich protein 1 |
| 3.157 | TCAP | XM_858636 | PREDICTED: Canis familiaris similar to Telethonin (Titin cap protein) |
| 3.109 | PPP1R12B | XM_843384 | PREDICTED: Canis familiaris similar to Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase targeting subunit 2) |
| 3.018 | DN756599 | DN756599 | GL-Cf-13395 GLGC-LIB0001-cf Canis familiaris Normalized Mixed Tissue cDNA |
| 2.949 | LOC488984 | XM_843847 | PREDICTED: Canis familiaris similar to Actin, alpha skeletal muscle |
| 2.865 | AB012223 | AB012223 | Canis familiaris LINE-1 element ORF2 mRNA |
| 2.855 | ENSCAFT00000034839 | ENSCAFT00000034839 | NADH-ubiquinone oxidoreductase chain 3 |
| 2.836 | PPP1R12B | XM_843384 | PREDICTED: Canis familiaris similar to Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase targeting subunit 2) |
| 2.812 | ENSCAFT00000034824 | ENSCAFT00000034824 | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2) |
| 2.812 | BM538907 | BM538907 | hb02c06.g1 Canis cDNA from testes cells Canis familiaris cDNA clone |
| 2.782 | LOC488984 | XM_843847 | PREDICTED: Canis familiaris similar to Actin, alpha skeletal muscle |
| 2.764 | TC55776 | TC55776 | Unknown |
| 2.761 | TAGLN | XM_856022 | PREDICTED: Canis familiaris similar to transgelin, transcript variant 3 |
| 2.734 | FHL1 | XM_549282 | PREDICTED: Canis familiaris similar to four and a half LIM domains 1 |
| 2.712 | LOC488984 | XM_843847 | PREDICTED: Canis familiaris similar to Actin, alpha skeletal muscle |
| 2.705 | OGN | XM_848247 | PREDICTED: Canis familiaris similar to Mimecan precursor (Osteoglycin) |
| 2.692 | DMD | NM_001003343 | Canis lupus familiaris dystrophin (muscular dystrophy, Duchenne and Becker types) |
| 2.673 | TC68286 | TC68286 | Unknown |
| 2.630 | ENSCAFT00000022802 | ENSCAFT00000022802 | Fibronectin (FN) (Fragment) |
| 2.626 | SLC22A3 | XM_533467 | PREDICTED: Canis familiaris similar to solute carrier family 22 member 3 |
| 2.617 | LOC488984 | XM_843847 | PREDICTED: Canis familiaris similar to Actin, alpha skeletal muscle |
| 2.598 | DN879822 | DN879822 | nae29h08.y1 Dog eye eye minus lens and cornea. Unnormalized (nae) |
| 2.594 | PTGS1 | NM_001003023 | Canis lupus familiaris prostaglandin-endoperoxide synthase 1 |
| 2.586 | LOC491592 | XM_548713 | PREDICTED: Canis familiaris similar to ankyrin repeat domain 26 |
| 2.567 | PPAP2B | XM_536696 | PREDICTED: Canis familiaris similar to phosphatidic acid phosphatase type 2B |
| 2.526 | PPAP2B | XM_536696 | PREDICTED: Canis familiaris similar to phosphatidic acid phosphatase type 2B |
| 2.489 | C1QTNF1 | XM_843609 | PREDICTED: Canis familiaris similar to C1q and tumor necrosis factor related protein 1 |
| 2.481 | TPM1 | ENSCAFT00000026876 | Tropomyosin (Fragment) |
| 2.460 | MYH11 | XM_857511 | PREDICTED: Canis familiaris similar to smooth muscle myosin heavy chain 11 isoform SM1 |
| 2.459 | ADCY5 | M88649 | Canis familiaris adenylyl cyclase type V |
| 2.458 | LOC480803 | XM_537918 | PREDICTED: Canis familiaris similar to ankyrin repeat domain 26 |
| 2.429 | TC48376 | TC48376 | MALAT_HUMAN (Q9UHZ2) Metastasis-associated lung adenocarcinoma transcript 1 |
| 2.407 | CF412534 | CF412534 | Canine heart normalized cDNA Library, pBluescript Canis familiaris cDNA clone CH3#080_C09 5' |
| 2.405 | DN368051 | DN368051 | LIB3731-002-Q6-K7-H5 LIB3731 Canis familiaris cDNA clone CLN2470649 |
| 2.393 | CN005552 | CN005552 | ip32c07.g1 Brain—Cerebellum Library (DOGEST8) Canis familiaris cDNA clone ip32c07 |
| 2.363 | CO586466 | CO586466 | DG2-130p9 DG2-brain Canis familiaris |
| 2.346 | ENSCAFT00000022799 | ENSCAFT00000022799 | Fibronectin (FN) (Fragment) |
| 2.331 | TCAP | XM_858636 | PREDICTED: Canis familiaris similar to Telethonin (Titin cap protein) |
| 2.322 | LOC488991 | XM_843985 | PREDICTED: Canis familiaris similar to Protein FAM13C1 |
| 2.309 | PDZRN4 | XM_543731 | PREDICTED: Canis familiaris similar to PDZ domain containing RING finger protein 4 (Ligand of Numb-protein X 4) |
| 2.301 | LOC607572 | XM_844308 | PREDICTED: Canis familiaris similar to ankyrin repeat domain 26 |
| 2.286 | PPP1R12A | XM_859971 | PREDICTED: Canis familiaris similar to Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase targeting subunit 1) |
| 2.286 | TC73854 | TC73854 | Unknown |
| 2.280 | A_11_P0000024197 | Unknown | TC47203 |
The pathways significantly implicated by the upregulated gene module (turquoise; Fig 1 D) include antigen processing and presentation, graft-versus-host disease, proteasome, and the lysosome. Specific genes from each upregulated pathway are detailed in Table 3. The pathways significantly implicated by the downregulated gene module (blue; Fig 1 E) include dilated cardiomyopathy, focal adhesion, and arrhythmogenic right ventricular cardiomyopathy and are detailed in Table 4. These identified pathways are consistent with pathways identified from prior expression arrays from MPS IIIb mouse brain [26, 30], MPS VII mouse brain [29] and liver [47].
Table 3. Upregulated Canine Arterial IDUA-/- Genes Sorted by Category.
Upregulated arterial genes in IDUA-/- canines fall into categories associated with lysosomal function, proteasomal function, and the immune system (graft-versus-host disease and antigen processing).
| Ontology Category | ||
|---|---|---|
| "LYSOSOMAL FUNCTION" | ||
| Gene Symbol | Gene Name | Entrez Gene ID |
| ATP6V0C | ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c | 479877 |
| ATP6V0B | ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b | 482528 |
| ATP6V0D1 | ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 | 479685 |
| ATP6V1H | ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H | 486953 |
| ATP6V0A1 | ATPase, H+ transporting, lysosomal V0 subunit a1 | 607705 |
| ATP6AP1 | ATPase, H+ transporting, lysosomal accessory protein 1 | 610922 |
| CD63 | CD63 molecule | 474391 |
| GM2A | GM2 ganglioside activator | 479324 |
| NAGA | N-acetylgalactosaminidase, alpha | 481226 |
| GNPTAB | N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits | 475443 |
| NAGPA | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase | 490019 |
| NAGLU | N-acetylglucosaminidase, alpha- | 490965 |
| ASAH1 | N-acylsphingosine amidohydrolase (acid ceramidase) 1 | 482897 |
| SGSH | N-sulfoglucosamine sulfohydrolase | 403707 |
| NPC1 | Niemann-Pick disease, type C1 | 403698 |
| TCIRG1 | T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3 | 483691 |
| AP3B2 | adaptor-related protein complex 3, beta 2 subunit | 479053 |
| AP3M1 | adaptor-related protein complex 3, mu 1 subunit | 489052 |
| AP3S2 | adaptor-related protein complex 3, sigma 2 subunit | 479042 |
| AP4B1 | adaptor-related protein complex 4, beta 1 subunit | 483125 |
| ARSA | arylsulfatase A | 474457 |
| AGA | aspartylglucosaminidase | 475638 |
| CTSA | cathepsin A | 611146 |
| CTSB | cathepsin B | 486077 |
| CTSC | cathepsin C | 403458; 478608 |
| CTSD | cathepsin D | 483662 |
| CTSG | cathepsin G | 608543 |
| CTSH | cathepsin H | 479065 |
| CTSK | cathepsin K | 608843 |
| CTSL2 | cathepsin L2 | 403708 |
| CTSS | cathepsin S | 403400 |
| CTSZ | cathepsin Z | 611983 |
| CLTA | clathrin, light chain (Lca) | 474765 |
| CTNS | cystinosis, nephropathic | 491220 |
| DNASE2 | deoxyribonuclease II, lysosomal | 476697 |
| GLB1 | galactosidase, beta 1 | 403873 |
| GBA | glucosidase, beta; acid (includes glucosylceramidase) | 612206 |
| GUSB | glucuronidase, beta | 403831 |
| GGA2 | golgi associated, gamma adaptin ear containing, ARF binding protein 2 | 608555 |
| HGSNAT | heparan-alpha-glucosaminide N-acetyltransferase | 482833 |
| HEXA | hexosaminidase A (alpha polypeptide) | 487633 |
| HEXB | hexosaminidase B (beta polypeptide) | 478100 |
| LGMN | legumain | 480232 |
| LIPA | lipase A, lysosomal acid, cholesterol esterase | 610650 |
| LAPTM5 | lysosomal multispanning membrane protein 5 | 487324 |
| LAPTM4A | lysosomal protein transmembrane 4 alpha | 475678 |
| LAMP1 | lysosomal-associated membrane protein 1 | 476995 |
| LAMP2 | lysosomal-associated membrane protein 2 | 481037 |
| M6PR | mannose-6-phosphate receptor (cation dependent) | 477700 |
| MAN2B1 | mannosidase, alpha, class 2B, member 1; similar to UPF0139 protein CGI-140 | 476703; 484932 |
| MANBA | mannosidase, beta A, lysosomal | 487883 |
| PPT1 | palmitoyl-protein thioesterase 1 | 475316 |
| PPT2 | palmitoyl-protein thioesterase 2 | 474856 |
| PSAP | prosaposin | 479240 |
| SCARB2 | scavenger receptor class B, member 2 | 478435 |
| NEU1 | sialidase 1 (lysosomal sialidase) | 481717 |
| AP1S2 | similar to Adapter-related protein complex 1 sigma 1B subunit (Sigma-adaptin 1B) | 611468 |
| SLC11A1 | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | 478909 |
| SLC17A5 | solute carrier family 17 (anion/sugar transporter), member 5 | 474969 |
| SORT1 | sortilin 1 | 479915 |
| TPP1 | tripeptidyl peptidase I | 485337 |
| "PROTEASOMAL FUNCTION" | ||
| Gene Symbol | Gene Name | Entrez Gene ID |
| PSMC1 | proteasome (prosome, macropain) 26S subunit, ATPase, 1 | 478703 |
| PSMC2 | proteasome (prosome, macropain) 26S subunit, ATPase, 2 | 475896 |
| PSMC3 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | 475980 |
| PSMC5 | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | 480478 |
| PSMD11 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | 480610 |
| PSMD14 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 | 478765 |
| PSMD2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 478654 |
| PSMD3 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | 491018 |
| PSMD6 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | 484700 |
| PSMD8 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 476470 |
| PSME1 | proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) | 480256 |
| PSME2 | proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | 480258 |
| PSME4 | proteasome (prosome, macropain) activator subunit 4 | 474594 |
| PSMA4 | proteasome (prosome, macropain) subunit, alpha type, 4 | 475132 |
| PSMA5 | proteasome (prosome, macropain) subunit, alpha type, 5 | 490123 |
| PSMA6 | proteasome (prosome, macropain) subunit, alpha type, 6 | 480290 |
| PSMA7 | proteasome (prosome, macropain) subunit, alpha type, 7 | 404305 |
| PSMB1 | proteasome (prosome, macropain) subunit, beta type, 1 | 475040 |
| PSMB10 | proteasome (prosome, macropain) subunit, beta type, 10 | 489749 |
| PSMB2 | proteasome (prosome, macropain) subunit, beta type, 2 | 475338 |
| PSMB3 | proteasome (prosome, macropain) subunit, beta type, 3 | 474987; 480537 |
| PSMB4 | proteasome (prosome, macropain) subunit, beta type, 4 | 475848 |
| PSMB5 | proteasome (prosome, macropain) subunit, beta type, 5 | 480246 |
| PSMB8 | proteasome (prosome, macropain) subunit, beta type, 8 | 474865 |
| PSMB9 | proteasome (prosome, macropain) subunit, beta type, 9 | 474867 |
| POMP | proteasome maturation protein | 477325 |
| PSMA1 | similar to Proteasome subunit alpha type 1 (Proteasome component C2) | 476867; 608388; 610188; 612667; 613000 |
| PSMA3 | similar to Proteasome subunit alpha type 3 (Proteasome component C8) | 480338; 607261 |
| "GRAFT-VS-HOST" | ||
| Gene Symbol | Gene Name | Entrez Gene ID |
| CD80 | CD80 molecule | 403765 |
| CD86 | CD86 molecule | 403764 |
| DLA-12 / DLA-64 | MHC class I DLA-12; MHC class I DLA-64 | 474838; 541592 |
| DLA88 | MHC class I DLA-88 | 474836 |
| DLA-DRB1 | MHC class II DLA DRB1 beta chain | 474860 |
| DLA-DRA1 | MHC class II DR alpha chain | 481731 |
| DLA-79 | MHC class Ib | 483594 |
| IL1A | interleukin 1, alpha | 403782 |
| IL1B | interleukin 1, beta | 403974 |
| IL2 | interleukin 2 | 403989 |
| IL6 | interleukin 6 (interferon, beta 2) | 403985 |
| HLA-DMA | major histocompatibility complex, class II, DM alpha | 481732 |
| HLA-DOB | major histocompatibility complex, class II, DO beta | 607786 |
| DLA-DQA1 | major histocompatibility complex, class II, DQ alpha 1 | 474861 |
| DLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 | 474862 |
| "ANTIGEN PROCESSING" | ||
| Gene Symbol | Gene Name | Entrez Gene ID |
| DLA-12 / DLA-64 | MHC class I DLA-12; MHC class I DLA-64 | 474838; 541592 |
| DLA88 | MHC class I DLA-88 | 474836 |
| DLA-DRB1 | MHC class II DLA DRB1 beta chain | 474860 |
| DLA-DRA1 | MHC class II DR alpha chain | 481731 |
| DLA-79 | MHC class Ib | 483594 |
| TAPBP | TAP binding protein (tapasin) | 481740 |
| B2M | beta-2-microglobulin | 478284 |
| CANX | calnexin | 403908 |
| CTSB | cathepsin B | 486077 |
| CTSS | cathepsin S | 403400 |
| HSPA1L | heat shock 70kDa protein 1-like | 474850 |
| HSP70 | heat shock protein 70 | 403612 |
| HSPA4 | heat shock protein Apg-2 | 474680 |
| KLRD1 | killer cell lectin-like receptor subfamily D, member 1 | 611360 |
| LGMN | legumain | 480232 |
| HLA-DMA | major histocompatibility complex, class II, DM alpha | 481732 |
| HLA-DOB | major histocompatibility complex, class II, DO beta | 607786 |
| DLA-DQA1 | major histocompatibility complex, class II, DQ alpha 1 | 474861 |
| DLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 | 474862 |
| NFYB | nuclear transcription factor Y, beta | 475450 |
| NFYC | nuclear transcription factor Y, gamma | 475312 |
| PSME1 | proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) | 480256 |
| PSME2 | proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | 480258 |
| RFXANK | regulatory factor X-associated ankyrin-containing protein | 476662 |
| LOC480726 | similar to 78 kDa glucose-regulated protein precursor (Endoplasmic reticulum lumenal Ca(2+) binding protein grp78) | 480726 |
| LOC476669 | similar to Gamma-interferon inducible lysosomal thiol reductase precursor (Gamma-interferon-inducible protein IP-30) | 476669 |
| LOC479329 | similar to HLA class II histocompatibility antigen, gamma chain (HLA-DR antigens associated invariant chain) (Ia antigen-associated invariant chain) | 479329 |
| LOC607470 | similar to Heat shock protein HSP 90-alpha (HSP 86) | 607470 |
| LOC490008 | similar to MHC class II transactivator | 490008 |
| HSPA8 | similar to heat shock 70kDa protein 8 isoform 2; heat shock 70kDa protein 8; similar to Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8); similar to heat shock protein 8 | 479406; 607182; 608802 |
| LOC480438 | similar to heat shock protein 1, alpha | 480438 |
| LOC608885 | similar to heat shock protein 8 | 608885 |
Table 4. Down-regulated Canine Arterial IDUA-/- Genes Sorted by Category.
Down-regulated arterial genes in IDUA-/- dogs fall into categories associated with cytoskeletal proteins, cellular adhesion, and ion channels (termed arrhythmogenic right ventricular cardiomyopathy, focal adhesion, and dilated cardiomyopathy).
| Ontology Category | ||
|---|---|---|
| "ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY" | ||
| Systematic Name | Gene Name | Entrez Gene ID |
| ACTB | actin, beta | 610787 |
| ACTN1 | actinin, alpha 1 | 480369 |
| ACTN2 | actinin, alpha 2 | 479191 |
| ACTN4 | actinin, alpha 4 | 484526 |
| LOC442937 | beta-actin | 442937 |
| CACNA1S | calcium channel, voltage-dependent, L type, alpha 1S subunit | 490244 |
| CACNB1 | calcium channel, voltage-dependent, beta 1 subunit | 491030 |
| CACNB2 | calcium channel, voltage-dependent, beta 2 subunit | 487113 |
| CACNB4 | calcium channel, voltage-dependent, beta 4 subunit | 609361 |
| CTNNA1 | catenin (cadherin-associated protein), alpha 2 | 483088 |
| CTNNA3 | catenin (cadherin-associated protein), alpha 3 | 489008 |
| DES | desmin | 497091 |
| DSC2 | desmocollin 2 | 403860 |
| DMD | dystrophin (muscular dystrophy, Duchenne and Becker types) | 606758 |
| GJA1 | gap junction protein, alpha 1, 43kDa | 403418 |
| ITGA5 | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | 486493 |
| ITGA6 | integrin, alpha 6 | 478800 |
| ITGA8 | integrin, alpha 8 | 487119 |
| ITGA9 | integrin, alpha 9 | 477021 |
| ITGB4 | integrin, beta 4 | 483318 |
| ITGB8 | integrin, beta 8 | 475253 |
| RYR2 | ryanodine receptor 2 (cardiac) | 403615 |
| SGCG | sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) | 486043 |
| LOC492249 | similar to Emerin | 492249 |
| "DILATED CARDIOMYOPATHY" | ||
| Systematic Name | Gene Name | Entrez Gene ID |
| GNAS | GNAS complex locus | 403943 |
| ACTB | actin, beta | 610787 |
| ADCY2 | adenylate cyclase 2 (brain) | 478624 |
| ADCY5 | adenylate cyclase 5 | 403859 |
| ADCY9 | adenylate cyclase 9 | 490031 |
| LOC442937 | beta-actin | 442937 |
| CACNA1S | calcium channel, voltage-dependent, L type, alpha 1S subunit | 490244 |
| CACNB1 | calcium channel, voltage-dependent, beta 1 subunit | 491030 |
| CACNB2 | calcium channel, voltage-dependent, beta 2 subunit | 487113 |
| CACNB4 | calcium channel, voltage-dependent, beta 4 subunit | 609361 |
| DES | desmin | 497091 |
| DMD | dystrophin (muscular dystrophy, Duchenne and Becker types) | 606758 |
| ITGA5 | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | 486493 |
| ITGA6 | integrin, alpha 6 | 478800 |
| ITGA8 | integrin, alpha 8 | 487119 |
| ITGA9 | integrin, alpha 9 | 477021 |
| ITGB4 | integrin, beta 4 | 483318 |
| ITGB8 | integrin, beta 8 | 475253 |
| PLN | phospholamban | 414755 |
| RYR2 | ryanodine receptor 2 (cardiac) | 403615 |
| SGCG | sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) | 486043 |
| LOC492249 | similar to Emerin | 492249 |
| TTN | titin | 478819 |
| TPM3 | tropomyosin 3; tropomyosin 1 (alpha); similar to tropomyosin 1 alpha chain isoform 4 | 478332; 480137; 484695; 491970 |
| TNF | tumor necrosis factor (TNF superfamily, member 2) | 403922 |
| "FOCAL ADHESION" | ||
| Systematic Name | Gene Name | Entrez Gene ID |
| RAP1A | RAP1A, member of RAS oncogene family | 483225 |
| RAPGEF1 | Rap guanine nucleotide exchange factor (GEF) 1 | 491286 |
| ARHGAP5 | Rho GTPase activating protein 5 | 490642 |
| ROCK1 | Rho-associated, coiled-coil containing protein kinase 1 | 480181 |
| ACTB | actin, beta | 610787 |
| ACTN1 | actinin, alpha 1 | 480369 |
| ACTN2 | actinin, alpha 2 | 479191 |
| ACTN4 | actinin, alpha 4 | 484526 |
| LOC442937 | beta-actin | 442937 |
| CAV1 | caveolin 1, caveolae protein, 22kDa | 403980 |
| CAV2 | caveolin 2 | 403611; 475294 |
| COL1A1 | collagen, type I, alpha 1 | 403651 |
| COL1A2 | collagen, type I, alpha 2 | 403824 |
| COL2A1 | collagen, type II, alpha 1 | 403826 |
| COL4A1 | collagen, type IV, alpha 1 | 403496 |
| COL5A1 | collagen, type V, alpha 1 | 480684 |
| COL6A3 | collagen, type VI, alpha 3 | 403582 |
| FLNC | filamin C, gamma (actin binding protein 280) | 482266 |
| ITGA5 | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | 486493 |
| ITGA6 | integrin, alpha 6 | 478800 |
| ITGA8 | integrin, alpha 8 | 487119 |
| ITGA9 | integrin, alpha 9 | 477021 |
| ITGB4 | integrin, beta 4 | 483318 |
| ITGB8 | integrin, beta 8 | 475253 |
| LAMA3 | laminin, alpha 3 | 480173 |
| LAMB2 | laminin, beta 2 (laminin S) | 476626 |
| MET | met proto-oncogene (hepatocyte growth factor receptor) | 403438 |
| MAPK8 | mitogen-activated protein kinase 8 | 477746 |
| MYLK | myosin light chain kinase | 488012 |
| MYLK2 | myosin light chain kinase 2 | 477187 |
| PAK4 | p21 protein (Cdc42/Rac)-activated kinase 4 | 484513 |
| PIK3CA | phosphoinositide-3-kinase, catalytic, alpha polypeptide | 488084 |
| PIK3R2 | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | 609956 |
| PGF | placental growth factor | 611916 |
| PDGFD | platelet derived growth factor D | 479460 |
| PDGFRB | platelet-derived growth factor receptor, beta polypeptide | 442985 |
| PPP1CB | protein phosphatase 1, catalytic subunit, beta isoform | 403558 |
| PPP1CC | protein phosphatase 1, catalytic subunit, gamma isoform | 403557 |
| PPP1R12A | protein phosphatase 1, regulatory (inhibitor) subunit 12A | 475411 |
| RHOA | ras homolog gene family, member A | 403954 |
| LOC479252 | similar to Vinculin (Metavinculin) | 479252 |
| LOC479790 | similar to mitogen activated protein kinase 3 | 479790 |
| TLN1 | talin 1 | 474759 |
| TNC | tenascin C | 481689 |
| AKT2 | v-akt murine thymoma viral oncogene homolog 2 | 449021 |
| CRKL | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | 608125 |
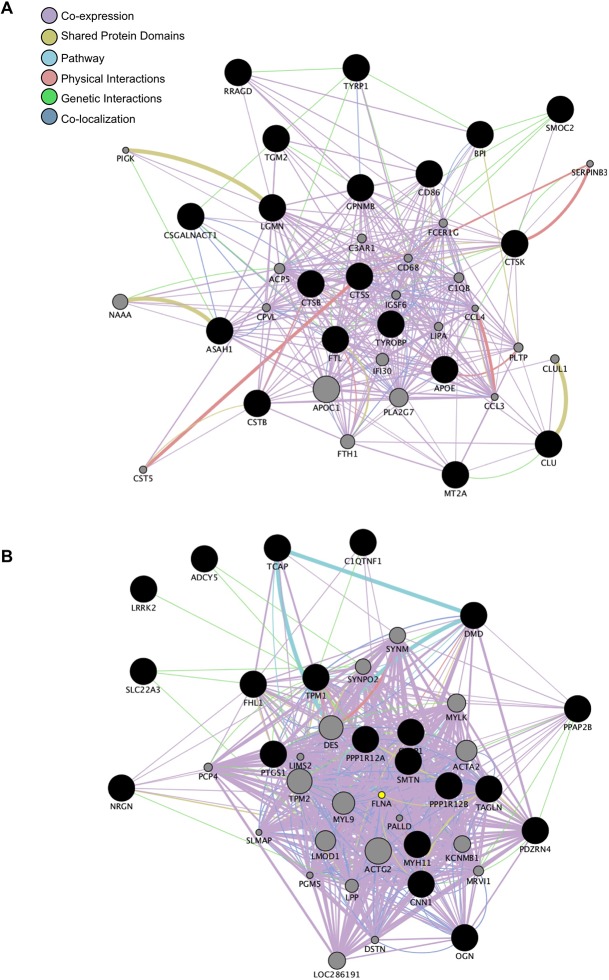
Next, gene networks were constructed, utilizing two methods to identify any potential interactions between genes with perturbed expression. The first was de novo using WGCNA, and the second utilized GeneMania. Signed networks were constructed in WGCNA using an adjacency function that incorporates gene correlation and connectivity across the dataset. GeneMania networks were constructed based on known interactions such as co-expression, shared protein domains, signaling pathways, physical interactions, genetic interactions, and co-localization. The results of the GeneMania-generated network analysis are shown in Fig 2, with correlated upregulated genes in the MPS I dogs in Fig 2A: notable genes exceeding 2-fold expression change in the upregulated network include lysosomal proteases (CTSB, CTSK, CTSS, LGMN); RRAGD, a GTPase involved with lysosomal energy sensing; immunity-related genes such as CD86, a macrophage-related signaling protein; BPI, a protein that binds bacterial lipopolysaccharide; and CLU, a marker previously identified in inflammatory processes and atherosclerosis. Fig 2B shows the correlated downregulated (reduced 2-fold) gene network in the MPS I dogs. Many of the downregulated genes are smooth muscle cytoskeletal elements, including TCAP (titin-cap), TPN1 (tropomyosin), DMD (dystrophin), SMTN (smoothelin), TAGLN (transgelin), MYH11 (smooth muscle myosin heavy chain 11), and CNN1 (smooth muscle calponin 1).
Fig 2. Up- and Downregulated Gene Networks in IDUA-/- Canine Arteries.
Gene networks formed using GeneMania using the top related genes and at most 20 attributes. Interactions observed include co-expression, co-localization, genetic interactions, pathway, and physical interactions. A. Network of the upregulated genes. Notable genes in the upregulated network include lysosomal proteases (CTSB, CTSK, CTSS, LGMN); RRAGD, a GTPase involved in lysosomal energy sensing; immunity-related genes such as CD86 and BPI; and clusterin, a protein associated with atherosclerotic cardiovascular disease. B. Network of the downregulated genes. The notable genes in the downregulated network are primarily smooth muscle cytoskeletal elements (CNN1, DMD, MYH11, SMTH, TAGLN, TCAP, and TPN1).
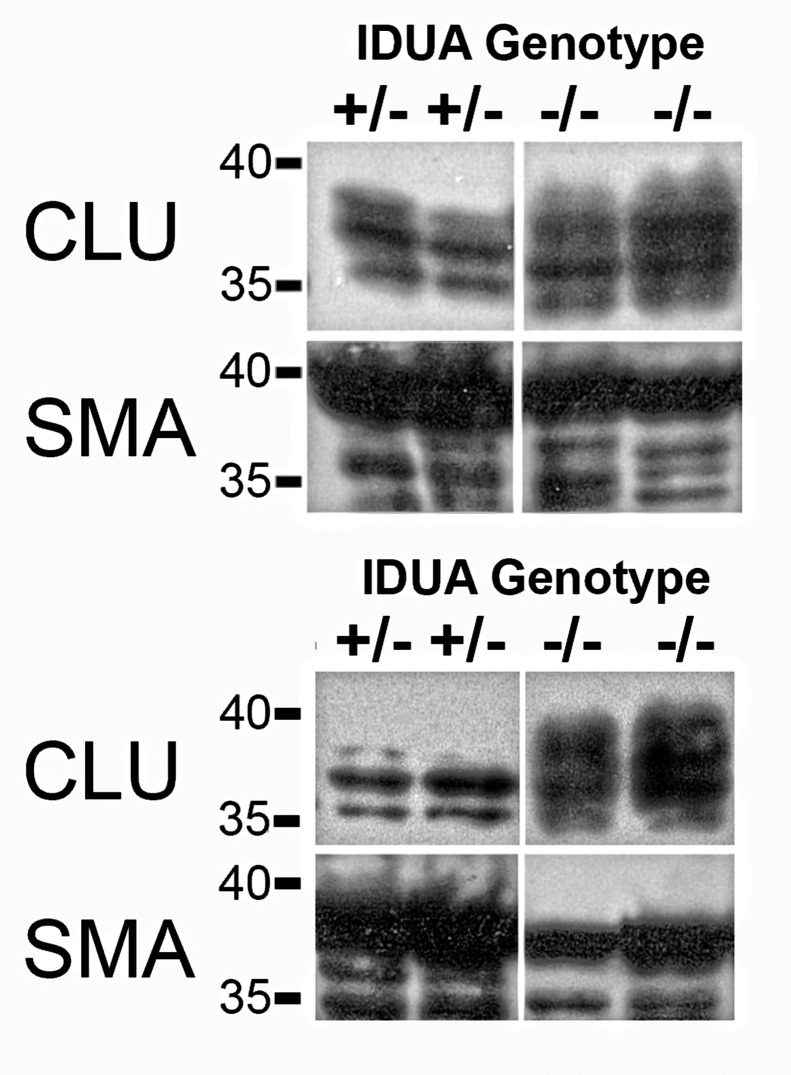
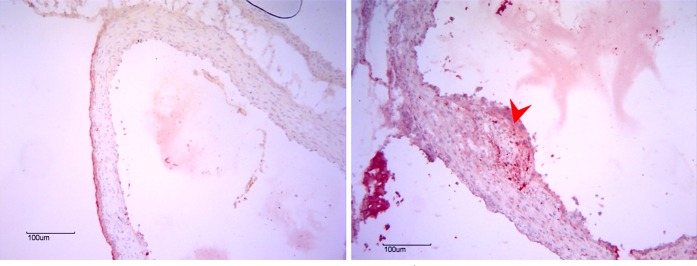
We further examined the expression pattern of clusterin, which was present in the upregulated gene network and the most significantly overexpressed gene (S3 Fig). The clusterin overexpression especially interested us, as it has been shown to be is involved in morphologic transformation of vascular smooth muscle cells [48] and present in human atherosclerotic plaques [49]. The MPS I dog aortas contained multiple eccentric plaques that intruded into the lumen, consistent with previous reports of vascular pathology in the model [23]. Importantly, we confirmed that clusterin protein is overexpressed in MPS I ascending (1.3-fold) and descending aortas (1.62-fold) compared to unaffected heterozygous dogs (Fig 3). The clusterin protein is localized primarily within the plaques of the MPS I aortas; little to no clusterin signal was seen in the control aortas (Fig 4). These results provide protein-level confirmation of clusterin gene overexpression within MPS I arterial vasculature. Additionally, Toll-like receptor 4 (TLR4) protein overexpression and increased phosphorylated STAT1 protein were seen only in canine MPS I arteries (S4 Fig).
Fig 3. Western Blot of Clusterin and Smooth Muscle Actin in Canine Aortas.
Clusterin is predicted to appear as a thicker band between 35–39 kDa owing to differential glycosylation. Alpha smooth muscle actin is predicted to appear at approximately 42 kDa. 20 μg of total protein was loaded in each lane. The upper set is from ascending aorta and the lower set is from descending aorta. Normalized mean clusterin band intensities from descending and ascending aortas are 1.62 and 1.3 fold greater in MPS I animals compared to unaffected animals.
Fig 4. Canine Arterial Clusterin Immunohistochemistry.
The MPS I aorta (right panel) has an overall increase in clusterin signal compared to unaffected aorta (left panel), and also contains an eccentric plaque (arrowhead) that stains strongly for clusterin.
Next, we assessed whether adult C57/Bl6 Idua-/- mice had similar cardiovascular findings to the IDUA-/- dogs, especially with regards to aortic histopathology. The mice had grossly evident hypertrophic cardiomyopathy compared to Idua+/- control mice, which is consistent with other assessments of cardiovascular phenotype [23]. While not as prominent as the canine MPS I model, the Idua-/- mice also demonstrated eccentric, sclerotic plaques and thickened proximal aortas compared to control mice. These plaques were visible only near the sinuses of Valsalva, a distribution different from the canine plaques, which were present throughout the ascending and descending aorta. Clusterin protein was absent in the control mice (Fig 5A), and prominently visible in the aortic plaques of the Idua-/- mice (Fig 5B). The full clusterin and smooth muscle actin blots can be viewed in S5 Fig.
Fig 5. Murine Aortic Clusterin Immunohistochemistry.
Taken at the level of the sinuses of Valsalva, the MPS I mouse aorta (right panel) is thicker than the wild-type mouse (left panel) and also contains an eccentric plaque (arrowhead) staining strongly for clusterin.
Discussion
We performed microarray analyses comparing arterial gene expression of untreated MPS I canines to unaffected heterozygous canines to gain insights into the gene perturbations caused by α-L-iduronidase deficiency and subsequent arterial glycosaminoglycan storage. Our study represents a comprehensive assessment of gene expression in the vasculature of any MPS model system, and identified upregulation of genes related to lysosomes, proteasomes, macrophages, and innate immunity. We also identified downregulation of cytoskeletal genes and calcium channel subunits. Our findings are consistent with results of gene expression studies from non-vascular tissues in differing MPS subtypes. This indicates the possibility that common mechanisms may be responsible for pathogenesis of MPS disease in multiple organ systems. We also identified evidence linking the innate immune system to pathogenesis of MPS I cardiovascular disease, as well as a biomarker for potentially following disease progression and treatment efficacy.
Overexpression of lysosomal hydrolases and housekeeping genes is consistent with the hypothesis that accumulation of primary substrate in lysosomal storage disorders results in a “traffic jam” with secondarily impaired degradation of other lysosomal substrates, and subsequent efforts by the cell to alleviate the secondary storage [50]. Our findings of cathepsin protease overexpression are consistent with aortic cathepsin B, D, L, and S overexpression from the mouse and canine models of MPS I and VII [31, 33, 34]. In addition, overexpression of lysosomal membrane proteins and enzymes has been observed in murine MPS VII liver [47] and brain [29]. An additional effect of the lysosomal traffic jam is impairment in organelle and protein degradation via a lysosome-dependent pathway known as autophagy [51]. Since the ubiquitin-proteasome system (UPS) is another prominent mechanism for recycling of accumulated proteins, the observed upregulation of proteasomal genes may indicate diversion of protein catabolism towards the UPS.
Downregulation of cellular adhesion and cytoskeletal genes (labeled as “Dilated Cardiomyopathy,” “Focal Adhesion,” and “Arrhythmogenic Right Ventricular Cardiomyopathy" in our study) has also been seen in MPS IIIb mouse brains [30]. These results support data suggesting that excess HS oligosaccharides impair function of cellular adhesion molecules and disrupt normal polarization and orientation of cultured MPSIIIB mouse astrocytes or neural stem cells [52].
The generalized upregulation of macrophage and immunity-related genes observed in the MPS I arteries indicate that inflammation may play a prominent role in the pathogenesis of MPS I cardiovascular disease. This immune-associated gene overexpression was also seen in several expression studies in MPS IIIb mice brains [26, 30] and is consistent with neuroinflammation observed in murine MPS IIIb and other neuropathic lysosomal storage disorders, including Sandhoff Disease and MPS I [53, 54]. While exploring potential etiologies of MPS I cardiovascular disease, significant overexpression of the Toll-like receptor 4 gene, in addition to numerous cathepsin proteases and matrix metalloproteinases was observed [34]. When MPS VII / cathepsin S and MPS VII / MMP-12 double knockout mice continued to develop arterial elastin degradation and aortic root dilatation, it was concluded that other proteases or inflammatory signals transduced by TLR4 activation could be responsible for the MPS VII cardiovascular phenotype [31].
TLR4 is a transmembrane protein that belongs to a family of proteins known as Pattern Recognition Receptors (PRRs). PRRs are expressed by cells of the innate immune system, bind foreign molecules that are typically associated with infections (pathogen-associated molecular patterns or PAMPs) or molecules released by damaged cells (damage-associated molecular patterns or DAMPs), and activate an immune response [55]. Lipopolysaccharide, the canonical PAMP for bacteria, activates TLR4 signaling. Acting via the MyD88 adaptor and STAT1 protein phosphorylation, activated TLR4 results in nuclear translocation of the transcription factor NF-κB and subsequent mRNA expression of proinflammatory cytokines.
Following the discovery that heparan sulfate, but neither chondroitin sulfate nor heparin, activates TLR4 signaling in dendritic cells and macrophages, evidence has been accumulating that heparan sulfate may play a role in the pathogenesis of MPS neurologic and orthopedic disease via TLR4-induced inflammation [56–58]. Wild-type mouse microglia respond in a dose-dependent fashion to HS-oligosaccharides isolated from MPS IIIB patients with overexpression of macrophage inflammatory protein 1α (Mip1a or Ccl3) and interleukin 1β mRNAs. There was significantly reduced cytokine mRNA expression when the same HS-oligosaccharides were administered to Tlr4-/- or MyD88-/- mouse microglia. MPS IIIb (Naglu-/-) mouse brains overexpress Mip1a mRNA as early as postnatal day 10 with concomitant microglial activation and neuroinflammation. Naglu / Tlr4 and Naglu / MyD88 double knockout mice did not overexpress Mip1a and demonstrated delayed neuroinflammation [33]. Similar neuroinflammatory findings have been reported in MPS IIIa, IIIb, and IIIc mice, all of which accumulate HS [28].
Our results suggest that HS–TLR4 mediated, monocyte/macrophage-induced inflammation contributes to the pathogenesis of cardiovascular disease in MPS I. Among the genes with highest overexpression in MPS I arteries was CD86 which, in conjunction with another overexpressed gene CD80, encode proteins found on the cell surface of activated monocytes. Other immunity-related genes with observed overexpression were members of the major histocompatibility complex II families, which are involved in antigen presentation and are found primarily on monocytes, macrophages, and dendritic cells. In addition, overexpression of the inflammatory cytokines interleukin-1α, interleukin-1β, interleukin 2, and interleukin 6 were observed. We also identified MPS I arterial overexpression of TLR4 protein and evidence of its activation via increased levels of phosphorylated STAT1.
Heparan sulfate-mediated inflammation cannot be the sole etiology of MPS cardiovascular disease pathogenesis. Patients who have MPS type VI store only DS GAG but often develop mitral or aortic valve dysfunction severe enough to necessitate surgical valve replacement, and also demonstrate coronary artery intimal thickening with arterial stiffness [2, 19,59]. Although the effects of DS GAG on TLR4 in MPS have not been directly investigated, there are studies that indicate it may also serve as a PAMP for TLR4 and promote inflammation like HS GAGs [60, 61]. Post-mortem studies of several MPS IVa patients, who store neither HS nor DS but accumulate keratan and chondroitin sulfate GAGs, have also demonstrated coronary sclerosis or aortic intimal thickening with macrophage infiltration and elastin fibril disruption [62, 63].
As our primary focus was canine MPS I arterial disease, mRNA and protein quantification of murine aorta clusterin or pSTAT1 were not performed. Despite this limitation, our study provides evidence from both canine and murine models confirming clusterin protein overexpression within MPS I arterial lesions. We believe that clusterin may have utility as an in vivo biomarker of MPS I cardiovascular disease for several reasons. First, overexpression of clusterin has been observed in many inflammatory conditions, including human atheromatous atherosclerosis, where it is observed only in vascular smooth muscle cells and stroma of atheromas and not in normal aorta [64, 65]. Second, while the effects of clusterin overexpression on human cardiovascular disease are unclear and may be pro-atherosclerotic [66] or protective [67, 68], clusterin serum levels correlate with severity of coronary artery stenosis and are highest during active myocardial infarction [69, 70]. Although lipid-mediated atheroma formation and atherosclerosis appear to proceed via pathways distinct from what occurs in MPS I, the roles of macrophages, inflammation, and Toll-like receptors in promoting atherosclerosis are well known [71]. Further studies are required to determine if circulating clusterin levels are indeed elevated in human MPS I patients, and if those levels correlate with severity of their cardiovascular disease.
Because GAGs are naturally present in healthy aortas [72], it will be important in the design of MPS cardiovascular disease therapeutics to determine if the arterial inflammation is caused solely by a quantitative increase of GAGs, or if the GAGs stored in MPS possess additional qualitative pro-inflammatory effects. The presence of storage-induced inflammation within MPS I canine arteries suggests several possible therapeutic strategies. Modified IDUA enzymes are being developed that have the potential to permeate deeper into the vascular parenchyma to alleviate GAG storage. An alternative strategy is the development of small molecule therapies that diffuse into arterial parenchyma and block the inflammatory response leading to MPS cardiovascular disease via inhibition of TLR4 signaling or pro-inflammatory cytokines. In fact, preclinical studies have been performed to investigate each of these modalities [73–76], and human clinical trials are in progress (European Union Clinical Trials Register # 2014-000350-11). Clusterin represents a promising biomarker to monitor efficacy of these therapeutic candidates in both preclinical and clinical studies for MPS I cardiovascular disease. The murine MPS I model system can be utilized to test efficacy of potential therapeutics targeted towards MPS cardiovascular disease.
Supporting Information
This protocol was utilized for identifying the Idua genotype of the mice utilized in this study.
(PDF)
The control samples cluster together and the IDUA-/- canine aorta samples cluster together. The same results are found if data from each tissue type is analyzed separately for up- and down-regulated genes, respectively.
(TIF)
Modules are designated on the left and the trait relationship is on the bottom (A for ascending aorta, C for carotid artery, and D for descending aorta). The table shows the correlation (top value) and the significance (bottom value) for each module-trait relationship. The turquoise and blue modules show the highest significance and thus were used for further analysis.
(TIF)
In the turquoise and blue modules, further assessment identified the top genes based on fold- change and p-value using Student’s t-test. A. Volcano plot of the–log10 of the p-value vs. log2 of fold change. B. Top up- and down-regulated genes in the module are represented in a bar graph with a log2 scale.
(TIF)
Immunohistochemistry for demonstrates overexpression of TLR4 in canine MPS I aorta, but not in unaffected canine aorta. The presence of increased pSTAT1 in canine MPS I aorta compared to unaffected canine aorta is evidence for TLR4 activation.
(TIF)
Scan of raw radiograph (Thermo Scientific CL-XPosure Film, ThermoFisher Scientific, Waltham, MA) exposure depicting enhanced chemiluminescence detection of canine aorta WB probed with anti-human clusterin alpha chain (EMD Millipore, Billerica, MA) and with anti-human smooth muscle alpha-actin (Dako North America, Inc., Carpinteria, CA). Hand written annotations refer to specific animal identifiers marking the lanes. Samples from descending or ascending aorta are grouped together and labeled as IDUA+/- (carrier) or IDUA-/- (MPS I). Handwritten markings indicate size and position of color coded protein ladder size markers (PageRuler Prestained Protein Ladder, Life Technologies, Grand Island, NY) traced by overlaying the film onto the transfer membrane. Clusterin is predicted to appear as a band between 35–39 kDa owing to differential glycosylation, while alpha smooth muscle actin is predicted to appear at approximately 42 kDa.
(TIF)
The full list of mRNA probes showing significant (p-value ≤ 0.05) and greater than 2-fold level of upregulation in IDUA-/- canine arteries, compared to controls. Note that some genes are represented more than once due to existence of multiple mRNA probes per gene.
(XLSX)
The full list of mRNA probes showing significant (p-value ≤ 0.05) and greater than 2-fold level of down-regulation in IDUA-/- canine arteries, compared to controls. Again, some genes are represented more than once due to existence of multiple mRNA probes per gene.
(XLSX)
Acknowledgments
We are grateful to Stanley Karsten, Ph.D., and Lili C. Kudo, Ph.D., for their technical assistance with the microarray setup and run. The Lysosomal Disease Network is a part of the National Institutes of Health (NIH) Rare Diseases Clinical Research Network (RDCRN), supported through collaboration between the NIH Office of Rare Diseases Research (ORDR) at the National Center for Advancing Translational Science (NCATS), the National Institute of Neurological Disorders and Stroke (NINDS) and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data Availability
In addition to data within paper and supporting information, expression data are uploaded to the NCBI GEO database (accession number: GSE78889).
Funding Statement
This work was supported by National Institutes of Health (NIH) grant T01NS05242-11 (NME), Children’s Hospital of Orange County Pediatric Subspecialty Faculty and the MPSI Research Foundation (RYW), Children’s Hospital of Orange County (OK and PHS), European Community FP7 Award, PIOF-GA-2010-273994 (PG), and the Lysosomal Disease Network U54-NS065768 (MV). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Neufeld EF, Muenzer J. The Online Metabolic and Molecular Bases of Inherited Diseases. Valle D, Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, et al. , editors. New York: McGraw-Hill; 2013. [Google Scholar]
- 2.Braunlin E, Orchard PJ, Whitley CB, Schroeder L, Reed RC, Manivel JC. Unexpected coronary artery findings in mucopolysaccharidosis. Report of four cases and literature review. Cardiovasc Pathol. 2014;23:145–151. 10.1016/j.carpath.2014.01.001 [DOI] [PubMed] [Google Scholar]
- 3.Brosius FC, Roberts WC. Coronary artery disease in the Hurler syndrome. Qualitative and quantitative analysis of the extent of coronary narrowing at necropsy in six children. Am J Cardiol. 1981;47:649–653. [DOI] [PubMed] [Google Scholar]
- 4.Cheitlin MD, McAllister HA, de Castro CM. Myocardial infarction without atherosclerosis. JAMA. 1975;231:951–959. [PubMed] [Google Scholar]
- 5.Leal GN, de Paula AC, Leone C, Kim CA. Echocardiographic study of paediatric patients with mucopolysaccharidosis. Cardiol Young. 2010;20:254–261. 10.1017/S104795110999062X [DOI] [PubMed] [Google Scholar]
- 6.Renteria VG, Ferrans VJ, Roberts WC. The heart in the Hurler syndrome: gross, histologic and ultrastructural observations in five necropsy cases. Am J Cardiol. 1976;38:487–501. [DOI] [PubMed] [Google Scholar]
- 7.Aldenhoven M, Wynn RF, Orchard PJ, O'Meara A, Veys P, Fischer A, et al. Long-term outcome of Hurler syndrome patients after hematopoietic cell transplantation: an international multicenter study. Blood. 2015;125:2164–2172. 10.1182/blood-2014-11-608075 [DOI] [PubMed] [Google Scholar]
- 8.Sifuentes M, Doroshow R, Hoft R, Mason G, Walot I, Diament M, et al. A follow-up study of MPS I patients treated with laronidase enzyme replacement therapy for 6 years. Mol Genet Metab. 2007;90:171–180. [DOI] [PubMed] [Google Scholar]
- 9.Wraith JE. Limitations of enzyme replacement therapy: current and future. J Inherit Metab Dis. 2006;29:442–447. [DOI] [PubMed] [Google Scholar]
- 10.Wraith JE, Clarke LA, Beck M, Kolodny EH, Pastores GM, Muenzer J, et al. Enzyme replacement therapy for mucopolysaccharidosis I: a randomized, double-blinded, placebo-controlled, multinational study of recombinant human alpha-L-iduronidase (laronidase). J Pediatr. 2004;144:581–588. [DOI] [PubMed] [Google Scholar]
- 11.Braunlin EA, Berry JM, Whitley CB. Cardiac findings after enzyme replacement therapy for mucopolysaccharidosis type I. Am J Cardiol. 2006;98:416–418. [DOI] [PubMed] [Google Scholar]
- 12.Fesslova V, Corti P, Sersale G, Rovelli A, Russo P, Mannarino S, et al. The natural course and the impact of therapies of cardiac involvement in the mucopolysaccharidoses. Cardiol Young. 2009;19:170–178. 10.1017/S1047951109003576 [DOI] [PubMed] [Google Scholar]
- 13.Lin HY, Lin SP, Chuang CK, Chen MR, Chen BF, Wraith JE. Mucopolysaccharidosis I under enzyme replacement therapy with laronidase—a mortality case with autopsy report. J Inherit Metab Dis. 2005;28:1146–1148. [DOI] [PubMed] [Google Scholar]
- 14.Yano S, Moseley K, Pavlova Z. Postmortem studies on a patient with mucopolysaccharidosis type I: histopathological findings after one year of enzyme replacement therapy. J Inherit Metab Dis. 2009;32:S53–57. 10.1007/s10545-009-1057-4 [DOI] [PubMed] [Google Scholar]
- 15.Kelly AS, Metzig AM, Steinberger J, Braunlin EA. Endothelial function in children and adolescents with mucopolysaccharidosis. J Inherit Metab Dis. 2013;36:221–225. 10.1007/s10545-011-9438-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemes A, Timmermans RG, Wilson JH, Soliman OI, Krenning BJ, ten Cate FJ, et al. The mild form of mucopolysaccharidosis type I (Scheie syndrome) is associated with increased ascending aortic stiffness. Heart Vessels. 2008;23:108–111. 10.1007/s00380-007-1013-x [DOI] [PubMed] [Google Scholar]
- 17.Wang RY, Braunlin EA, Rudser KD, Dengel DR, Metzig AM, Covault KK, et al. Carotid intima-media thickness is increased in patients with treated mucopolysaccharidosis types I and II, and correlates with arterial stiffness. Mol Genet Metab. 2014;111:128–132. 10.1016/j.ymgme.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang RY, Covault KK, Halcrow EM, Gardner AJ, Cao X, Newcomb RL, et al. Carotid intima-media thickness is increased in patients with mucopolysaccharidoses. Mol Genet Metab. 2011;104:592–596. 10.1016/j.ymgme.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang RY, Rudser KD, Dengel DR, Braunlin EA, Steinberger J, Jacobs DR, et al. Carotid intima-media thickness and arterial stiffness of pediatric mucopolysaccharidosis patients are increased compared to both pediatric and adult populations. Mol Genet Metab. 2015;114:S123–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gompf RE, Shull RM, Breider MA, Scott JA, Constantopoulos GC. Cardiovascular changes after bone marrow transplantation in dogs with mucopolysaccharidosis I. Am J Vet Res. 1990;51:2054–2060. [PubMed] [Google Scholar]
- 21.Kakkis ED, McEntee MF, Schmidtchen A, Neufeld EF, Ward DA, Gompf RE, et al. Long-term and high-dose trials of enzyme replacement therapy in the canine model of mucopolysaccharidosis I. Biochem Mol Med. 1996;58:156–167. [DOI] [PubMed] [Google Scholar]
- 22.Shull RM, Helman RG, Spellacy E, Constantopoulos G, Munger RJ, Neufeld EF. Morphologic and biochemical studies of canine mucopolysaccharidosis I. Am J Pathol. 1984;114:487–495. [PMC free article] [PubMed] [Google Scholar]
- 23.Lyons JA, Dickson PI, Wall JS, Passage MB, Ellinwood NM, Kakkis ED, et al. Arterial pathology in canine mucopolysaccharidosis-I and response to therapy. Lab Invest. 2011;91:665–674. 10.1038/labinvest.2011.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braunlin E, Mackey-Bojack S, Panoskaltsis-Mortari A, Berry JM, McElmurry RT, Riddle M, et al. Cardiac functional and histopathologic findings in humans and mice with mucopolysaccharidosis type I: implications for assessment of therapeutic interventions in hurler syndrome. Pediatr Res. 2006;59:27–32. [DOI] [PubMed] [Google Scholar]
- 25.Jordan MC, Zheng Y, Ryazantsev S, Rozengurt N, Roos KP, Neufeld EF. Cardiac manifestations in the mouse model of mucopolysaccharidosis I. Mol Genet Metab. 2005;86:233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiRosario J, Divers E, Wang C, Etter J, Charrier A, Jukkola P, et al. Innate and adaptive immune activation in the brain of MPS IIIB mouse model. J Neurosci Res. 2009;87:978–990. 10.1002/jnr.21912 [DOI] [PubMed] [Google Scholar]
- 27.Killedar S, Dirosario J, Divers E, Popovich PG, McCarty DM, Fu H. Mucopolysaccharidosis IIIB, a lysosomal storage disease, triggers a pathogenic CNS autoimmune response. J Neuroinflammation. 2010;7:39 10.1186/1742-2094-7-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martins C, Hulkova H, Dridi L, Dormoy-Raclet V, Grigoryeva L, Choi Y, et al. Neuroinflammation, mitochondrial defects and neurodegeneration in mucopolysaccharidosis III type C mouse model. Brain. 2015;138:336–355. 10.1093/brain/awu355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parente MK, Rozen R, Cearley CN, Wolfe JH. Dysregulation of gene expression in a lysosomal storage disease varies between brain regions implicating unexpected mechanisms of neuropathology. PLoS One. 2012;7:e32419 10.1371/journal.pone.0032419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villani GR, Gargiulo N, Faraonio R, Castaldo S, Gonzalez YRE, Di Natale P. Cytokines, neurotrophins, and oxidative stress in brain disease from mucopolysaccharidosis IIIB. J Neurosci Res. 2007;85:612–622. [DOI] [PubMed] [Google Scholar]
- 31.Baldo G, Wu S, Howe RA, Ramamoothy M, Knutsen RH, Fang J, et al. Pathogenesis of aortic dilatation in mucopolysaccharidosis VII mice may involve complement activation. Mol Genet Metab. 2011;104:608–619. 10.1016/j.ymgme.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Xu L, Hennig AK, Kovacs A, Fu A, Chung S, et al. Liver-directed neonatal gene therapy prevents cardiac, bone, ear, and eye disease in mucopolysaccharidosis I mice. Mol Ther. 2005;11:35–47. [DOI] [PubMed] [Google Scholar]
- 33.Ausseil J, Desmaris N, Bigou S, Attali R, Corbineau S, Vitry S, et al. Early neurodegeneration progresses independently of microglial activation by heparan sulfate in the brain of mucopolysaccharidosis IIIB mice. PLoS One. 2008;3:e2296 10.1371/journal.pone.0002296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metcalf JA, Linders B, Wu S, Bigg P, O'Donnell P, Sleeper MM, et al. Upregulation of elastase activity in aorta in mucopolysaccharidosis I and VII dogs may be due to increased cytokine expression. Mol Genet Metab. 2010;99:396–407. 10.1016/j.ymgme.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menon KP, Tieu PT, Neufeld EF. Architecture of the canine IDUA gene and mutation underlying canine mucopolysaccharidosis I. Genomics. 1992;14:763–768 [DOI] [PubMed] [Google Scholar]
- 36.Wang RY, Aminian A, McEntee MF, Kan SH, Simonaro CM, Lamanna WC, et al. Intra-articular enzyme replacement therapy with rhIDUA is safe, well-tolerated, and reduces articular GAG storage in the canine model of mucopolysaccharidosis type I. Mol Genet Metab. 2014;112:286–293. 10.1016/j.ymgme.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daugherty A, Whitman SC. Quantification of atherosclerosis in mice. Methods Mol Biol. 2003;209:293–309. [DOI] [PubMed] [Google Scholar]
- 38.Khalid O, Kim JJ, Duan L, Hoang M, Elashoff D, Kim Y. Genome-wide transcriptomic alterations induced by ethanol treatment in human dental pulp stem cells (DPSCs). Genom Data. 2014;2:127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khalid O, Kim JJ, Kim HS, Hoang M, Tu TG, Elie O, et al. Gene expression signatures affected by alcohol-induced DNA methylomic deregulation in human embryonic stem cells. Stem Cell Res. 2014;12:791–806. 10.1016/j.scr.2014.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim JJ, Khalid O, Namazi A, Tu TG, Elie O, Lee C, et al. Discovery of consensus gene signature and intermodular connectivity defining self-renewal of human embryonic stem cells. Stem Cells. 2014;32:1468–1479. 10.1002/stem.1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JJ, Khalid O, Vo S, Sun HH, Wong DT, Kim Y. A novel regulatory factor recruits the nucleosome remodeling complex to wingless integrated (Wnt) signaling gene promoters in mouse embryonic stem cells. J Biol Chem. 2012;287:41103–41117. 10.1074/jbc.M112.416545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, et al. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics. 2009;Chapter 13:Unit 13 1. [DOI] [PubMed] [Google Scholar]
- 43.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 44.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20 10.1093/nar/gkq537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woloszynek JC, Roberts M, Coleman T, Vogler C, Sly W, Semenkovich CF, et al. Numerous transcriptional alterations in liver persist after short-term enzyme-replacement therapy in a murine model of mucopolysaccharidosis type VII. Biochem J. 2004;379:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diemer V, Hoyle M, Baglioni C, Millis AJ. Expression of porcine complement cytolysis inhibitor mRNA in cultured aortic smooth muscle cells. Changes during differentiation in vitro. J Biol Chem. 1992;267:5257–5264. [PubMed] [Google Scholar]
- 49.Jordan-Starck TC, Lund SD, Witte DP, Aronow BJ, Ley CA, Stuart WD, et al. Mouse apolipoprotein J: characterization of a gene implicated in atherosclerosis. J Lipid Res. 1994;35:194–210. [PubMed] [Google Scholar]
- 50.Schulze H, Sandhoff K. Lysosomal lipid storage diseases. Cold Spring Harb Perspect Biol. 2011;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–129. [DOI] [PubMed] [Google Scholar]
- 52.Bruyere J, Roy E, Ausseil J, Lemonnier T, Teyre G, Bohl D, et al. Heparan sulfate saccharides modify focal adhesions: implication in mucopolysaccharidosis neuropathophysiology. J Mol Biol. 2015;427:775–791. 10.1016/j.jmb.2014.09.012 [DOI] [PubMed] [Google Scholar]
- 53.Ohmi K, Greenberg DS, Rajavel KS, Ryazantsev S, Li HH, Neufeld EF. Activated microglia in cortex of mouse models of mucopolysaccharidoses I and IIIB. Proc Natl Acad Sci U S A. 2003;100:1902–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wada R, Tifft CJ, Proia RL. Microglial activation precedes acute neurodegeneration in Sandhoff disease and is suppressed by bone marrow transplantation. Proc Natl Acad Sci U S A. 2000;97:10954–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. 10.1016/j.cyto.2008.01.006 [DOI] [PubMed] [Google Scholar]
- 56.Johnson GB, Brunn GJ, Kodaira Y, Platt JL. Receptor-mediated monitoring of tissue well-being via detection of soluble heparan sulfate by Toll-like receptor 4. J Immunol. 2002;168:5233–5239. [DOI] [PubMed] [Google Scholar]
- 57.Archer LD, Langford-Smith KJ, Bigger BW, Fildes JE. Mucopolysaccharide diseases: a complex interplay between neuroinflammation, microglial activation and adaptive immunity. J Inherit Metab Dis. 2014;37:1–12. 10.1007/s10545-013-9613-3 [DOI] [PubMed] [Google Scholar]
- 58.Simonaro CM, Ge Y, Eliyahu E, He X, Jepsen KJ, Schuchman EH. Involvement of the Toll-like receptor 4 pathway and use of TNF-alpha antagonists for treatment of the mucopolysaccharidoses. Proc Natl Acad Sci U S A. 2010;107:222–227. 10.1073/pnas.0912937107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kampmann C, Lampe C, Whybra-Trumpler C, Wiethoff CM, Mengel E, Arash L, et al. Mucopolysaccharidosis VI: cardiac involvement and the impact of enzyme replacement therapy. J Inherit Metab Dis. 2014;37:269–276. 10.1007/s10545-013-9649-4 [DOI] [PubMed] [Google Scholar]
- 60.Moreth K, Iozzo RV, Schaefer L. Small leucine-rich proteoglycans orchestrate receptor crosstalk during inflammation. Cell Cycle. 2012;11:2084–2091. 10.4161/cc.20316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaefer L, Babelova A, Kiss E, Hausser HJ, Baliova M, Krzyzankova M, et al. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. J Clin Invest. 2005;115:2223–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Factor SM, Biempica L, Goldfischer S. Coronary intimal sclerosis in Morquio's syndrome. Virchows Arch A Pathol Anat Histol. 1978;379:1–10. [DOI] [PubMed] [Google Scholar]
- 63.Yasuda E, Fushimi K, Suzuki Y, Shimizu K, Takami T, Zustin J, et al. Pathogenesis of Morquio A syndrome: an autopsied case reveals systemic storage disorder. Mol Genet Metab. 2013;109:301–311. 10.1016/j.ymgme.2013.04.009 [DOI] [PubMed] [Google Scholar]
- 64.Ishikawa Y, Akasaka Y, Ishii T, Komiyama K, Masuda S, Asuwa N, et al. Distribution and synthesis of apolipoprotein J in the atherosclerotic aorta. Arterioscler Thromb Vasc Biol. 1998;18:665–672. [DOI] [PubMed] [Google Scholar]
- 65.Ishikawa Y, Ishii T, Akasaka Y, Masuda T, Strong JP, Zieske AW, et al. Immunolocalization of apolipoproteins in aortic atherosclerosis in American youths and young adults: findings from the PDAY study. Atherosclerosis. 2001;158:215–225. [DOI] [PubMed] [Google Scholar]
- 66.Hamada N, Miyata M, Eto H, Ikeda Y, Shirasawa T, Akasaki Y, et al. Loss of clusterin limits atherosclerosis in apolipoprotein E-deficient mice via reduced expression of Egr-1 and TNF-alpha. J Atheroscler Thromb. 2011;18:209–216. [DOI] [PubMed] [Google Scholar]
- 67.Kim HJ, Yoo EK, Kim JY, Choi YK, Lee HJ, Kim JK, et al. Protective role of clusterin/apolipoprotein J against neointimal hyperplasia via antiproliferative effect on vascular smooth muscle cells and cytoprotective effect on endothelial cells. Arterioscler Thromb Vasc Biol. 2009;29:1558–1564. 10.1161/ATVBAHA.109.190058 [DOI] [PubMed] [Google Scholar]
- 68.Schwarz M, Spath L, Lux CA, Paprotka K, Torzewski M, Dersch K, et al. Potential protective role of apoprotein J (clusterin) in atherogenesis: binding to enzymatically modified low-density lipoprotein reduces fatty acid-mediated cytotoxicity. Thromb Haemost. 2008;100:110–118. 10.1160/TH07-12-0737 [DOI] [PubMed] [Google Scholar]
- 69.Poulakou MV, Paraskevas KI, Wilson MR, Iliopoulos DC, Tsigris C, Mikhailidis DP, et al. Apolipoprotein J and leptin levels in patients with coronary heart disease. In Vivo. 2008;22:537–542. [PubMed] [Google Scholar]
- 70.Trougakos IP, Poulakou M, Stathatos M, Chalikia A, Melidonis A, Gonos ES. Serum levels of the senescence biomarker clusterin/apolipoprotein J increase significantly in diabetes type II and during development of coronary heart disease or at myocardial infarction. Exp Gerontol. 2002;37:1175–1187. [DOI] [PubMed] [Google Scholar]
- 71.Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145:341–355. 10.1016/j.cell.2011.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yla-Herttuala S, Solakivi T, Hirvonen J, Laaksonen H, Mottonen M, Pesonen E, et al. Glycosaminoglycans and apolipoproteins B and A-I in human aortas. Chemical and immunological analysis of lesion-free aortas from children and adults. Arteriosclerosis. 1987;7:333–340. [DOI] [PubMed] [Google Scholar]
- 73.Esko JD, Tong W, Hamil K, Tor Y. Intranasal enzyme replacement therapy in mice. Mol Genet Metab. 2015;114:S42–3. [Google Scholar]
- 74.Frohbergh M, Ge Y, Meng F, Karabul N, Solyom A, Lai A, et al. Dose responsive effects of subcutaneous pentosan polysulfate injection in mucopolysaccharidosis type VI rats and comparison to oral treatment. PLoS One. 2014;9:e100882 10.1371/journal.pone.0100882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schuchman EH, Ge Y, Lai A, Borisov Y, Faillace M, Eliyahu E, et al. Pentosan polysulfate: a novel therapy for the mucopolysaccharidoses. PLoS One. 2013;8:e54459 10.1371/journal.pone.0054459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simonaro CM, Tomatsu S, Ge Y, Meng F, Frohbergh M, Haskins ME, et al. Pentosan polysufate: New mechanistic insights and treatment of the mucopolysaccharidosis. Mor Genet Metab. 2015;114:S106–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This protocol was utilized for identifying the Idua genotype of the mice utilized in this study.
(PDF)
The control samples cluster together and the IDUA-/- canine aorta samples cluster together. The same results are found if data from each tissue type is analyzed separately for up- and down-regulated genes, respectively.
(TIF)
Modules are designated on the left and the trait relationship is on the bottom (A for ascending aorta, C for carotid artery, and D for descending aorta). The table shows the correlation (top value) and the significance (bottom value) for each module-trait relationship. The turquoise and blue modules show the highest significance and thus were used for further analysis.
(TIF)
In the turquoise and blue modules, further assessment identified the top genes based on fold- change and p-value using Student’s t-test. A. Volcano plot of the–log10 of the p-value vs. log2 of fold change. B. Top up- and down-regulated genes in the module are represented in a bar graph with a log2 scale.
(TIF)
Immunohistochemistry for demonstrates overexpression of TLR4 in canine MPS I aorta, but not in unaffected canine aorta. The presence of increased pSTAT1 in canine MPS I aorta compared to unaffected canine aorta is evidence for TLR4 activation.
(TIF)
Scan of raw radiograph (Thermo Scientific CL-XPosure Film, ThermoFisher Scientific, Waltham, MA) exposure depicting enhanced chemiluminescence detection of canine aorta WB probed with anti-human clusterin alpha chain (EMD Millipore, Billerica, MA) and with anti-human smooth muscle alpha-actin (Dako North America, Inc., Carpinteria, CA). Hand written annotations refer to specific animal identifiers marking the lanes. Samples from descending or ascending aorta are grouped together and labeled as IDUA+/- (carrier) or IDUA-/- (MPS I). Handwritten markings indicate size and position of color coded protein ladder size markers (PageRuler Prestained Protein Ladder, Life Technologies, Grand Island, NY) traced by overlaying the film onto the transfer membrane. Clusterin is predicted to appear as a band between 35–39 kDa owing to differential glycosylation, while alpha smooth muscle actin is predicted to appear at approximately 42 kDa.
(TIF)
The full list of mRNA probes showing significant (p-value ≤ 0.05) and greater than 2-fold level of upregulation in IDUA-/- canine arteries, compared to controls. Note that some genes are represented more than once due to existence of multiple mRNA probes per gene.
(XLSX)
The full list of mRNA probes showing significant (p-value ≤ 0.05) and greater than 2-fold level of down-regulation in IDUA-/- canine arteries, compared to controls. Again, some genes are represented more than once due to existence of multiple mRNA probes per gene.
(XLSX)
Data Availability Statement
In addition to data within paper and supporting information, expression data are uploaded to the NCBI GEO database (accession number: GSE78889).