Abstract
The SNARE complex composed of VAMP727, SYP22, VTI11 and SYP51 is critical for protein trafficking and PSV biogenesis in Arabidopsis. This SNARE complex directs the fusion between the prevacuolar compartment (PVC) and the vacuole, and thus mediates protein trafficking to the vacuole. In this study, we examined the role of AtNHX5 and AtNHX6 in regulating this SNARE complex and its function in protein trafficking. We found that AtNHX5 and AtNHX6 were required for seed production, protein trafficking and PSV biogenesis. We further found that the nhx5 nhx6 syp22 triple mutant showed severe defects in seedling growth and seed development. The triple mutant had short siliques and reduced seed sets, but larger seeds. In addition, the triple mutant had numerous smaller protein storage vacuoles (PSVs) and accumulated precursors of the seed storage proteins in seeds. The PVC localization of SYP22 and VAMP727 was repressed in nhx5 nhx6, while a significant amount of SYP22 and VAMP727 was trapped in the Golgi or TGN in nhx5 nhx6. AtNHX5 and AtNHX6 were co-localized with SYP22 and VAMP727. Three conserved acidic residues, D164, E188, and D193 in AtNHX5 and D165, E189, and D194 in AtNHX6, were essential for the transport of the storage proteins, indicating the importance of exchange activity in protein transport. AtNHX5 or AtNHX6 did not interact physically with the SNARE complex. Taken together, AtNHX5 and AtNHX6 are required for the PVC localization of the SNARE complex and hence its function in protein transport. AtNHX5 and AtNHX6 may regulate the subcellular localization of the SNARE complex by their transport activity.
Introduction
Seeds of crop plants are the principle source of proteins for humans and livestock [1–3]. Seed storage proteins are primarily accumulated in the protein storage vacuoles (PSVs) [4–7]. Thus, understanding the mechanism underlying the trafficking of the storage proteins to the PSVs is critical for improvements in the protein yield as well as protein quality of the crop seeds [2, 3].
Seed storage proteins are synthesized as precursors in the endoplasmic reticulum (ER). They are then transported into the PSVs where the precursors are converted to mature forms [8–10]. Two distinct pathways have been identified for the trafficking of storage proteins to the PSVs: the Golgi-dependent and -independent pathways [11–14]. In the Golgi-dependent pathway, proteins are transported through the Golgi and then dense vesicles before being delivered into the PSVs [15–19]. In the Golgi-independent pathway, however, proteins are transported from the ER to the PSVs through the precursor-accumulating (PAC) vesicles [11, 13].
The molecular machinery involved in protein trafficking to the PSVs is beginning to be explored. VSR1, VSR3 and VSR4 have been shown to function as the vacuolar sorting receptors (VSR) of the PSVs in Arabidopsis [20–22]. RMR1, the receptor homology region transmembrane domain ring H2 motif protein, is another type of sorting receptors for the trafficking to the PSVs in Arabidopsis [19, 23]. PV72 was reported to be the pumpkin VSR on the membranes of the PAC vesicles [24]. AtVPS29, a member of the retromer complex that recycles VPS10 from the prevacuolar compartment (PVC) to the Golgi, is involved in recycling VSRs for the sorting of storage proteins [7, 25]. In addition, the SNARE complex composed of VAMP727, SYP22, VTI11, and SYP51, which mediates the fusion between the PVC and the vacuole, is crucial for protein transport into the PSVs in Arabidopsis [26].
Cellular pH is a key regulatory factor of protein trafficking in both the secretory and endocytic pathways [27–30]. Studies have shown that the organelles become more acidic along the process of maturation in the exocytic or endocytic pathways in both the plant and animal cells [27, 28, 31]. In yeast and animals, the acidity of the organelles is generated by the vacuolar-type H+-ATPases (V-ATPases) [27, 32]. Moreover, Na+/H+ antiporters (NHXs) in animals conduct proton leak to counter organelle acidification in order to maintain an optimal pH [33, 34]. In plants, however, the acidic pH of the organelles is maintained by the proton pumps V-ATPases and pyrophosphatase [30, 31]. Similar to their animal counterparts, the Golgi/TGN-localized plant NHX antipoters, AtNHX5 and AtNHX6, may act as a H+-leak system to counter the luminal acidification [31].
Studies have shown that protein trafficking to the PSVs is controlled by pH and requires V-ATPases and NHX antiporters: (1) The binding of the VSR to its ligand is pH-dependent; the binding occurs from pH 4.0 to 7.0, with an optimal binding at pH 6.0 [35]; (2) V-ATPase is required for the sorting of soluble vacuolar protein precursors in tobacco cells [36]; (3) The antibiotic Na+/H+ antiporter monensin, which causes the acidification of the TGN, affects the vacuolar transport of the seed storage proteins [37, 38]; and (4) AtNHX5 and AtNHX6 are required for the transport of the seed storage proteins into the vacuole as well as processing of the seed storage proteins in Arabidopsis [39,40].
Plant NHX antiporters are membrane proteins that transport protons (H+) across a membrane in exchange for Na+ or K+. Studies show that plant NHX antiporters are critical for cellular ion homeostasis and pH regulation, and play significant roles in diverse cellular processes, including pH homeostasis, Na+ and K+ movement, vesicle trafficking and fusion, growth and development, and salt tolerance [41–45]. The Arabidopsis NHX gene family contains 8 members that are divided into three distinct classes: vacuolar (AtNHX1-AtNHX4), endosomal (AtNHX5 and AtNHX6), and plasma membrane (AtNHX7/SOS1and AtNHX8) [41, 43, 46]. AtNHX5 and AtNHX6 are localized in the Golgi, TGN, and PVC, and share high sequence similarity (78.7%) [39, 41, 47]. AtNHX5 and AtNHX6 play an important role in plant growth and development [48, 49]. nhx5 nhx6 double mutant showed profound defects in growth and development. nhx5 nhx6 had smaller rosettes and shorter seedlings, and was flowering and bolting late [48, 49]. These results demonstrate that AtNHX5 and AtNHX6 are essential for the growth and development in Arabidopsis.
How the trafficking of seed storage proteins is controlled by AtNHX5 and AtNHX6, however, remains unclear. Reguera et al. (2015) found that the binding of VSR to its cargoes was reduced in nhx5 nhx6 [39]. This discovery suggested that AtNHX5 and AtNHX6 might function in controlling the interaction between VSR and its cargoes [39]. In addition, Ashnest et al. (2015) showed that AtNHX6 interacted with SNX1, a component of the Retromer. Retromer is the cellular sorting machinery that recycles VSRs back to the ER from the TGN [40]. Therefore, these findings from Ashnest et al. (2015) suggest that AtNHX5 and AtNHX6 might regulate the recycling of VSRs. Considering that AtNHX5 and AtNHX6 are localized in Golgi, TGN, and PVC, key players of the protein trafficking pathway, their function may not be limited to regulating VSR binding activity [4, 39, 50]. They may have diversified function. As mentioned above, the four SNAREs, VAMP727, SYP22, VTI11, and SYP51, play a crucial role in vacuolar transport, seed maturation, and vacuole biogenesis [26]. They form a complex that mediates the fusion between the PVC and the vacuole, and through which proteins are delivered into the vacuole [26]. Notably, these four SNAREs are localized to the PVC, the same organelle where AtNHX5 and AtNHX6 reside. Then, it is interesting to understand whether this SNARE complex is regulated by AtNHX5 and AtNHX6.
In this study, we aim to examine the role of AtNHX5 and AtNHX6 in regulating the SNARE complex of VAMP727, SYP22, VTI11, and SYP51. We started with genetic analysis for the double mutant nhx5 nhx6 and the triple mutant nhx5 nhx6 syp22. We found that AtNHX5 and AtNHX6 were required for seed production, protein trafficking and PSV biogenesis. We further found that the triple mutant showed severe defects in seedling growth and seed development. The PSV was smaller but its number was increased in the triple mutant. The precursors of the seed storage proteins were accumulated in the triple mutant. We further found that the PVC localization of the SNARE proteins SYP22 and VAMP727 was repressed in nhx5 nhx6. We identified three conserved acidic residues that were essential for the transport of the storage proteins. But AtNHX5 and AtNHX6 did not interact physically with these SNARE molecules. These results suggest that AtNHX5 and AtNHX6 regulate the subcellular localization of the SNARE complex and thus its function in protein transport.
Results
Seed production is altered in nhx5 nhx6
In order to investigate the role of AtNHX5 and AtNHX6 in controlling the trafficking of the seed storage proteins, we examined silique growth and seed production of the nhx5 nhx6 double mutant. The nhx5 nhx6 double mutants were generated in our previous study [49]. Briefly, we obtained one T-DNA line for the AtNHX5 gene (nhx5-1) and two separate T-DNA lines for the AtNHX6 gene (nhx6-1 and nhx6-2) [49]. The double knockout lines were produced by crossing nhx5-1 with nhx6-1 or nhx6-2, respectively, to obtain two independent double knockout lines, nhx5-1 nhx6-1 and nhx5-1 nhx6-2. The absence of the AtNHX5 and AtNHX6 transcripts in these double knockout lines was confirmed by RT-PCR [49]. These two double knockout lines had identical growth phenotypes [49]. The nhx5-1 nhx6-1 double mutant line was used in the following experiments.
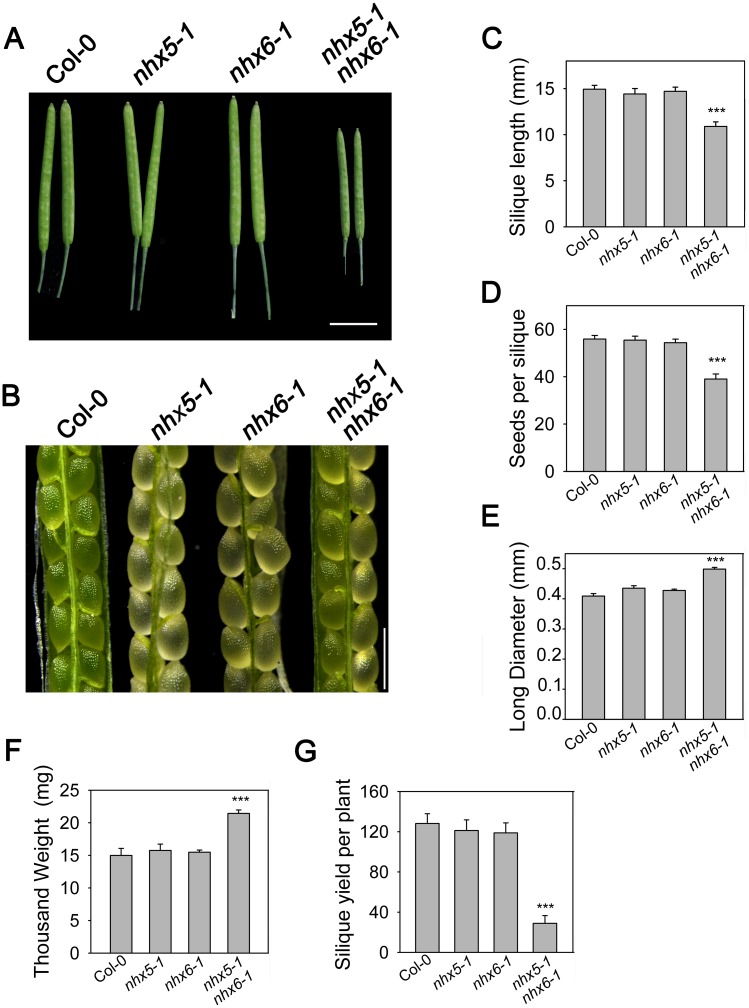
Interestingly, besides the profound defects in growth and development, nhx5 nhx6 double mutants had smaller siliques (Fig 1A). The siliques of nhx5 nhx6 were 26% shorter than that of the wild-type plants (Fig 1C). Additionally, nhx5 nhx6 produced less siliques (Fig 1G) and contained less seeds (Fig 1D). The seed yield per silique of nhx5 nhx6 was reduced 29% compared to the wild-type plants (Fig 1D). Similar to what Reguera et al. (2015) reported, we found that nhx5 nhx6 produced larger seeds (Fig 1B and 1E). The seeds of nhx5 nhx6 were 21% longer than that of the wild-type plants (Fig 1E). The thousand grain weight of nhx5 nhx6 was increased by 43% compared with the wide-type plants (Fig 1F). But the morphology of the mature seeds still looked the same as the wild type plants (Fig 1B). These results suggest that AtNHX5 and AtNHX6 play an important role in seed growth and development in Arabidopsis. However, there was no significant difference between the single mutant nhx5 or nhx6 and the wild type plants in silique growth and seed production (Fig 1), suggesting that AtNHX5 and AtNHX6 are functionally redundant.
Fig 1. Seed production was altered in nhx5 nhx6.
(A) Silique phenotype. Photos were taken for siliques on the main stem at 22 DAF. Bar = 0.5 cm. (B) Embryo morphology at 14 DAF. Bars = 1mm. (C) Silique length. Length of siliques on the main stem was measured (average length of 6 siliques) at 22 DAF. Error bars represent SD; n = 15. (D) Seed yield per silique. The total seeds per silique were counted (average number of 6 siliques). Error bars represent SD, n = 15. (E) Long diameter of dry seeds. Error bars represent SD, n = 250. (F) Thousand grain weight of seeds. Error bars represent SD, n = 8. (G) Silique yield per plant. The total siliques of each plant were counted. Error bars represent SD, n = 15. ***P≤0.001, by t test.
The artificial seed protein GFP-CT24 is missorted in nhx5 nhx6
We then examined if AtNHX5 and AtNHX6 controlled the transport of seed storage proteins to the PSVs. We used the artificial protein GFP-CT24 as a marker to track protein trafficking toward the PSVs in the embryo of the mature seeds. The artificial protein GFP-CT24 is produced by expressing the construct SP-GFP-CT24 in Arabidopsis. SP-GFP-CT24 consists of a signal peptide, GFP, and the C-terminal 24 amino acids (CT24) of the α’ subunit of β-conglycinin [10, 51]. β-conglycinin, a 7S globulin, is one of the major storage proteins in soybean seeds. CT24 contains the vacuolar sorting determinant of α’ subunit. Studies have shown that the artificial protein GFP-CT24 can be expressed and transported into the PSVs in the seeds of Arabidopsis and soybean [10, 51].
To track protein trafficking with GFP-CT24, we generated the transgenic seedlings expressing the artificial seed protein GFP-CT24. The transformants were produced in the backgrounds of wild-type, single mutant (nhx5 or nhx6) and double mutant (nhx5 nhx6) plants (Fig 2). We then compared the expression and transport of GFP-CT24 in the seeds of the mutant plants with that of the wild-type plants. GFP-CT24 fluorescent signals were visualized with a confocal laser scanning microscope. In the seeds of wild-type plants, GFP fluorescence predominantly appeared in the PSVs (Fig 2), indicating that the artificial seed protein GFP-CT24 was expressed and transported effectively into the PSVs in the wild-type plants. However, in either nhx5 or nhx6 single mutants, a strong GFP fluorescence signal was observed in the extracellular spaces, while a relatively faint signal of GFP fluorescence appeared in the PSVs, indicating that protein transport is impaired in nhx5 or nhx6 single mutants (Fig 2). Interestingly, GFP fluorescence predominantly appeared in the extracellular spaces in nhx5 nhx6, indicating that the artificial seed protein GFP-CT24 is missorted out of the cells in nhx5 nhx6 (Fig 2). These results suggest that AtNHX5 and AtNHX6 may control the transport of the seed storage proteins to the PSVs in Arabidopsis.
Fig 2. The artificial protein GFP-CT24 is missorted out of the cells of the mature seeds in nhx5 nhx6.
The artificial seed protein GFP-CT24 was used as a marker to track protein trafficking to the PSVs in the embryos of the mature seeds. GFP-CT24 was expressed in Arabidopsis seedlings and GFP-CT24 fluorescent signal was visualized with a confocal laser scanning microscope. Bars = 10 μm.
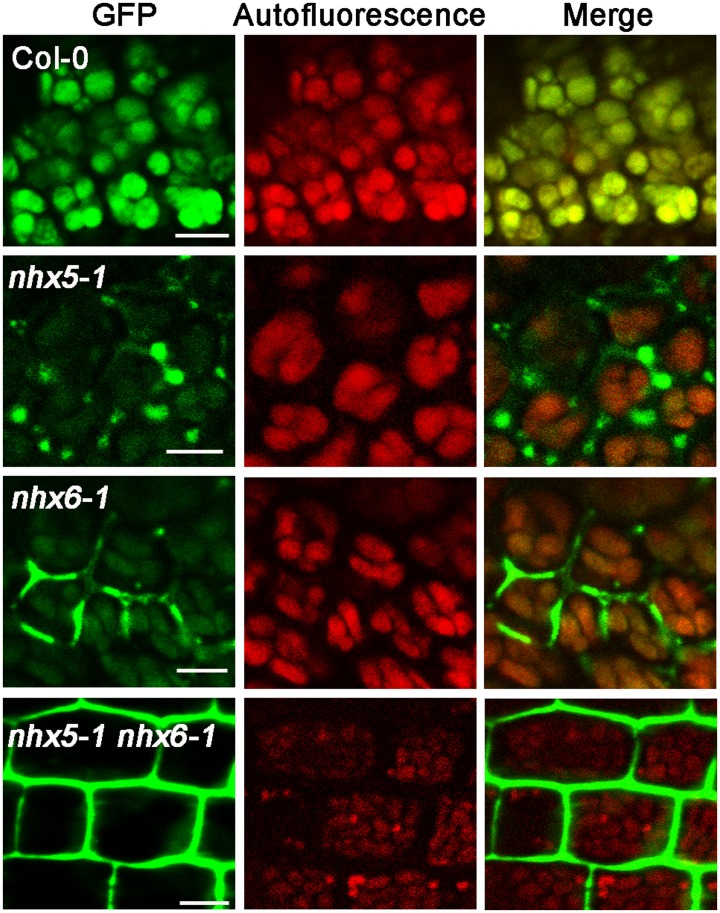
We next examined the profile of seed storage proteins using SDS-PAGE and immunoblot. We found that nhx5 nhx6 accumulated a significant amount of the precursor proteins of both 12S globulin and 2S albumin (S1 Fig). This result was verified by examining the distribution of 12S globulin in the embryo of the mature seeds using a transmission electron microscopy. The immune EM showed that a striking amount of the unprocessed precursors of 12S globulin were detected in the intercellular space of nhx5 nhx6 (S2 Fig). In addition, we found that PSV’s size is reduced but its number is increased in nhx5 nhx6, suggesting that AtNHX5 and AtNHX6 may involve in the biogenesis of the PSVs in Arabidopsis (S3 Fig).
All these results are similar to the findings of Reguera et al. (2015) [39]. These results confirmed the notion that AtNHX5 and AtNHX6 are required for the transport of the seed storage proteins into the PSVs as well as the biogenesis of the PSVs in Arabidopsis [39].
Three conserved acidic residues in AtNHX5 and AtNHX6 are critical for the transport of seed storage proteins into the PSVs in Arabidopsis
Our previous study has shown that AtNHX5 and AtNHX6 contain the conserved acidic amino acid residues in transmembrane domains that align with the yeast Na+/H+ antiporter ScNhx1p [49]. These conserved acidic residues of AtNHX5 (D164, E188, D193 and E320) and AtNHX6 (D165, E189, D194 and E320) line up with the corresponding residues in ScNhx1p (D201, E225, D230 and E355) [49, 52]. We have shown that three of these conserved acidic residues of AtNHX5 (D164, E188, and D193) and AtNHX6 (D165, E189, and D194) are essential for the ion transport activity of AtNHX5 and AtNHX6, and thus play an important role in growth and development in Arabidopsis. We are then asking whether the transport activity of AtNHX5 and AtNHX6 is required for the transport of seed storage proteins.
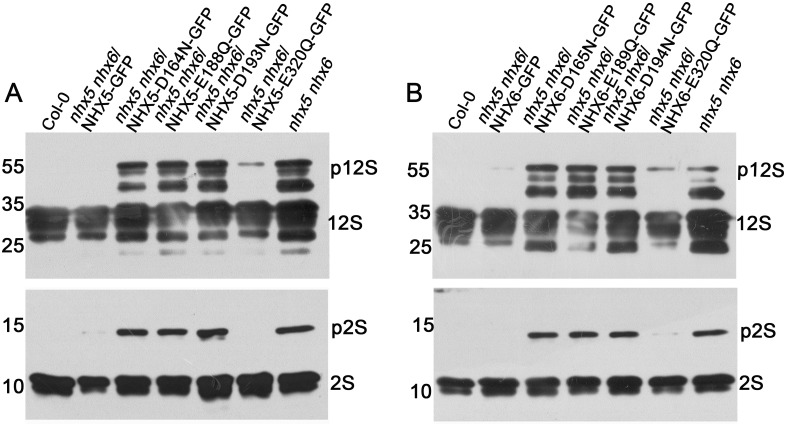
To this end, we expressed AtNHX5 and AtNHX6 genes mutated in these four conserved residues in nhx5 nhx6. The mutations were made by replacing the acidic residues with the uncharged polar residues. In AtNHX5, D164 was mutated to N, E188 to Q, D193 to N and E320 to Q; in AtNHX6, D165 to N, E189 to Q, D194 to N and E320 to Q. Protein profiles of the mature seeds were analyzed by SDS-PAGE and immunoblot (Fig 3). No precursors were detected for the control plants expressing the wild type AtNHX5 or AtNHX6 gene (Fig 3A and 3B), indicating that the transformed AtNHX5 or AtNHX6 gene functions efficiently in the transport of the seed storage proteins. However, the precursor proteins of both 12S globulin and 2S albumin were detected for three of the point mutations in both AtNHX5 and AtNHX6 genes (D164N, E188Q and D193N of AtNHX5, and D165N, E189Q and D194N of AtNHX6) (Fig 3A and 3B), indicating that these mutant genes were malfunctioning in protein transport. These results suggest that the transport activity of AtNHX5 and AtNHX6 is required for the transport of seed storage proteins.
Fig 3. Three conserved acidic residues in AtNHX5 and AtNHX6 are critical for the transport of the storage proteins into the PSVs in Arabidopsis.
(A) and (B) Immunoblot analysis for the point mutations in AtNHX5 (A) and AtNHX6 (B). The point mutated genes of the four conserved acidic residues of AtNHX5 and AtNHX6 were introduced into the nhx5 nhx6 background, respectively. Total proteins were extracted from the mature seeds. 10 μg proteins were loaded in each lane. Proteins were probed with anti-12S globulin and anti-2S albumin antibodies, respectively.
Our previous study has shown that E320 in both AtNHX5 and AtNHX6 genes may not be involved in exchange activity and cellular functions, since E320Q mutant remained its activity in conferring yeast growth under both high K+ and hygromicin B, and E320Q mutant fully recovered the growth of the nhx5 nhx6 seedlings [49]. As expected, E320Q mutant accumulated no or little precursors for both AtNHX5 and AtNHX6 genes (Fig 3A and 3B), indicating that protein transport was not or partially impaired in E320Q.
Seedling growth and seed development are altered in nhx5 nhx6 syp22
Since AtNHX5, AtNHX6 and the SNAREs VAMP727, SYP22, VTI11 and SYP51 are all localized to the same organelle PVC, we reason that AtNHX5 and AtNHX6 may control the trafficking of seed storage proteins by regulating the function of this SNARE complex.
To test this hypothesis, we generated the nhx5 nhx6 syp22 triple mutant (Fig 4). We obtained two separate T-DNA lines for the SYP22 gene (syp22-1 and syp22-3) (S4A Fig). The nhx5-1 nhx6-1 double mutant line was generated in our previous study [49]. The double mutant line nhx5-2 nhx6-2 was generated by crossing nhx5-2 and nhx6-2. The triple mutant lines were generated by crossing nhx5-1 nhx6-1 with syp22-3, and nhx5-2 nhx6-2 with syp22-1, respectively, to obtain two independent triple knockout lines, nhx5-1 nhx6-1 syp22-3 and nhx5-2 nhx6-2 syp22-1. The absence of AtNHX5 and AtNHX6 transcripts in the double knockout lines was confirmed by RT-PCR (S4C Fig; 49). The absence of AtNHX5, AtNHX6 and SYP22 transcripts in triple knockout lines was confirmed by RT-PCR (S4D Fig). These two triple knockout lines had identical growth phenotypes (S4B Fig). The nhx5-1 nhx6-1 syp22-3 triple mutant line was used in the following experiments.
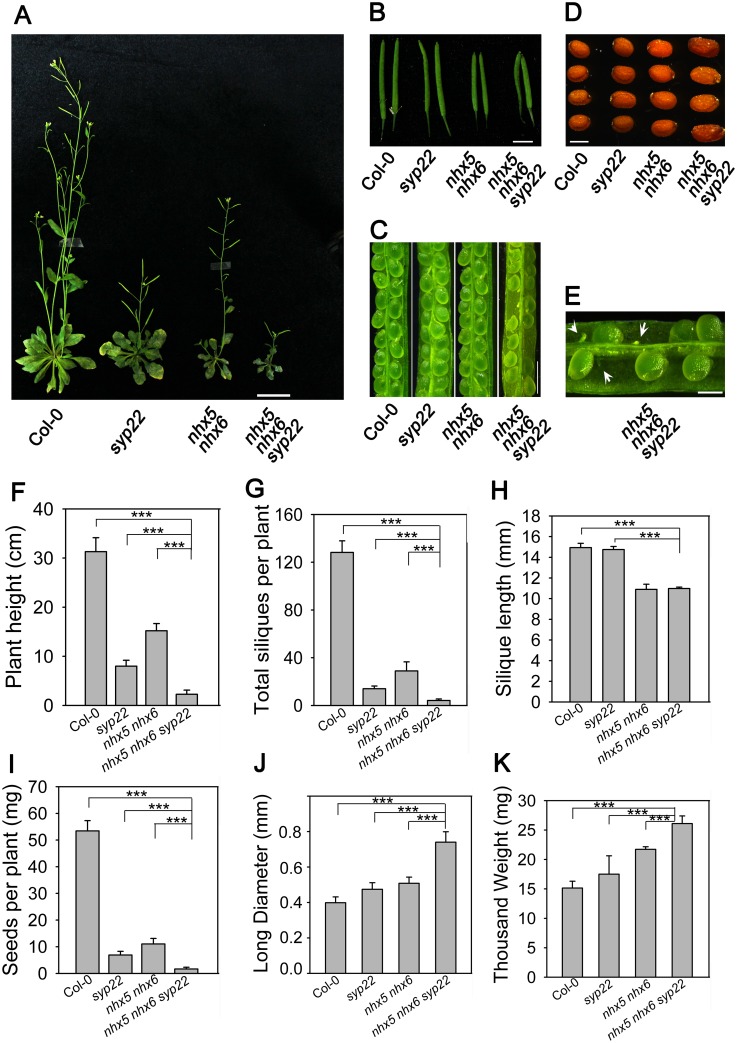
Fig 4. Seedling growth and seed development are altered in nhx5 nhx6 syp22.
(A) Phenotype of the triple mutant grown on soil for 60 days. (B) Silique phenotype. Photos were taken at 22 DAF. Bar = 0.5 cm. (C) The embryo morphology at 14 DAF. Bars = 1mm. (D) The mature seeds. Photos were taken when the seeds were dehydrated for 30 days after harvest. Bars = 0.5mm. (E) Embryo morphology of the triple mutant. Bars = 0.5mm. (F) Plant height. Error bars represent SD; n = 30. (G) Silique yield per plant. Error bars represent SD; n = 30. (H) Silique length. Silique length was measured on the main stem at 22 DAF. Error bars represent SD; n = 15. (I) Seed yield per silique. The total seeds per silique were counted on the main stem. Error bars represent SD, n = 15. (J) Long diameter of the dry seeds. Error bars represent SD. n = 300. (K) Thousand weight of the seeds. Error bars represent SD, n = 8. ***P≤0.001, by t test.
Studies have found that the nhx5 nhx6 double mutant showed profound defects in growth and development (Fig 4A) [48, 49]. nhx5 nhx6 had smaller rosettes and shorter seedlings, was flowering and bolting late, and produced less seeds (Fig 4A) [48, 49]. In addition, the syp22 mutant is impaired in growth and development (Fig 4A) [53, 54]. syp22 showed multiple abnormal phenotypes, including semi-dwarfism, serrated wavy leaves, late flowering, and resistance to salt stress (Fig 4A) [53, 54]. Interestingly, the nhx5 nhx6 syp22 triple mutant showed much more severe defects in growth and development (Fig 4A). The triple mutant was smaller than nhx5 nhx6 or syp22 (Fig 4A). The height of the triple mutant seedlings was 13%, 40% and 26% of the wild type, syp22, and nhx5 nhx6 seedlings, respectively, when grown in soil for 60 days (Fig 4A and 4F). The triple mutant had short siliques and reduced seed sets, and contained approximately 24% aborted seeds (Fig 4B, 4C and 4E). The triple mutant produced less siliques and seeds (Fig 4G and 4I). However, the triple mutant produced much larger seeds (Fig 4D). The seed of the triple mutant was 85%, 56%, and 45% larger in length than the wild type, syp22, and nhx5 nhx6, respectively (Fig 4D and 4J). The thousand grain weight of the triple mutant was increased by 73%, 53% and 23%, respectively, comparing with the wild type, syp22 and nhx5 nhx6 (Fig 4K). The severe defects in seedling growth and seed production of the triple mutant suggest that AtNHX5, AtNHX6 and SYP22 have an overlapped function in growth and development, particularly in silique growth and seed development.
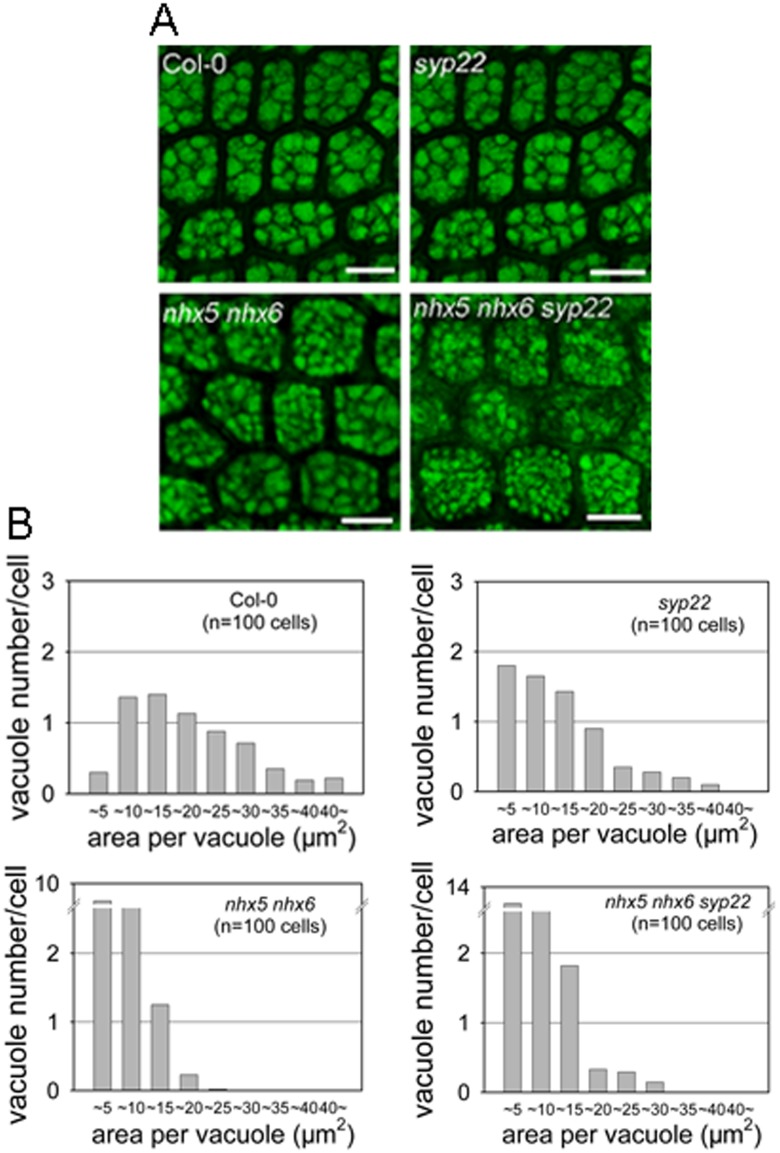
PSV’s size is reduced but its number is increased in nhx5 nhx6 syp22
In order to understand whether AtNHX5, AtNHX6 and SYP22 control the biogenesis of the PSVs in Arabidopsis, we analyzed the morphology of the PSVs (Fig 5). The autofluorescence of the PSVs was visualized under a confocal microscope (Fig 5A). For the wild-type plants, the average section area per vacuole is 17.55 ± 6.38 μm2 (mean ± SD; n = 654 PSVs in 100 cells) (Fig 5A and 5B). The PSVs in syp22 was slightly reduced: 10.42 ± 8.5 μm2 (mean ± SD; n = 665 PSVs in 100 cells) (Fig 5A and 5B). The PSVs in nhx5 nhx6 were fragmented and showed a significant reduction in average vacuole size: 5.5 ± 3.12 μm2 (mean ± SD; n = 1647 PSVs in 100 cells) (Fig 5A and 5B). Interestingly, the triple mutant had a much smaller PSV with an average size of 4.5±2.12 μm2 (mean ± SD; n = 2400 PSVs in 100 cells) (Fig 5A and 5B).
Fig 5. AtNHX5, AtNHX6 and AtSYP22 play an important role in the biogenesis of the PSVs in Aabidopsis.
(A) Morphology of the PSVs. Autofluorescence of the PSVs was visualized with a confocal laser scanning microscope. Bars = 10 μm. (B) Histograms representing a distribution of size and number of vacuoles within a single cell. The X axis indicates the areas of the vacuole. Area of each vacuole was measured from the embryo cells of Col-0 and mutants. 100 cells were measured for PSV analysis.
In addition, the number of the PSVs in the triple mutant was increased significantly. While the average number of the PSVs is 6.5 in wild-type plants, syp22 and nhx5 nhx6 had an average of 6.6 and 16 PSVs, respectively (Fig 5A and 5B). However, the triple mutant had a significantly increased number of PSVs: 24 PSVs per embryo cell (Fig 5A and 5B). These results suggest that AtNHX5, AtNHX6 and SYP22 may involve in the biogenesis of the PSVs in Arabidopsis.
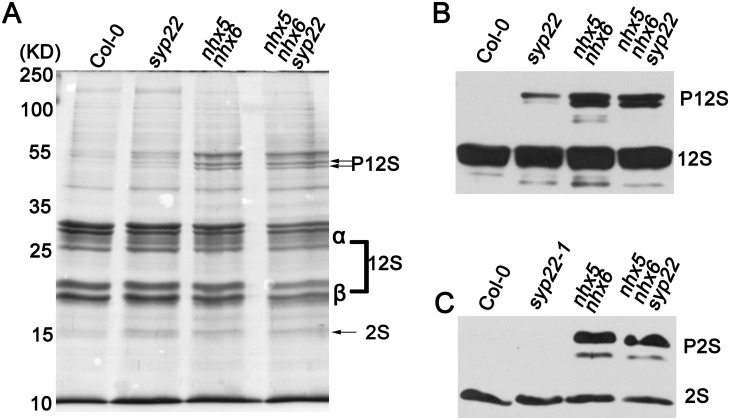
Precursors of seed storage proteins are accumulated in nhx5 nhx6 syp22
12S globulin and 2S albumin are the two major storage proteins of the PSVs in Arabidopsis [10, 20]. They are synthesized as precursors in endoplasmic reticulum and sorted to the PSVs to convert to the mature proteins [20]. It is interesting to ask whether AtNHX5, AtNHX6 and SYP22 control the transport of 12S globulin and 2S albumin into the PSVs in Arabidopsis.
To this end, we examined the profile of seed storage proteins using SDS-PAGE and immunoblot. Total proteins were extracted from the mature seeds of wild-type, syp22, nhx5 nhx6, and nhx5 nhx6 syp22 seedlings, respectively. The proteins were separated on SDS-PAGE, stained with coomassie blue, and blotted with anti-12S or anti-2S antibodies (Fig 6). A high amount of the mature forms of 12S globulin and 2S albumin were detected in the seeds of wild-type plants; but no precursor proteins were detected (Fig 6A to 6C), indicating that 12S globulin and 2S albumin were transported into the PSVs and converted to their mature forms in wild-type plants. A faint band of the precursor proteins of 12S globulin was detected in the seeds of syp22; but the precursors of 2S albumin was not detected in syp22 (Fig 6A to 6C). Interestingly, nhx5 nhx6 and nhx5 nhx6 syp22 accumulated a significant amount of the precursor proteins of both 12S globulin and 2S albumin, while the mature forms of 12S globulin and 2S albumin were slightly reduced (Fig 6A to 6C). These results suggest that the transport of 12S globulin and 2S albumin was partially impaired, and 12S globulin and 2S albumin were missorted out of the cells in nhx5 nhx6 syp22.
Fig 6. Precursors of the storage proteins are accumulated in the seeds of nhx5 nhx6 syp22.
(A) SDS-PAGE stained with the Coomassie blue. Total proteins were extracted from the mature seeds. 10 μg proteins were loaded in each lane. (B) and (C) Immunoblot analysis of 12S globulin (B) and 2S albumin (C).
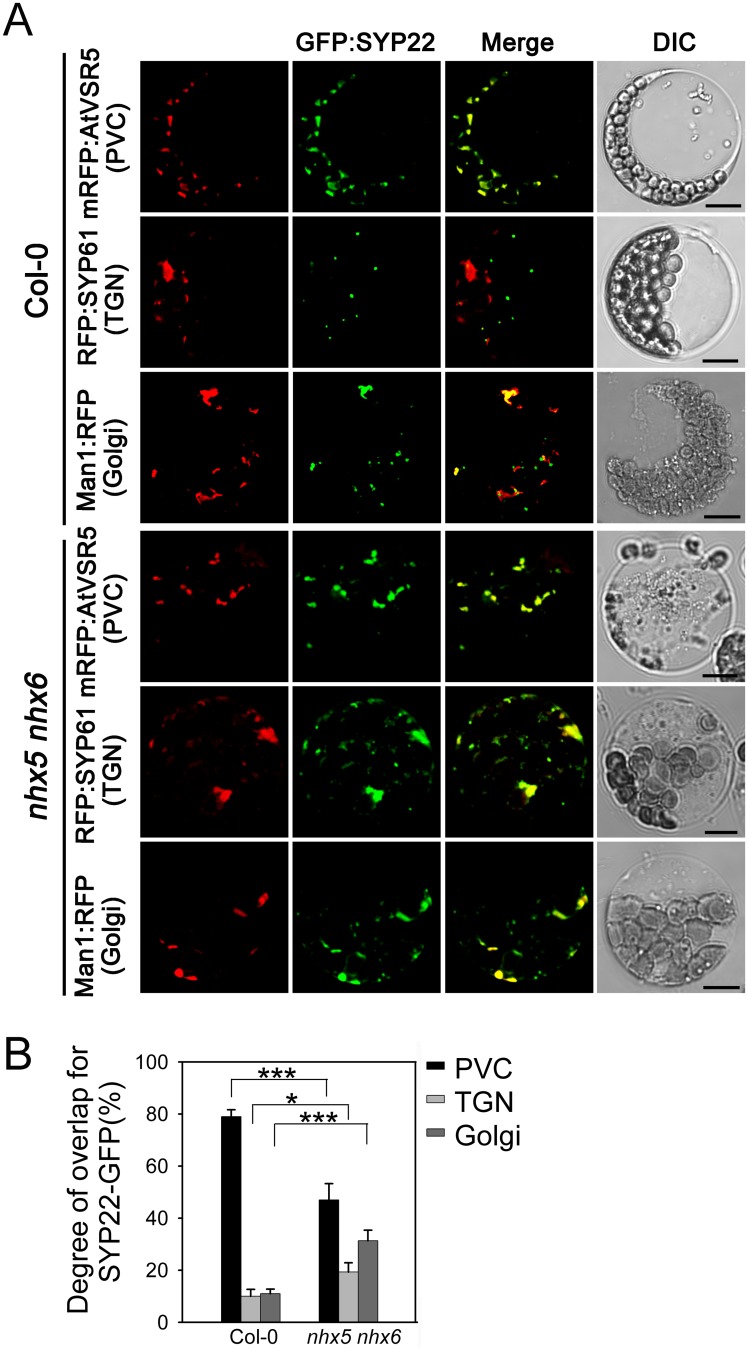
Subcellular localization of SYP22 and VAMP727 is altered in nhx5 nhx6
In plant cells, proteins are transported to the vacuole through a vesicle-mediated trafficking pathway that includes the ER, Golgi, TGN, and MVB/PVC [50]. Thus, the Golgi, TGN and MVB/PVC are major protein sorting stations in vacuolar transport [55, 56]. Since AtNHX5 and AtNHX6 are localized to the Golgi, TGN and PVC, these two antiporters may involve in protein trafficking towards the vacuole [39]. Studies have shown that the SNARE complex composed of VAMP727, SYP22, VTI11 and SYP51 was localized to PVC to mediate the fusion between the PVC and vacuole [26]. Then, it is interesting to ask whether AtNHX5 and AtNHX6 are required for the trafficking and localization of the SNAREs VAMP727, SYP22, VTI11 and SYP51.
To this end, we examined the trafficking and subcellular distribution of SYP22 and VAMP727 by transient expression in Arabidopsis protoplasts (Figs 7 and 8).
Fig 7. Subcellular localization of AtSYP22 is altered in nhx5 nhx6.
(A) Subcellular localization of GFP:SYP22. The SYP22:GFP plasmid was co-transformed with the PVC marker mRFP:AtVSR5, TGN marker RFP:SYP61 or Golgi marker Man1:RFP into the leaf protoplasts of Arabidopsis, respectively. Fluorescence was visualized by a confocal laser scanning microscope. Bar = 10μm. (B) Quantification of the subcellular localization pattern of AtSYP22. The overlapping percentage of GFP:SYP22 signal with mRFP:AtVSR5, RFP:SYP61 or Man1:RFP, respectively, was determined from more than 100 protoplasts obtained from three independent experiments. Error bars represent SD, n = 3. ***P≤0.001, *P≤0.05, by t test.
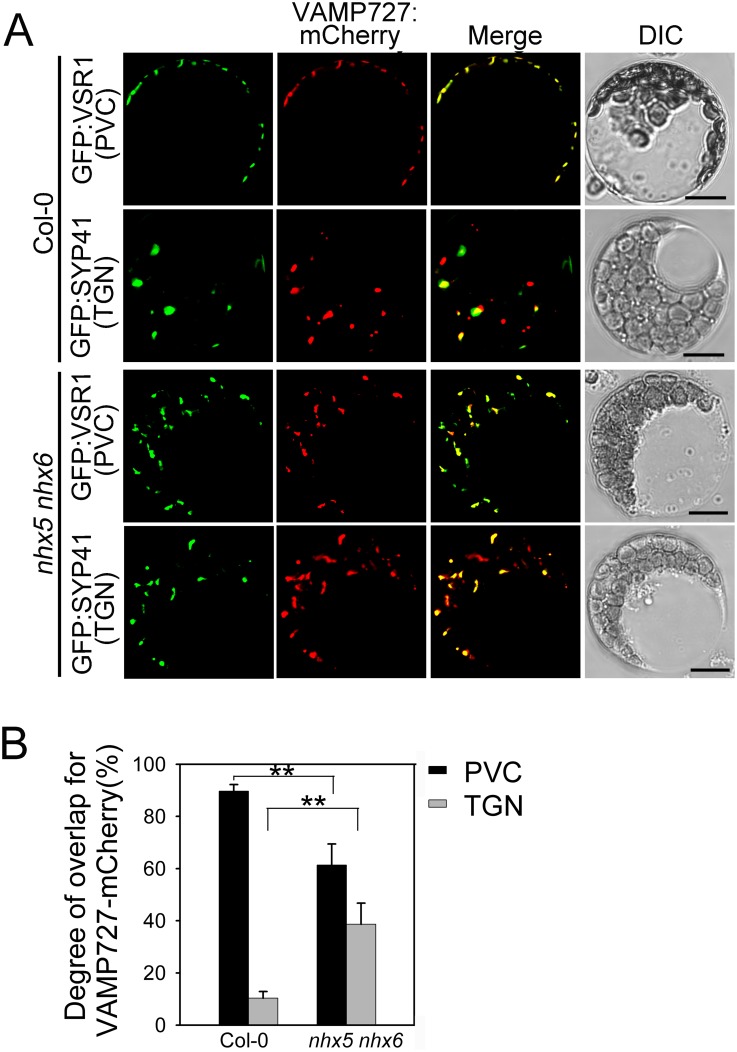
Fig 8. Subcellular localization of AtVAMP727 is altered in nhx5 nhx6.
(A) Subcellular localization of VAMP727:mCherry. The VAMP727:mCherry plasmid was co-transformed with the PVC marker GFP:AtVSR1 and TGN marker GFP:SYP41, respectively, into the leaf protoplasts of Arabidopsis. Fluorescence was visualized by a confocal laser scanning microscope. Bar = 10μm. (B) Quantification of the subcellular localization pattern of AtVAMP727. The overlapping percentage of VAMP727:mCherry signal with GFP:AtVSR1 or GFP:SYP41, respectively, was determined from more than 50 protoplasts obtained from three independent experiments. Error bars represent SD, n = 3. **P≤0.01, by t test.
For SYP22 assay, GFP gene was fused to the N-terminal end of SYP22, driven by the 35S promoter. The SYP22:GFP plasmid was transiently co-expressed in Arabidopsis leaf protoplasts with the PVC marker mRFP:AtVSR5, TGN marker RFP:SYP61 or Golgi marker Man1:RFP, respectively. SYP22:GFP fluorescence appeared on the punctate structures in the cytosol (Fig 7A). In Col-0, 79% of the SYP22:GFP fluorescent signals were co-localized with mRFP:AtVSR5, but only 10% with RFP:SYP61 and 11% with Man1:RFP, suggesting that the majority of SYP22 is localized to the PVC in Col-0 (Fig 7A and 7B). In nhx5 nhx6, however, the cellular distribution pattern of AtSYP22 was altered: 47% of SYP22:GFP overlapped with mRFP:AtVSR5, whereas 19% with SYP61and 31% with Man1:RFP, respectively (Fig 7A and 7B). These results suggest that a significant amount of SYP22 is trapped in the Golgi or TGN on the way to the PVC in the absence of AtNHX5 and AtNHX6. AtNHX5 and AtNHX6 are required for the localization of SYP22 in the PVC.
For VAMP727 assay, mCherry gene was fused to the C-terminal end of VAMP727, driven by the 35S promoter. The VAMP727:mCherry plasmid was transiently co-expressed in Arabidopsis leaf protoplasts with the PVC marker GFP:AtVSR1 and TGN marker GFP:SYP41, respectively. VAMP727:mCherry fluorescence appeared on the punctate structures in the cytosol (Fig 8A). In Col-0, 91% of VAMP727:mCherry overlapped with the PVC marker GFP:AtVSR1, only 9% with the TGN marker GFP:SYP41, suggesting that the majority of VAMP727 is localized to the PVC in Col-0 (Fig 8A and 8B). By contrast, in nhx5 nhx6, 61% of VAMP727:mCherry overlapped with the PVC marker GFP:AtVSR1, while 38% of VAMP727:mCherry overlapped with the TGN marker GFP:SYP41 (Fig 8A and 8B). These results suggest that the PVC localization of VAMP727:mCherry is considerately repressed in nhx5 nhx6. A significant amount of VAMP727 is trapped in the TGN on the way to the PVC in the absence of AtNHX5 and AtNHX6.
AtNHX5 and AtNHX6 are co-localized with SYP22 and VAMP727
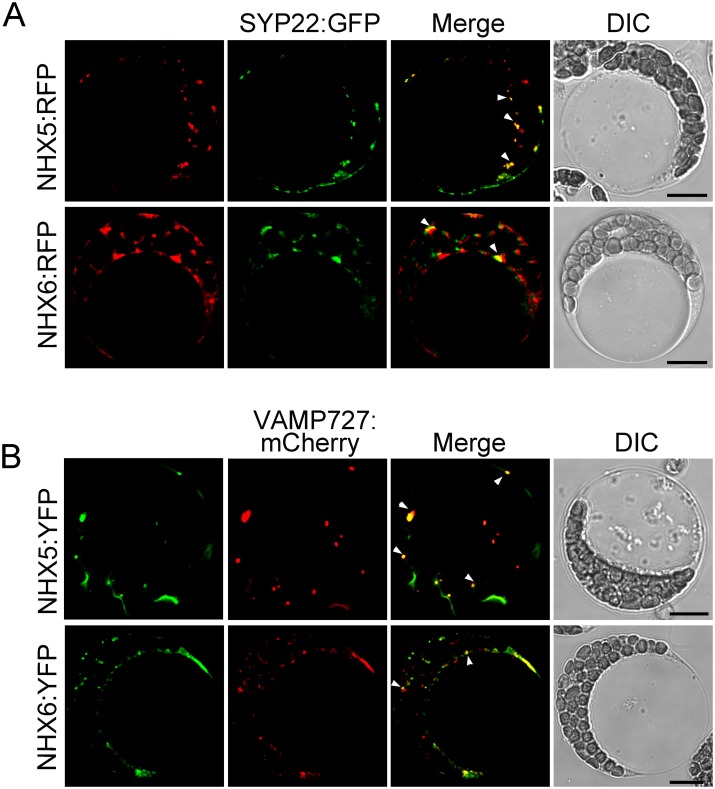
We examined the colocalization of AtNHX5 and AtNHX6 with the SNAREs by transient expression in Arabidopsis protoplasts. AtNHX5 and AtNHX6 were tagged at the N-terminal ends with RFP and YFP, respectively, driven by the 35S promoter. The RFP:AtNHX5 or RFP:AtNHX6 plasmids were transiently co-expressed in Arabidopsis leaf protoplasts with SYP22:GFP, while the YFP:AtNHX5 or YFP:AtNHX6 plasmids were co-expressed with VAMP727:mCherry. As shown in Fig 9, RFP:AtNHX5 and RFP:AtNHX6 fluorescent signals partially co-localized with SYP22-GFP. In addition, YFP:AtNHX5 and YFP:AtNHX6 fluorescent signals were partially co-localized with VAMP727:mCherry. These results suggest that AtNHX5 and AtNHX6 are co-localized with SYP22 and VAMP727.
Fig 9. AtNHX5 and AtNHX6 are co-localized with SYP22 and VAMP727.
(A) Subcellular localization of RFP:NHX5 or RFP:NHX6 with SYP22:GFP. The RFP:AtNHX5 or RFP:AtNHX6 plasmids were transiently co-expressed in Arabidopsis leaf protoplasts with SYP22:GFP, respectively. Fluorescence was visualized by a confocal laser scanning microscope. (B) Subcellular localization of YFP:NHX5 or YFP:NHX6 with VAMP727:mCherry. The YFP:AtNHX5 or YFP:AtNHX6 plasmids were transiently co-expressed in Arabidopsis leaf protoplasts with VAMP727:mCherry, respectively. Fluorescence was visualized by a confocal laser scanning microscope. Bar = 10 μm in (A) and (B).
AtNHX5 and AtNHX6 do not interact physically with the SNARE complex
We next examined whether AtNHX5 or AtNHX6 interacted physically with the SNARE complex by coimmunoprecipitation (Fig 10).
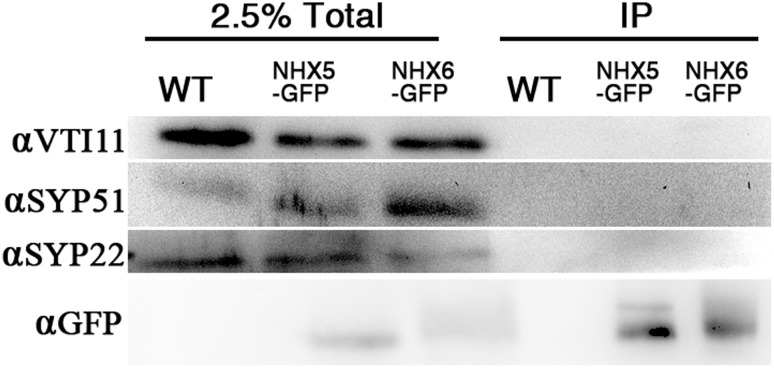
Fig 10. AtNHX5 and AtNHX6 do not interact physically with the SNARE complex.
The transgenic Arabidopsis seedlings expressing GFP-tagged NHX5 or NHX6 were generated in our previous study (Wang et al., 2015). Anti-GFP monoclonal antibody was added to the plant lysate to precipitate NHX5-GFP or NHX6-GFP and their binding proteins. Coimmuoprecipitation of the SNARE proteins was analyzed with immunoblot with antibodies against SYP22, VTI11, or SYP51.
We performed the coimmunoprecipitation using the transgenic Arabidopsis seedlings expressing GFP-tagged NHX5 or NHX6 that were generated in our previous study [49]. GFP was fused with the C-terminus of the AtNHX5 or AtNHX6 genes under the control of 35S promoters. The constructs were introduced into the nhx5 nhx6 and wild-type plants. The chimeric genes coding for NHX5-GFP or NHX6-GFP functioned well since transformation of nhx5 nhx6 with these constructs rescued the mutant phenotype [49]. The wild-type plants expressing GFP-tagged NHX5 or NHX6 were used to conduct the assay. Anti-GFP monoclonal antibody was added to the plant lysate to precipitate NHX5-GFP or NHX6-GFP and their binding proteins, followed by immunoblotting with antibodies against SYP22, VTI11, or SYP51. As shown in Fig 10, SYP22, VTI11 and SYP51 were not co-immunoprecipitated by the anti-GFP antibody, indicating that there was no physical interreaction between the SNARE complex and AtNHX5 or AtNHX6.
Discussion
AtNHX5 and AtNHX6 regulate the subcellular localization of the SNARE complex in Arabidopsis
In this report, we examined the role of AtNHX5 and AtNHX6 in the trafficking of the seed storage proteins to the PSVs. Using genetics, cellular and molecular techniques, we found that: (1) seed growth and production were altered in nhx5 nhx6 double mutants (Fig 1); (2) transport of seed storage proteins into the PSVs was impaired in nhx5 nhx6 (Fig 2, S1 and S2 Figs); (3) PSV biogenesis was changed in nhx5 nhx6 (S3 Fig). These results indicate that AtNHX5 and AtNHX6 are involved in seed production, protein trafficking and PSV biogenesis, which are similar to what Reguera et al. (2015) had reported [39]. Together, both Reguera et al. (2015) and our results demonstrate that AtNHX5 and AtNHX6 are required for the transport of the seed storage proteins into the PSVs as well as the biogenesis of the PSVs in Arabidopsis [39].
It has been reported that the SNARE complex, which is composed of VAMP727, SYP22, VTI11 and SYP51, is critical for protein trafficking and PSV biogenesis in Arabidopsis [26]. This SNARE complex is localized to PVC to mediate the fusion between the PVC and the vacuole, through which proteins are delivered into the vacuole [26]. It is interesting to mention that AtNHX5 and AtNHX6 are localized to the Golgi, TGN and PVC, and these organelles are critical for protein trafficking to the vacuole in plants [39, 48, 55, 56]. There is great potential, therefore, for AtNHX5 and AtNHX6 to regulate the trafficking and subcellular distribution of this SNARE complex and hence its function in protein transport. We examined the role of AtNHX5 and AtNHX6 in regulating this SNARE complex. We found that: (1) seedling growth and seed development were altered in nhx5 nhx6 syp22 (Fig 4); (2) PSV biogenesis was changed in nhx5 nhx6 syp22 (Fig 5); (3) protein trafficking into the PSVs was impaired in nhx5 nhx6 syp22 (Fig 6); (4) the localization of this SNARE complex in the PVC was repressed in the nhx5 nhx6 (Figs 7 and 8). Our results indicate that AtNHX5 and AtNHX6 are required for the subcellular localization of this SNARE complex and hence its function in protein transport. Collectively, these results indicate that AtNHX5 and AtNHX6 may control the trafficking of the seed storage proteins to the PSVs through regulating the subcellular localization of the SNARE complex in Arabidopsis.
The transport activity of AtNHX5 and AtNHX6 is required for the trafficking and subcellular localization of the SNARE complex
We further examined how AtNHX5 and AtNHX6 regulated the subcellular localization of the SNARE complex: via transport activity, physical interaction, or both. Studies from bacteria, yeast and mammals show that the acidic amino acid residues in transmembrane domains of Na+/H+ antiporters are critical for exchange activity [57–59]. Mutation of these conserved acidic residues in yeast ScNhx1p blocked protein trafficking in yeast. Our previous study has shown that AtNHX5 and AtNHX6 contain the conserved acidic amino acid residues in transmembrane domains that align with the yeast Na+/H+ antiporter ScNhx1p [49]. We have shown that three of these conserved acidic residues of AtNHX5 (D164, E188, and D193) and AtNHX6 (D165, E189, and D194) are essential for the ion transport activity of AtNHX5 and AtNHX6, and play an important role in growth and development in Arabidopsis [49]. In this study, we found that the precursor proteins of both 12S globulin and 2S albumin were detected for three of the point mutations in both AtNHX5 and AtNHX6 genes (D164N, E188Q and D193N of AtNHX5, and D165N, E189Q and D194N of AtNHX6) (Fig 3A and 3B), indicating that the transport activity of AtNHX5 and AtNHX6 is critical for protein transport into the PSVs. These results suggest that AtNHX5 and AtNHX6 may regulate the subcellular localization of the SNARE complex by their transport activity.
AtNHX5 and AtNHX6 are H+-coupled cotransporters whose biochemical activity is transferring the Na+ or K+ across a membrane in exchange for protons (H+). Thus, AtNHX5 and AtNHX6 play an important role in pH and ion homeostasis in Arabidopsis [39, 48, 49]. Our finding that the localization of the SNARE complex was regulated by the transport activity of AtNHX5 and AtNHX6, therefore, suggests that the localization of the SNARE complex was regulated by the pH and/or ion homeostasis maintained by AtNHX5 and AtNHX6. This is supported by the recent studies which show that AtNHX5 and AtNHX6 are vital for the endosomal pH homeostasis and this pH homeostasis is required for protein trafficking to the vacuole in Arabidopsis [31, 39].
To determine whether AtNHX5 or AtNHX6 physically interact with the SNARE complex, we performed coimmunoprecipitation assay. We found that SYP22, VTI11 and SYP51 were not co-immunoprecipitated by the anti-GFP antibody, suggesting that there is no physical interrelation between the SNARE complex and AtNHX5 or AtNHX6 (Fig 10).
AtNHX5 and AtNHX6 have diversified roles in protein trafficking
In this report, we demonstrate that AtNHX5 and AtNHX6 control the trafficking of seed storage proteins by regulating the subcellular localization of the SNARE complex (Figs 7 and 8). On the other hand, Reguera et al. (2015) found that AtNHX5 and AtNHX6 controlled the trafficking of seed storage proteins by regulating the interaction between VSR and its cargoes [39]. Ashnest et al. (2015) found that AtNHX5 and AtNHX6 might regulate the recycling of VSRs by interacting with SNX1, a component of the Retromer [40]. Therefore, the results of Reguera et al. (2015), Ashnest et al. (2015) and ours revealed three distinct mechanisms for the role of AtNHX5 and AtNHX6 in protein trafficking: regulating the SNARE complex that mediates the fusion between the PVC and the vacuole, regulating the binding of VSR with its cargoes at the TGN, and regulating the recycling of VSRs from the TGN back to the ER. AtNHX5 and AtNHX6, thus, regulate three distinct steps of the protein trafficking pathway. In other words, AtNHX5 and AtNHX6 may have diversified roles in protein trafficking.
Why AtNHX5 and AtNHX6, then, have such diversified roles in protein trafficking? The short answer is that they have unique subcellular localizations in cells. We know that, in plant cells, proteins are transported to the vacuole through a vesicle-mediated trafficking pathway that includes the ER, Golgi, TGN, and MVB/PVC [50]. Thus, the Golgi, TGN and MVB/PVC are major protein sorting stations in vacuolar transport [55, 56]. Since AtNHX5 and AtNHX6 are localized to the Golgi, TGN and PVC [39], these two antiporters may regulate the protein trafficking activities carried out in these organelles, including the function of the SNARE complex and the receptor protein VSR. Therefore, AtNHX5 and AtNHX6 have such diversified roles in protein trafficking.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana ecotypes Columbia (Col-0), mutant and transgenic lines were used in this study. In the growth chamber, plants were grown on compost (Pindstrup Substrate, Latvia) and subirrigated with tap water. Greenhouse conditions were as follows: 16-h-light /8-h-dark cycles, light intensity 100 μmol s-1 m-2 photosynthetically active radiation, temperature 22°C, and relative humidity 50±10%.
For plate-grown plants, Arabidopsis thaliana seeds were surface sterilized with 20% (v/v) bleach. After cold treatment at 4°C for 3 days in the dark, the seeds were germinated on plates with Murashige and Skoog (MS) medium containing 1.0% agar, pH 5.8.
For silique phenotype observation, photos were taken at 22 DAF (DAF, days after flowering). The length of siliques on the main stem was measured, where the fifth to eighth siliques were measured.
To observe the developing embryo, the embryos were stripped from siliques at 14 DAF. For morphology observation of the mature seeds, photos were taken when the seeds were dehydrated for 30 days after harvest.
Generation of the nhx5 nhx6 syp22 triple mutant
T-DNA insertion lines were obtained from the SALK collection and GABI collection. T-DNA lines used in this work were Wisc-DsLox345-348M8 (nhx5-1), GABI_363E02 (nhx5-2), SALK_113129C (nhx6-1), SALK_100042 (nhx6-2), SALK_ 060946C (syp22-1) and 075924 (syp22-3). Insertion mutant information was obtained from the SIGnAL website (http://signal.salk.edu/cgi-bin/tdnaexpress) and confirmed experimentally. Positions of nhx5-1, nhx6-1 and nhx6-2 T-DNA insertion sites have been shown in Wang et al. (2015) [49]. The T-DNA insertion sites of syp22-1, syp22-3 and nhx5-2 are shown in S4A Fig online. The T-DNA insertion in line syp22-1 occurred at nucleotide +1058 relative to the start codon, whereas mutant syp22-3 carries the insertion at nucleotide +154 (S4A Fig). T-DNA insertion in line nhx5-2 occurred at nucleotide +1371bp relative to the start codon (S4A Fig). Homozygous mutant lines were identified by PCR screening with allele-specific primers designed to amplify wild-type or mutated loci. The primers used for homozygous identification were as follows: syp22-1 (SALK_060946C LP: 5’- TTTGAGCTCAAAGGGTGAATC-3’; SALK_060946C RP: 5’- TGCTGCTGAAAGAGAAACCAC-3’; BP LB1.3: 5’- ATTTTGCCGATTTCGGAAC-3’); syp22-3 (SALK_075924 LP: 5’- TTAAACTAAGGGCACCGAACC-3’; SALK_075924 RP: 5’- AAAGTCCCTTGCAAGCTTAGC-3’; BP LB1.3: 5’- ATTTTGCCGATTTCGGAAC-3’); nhx5-2 (GABI_363E02 LP: 5’- CAATTGCTCCGAAGACTTCAG-3’; GABI_363E02 RP: 5’- TTGGCACATCGATAGGACTTC -3’; GABI BP: 5’- ATAATAACGCTGCGGACATCTACAT -3’)
The nhx5 nhx6 double mutant line, nhx5-1 nhx6-1 was generated in our previous study [49]. The nhx5-2 nhx6-2 was generated by crossing nhx5-2 and nhx6-2 (S4B Fig).
The triple mutant lines were generated by crossing nhx5-1 nhx6-1 with syp22-3, or crossing nhx5-2 nhx6-2 with syp22-1, using nhx5-1 nhx6-1 and nhx5-2 nhx6-2 as pollen donors. The homozygous triple mutant lines were identified by PCR screening with allele-specific primers designed to amplify wild-type or mutated loci.
Transformation of SP-GFP-CT24 into Arabidopsis
The plasmid SP-GFP-CT24 was kindly provided by Dr. Shigeru Utsumi of Kyoto University, Japan. Transformation of Arabidopsis was performed by floral dipping using Agrobacterium tumefaciens (strain GV3101) [60]. The transformed seedlings (T1 plants) were selected on MS agar plates containing 200 μg ml-1 carbenicillin and 30 μg ml-1 kanamycin. The antibiotic positive seedlings were re-confirmed with PCR amplification of the GFP fragment using the following primers: GFP F: 5’- GTGAGCAAGGGCGAGGAGCTGTTC -3’, and GFP R: 5’- CAGCTCGTCCATGCCGAGAGTGATC -3’. The T2 seeds were collected and used for the experiments.
Confocal laser scanning microscopy
The confocal laser scanning analysis was performed as described [10]. T2 seeds of the transformation plants were soaked for 20 minutes in PBS or distilled water. Then the cotyledons were peeled off from the seed coat. To observe the fluorescence, the cotyledons were placed on the glass slides, added a drop of glycerol, and covered with glass strips. Fluorescence was visualized by a confocal laser scanning microscope (FV1000, Olympus). The excitation wavelength was 488 nm for GFP and 594 nm for autofluorescence of PSVs, and emission was 500–530 nm for GFP and 590–630 nm for autofluorescence of PSVs. Images were processed with the Adobe Photoshop 3.0 (Adobe System). For quantitative analysis, 100 cells of the hypocotyls were measured in indicated genotypes seeds. Images were processed with Image J software to measure the size and numbers of the PSVs.
Seed protein extraction
Seed proteins were extracted according to Nishizawa et al. (2003) with minor modifications [51]. Seeds were harvested and dehydrated for 30 days before protein extraction. Seeds were homogenized at 4°C in extraction buffer (150–200 μL extraction buffer/mg seeds). The extraction buffer contained 35 mM sodium phosphate buffer (pH 7.6), 0.4 M NaCl, 1 mM EDTA, 0.02% (w/v) NaN3, 0.1 mM pepstatin A, 10 mM 2-mercaptoethanol, and 0.1 mM (p-amidinophenyl) methanesulfonyl fluoride (p-APMSF). The homogenates were centrifuged at 13,000 × g for 15 min at 4°C to remove the cellular debris. The supernatants were collected as the crude protein extracts. The total protein of the crude extracts was determined with the Bradford protein assay kit (Sangon Biotech, China).
SDS-PAGE and immunoblot analysis
Total protein (10 μg) of the supernatants was mixed with 2 μL 5× loading buffer composed of 250 mM Tris-HCl (pH = 6.8), 10% (W/V) SDS, 0.5% (W/V) Bromophenol Blue, 50% (V/V) glycerol and 5% (V/V) β-mercaptoethanol, then added protein extraction buffer to a total volume of 15 μL. The samples were boiled for 5 minutes and subjected to SDS-PAGE on 12% polyacrylamide gel. The separated proteins on gels were transferred electrophoretically to a polyvinylidene difluoride filter (0.45 μm) in a wet-electroblotting system, the membrane were blocked at room temperature for an hour with blocking solution containing 5% (w/v) skim milk dissolved in TBST (0.1 M Tris-HCl, pH 8.0, 0.15 M NaCl, 0.05% (w/w) Tween-20). Then the membrane was incubated with the primary antibodies against 12S albumin (diluted 10000-fold) or 2S globulin (diluted 10000-fold) in 1% (w/v) skim milk dissolved in TBST at 4°C overnight. After rinsed several times with TBST, the membrane was incubated in the horseradish peroxidase-conjugated goat antibody against rabbit IgG which was diluted 10000-fold in TBST plus 1% skim milk. Signals were detected with an ECL kit (Thermo Scientific).
The anti-12S globulin and anti-2S albumin antibodies were kindly provided by Dr. Shigeru Utsumi of Kyoto University, Japan. The second antibody (the horseradish peroxidase-conjugated goat antibody against rabbit IgG) was purchased from ZSGB-BIO, China.
Generation of the point mutants of the conserved acidic residues in AtNHX5 and AtNHX6
The point mutants of the conserved acidic residues of AtNHX5 and AtNHX6 were generated from our previous study [49]. Briefly, pDONR-NHX5 and pDONR-NHX6 were used as templates to generate the point mutants. The site-directed mutagenesis was performed by Quikchange mutagenesis. For AtNHX5, the mutations were GAC to AAT (pDONR-NHX5-D164N), GAA to CAA (pDONR-NHX5-E188Q), GAT to AAT (pDONR-NHX5-D193N), and GAA to CAA (pDONR-NHX5-E320Q). The mutations of AtNHX6 were GAT to AAT (pDONR-NHX6-D165N), GAA to CAA (pDONR-NHX6-E189Q), GAT to AAT (pDONR-NHX6-D194N), and GAG to CAA (pDONR-NHX6-E320Q). These point mutants of AtNHX5 or AtNHX6 were recombined into pUBC-GFP using Gateway technology. The C-termini of these genes was fused with GFP, driven by Ubiquitin-10 (Ub10) promoter. These plasmids were transformed into the GV3101 A. tumefaciens strain, and the resulting bacterial clones were used to transform the A. thaliana (ecotype Columbia) by the floral dip procedure [60]. Transgenic plants were screened in vitro on MS medium supplemented with 0.0015% Basta. The homozygous lines of T3 progeny were selected for experiments.
Transient expression in Arabidopsis protoplasts and confocal microscopy
The PVC makers mRFP:AtVSR5 and GFP:AtVSR1, the TGN marker RFP:SYP61, and the Golgi marker Man1:RFP were generously provided by Dr. Liwen Jiang of The Chinese University of Hong Kong, China. The PVC/vacuoloar membrane marker GFP:SYP22 and the TGN marker GFP:SYP41 were kindly provided by Dr. Masa H. Sato of Kyoto University, Japan.
VAMP727 gene was constructed into the plasmid pSAT4A-mCherry-N1. mCherry gene was fused in frame to VAMP727 at its C-terminus. The VAMP727 gene was amplified using the following primers: VAMP727 (5’-CGCGGATCCATGAGTCAAAAGGGT-3’ and 5’- TGCTCTAGATGATGAGCATTTGAAACCTC-3’). Then the amplified production was cloned into the BamH1 and Xba1 sites of the pSAT4A-mCherry-N1 vector to yield the final plasmid VAMP727: mCherry. The positive gene fragment was verified by sequencing.
RFP gene and YFP gene were fused in frame to the N-termini of AtNHX5 and AtNHX6. The AtNHX5 and AtNHX6 were amplified using the following primers: AtNHX5 (5’-TCGCGGATCCATGGAGGAAGTGATGATT-3’ and 5’- CCCGGAATTCCTACTCCCCATCTCCATC-3’), AtNHX6 (5’- ATCGCGGATCCATGTCGTCGGAGCT-3’ and 5’- TCCGGAATTCTTAGCCGCGGTTATTTAGAT-3’). The amplified fragments were cloned into the EcoRI and BamHI sites of the pSAT6-C1-red fluorescent protein (RFP) and pSAT6-C1-yellow fluorescent protein (YFP) vector to yield the final plasmids RFP:AtNHX5, RFP:AtNHX6, YFP:AtNHX5 and YFP:AtNHX6.
The transient expression in protoplasts was performed as described [61, 62]. Arabidopsis seedlings of 4 weeks old were used for protoplast isolation. The protoplasts were derived from the leaf mesophyll cells of Arabidopsis. For SYP22 analysis, the plasmid GFP:SYP22 was transiently co-expressed in leaf protoplasts with the PVC marker mRFP:AtVSR5, TGN marker RFP:SYP61 and Golgi marker Man1:RFP. For VAMP727, VAMP727:mCherry was transiently co-expressed in leaf protoplasts with the PVC marker GFP:AtVSR1 and TGN marker GFP:SYP41. For co-localization assay between AtNHX5 or AtNHX6 and SYP22, the RFP:AtNHX5 or RFP:AtNHX6 plasmids were transiently co-expressed in Arabidopsis leaf protoplasts with SYP22:GFP. For co-localization assay between AtNHX5 or AtNHX6 and VAMP727, the YFP:AtNHX5 or YFP:AtNHX6 plasmids were transiently co-expressed in Arabidopsis leaf protoplasts with VAMP727:mCherry.
For confocal laser scanning microscopy, fixed cell were examined using object UPLAPO40×O13 (Olympus FV1000). The excitation/emission wavelengths were 488 /500-530 nm for GFP, 515/ 515–550 nm for YFP, and 559/590-630 for RFP, respectively. The colocalization was examined by directly monitoring the red, green or yellow fluorescence signal. To compare the images between Col-0 and nhx5 nhx6, the same conditions for image-taking and data analysis, including laser intensity, gray level, line average, and image processing, were used for Col-0 and nhx5 nhx6.
Quantitation of the colocalization of different fluorescence signals was performed as described [7,12]. The images of single frames from projection of the Z-stack were used for quantitation analysis. The images were disposed by Adobe Photoshop before quantitating the colocalization area with colocalization plugin in Image J (http://imagej.nih.gov/ij/plugins/).
Crosslinking and immunoprecipitation
1.5 g of 12-day-old wild-type seedlings and transgenic seedlings expressing GFP-tagged NHX5 or NHX6 were incubated in crosslinking buffer (PBS pH 7.4 and 1 mM dithiobis (succinimidyl propionate))for 1 hr at room temperature (RT), and then 1M Tris-HCl (pH 7.5) was added (final concentration, 20 mM) for blocking and incubated for 30 min at RT. Samples were then grinded in 1 ml of grinding buffer (50 mM HEPES-KOH pH7.5, 5 mM MgCl2, 2 mM EGTA, 250 mM sorbitol, and protease inhibitor cocktail Complete, EDTA-free (Roche)), and centrifuged at 1,000 × g for 10 min to remove debris. Supernatants were centrifuged at 20,000 ×g for 10 min to collect membrane fraction, and pellets were applied to immunoprecipitation. Immunoprecipitation from detergent extracts was conducted using the micro-MACS GFP-tagged protein isolation kit (Miltenyi Biotec) according to the manufacturer's instructions.
Electron microscopy
Sections were carried out as described with modifications [26, 50]. The Arabidopsis thaliana dry seeds were soaked in 50 mM PBS (pH 7.2) for 30 min, then, dozens of whole cotyledons were striped from the testa and fixed with buffer containing PBS (50 mM, pH 7.2) and 0.5% (w/v) glutaraldehyde for less than two hours, followed by dehydration in an ethanol series, and embedded in epoxy resin (Quetol-812; China).
For immunolabeling, sections were mounted on formvar-coated nickel grids etched with 3% (v/v) aqueous hydrogen peroxide for 10 min. After rinsing with distilled water several times, the ultrathin sections blocked for 30 min with blocking solution containing 5% (w/v) BSA and 0.1% Tween-20 in PBS (pH = 7.4). Then sections were incubated with antiserum against 12S globulin (1:50) diluted with diluting solution composed of PBS containing 0.25% Tween-20 and 1% (w/v) BSA at 4°C for overnight. After washing in diluting solution for three times, sections were incubated for 1hour at room temperature with antirabbit IgG secondary antibody conjugated to 15 nm colloidal gold (Sangon Biotech, China) diluted 1:20 in diluting solution. The sections were rinsed with PBS or distilled water and aired without stain. The sections were examined with a transmission electron microscope. Control experiment was performed by omitting the primary antibody.
Accession numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are, NHX5 (At1g54370), NHX6 (At1g79610), VAMP727 (At3g54300), SYP22 (At5g46860), VSR1 (AT3G52850), VSR5 (AT2G34940), SYP41 (AT5G26980), SYP61 (AT1G28490) and Man1 (AT1G02310).
Supporting Information
(A) SDS-PAGE stained with the Coomassie blue. Total proteins were extracted from the mature seeds. 10 μg Proteins were loaded in each lane. (B) and (C) Immunoblot analysis of 12S globulin (B) and 2S albumin (C).
(TIF)
(A) and (B) Immune EM assay. Embryos of Col-0 (A) and nhx5 nhx6 (B) were labeled with 12S albumin antibody. Bars = 250 nm. (C) and (D) Enlarged photos showing the extracellular spaces in embryos. The 12S antibody labeled the extracellular space in the double mutant (arrowheads). CW, cell wall; L, lipid body. Bars = 1μm.
(TIF)
(A) Morphology of the PSVs. Autofluorescence of the PSVs was visualized with a confocal laser scanning microscope. Bars = 10 μm. (B) Histograms representing a distribution of size and number of vacuoles within a single cell. Area of each vacuole was measured in cells of Col-0 and mutant embryos.
(TIF)
(A) T-DNA insertion sites in mutant alleles nhx5-2, syp22-1 and syp22-3. Black boxes and lines represent the exons and introns, respectively, in the coding region. (B) The growth phenotype of nhx5-2 nhx6-2 syp22-1 triple mutant. Seedlings were grown on soil for 30 d. (C) RT-PCR analysis of the mRNA expression level in the nhx5-2 nhx6-2 double mutant line. ACT2 was used as a control. (D) RT-PCR analysis of the mRNA expression level in the triple mutant lines. ACT2 was used as a control. In the nhx5-2 nhx6-2 syp22-1 triple mutant line, a cDNA fragment of SYP22, which was short than the WT, was detected by RT-PCR. This is consistent with the report of Ohtomo et al. (2005), where they showed that a 63 bp deletion was generated in syp22 mutant. Both Ohtomo et al. (2005) and us used the same T-DNA insertion line SALK_ 060946C, which has a T-DNA insertion in the fifth exon of SYP22.
(TIF)
Acknowledgments
We thank Dr. Shigeru Utsumi for sending the plasmid SP-GFP-CT24 and the anti-12S globulin and anti-2S albumin antibodies, Dr. Liwen Jiang for the vectors mRFP-AtVSR5, GFP-AtVSR1, RFP-SYP61 and the Man1-RFP, and Dr. Masa H. Sato for the vectors GFP-SYP22 and GFP-SYP41.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Basic Research Program of China (973 project, 2013CB429904 to QSQ), National Natural Science Foundation of China (NSFC,31070222 to QSQ), National Natural Science Foundation of China (NSFC,31371438 to QSQ), National Natural Science Foundation of China (NSFC,31571464 to QSQ), Doctoral Fund of Ministry of Education of China (RFDP, 20130211110001 to QSQ), Fundamental Research Funds for the Central Universities (lzujbky-2013-k10 to QSQ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Shewry PR, Napier JA, Tatham AS. Seed storage proteins: structures and biosynthesis. The Plant cell. 1995;7(7):945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herman EM, Larkins BA. Protein storage bodies and vacuoles. The Plant cell. 1999;11(4):601–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shewry PR, Halford NG. Cereal seed storage proteins: structures, properties and role in grain utilization. J Exp Bot. 2002;53(370):947–58. [DOI] [PubMed] [Google Scholar]
- 4.Robinson DG, Oliviusson P, Hinz G. Protein sorting to the storage vacuoles of plants: a critical appraisal. Traffic. 2005;6(8):615–25. [DOI] [PubMed] [Google Scholar]
- 5.Vitale A, Hinz G. Sorting of proteins to storage vacuoles: how many mechanisms? Trends Plant Sci. 2005;10(7):316–23. [DOI] [PubMed] [Google Scholar]
- 6.Sohn EJ, Rojas-Pierce M, Pan S, Carter C, Serrano-Mislata A, Madueno F, et al. The shoot meristem identity gene TFL1 is involved in flower development and trafficking to the protein storage vacuole. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(47):18801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang H, Kim SY, Song K, Sohn EJ, Lee Y, Lee DW, et al. Trafficking of vacuolar proteins: the crucial role of Arabidopsis vacuolar protein sorting 29 in recycling vacuolar sorting receptor. The Plant cell. 2012;24(12):5058–73. 10.1105/tpc.112.103481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hara-Hishimura I, Takeuchi Y, Inoue K, Nishimura M. Vesicle transport and processing of the precursor to 2S albumin in pumpkin. Plant J. 1993;4(5):793–800. [DOI] [PubMed] [Google Scholar]
- 9.Shimada T, Fuji K, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I. Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):16095–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuji K, Shimada T, Takahashi H, Tamura K, Koumoto Y, Utsumi S, et al. Arabidopsis vacuolar sorting mutants (green fluorescent seed) can be identified efficiently by secretion of vacuole-targeted green fluorescent protein in their seeds. The Plant cell. 2007;19(2):597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara-Nishimura I, Shimada T, Hatano K, Takeuchi Y, Nishimura M. Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. The Plant cell. 1998;10(5):825–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, Rogers JC. Integral membrane protein sorting to vacuoles in plant cells: evidence for two pathways. J Cell Biol. 1998;143(5):1183–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toyooka K, Okamoto T, Minamikawa T. Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. J Cell Biol. 2000;148(3):453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park M, Kim SJ, Vitale A, Hwang I. Identification of the protein storage vacuole and protein targeting to the vacuole in leaf cells of three plant species. Plant Physiol. 2004;134(2):625–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chrispeels MJ. The Golgi apparatus mediates the transport of phytohemagglutinin to the protein bodies in bean cotyledons. Planta. 1983;158(2):140–51. 10.1007/BF00397707 [DOI] [PubMed] [Google Scholar]
- 16.Hohl I, Robinson DG, Chrispeels MJ, Hinz G. Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. J Cell Sci. 1996;109(Pt 10):2539–50. [DOI] [PubMed] [Google Scholar]
- 17.Hinz G, Hillmer S, Baumer M, Hohl II. Vacuolar storage proteins and the putative vacuolar sorting receptor BP-80 exit the golgi apparatus of developing pea cotyledons in different transport vesicles. The Plant cell. 1999;11(8):1509–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillmer S, Movafeghi A, Robinson DG, Hinz G. Vacuolar storage proteins are sorted in the cis-cisternae of the pea cotyledon Golgi apparatus. J Cell Biol. 2001;152(1):41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park M, Lee D, Lee GJ, Hwang I. AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J Cell Biol. 2005;170(5):757–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimada T, Yamada K, Kataoka M, Nakaune S, Koumoto Y, Kuroyanagi M, et al. Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J Biol Chem. 2003;278(34):32292–9. [DOI] [PubMed] [Google Scholar]
- 21.Zouhar J, Munoz A, Rojo E. Functional specialization within the vacuolar sorting receptor family: VSR1, VSR3 and VSR4 sort vacuolar storage cargo in seeds and vegetative tissues. Plant J. 2010;64(4):577–88. 10.1111/j.1365-313X.2010.04349.x [DOI] [PubMed] [Google Scholar]
- 22.Lee Y, Jang M, Song K, Kang H, Lee MH, Lee DW, et al. Functional identification of sorting receptors involved in trafficking of soluble lytic vacuolar proteins in vegetative cells of Arabidopsis. Plant Physiol. 2013;161(1):121–33. 10.1104/pp.112.210914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang L, Phillips TE, Rogers SW, Rogers JC. Biogenesis of the protein storage vacuole crystalloid. J Cell Biol. 2000;150(4):755–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimada T, Kuroyanagi M, Nishimura M, Hara-Nishimura I. A pumpkin 72-kDa membrane protein of precursor-accumulating vesicles has characteristics of a vacuolar sorting receptor. Plant Cell Physiol. 1997;38(12):1414–20. [DOI] [PubMed] [Google Scholar]
- 25.Shimada T, Koumoto Y, Li L, Yamazaki M, Kondo M, Nishimura M, et al. AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 2006;47(9):1187–94. [DOI] [PubMed] [Google Scholar]
- 26.Ebine K, Okatani Y, Uemura T, Goh T, Shoda K, Niihama M, et al. A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. The Plant cell. 2008;20(11):3006–21. 10.1105/tpc.107.057711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshansky V, Futai M. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol. 2008;20(4):415–26. 10.1016/j.ceb.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11(1):50–61. 10.1038/nrm2820 [DOI] [PubMed] [Google Scholar]
- 29.Shen J, Zeng Y, Zhuang X, Sun L, Yao X, Pimpl P, et al. Organelle pH in the Arabidopsis endomembrane system. Mol Plant. 2013;6(5):1419–37. 10.1093/mp/sst079 [DOI] [PubMed] [Google Scholar]
- 30.Schumacher K. pH in the plant endomembrane system-an import and export business. Curr Opin Plant Biol. 2014;22:71–6. 10.1016/j.pbi.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 31.Martiniere A, Bassil E, Jublanc E, Alcon C, Reguera M, Sentenac H, et al. In vivo intracellular pH measurements in tobacco and Arabidopsis reveal an unexpected pH gradient in the endomembrane system. The Plant cell. 2013;25(10):4028–43. 10.1105/tpc.113.116897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paroutis P, Touret N, Grinstein S. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology. 2004;19:207–15. [DOI] [PubMed] [Google Scholar]
- 33.Demaurex N. pH Homeostasis of cellular organelles. News Physiol Sci. 2002;17:1–5. [DOI] [PubMed] [Google Scholar]
- 34.Orlowski J, Grinstein S. Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Curr Opin Cell Biol. 2007;19(4):483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirsch T, Paris N, Butler JM, Beevers L, Rogers JC. Purification and initial characterization of a potential plant vacuolar targeting receptor. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(8):3403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuoka K, Higuchi T, Maeshima M, Nakamura K. A Vacuolar-Type H+-ATPase in a Nonvacuolar Organelle Is Required for the Sorting of Soluble Vacuolar Protein Precursors in Tobacco Cells. The Plant cell. 1997;9(4):533–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boss WF, Morre DJ, Mollenhauer HH. Monensin-induced swelling of Golgi apparatus cisternae mediated by a proton gradient. Eur J Cell Biol. 1984;34(1):1–8. [PubMed] [Google Scholar]
- 38.Matsuoka K, Matsumoto S, Hattori T, Machida Y, Nakamura K. Vacuolar targeting and posttranslational processing of the precursor to the sweet potato tuberous root storage protein in heterologous plant cells. J Biol Chem. 1990;265(32):19750–7. [PubMed] [Google Scholar]
- 39.Reguera M, Bassil E, Tajima H, Wimmer M, Chanoca A, Otegui MS, et al. pH Regulation by NHX-Type Antiporters Is Required for Receptor-Mediated Protein Trafficking to the Vacuole in Arabidopsis. The Plant cell. 2015;27(4):1200–17. 10.1105/tpc.114.135699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashnest JR, Huynh DL, Dragwidge JM, Ford BA, Gendall AR. Arabidopsis Intracellular NHX-Type Sodium-Proton Antiporters are Required for Seed Storage Protein Processing. Plant Cell Physiol. 2015;56(11):2220–33. 10.1093/pcp/pcv138 [DOI] [PubMed] [Google Scholar]
- 41.Pardo JM, Cubero B, Leidi EO, Quintero FJ. Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J Exp Bot. 2006;57(5):1181–99. [DOI] [PubMed] [Google Scholar]
- 42.Bassil E, Coku A, Blumwald E. Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J Exp Bot. 2012;63(16):5727–40. 10.1093/jxb/ers250 [DOI] [PubMed] [Google Scholar]
- 43.Chanroj S, Wang G, Venema K, Zhang MW, Delwiche CF, Sze H. Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front Plant Sci. 2012;3(25). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu QS. Plant and yeast NHX antiporters: roles in membrane trafficking. J Integr Plant Biol. 2012;54(2):66–72. 10.1111/j.1744-7909.2012.01097.x [DOI] [PubMed] [Google Scholar]
- 45.Reguera M, Bassil E, Blumwald E. Intracellular NHX-type cation/H+ antiporters in plants. Mol Plant. 2014;7(2):261–3. 10.1093/mp/sst091 [DOI] [PubMed] [Google Scholar]
- 46.Brett CL, Tukaye DN, Mukherjee S, Rao R. The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol Biol Cell. 2005;16(3):1396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, et al. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2002;30(5):529–39. [DOI] [PubMed] [Google Scholar]
- 48.Bassil E, Ohto MA, Esumi T, Tajima H, Zhu Z, Cagnac O, et al. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. The Plant cell. 2011;23(1):224–39. 10.1105/tpc.110.079426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, Wu X, Liu Y, Qiu QS. AtNHX5 and AtNHX6 Control Cellular K+ and pH Homeostasis in Arabidopsis: Three Conserved Acidic Residues Are Essential for K+ Transport. PLoS One. 2015;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bassham DC, Raikhel NV. Unique features of the plant vacuolar sorting machinery. Curr Opin Cell Biol. 2000;12(4):491–5. [DOI] [PubMed] [Google Scholar]
- 51.Nishizawa K, Maruyama N, Satoh R, Fuchikami Y, Higasa T, Utsumi S. A C-terminal sequence of soybean beta-conglycinin alpha' subunit acts as a vacuolar sorting determinant in seed cells. Plant J. 2003;34(5):647–59. [DOI] [PubMed] [Google Scholar]
- 52.Bowers K, Levi BP, Patel FI, Stevens TH. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2000;11(12):4277–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohtomo I, Ueda H, Shimada T, Nishiyama C, Komoto Y, Hara-Nishimura I, et al. Identification of an allele of VAM3/SYP22 that confers a semi-dwarf phenotype in Arabidopsis thaliana. Plant Cell Physiol. 2005;46(8):1358–65. [DOI] [PubMed] [Google Scholar]
- 54.Uemura T, Morita MT, Ebine K, Okatani Y, Yano D, Saito C, et al. Vacuolar/pre-vacuolar compartment Qa-SNAREs VAM3/SYP22 and PEP12/SYP21 have interchangeable functions in Arabidopsis. Plant J. 2010;64(5):864–73. 10.1111/j.1365-313X.2010.04372.x [DOI] [PubMed] [Google Scholar]
- 55.Otegui MS, Spitzer C. Endosomal functions in plants. Traffic. 2008;9(10):1589–98. 10.1111/j.1600-0854.2008.00787.x [DOI] [PubMed] [Google Scholar]
- 56.Richter S, Voss U, Jurgens G. Post-Golgi traffic in plants. Traffic. 2009;10(7):819–28. 10.1111/j.1600-0854.2009.00916.x [DOI] [PubMed] [Google Scholar]
- 57.Fafournoux P, Noel J, Pouyssegur J. Evidence that Na+/H+ exchanger isoforms NHE1 and NHE3 exist as stable dimers in membranes with a high degree of specificity for homodimers. J Biol Chem. 1994;269(4):2589–96. [PubMed] [Google Scholar]
- 58.Inoue H, Noumi T, Tsuchiya T, Kanazawa H. Essential aspartic acid residues, Asp-133, Asp-163 and Asp-164, in the transmembrane helices of a Na+/H+ antiporter (NhaA) from Escherichia coli. FEBS Lett. 1995;363(3):264–8. [DOI] [PubMed] [Google Scholar]
- 59.Dibrov P, Young PG, Fliegel L. Functional analysis of amino acid residues essential for activity in the Na+/H+ exchanger of fission yeast. Biochemistry. 1998;37(23):8282–8. [DOI] [PubMed] [Google Scholar]
- 60.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–43. [DOI] [PubMed] [Google Scholar]
- 61.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–72. [DOI] [PubMed] [Google Scholar]
- 62.Li X, Chanroj S, Wu Z, Romanowsky SM, Harper JF, Sze H. A distinct endosomal Ca2+/Mn2+ pump affects root growth through the secretory process. Plant Physiol. 2008;147(4):1675–89. 10.1104/pp.108.119909 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) SDS-PAGE stained with the Coomassie blue. Total proteins were extracted from the mature seeds. 10 μg Proteins were loaded in each lane. (B) and (C) Immunoblot analysis of 12S globulin (B) and 2S albumin (C).
(TIF)
(A) and (B) Immune EM assay. Embryos of Col-0 (A) and nhx5 nhx6 (B) were labeled with 12S albumin antibody. Bars = 250 nm. (C) and (D) Enlarged photos showing the extracellular spaces in embryos. The 12S antibody labeled the extracellular space in the double mutant (arrowheads). CW, cell wall; L, lipid body. Bars = 1μm.
(TIF)
(A) Morphology of the PSVs. Autofluorescence of the PSVs was visualized with a confocal laser scanning microscope. Bars = 10 μm. (B) Histograms representing a distribution of size and number of vacuoles within a single cell. Area of each vacuole was measured in cells of Col-0 and mutant embryos.
(TIF)
(A) T-DNA insertion sites in mutant alleles nhx5-2, syp22-1 and syp22-3. Black boxes and lines represent the exons and introns, respectively, in the coding region. (B) The growth phenotype of nhx5-2 nhx6-2 syp22-1 triple mutant. Seedlings were grown on soil for 30 d. (C) RT-PCR analysis of the mRNA expression level in the nhx5-2 nhx6-2 double mutant line. ACT2 was used as a control. (D) RT-PCR analysis of the mRNA expression level in the triple mutant lines. ACT2 was used as a control. In the nhx5-2 nhx6-2 syp22-1 triple mutant line, a cDNA fragment of SYP22, which was short than the WT, was detected by RT-PCR. This is consistent with the report of Ohtomo et al. (2005), where they showed that a 63 bp deletion was generated in syp22 mutant. Both Ohtomo et al. (2005) and us used the same T-DNA insertion line SALK_ 060946C, which has a T-DNA insertion in the fifth exon of SYP22.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.