Abstract
The contractile protein myosin II is ubiquitous in muscle. It is widely accepted that animals express tissue-specific myosin isoforms that differ in amino acid sequence and ATPase activity in order to tune muscle contractile velocities. Recent studies, however, suggested that the squid Doryteuthis pealeii might be an exception; members of this species do not express muscle-specific myosin isoforms, but instead alter sarcomeric ultrastructure to adjust contractile velocities. We investigated whether this alternative mechanism of tuning muscle contractile velocity is found in other coleoid cephalopods. We analyzed myosin heavy chain transcript sequences and expression profiles from muscular tissues of a cuttlefish, Sepia officinalis, and an octopus, Octopus bimaculoides, in order to determine if these cephalopods express tissue-specific myosin heavy chain isoforms. We identified transcripts of four and six different myosin heavy chain isoforms in S. officinalis and O. bimaculoides muscular tissues, respectively. Transcripts of all isoforms were expressed in all muscular tissues studied, and thus S. officinalis and O. bimaculoides do not appear to express tissue-specific muscle myosin isoforms. We also examined the sarcomeric ultrastructure in the transverse muscle fibers of the arms of O. bimaculoides and the arms and tentacles of S. officinalis using transmission electron microscopy and found that the fast contracting fibers of the prey capture tentacles of S. officinalis have shorter thick filaments than those found in the slower transverse muscle fibers of the arms of both species. It thus appears that coleoid cephalopods, including the cuttlefish and octopus, may use ultrastructural modifications rather than tissue-specific myosin isoforms to adjust contractile velocities.
Additional key words: octopus, cuttlefish, contraction, myofilament
Myosin II is the major contractile protein found in both vertebrate and invertebrate muscle. It is composed of six subunits: two heavy chains in a helical dimer that form the rod, the flexible myosin S2 hinge region, and the myosin S1 heads, and four light chains that modulate myosin function by binding to myosin S2 and S1. The myosin S1 heads contain binding sites for ATP and for the thin filament protein actin, which positions myosin to initiate the cross bridge cycle of muscle contraction through the hydrolysis of ATP and interactions with actin.
The various myosin isoforms show a wide range of ATPase activity. For example, rat fast skeletal myosin (type II) hydrolyzes ATP three to five times faster than rat slow skeletal myosin (type I) (Schiaffino & Reggiani 1994). The difference in ATPase activity is due to differences in amino acid composition in two major regions of the myosin S1 heads: surface loop 1, which binds ATP (Murphy & Spudich 1998), and surface loop 2, which binds actin (Uyeda et al. 1994). Differences in ATPase activity in scallop striated and catch muscle myosin heavy chain isoforms have been attributed to sequence differences in surface loop 1 (Perreault-Micale et al. 1996).
Many animals tune muscle fiber contractile velocity largely through the expression of tissue-specific myosin isoforms that vary in ATPase activity. For example, rat fast skeletal muscle fibers shorten two to three times faster than rat slow skeletal muscle fibers (Bottinelli et al. 1991), primarily due to the higher ATPase activity of the rat fast skeletal myosin isoform. As demonstrated by Barany (1967), the shortening velocity of a muscle fiber is directly proportional to the ATPase activity of the myosin isoform expressed in that fiber; higher ATPase activity means that cross-bridge cycling rate and thus interfilamentary sliding velocity are higher, with a consequent direct effect on muscle fiber shortening velocity.
This mechanism for the modulation of shortening velocity was thought to be ubiquitous, but recent studies of the long-finned squid, Doryteuthis pealeii (Lesueur 1821), suggest that an alternative mechanism may be used by cephalopod molluscs. Squids are excellent models for the study of shortening velocities because a range of shortening velocities are found in the musculature of the appendages. Squid possess two tentacles that are rapidly elongated to capture prey, and eight arms that are used for slower bending movements such as manipulation of prey and steering during locomotion (Kier 1982; van Leeuwen & Kier 1997). The transverse muscle fibers of the tentacle contain short, cross-striated sarcomeres, whereas the transverse muscle of the arm contains obliquely-striated muscle fibers with long myosin filaments (Kier 1985). Shortening velocities of the tentacle cross-striated muscle fibers were found to be ten times higher than fibers from the arm transverse musculature (Kier & Curtin 2002). If the relationship demonstrated by Barany (1967) was true in squid, then distinct myosin heavy chain isoforms should be found in the tentacle and arm muscular tissues. However, Shaffer & Kier (2012) isolated and sequenced myosin heavy chain transcripts and found that D. pealeii does not express tissue-specific myosin heavy chain isoforms. Although three isoforms were identified (termed A, B, and C), all were found in all muscle tissues studied, and isoforms A and B were observed to be expressed at similar levels in the arm and tentacle musculature (Kier & Schachat 2008). These results suggest that squid use an alternative mechanism to tune contractile velocities. Comparison of thick filament lengths in the two muscle types revealed that the thick filaments of the fast contracting tentacle fibers are approximately 0.8 μm long while the slower fibers of the arms have thick filaments that are approximately 7.4 μm. The short myofilaments and sarcomeres of the tentacle fibers result in more elements in series per unit length of fiber, and since shortening velocities of elements in series are additive (Huxley & Simmons 1972; Josephson 1975; van Leeuwen 1991), this ultrastructural specialization is likely responsible for the observed differences in shortening velocity (Kier & Curtin 2002).
While D. pealeii uses this alternative mechanism to tune contractile velocities, it is unknown whether other coleoid cephalopods employ it as well (the cephalopod subclass Coleoidea includes all of the living cephalopods except the chambered nautiluses). In this study we examined an additional representative from each of the two superorders of coleoids, a cuttlefish species from the Decapodiformes (squids, cuttlefish, bobtail squid, and pygmy squids) and an octopus species from the Octopodiformes (octopuses, vampire squid, and argonauts). The cuttlefish Sepia officinalis Linnaeus 1758 studied here is a particularly relevant comparison since, like D. pealeii, it possesses two tentacles that are elongated rapidly during prey capture and eight arms that undergo slower bending movements (Messenger 1968, 1977). The octopus Octopus bimaculoides Pickford & McConnaughey 1949 was also examined since it is a representative of the other coleoid superorder. Octopus bimaculoides lacks the rapidly elongating prey capture tentacles of cuttlefish and squid, and uses its more slowly moving but highly manipulative eight arms for tasks such as locomotion, prey capture, and manipulation of objects.
The goal of this study was to determine if the mechanism employed by squid to tune contractile velocity may be more widespread in coleoid cephalopods. We sequenced myosin heavy chain transcripts from a number of tissues in S. officinalis and O. bimaculoides. In addition, we examined sarcomeric ultrastructure from selected muscles of these species using transmission electron microscopy.
Methods
Experimental animals
Live specimens of Octopus bimaculoides were purchased from Aquatic Research Consultants (San Pedro, CA), maintained in a recirculating artificial sea water system, and fed daily until used. Animals were anesthetized in chilled sea water (O’Dor & Shadwick 1989) and sacrificed. Tissue samples from the mantle, arm, funnel retractor, and buccal mass musculature were removed, frozen in RNAlater (Ambion, Life Technologies, Grand Island, NY) according to manufacturer’s instructions, and stored at −80°C until use.
Tissue samples of Sepia officinalis were purchased from Marine Biomedical Technologies (Bohemia, NY). Tissue samples from the mantle, arm, tentacular stalk, fin, and funnel retractor musculature were removed, flash frozen in liquid nitrogen, and shipped on dry ice. Samples were stored at −80°C until use.
Molecular cloning and sequencing methods
Myosin heavy chain transcripts were isolated, used as a template to produce cDNA, amplified with PCR, cloned and transformed into E. coli, and sequenced as described previously (Shaffer & Kier 2012). Briefly, total RNA was isolated from tissue samples using the GeneJET RNA Purification Kit (Fermentas, Glen Burnie, MD) according to manufacturer’s instructions. cDNA was synthesized with the Phusion RT-PCR kit (Finnzymes, Espoo, Finland) using an oligo(dT)15 primer for conventional PCR, an anchor primer (Q_total) for 3′RACE (rapid amplification of cDNA ends), or gene specific primers (Sepia_5′RACE_RT and Oct_5′RACE_RT) for 5′RACE (Table 1). The protocols for RACE were based on those described previously (Scotto-Lavino et al. 2006a, 2006b). For additional details, see Shaffer and Kier (2012).
Table 1.
Primers used for 5′ and 3′ RACE to clone myosin heavy chain sequences from cuttlefish (Sepia officinalis) and octopus (Octopus bimaculoides). All primers are written 5′ to 3′. Primers without a genus prefix (“Sepia” or “Oct”) were used for both cuttlefish and octopus PCR reactions.
| Primer name | Primer sequence |
|---|---|
| Q_total | CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGC(T)17 |
| Q_outer | CCAGTGAGCAGAGTGACG |
| Q_inner | GAGGACTCGAGCTCAAGC |
| 3′RACE_1 | GGAACACTACCAGATGGCTG |
| 3′RACE_2 | AGACCCAGAAGAGCATGC |
| Sepia_5′RACE_RT | ATCAGCTTCAGCCTGTTCA |
| Sepia_5′RACE_1 | GGTGACACGGGATTTCTC |
| Sepia_5′RACE_2 | TAGGCGTTGTCAGCCAC |
| Oct_5′RACE_RT | TGTTCAAAGCTGTTGAAGTCA |
| Oct_5′RACE_1 | CAGTGAAGCCCAACACATC |
| Oct_5′RACE_2 | GCAACGGAGAACAAATGG |
Conventional PCR and 5′ and 3′RACE reactions were carried out with the Phusion Hot Start II High-Fidelity DNA Polymerase (Finnzymes) according to manufacturer’s instructions in order to clone the myosin heavy chain cDNA in segments (see Shaffer & Kier [2012] for details). The primers used in these reactions are listed in Tables 1, 2, and 3. These primers were designed by aligning known cephalopod myosin heavy chain DNA sequences and locating degenerate regions. The sequences used were Doryteuthis pealeii isoform A (UniProt accession number G4V4Y6); Heterololigo bleekeri (Keferstein 1866) (as Doryteuthis bleekeri; B2ZTQ5); Todarodes pacificus (Steenstrup 1880) (F1ADJ4); Sepia esculenta Hoyle 1885 (B2ZTQ6); Argopecten irradians (Lamarck 1819) (P24733); Pecten maximus (Linnaeus 1758) (Q9U7E3); Mizuhopecten yessoensis (Jay 1857) (Q9BLD0); and both catch and striated myosin heavy chain isoforms from Placopecten magellanicus (Gmelin 1791) (Q26080 and Q26079, respectively). Sequences were aligned with ClustalX2 (version 2.1) (Larkin et al. 2007) and degenerate regions were identified by eye.
Table 2.
Primers used for conventional PCR to clone myosin heavy chain sequences from cuttlefish (Sepia officinalis) and octopus (Octopus bimaculoides). All primers are written 5′ to 3′. Primers without the genus prefix “Sepia” were used for both cuttlefish and octopus PCR reactions.
| Primer name | Primer sequence |
|---|---|
| 368_F | TCAACCCCTACAGACGTCTT |
| 2166_R | GTATCTCTGCTTGAACTCAGAGTAGA |
| 2071_F | CAGCTCCGTTGTAACGG |
| 4405_R | TTCTCCAATTCAGCCTGG |
| 4361_F | AATGGCAACACAAATGCAACG |
| 5320_R | GGTCTTGTTCTTGTCTAAGC |
| Sepia_1_F | ATGTCGTCCTACGATCCAAG |
| Sepia_540_R | TCCAGACTCTCCAGTAATCAACA |
| Sepia_3781_F | ATGCTCCGAGAAGATGAGC |
| Sepia_5814_R | TTACATGGGACTGGCAGC |
Table 3.
Primers used for conventional PCR to detect specific myosin heavy chain isoforms in cuttlefish (Sepia officinalis) and octopus (Octopus bimaculoides) tissues. All primers are written 5′ to 3′.
| Primer name | Primer sequence |
|---|---|
| Sepia_A-B_F | GCAGGTAAAACTGAGAACACG |
| Sepia_A-B_R | TGTGTGCCGAAGTGGAT |
| Sepia_A-C_F | AGACCCAGAAGAGCATGC |
| Sepia_A_R | TACATGGGACTGGCACG |
| Sepia_C_R | TCATTGATTCGATGGAGCT |
| Oct_A-B_F | AGACGAAAAGAAGGGTACCC |
| Oct_A-B_R | TGGGAAGGTAGGTGTAAGGA |
| Oct_A-C-D-E_F | AGACCCAGAAGAGCATGC |
| Oct_A_R | TTACAAAGGACTAGCGCGAG |
| Oct_C_R | TCATTGTGAGACAGAACTGGC |
| Oct_D_R | TCACTGAGAAGATGACGCG |
| Oct_E_R | TTAGAAGTGGTCATCTTCTCCAC |
Once PCR was completed, samples were separated using agarose gel electrophoresis, and bands were excised and purified using the GeneJET Gel Extraction Kit (Fermentas). Purified PCR products were cloned and ligated into the pJET1.2/blunt vector (Fermentas) and transformed into competent JM107 E. coli bacteria prepared using the TransformAid Bacterial Transformation Kit (Fermentas). Colonies were selected, grown overnight, and plasmid DNA was isolated using the GeneJET Plasmid Miniprep Kit (Fermentas). Plasmid DNA was sequenced by Eton Biosciences (Durham, NC) using the Fermentas pJET forward and reverse primers, as well as myosin-specific primers in some cases. Sequence analysis was performed using Sequence Scanner software (v1.0, Applied Biosystems, Carlsbad, CA) and tools on the ExPASy web server (Gasteiger et al. 2003).
Conventional PCR was also used to determine if specific myosin heavy chain isoforms were expressed in tissues of S. officinalis and O. bimaculoides. Once Sepia isoform C and Octopus isoforms C, D, and E were found using the methods described above, specific primers were designed for these isoforms (Table 3) and PCR was completed with all tissue cDNA samples in order to search for the expression of these isoforms. In order to minimize chances of cross-reactions with other isoforms, primers were design with significant overlap with the specific isoforms they were designed to amplify; in addition, the annealing temperatures used in the PCR reactions were within 1 to 2°C of the Tm of the primers (see Shaffer and Kier [2012] for additional details). Upon completion of PCR, the samples were separated using agarose gel electrophoresis and PCR products were analyzed based on size.
Transmission electron microscopy
Cross-sectional slices 2–3 mm thick of the tentacles and arms of three adult specimens of S. officinalis and of the arms of four adult specimens of O. bimaculoides were fixed in 3.0% glutaraldehyde, 0.065M phosphate buffer, 0.5% tannic acid, and 6% sucrose for 6–8 h at 4°C. Following fixation, small (~1×1×2 mm) blocks of tissue were cut from the slices, rinsed overnight in chilled 0.065 M phosphate buffer, and post-fixed for 40 min at 4°C in a 1:1 mixture of 2% osmium tetroxide and 2% potassium ferrocyanide. The blocks were rinsed in chilled 0.065 M cacodylate buffer for 15 min, and then dehydrated and cleared in a graded acetone series. Acetone was used in place of ethanol and propylene oxide in order to minimize dimensional changes in the myofilaments (Page & Huxley 1963). The tissue blocks were embedded in epoxy resin (Epox 812, Ernest R. Fullam, Latham, NY). During sectioning, we attempted to align the section plane with the longitudinal axis of the fibers by examining 0.5–1.0 μm sections in the light microscope and reorienting the block until individual fibers in the area of interest remained in the section plane. The block was then trimmed and sectioned with a diamond knife. Sections of silver to gold interference color were stained with saturated aqueous uranyl acetate and Reynolds lead citrate (Reynolds 1963) and photographed in Zeiss EM 10CA and Philips Tecnai 12 electron microscopes. Thick filament lengths were measured on micrographs using morphometrics software (Sigma-Scan Pro, Systat Sofware, Inc., San Jose, CA).
Results
Myosin heavy chain sequences in Sepia officinalis
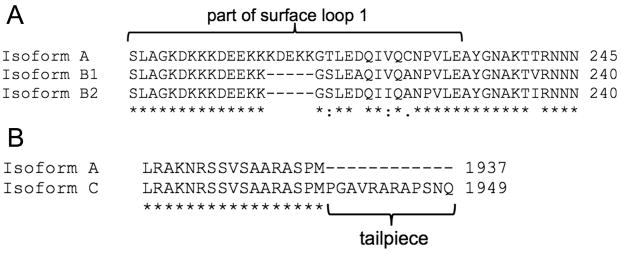
The complete myosin heavy chain transcript sequences for five muscular tissues (mantle, arm, tentacle, fin, and funnel retractor) were obtained from Sepia officinalis. Four isoforms were identified; these are homologous to those found in Doryteuthis pealeii (Shaffer & Kier 2012). The coding region (CDS) of isoform A was composed of 5814 nucleotides and was translated to 1937 amino acids. Isoform A was 95.3% identical to isoform A from D. pealeii (UniProt G4V4Y6). Partial sequences of three additional isoforms (referred to as isoforms B1, B2, and C) were found by analyzing the PCR products from the different tissue cDNAs. Figure 1 displays partial sequences of the four isoforms and how they differ from one another (full sequences can be found through GenBank or EMBL, Table 4). Isoforms B1 and B2 differ from isoform A due to the absence of a five amino acid sequence in surface loop 1 near the N-terminus, and there are also four amino acid substitutions in surface loop 1 (Fig. 1A). Isoform B1 differs from isoform B2 by three amino acid substitutions in surface loop 1. Isoform C contains a twelve amino-acid tailpiece at the C-terminus compared with isoform A (Fig. 1B). These isoforms are homologous to isoforms B and C described previously (Matulef et al. 1998; Shaffer & Kier 2012).
Fig. 1.
Comparisons of Sepia officinalis myosin isoforms A, B1, B2, and C. A. Translated amino acid sequences of myosin heavy chain isoforms A, B1, and B2. Isoforms B1 and B2 contain a five amino acid deletion and four amino acid substitutions in surface loop 1 compared to isoform A. Isoform B1 differs from isoform B2 by three amino acids. B. Translated amino acid sequences of myosin heavy chain isoforms A and C. Isoform C contains a 12 amino acid extension compared to isoform A. Asterisks (*) denote identical amino acids, colons (:) denote well conserved amino acids, periods (.) denote somewhat conserved amino acids, and blank spaces denote no conservation.
Table 4.
Summary of differences among Sepia officinalis muscle myosin heavy chain isoforms, relative to isoform A.
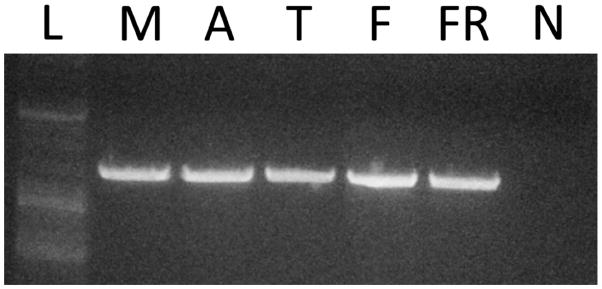
All four myosin heavy chain isoforms were found to be expressed in all five S. officinalis tissues studied. Isoforms A, B1, and B2 were observed by sequencing short PCR products that contained the surface loop 1 sequence (Sepia_A-B_F and Sepia_A-B_R primers, Table 3) prepared from the five tissues studied. Isoform C was first detected using 3′RACE with cDNA from tentacle tissue, and then a reverse primer specific to the isoform C sequence was created (Table 3) and used in PCR to determine if isoform C was expressed in the other four S. officinalis tissues. As shown in Figure 2, a band for isoform C was evident in all five cuttlefish tissues studied, suggesting that isoform C was expressed in all five tissues.
Fig. 2.
Expression of isoform C in muscular tissues of Sepia officinalis. PCR was used with a specific isoform C reverse primer to detect isoform C in S. officinalis tissues. L, ladder; M, mantle; A, arm; T, tentacle; F, fin; FR, funnel retractor; N, non-template control.
Ultrastructure of arm and tentacle transverse muscle in Sepia officinalis
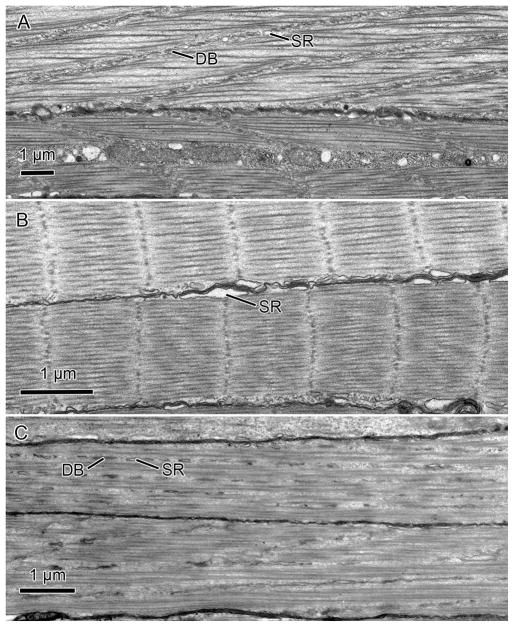
The ultrastructure of transverse muscles we observed in S. officinalis was indistinguishable from that described previously for squid arms and tentacles (Kier 1982, 1991; Kier & Curtin 2002), so we provide only a summary here. The fibers of the transverse muscle mass of the arms of S. officinalis were obliquely striated, the typical striation pattern found in the musculature of cephalopods (for a review see Budelmann et al. 1997) (Fig. 3A). The myofilaments of this fiber type were oriented parallel to the long axis of the fiber but they were not aligned in register across the cell. Instead, they were arranged in a staggered array so that the A and I bands followed oblique or helical courses of low angle relative to the longitudinal axis of the fiber. This was most easily observed in longitudinal sections, where the dense bodies or Z elements, which anchor the thin myofilaments, could be seen to be aligned at a small angle to the longitudinal axis of the fiber (Fig. 3A). It was challenging to obtain accurate measurements of thick filament length in these fibers because of the difficulty of obtaining precisely longitudinal sections. In some sections we were able to trace thick filaments for 7.6 μm, but this may not represent their total length. The sarcoplasmic reticulum appeared similar to that described previously for the homologous muscle in squids and included a peripheral zone present in the subsarcolemmal cytoplasm, a zone in the plane of the Z elements, and a central zone surrounding the mitochondria that occupied the core of the fibers. The fibers lacked invaginated tubules and the sarcoplasmic reticulum made specialized peripheral couplings with the sarcolemma.
Fig. 3.
Transmission electron micrographs of transverse muscle fibers of Sepia officinalis and Octopus bimaculoides. A. Transverse fibers from the arm of S. officinalis. B. Transverse fibers from the tentacle of S. officinalis. C. Transverse fibers from the arm of O. bimaculoides. DB, dense body; SR, sarcoplasmic reticulum.
In contrast to the fibers of the arms, muscle cells from the transverse muscle mass of the tentacles were cross-striated with similar ultrastructure to that described previously for the fast-contracting transverse muscle mass of the tentacles of loliginid and ommastrephid squids (Kier 1985, 1991; Kier & Curtin 2002) (Fig. 3B). The A-band of these fibers, as in the squids, was quite short (mean=0.81 μm, S.D.=0.18 μm). The material of the Z-discs was not the organized network that has been described for vertebrate skeletal muscle fibers, but instead appeared to be a loosely arranged grouping of electron-dense material. The sarcomeres appeared to be susceptible to shearing, and the Z discs were often not precisely perpendicular to the long axis of the cell. As in the arm fibers, the tentacle cells lacked invaginated tubules and the sarcoplasmic reticulum formed specialized couplings directly with the sarcolemma.
Myosin heavy chain sequences in Octopus bimaculoides
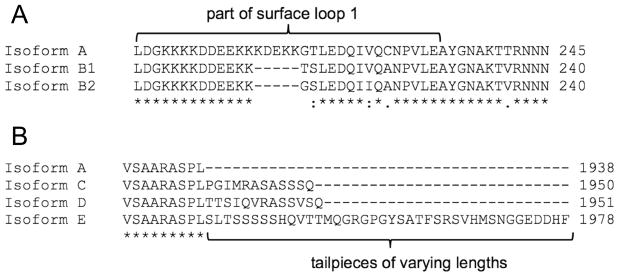
We identified six different myosin heavy chain isoforms in four muscular tissues of Octopus bimaculoides (mantle, arm, funnel retractor, and buccal mass). We identified myosin heavy chain isoform A (CDS of 5817 nucleotides, 1938 amino acids), homologous to isoform A found in S. officinalis (89.4% identity) and squid (89.4% identity), and also partial sequences for two variants of isoform B that were homologous to the B isoforms from S. officinalis and D. pealeii (Table 5). Both isoform B1 and B2 contained a five-amino acid deletion of surface loop 1 compared to isoform A, but they differed from one another by virtue of the specific substitutions (Fig. 4A). Partial sequences for three C-terminal isoforms were also identified and are summarized in Table 5 and Figure 4B. Isoform C contained a twelve-amino acid tailpiece, isoform D contained a thirteen-amino acid tailpiece, and isoform E contained a 48-amino acid tailpiece. All tailpiece sequences of isoforms C, D, and E were distinct from one another.
Table 5.
Summary of differences among Octopus bimaculoides muscle myosin heavy chain isoforms, relative to isoform A.
Fig. 4.
Comparisons of Octopus bimaculoides myosin isoforms A, B1, B2, C, D, and E. A. Translated amino acid sequences of myosin heavy chain isoforms A, B1, and B2. Isoform B1 and B2 contain five amino acid deletions and four amino acid substitutions in surface loop 1 compared to isoform A. Isoform B1 differs from isoform B2 by two amino acids. B. Translated amino acid sequences of myosin heavy chain isoforms A, C, D, and E. Isoforms C, D, and E contain 12, 13, and 40 amino acid extensions, respectively, compared to isoform A. Asterisks (*) denote identical amino acids, colons (:) denote well conserved amino acids, periods (.) denote somewhat conserved amino acids, and blank spaces denote no conservation.
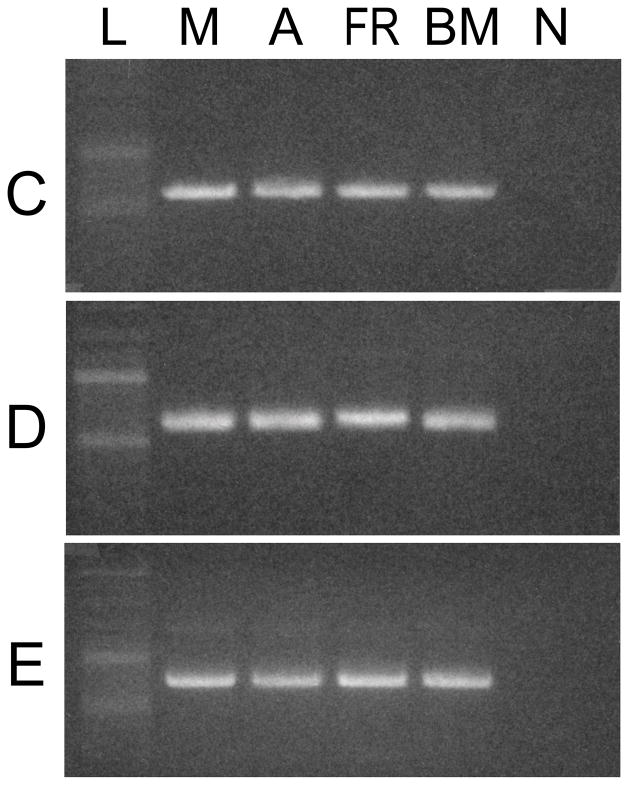
All six isoforms were found in all four O. bimaculoides tissues studied. Isoforms A, B1, and B2 were identified by sequencing short PCR products that contained the surface loop 1 sequence (Oct_A-B_F and Oct_A-B_R primers, Table 3) prepared from each of the four O. bimaculoides tissues studied. Isoforms C, D, and E were first detected with 3′RACE from the buccal mass, mantle, and funnel retractor cDNA, respectively. Once these isoforms were identified, specific reverse primers were developed and used with a common forward primer (Oct_A-C-D-E_F, Table 3) in PCR reactions to determine if these isoforms were expressed in all four tissues. Bands were observed for isoforms C, D, and E in all four tissues (Fig. 5).
Fig. 5.
Expression of isoforms C, D, and E in muscular tissues of Octopus bimaculoides. PCR was used with specific isoform C, D, and E reverse primers to detect isoforms in O. bimaculoides tissues. The top panel shows the distribution of isoform C; the middle panel that of isoform D; and the bottom panel that of isoform E. L, ladder; M, mantle; A, arm; FR, funnel retractor; BM, buccal mass; N, non-template control.
Ultrastructure of the transverse muscle of the arms of Octopus bimaculoides
The transverse muscle fibers of the arms of O. bimaculoides were observed to be obliquely striated with similar ultrastructure (Fig. 3C) to that described above for the transverse muscle of the arms of the cuttlefish S. officinalis. The thick myofilaments were quite long in these fibers as well; we were able to trace thick filaments for 7.9 μm, but this may not represent their full length, given the challenges of obtaining perfectly aligned longitudinal sections.
Discussion
The major finding from this study is that coleoid cephalopods, including squid, cuttlefish, and octopus, appear to use sarcomeric ultrastructural differences, rather than the expression of tissue-specific myosin heavy chain isoforms, to tune contractile velocity. This is also the first time that the myosin heavy chain has been sequenced from octopus muscular tissues. The results from this study provide important insight into the diversity of mechanisms of muscle regulation.
While it is widely accepted that animals express tissue-specific myosin heavy chain isoforms to tune muscle contractile velocities (Barany 1967), squids appear to be an exception. Shaffer & Kier (2012) showed that the muscular tissues of the squid Doryteuthis pealeii do not express tissue-specific myosin heavy chain isoforms. The results from the current study are consistent with this finding and provide preliminary support for the hypothesis that cephalopod molluscs in general use ultrastructural differences rather than differences in myosin heavy chain isoforms to tune contractile velocity. Similar to D. pealeii, the cuttlefish Sepia officinalis analyzed in the present study has two tentacles that rapidly elongate and eight more slowly moving arms, yet the arms and tentacles express identical transcripts for myosin heavy chain isoforms. Transcripts of these isoforms were also expressed in the mantle, fin, and funnel retractor musculature. While Octopus bimaculoides does not possess musculature that varies so widely in terms of contractile velocity, we also found that this species expresses identical myosin heavy chain isoforms in the mantle, arm, funnel retractor, and buccal mass tissues. Thus, none of the representatives from three major coleoid cephalopod groups (squids, cuttlefishes, and octopuses) that have been studied express tissue-specific transcripts of myosin heavy chain isoforms in their muscular tissues.
Previous research on loliginid and ommastrephid squids has shown that instead of expressing tissue-specific myosin heavy chain isoforms, the modulation of shortening velocity occurs through variation in the ultrastructure of the sarcomeres (Kier 1985, 1991; Kier & Schachat 1992, 2008; Shaffer & Kier 2012; van Leeuwen & Kier 1997). The most important factors are the thick filament and sarcomere length. For example, in the squid D. pealeii, the thick filaments of the fast-contracting cross-striated tentacle fibers were found to be ~0.8 μm long, while those of the more slowly contracting obliquely striated muscle of the arms were ~7.5 μm long (Kier & Curtin 2002). Because shortening velocities of elements in series are additive, muscles with shorter myofilaments and sarcomeres show higher unloaded shortening velocities, assuming other factors are constant (Huxley & Simmons 1972; Josephson 1975). Since the difference in thick filament length in D. pealeii was approximately ten fold, the shortening velocity of the tentacle fibers was predicted to be approximately ten times higher. Measurement of the contractile properties of these fibers was consistent with this prediction; the unloaded shortening velocity of the tentacle fibers was found to be approximately 15 L0s−1 while that of the arm fibers was approximately 1.5 L0s−1 (Kier & Curtin 2002).
The results of the preliminary ultrastructural analysis of the S. officinalis and O. bimaculoides muscle fibers described in the current study are consistent with the previous work on squids, and show that the fast contracting tentacle fibers of S. officinalis possess shorter myofilaments and sarcomeres, compared with the more slowly contracting fibers of the arms of S. officinalis and O. bimaculoides. Although measurements of contractile properties are needed, along with a more extensive morphometric analysis of the ultrastructure, the results presented here are consistent with the hypothesis that cephalopods may tune contractile properties primarily through variation in ultrastructure.
Four myosin heavy chain isoforms were identified in muscular tissues of S. officinalis in this study, whereas six isoforms were identified in muscular tissues of O. bimaculoides. Previously, three isoforms were identified in the muscular tissues of squid (Shaffer & Kier 2012). Why then, do S. officinalis and O. bimaculoides express more myosin isoforms than the squid? One possibility is that these additional isoforms perform functional roles in the cuttlefish and octopus not found in squid. This seems unlikely to be the case for isoforms B1 and B2 because they only differ from each other at two positions for the octopus (T213G and V220I) and three positions for the cuttlefish (A219D, V221I, and V236I). Both of thee substitutions in the octopus myosin heavy chain are conserved small hydrophobic amino acids, so functional differences are unlikely to result. Two of the three substitutions between cuttlefish isoforms B1 and B2 are small hydrophobic amino acids (V221I and V236I) so are unlikely to result in functional differences, however the substitution A219D is a small hydrophobic alanine to a negatively charged aspartic acid, which may affect function. Functional differences are also more likely to occur within the variable C-terminal isoforms C, D, and E, which have C-terminal tails of 12, 13, and 40 amino acids, respectively, compared with isoform A (Fig. 4B). Prior work has shown that scallop catch myosin heavy chain contains a 10-amino-acid C-terminal tailpiece (Perreault-Micale et al. 1996) of unknown significance. Rabbit smooth muscle myosin is alternatively spliced to produce two C-terminal isoforms, one with a 43 amino acid tail (SM1) and the other with a nine amino acid tail (SM2) (Rovner et al. 2002), and thick filament assembly differed depending on whether the SM1 or SM2 myosin isoform was involved. In Drosophila indirect flight muscle, expression of a myosin heavy chain isoform with a C-terminal tailpiece may contribute to thick filament assembly and stability and A-band development (Orfanos & Sparrow 2013). Based on these studies, it could therefore be possible that thick filament assembly differs with the expression of isoforms C, D, and E described in this study. While a functional difference is possible between the shorter C-terminal isoforms C and D and the longer C-terminal isoform E, all three of these isoforms were expressed in all four O. bimaculoides tissues studied (Fig. 5), so a functional difference, if any, would not affect function in a tissue-specific manner. A final possibility is that the squid also expresses more than three myosin heavy chain isoforms, but that they were missed in the previous study by Shaffer & Kier (2012). While possible, we think that it is unlikely due to the large number of samples prepared and sequenced over the course of the study.
In this study we identified myosin heavy chain isoforms that vary in sequence at surface loop 1 near the N-terminus, and also isoforms that vary in length at the C-terminus. Because we sequenced these isoforms from PCR products that did not encompass the full-length cDNA, it is possible that various combinations of isoforms exist. As we and others have described earlier (Matulef et al. 1998; Shaffer & Kier 2012), isoform A contains a five amino acid insertion in surface loop 1 compared with isoform B and has a shorter C-terminus than isoforms C, D, or E. It is possible, however, that the isoform A characteristics of surface loop 1 (the five amino acid addition) could be found with a longer C-terminus, thus resulting in a hybrid of isoforms A and C (or D or E in the octopus). Additionally, isoform B could be paired with either the short isoform A C-terminus or the longer C-termini of isoforms C, D, and E. In total, there are four potential isoform combinations in the squid and cuttlefish, and 12 potential combinations in the octopus. In addition, differential post-translation modifications of the myosin heavy chain protein isoforms could affect sarcomeric function in S. officinalis and O. bimaculoides. Molluscan catch muscle myosin can be phosphorylated in the rod region (Castellani & Cohen 1987) to regulate the catch state and ATPase activity. The tailpieces identified in this study contain potentially phosphorylatable serine residues that could participate in similar regulation of ATPase activity. Additional investigation is needed to determine what potential combinations of myosin heavy chain isoforms are expressed in cephalopod tissues and how potential post-translational modifications may affect function.
The existence of multiple myosin isoforms in both the cuttlefish and octopus also raises that possibility that they are expressed at unequal levels in the tissues. Quantitative PCR analysis was beyond the scope of the present study so we do not know whether or not the isoforms are expressed at different levels in the various tissues studied. This is of interest since differences in the relative levels of transcripts could potentially affect ATPase activity and thereby the shortening velocities of the various muscle groups. Kier & Schachat (2008) used semi-quantitative PCR and demonstrated that in both the squid arm and tentacle transverse musculature, ~90% of the myosin heavy chain expressed is isoform A, while the other 10% is isoform B. We found the homologous isoforms A and B in S. officinalis and O. bimaculoides, and A may prove to be the dominant form in these species as well, but future work employing quantitative methods is needed in order to assess any potential variation in expression in the various muscular tissues.
In conclusion, we have shown that S. officinalis and O. bimaculoides, like the squid D. pealeii (Shaffer & Kier 2012), do not appear to express tissue-specific myosin heavy chain isoforms. Although additional work is needed, these results suggest that coleoid cephalopods in general may use ultrastructural differences, primarily variation in thick filament length, to tune contractile velocity. This unusual form of regulation of contractile velocity warrants additional investigation to determine if it is more widespread than currently recognized.
Acknowledgments
The authors thank Allison Sarfati, Dylan Catlett, and Sonia Guarda for technical assistance, and Kathleen K. Smith for use of equipment. This work was supported by the Seeding Postdoctoral Innovators in Research and Education (SPIRE) program funded by the National Institutes of Health, Minority Opportunities in Research division of the National Institute of General Medical Sciences [grant K12GM000678 to J.S.] and the National Science Foundation [grant IOS-0951067 to W.M.K.].
References
- Barany M. ATPase activity of myosin correlated with speed of muscle shortening. J Gen Physiol. 1967;50:197–218. doi: 10.1085/jgp.50.6.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottinelli R, Schiaffino S, Reggiani C. Force-velocity relations and myosin heavy-chain isoform compositions of skinned fibers from rat skeletal-muscle. J Physiol-London. 1991;437:655–672. doi: 10.1113/jphysiol.1991.sp018617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budelmann BU, Schipp R, Boletsky SV. Cephalopoda. In: Harrison FW, Kohn AJ, editors. Microscopic Anatomy of Invertebrates, Vol.2. Mollusca II. Wiley-Liss; New York: 1997. pp. 119–414. [Google Scholar]
- Castellani L, Cohen C. Myosin rod phosphorylation and the catch state of molluscan muscles. Science. 1987;235:334–337. doi: 10.1126/science.3026049. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley AF, Simmons RM. Mechanical transients and the origin of muscular force. Cold Spring Harb Sym. 1972;37:669–680. [Google Scholar]
- Josephson RK. Extensive and intensive factors determining the performance of striated muscle. J Exp Zool. 1975;194:135–154. doi: 10.1002/jez.1401940109. [DOI] [PubMed] [Google Scholar]
- Kier WM. The functional-morphology of the musculature of squid (Loliginidae) arms and tentacles. J Morphol. 1982;172:179–192. doi: 10.1002/jmor.1051720205. [DOI] [PubMed] [Google Scholar]
- Kier WM. The musculature of squid arms and tentacles: ultrastructural evidence for functional differences. J Morphol. 1985;185:223–239. doi: 10.1002/jmor.1051850208. [DOI] [PubMed] [Google Scholar]
- Kier WM. Squid cross-striated muscle: the evolution of a specialized muscle fiber type. Bull Mar Sci. 1991;49:389–403. [Google Scholar]
- Kier WM, Curtin NA. Fast muscle in squid (Loligo pealei): contractile properties of a specialized muscle fibre type. J Exp Biol. 2002;205:1907–1916. doi: 10.1242/jeb.205.13.1907. [DOI] [PubMed] [Google Scholar]
- Kier WM, Schachat FH. Biochemical comparison of fast- and slow- contracting squid muscle. J Exp Biol. 1992;168:41–56. doi: 10.1242/jeb.168.1.41. [DOI] [PubMed] [Google Scholar]
- Kier WM, Schachat FH. Muscle specialization in the squid motor system. J Exp Biol. 2008;211:164–169. doi: 10.1242/jeb.008144. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Matulef K, Sirokman K, Perreault-Micale CL, Szent-Gyorgyi AG. Amino-acid sequence of squid myosin heavy chain. J Muscle Res Cell M. 1998;19:705–712. doi: 10.1023/a:1005341416989. [DOI] [PubMed] [Google Scholar]
- Murphy CT, Spudich JA. Dictyostelium myosin 25–50K loop substitutions specifically affect ADP release rates. Biochemistry. 1998;37:6738–6744. doi: 10.1021/bi972903j. [DOI] [PubMed] [Google Scholar]
- O’Dor RK, Shadwick RE. Squid, the olympian cephalopods. J Ceph Biol. 1989;1:33–55. [Google Scholar]
- Orfanos Z, Sparrow JC. Myosin isoform switching during assembly of the Drosophila flight muscle thick filament lattice. J Cell Sci. 2013;126:139–148. doi: 10.1242/jcs.110361. [DOI] [PubMed] [Google Scholar]
- Page SG, Huxley HE. Filament lengths in striated muscle. J Cell Biol. 1963;19:369–390. doi: 10.1083/jcb.19.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault-Micale CL, Kalabokis VN, Nyitray L, Szent-Gyorgyi AG. Sequence variations in the surface loop near the nucleotide binding site modulate the ATP turnover rates of molluscan myosins. J Muscle Res Cell M. 1996;17:543–53. doi: 10.1007/BF00124354. [DOI] [PubMed] [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovner AS, Fagnant PM, Lowey S, Trybus KM. The carboxyl-terminal isoforms of smooth muscle myosin heavy chain determine thick filament assembly properties. J Cell Biol. 2002;156:113–123. doi: 10.1083/jcb.200107131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Myosin isoforms in mammalian skeletal muscle. J Appl Physiol. 1994;77:493–501. doi: 10.1152/jappl.1994.77.2.493. [DOI] [PubMed] [Google Scholar]
- Scotto-Lavino E, Du G, Frohman MA. 3′ end cDNA amplification using classic RACE. Nat Protoc. 2006a;1:2742–2745. doi: 10.1038/nprot.2006.481. [DOI] [PubMed] [Google Scholar]
- Scotto-Lavino E, Du G, Frohman MA. 5′ end cDNA amplification using classic RACE. Nat Protoc. 2006b;1:2555–2562. doi: 10.1038/nprot.2006.480. [DOI] [PubMed] [Google Scholar]
- Shaffer JF, Kier WM. Muscular tissues of the squid Doryteuthis pealeii express identical myosin heavy chain isoforms: an alternative mechanism for tuning contractile speed. J Exp Biol. 2012;215:239–46. doi: 10.1242/jeb.064055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda TQP, Ruppel KM, Spudich JA. Enzymatic-activities correlate with chimeric substitutions at the actin-binding face of myosin. Nature. 1994;368:567–569. doi: 10.1038/368567a0. [DOI] [PubMed] [Google Scholar]
- van Leeuwen JL. Optimum power output and structural design of sarcomeres. J Theor Biol. 1991;149:229–256. doi: 10.1016/s0022-5193(05)80279-6. [DOI] [PubMed] [Google Scholar]
- van Leeuwen JL, Kier WM. Functional design of tentacles in squid: linking sarcomere ultrastructure to gross morphological dynamics. Philos T Roy Soc B. 1997;352:551–571. [Google Scholar]