Abstract
The conserved Piwi family of proteins and piwi-interacting RNAs (piRNAs) play a central role in genomic stability, which is inextricably tied with germ cell formation, by forming ribonucleoproteins (piRNPs) that silence transposable elements (TEs)1. In Drosophila melanogaster and other animals, primordial germ cell (PGC) specification in the developing embryo is driven by maternal mRNAs and proteins that assemble into specialized mRNPs localized in the germ (pole) plasm at the posterior of the oocyte2,3. Maternal piRNPs, especially those loaded on Aubergine (Aub), a Piwi protein, are transmitted to the germ plasm to initiate transposon silencing in the offspring germline4–7. Transport of mRNAs to the oocyte by midoogenesis is an active, microtubule-dependent process8; mRNAs necessary for PGC formation are enriched in the germ plasm at late oogenesis via a diffusion and entrapment mechanism, whose molecular identity remains unknown8,9. Aub is a central component of germ granule RNPs, which house mRNAs in the germ plasm10–12 and interactions between Aub and Tudor are essential for the formation of germ granules13–16. Here we show that Aub-loaded piRNAs use partial base pairing characteristic of Argonaute RNPs to bind mRNAs randomly, acting as an adhesive trap that captures mRNAs in the germ plasm, in a Tudor-dependent manner. Strikingly, germ plasm mRNAs in Drosophilids are generally longer and more abundant than other mRNAs, suggesting that they provide more target sites for piRNAs to promote their preferential tethering in germ granules. Thus complexes containing Tudor, Aub piRNPs and mRNAs couple piRNA inheritance with germline specification. Our findings reveal an unexpected function for Piwi ribonucleoprotein complexes in mRNA trapping that may be generally relevant to the function of animal germ granules.
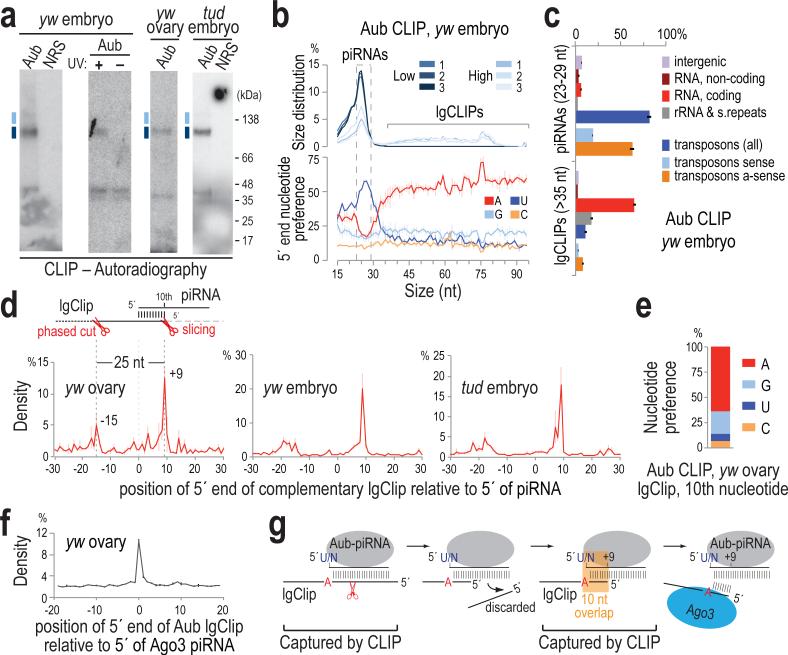
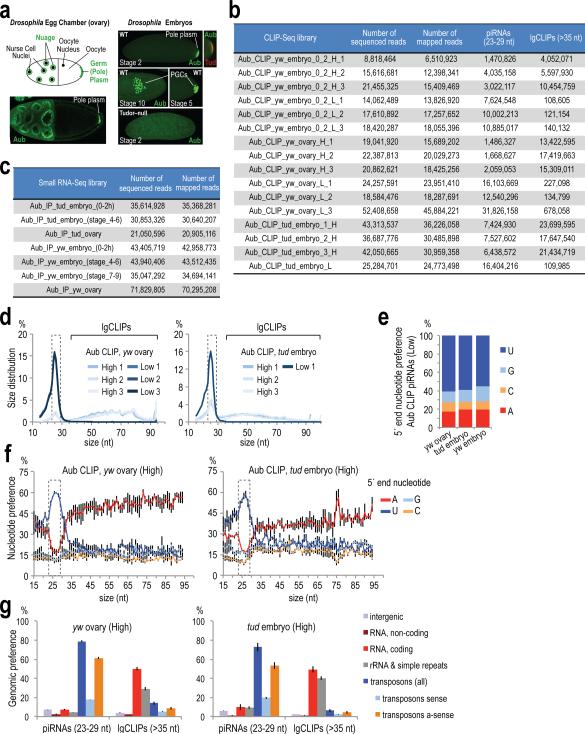
We performed stringent immunoprecipitations for Aub after ultraviolet crosslinking (UV CLIP)17 (Fig. 1a) and standard small RNA immunoprecipitations (IP) employing a highly specific antibody that we generated (Extended Data Fig. 1a) from wild-type (yw) ovaries and from yw and Tudor null (tud) embryos collected up to 2 h post-laying (0-2 h embryos); this is prior to zygotic transcription and degradation of maternal mRNAs. Crosslinked RNA-Aub complexes yielded strong, specific signals that were absent from non-immune serum (NRS) and no-UV controls (Fig. 1a). CLIP and IP libraries contained essentially identical 23-29 nt piRNAs (Fig. 1b, Extended Data Figs. 1b-g, 2a-f, Extended Data Table 1). We verified minimal changes in the piRNA load of Aub in tud versus yw ovaries (Extended Data Fig. 2g)13, and found no changes in the piRNA load of 0-2 h embryos compared to ovaries in both genotypes (Extended Data Fig. 2h, i). Larger CLIP tags (lgClips, ≥36 nt) are present in libraries prepared from larger RNP complexes (Fig. 1a-c, Extended Data Fig. 1d, Supplementary Results).
Figure 1. Transcriptome-wide identification of RNAs bound by Aubergine and in vivo retrotransposon targeting and slicing captured by CLIP.
a. Aub CLIPs; separate libraries were prepared from RNA extracted from indicated positions; uncropped gels can be found in Supplementary Figure 1.
b. Size distribution and 5′ end nucleotide composition per size of CLIP tag. Error bars represent one standard deviation (±S.D.; n=3; same applies to c, e, g).
c. Genomic distribution of CLIP tags for three High yw embryo (0-2 h) Aub CLIPs.
d. Position of 5′ ends of retrotransposon lgClips relative to 5′ ends of complementary piRNAs (0, x-axis).
e. Nucleotide composition at +9 of retrotransposon-derived lgClips with 10-nt overlap to complementary piRNAs.
f. yw ovary Aub lgClip 5′ end positions relative to the 5′ ends of Ago3-loaded piRNAs (0, x-axis).
g. Schematic of processing fragments captured by Aub CLIP.
We observe considerable overlap of retrotransposon lgClips with complementary piRNAs (Extended Data Fig. 3a, Supplementary Table 1) and strong positive correlation of their abundances (Extended Data Fig. 3b, c). Relative distance analysis reveals high occurrence of lgClips with a 10-nucleotide (nt) overlap to complementary piRNAs (Fig. 1d, peak at position +9) for all three genotypes. The majority of such lgClips bear an adenine at the tenth position (Fig. 1e) and show prominent 5′-5′ end coincidence with Ago3 piRNAs (Fig. 1f), indicating that they correspond to ping-pong intermediate fragments produced by Aub slicing1. Furthermore, a second peak at position −15 (Fig. 1d), which is 25 nt (the median Aub piRNA length) from position +9, represents 5′ ends of fragments of trigger piRNA targets undergoing phased piRNA biogenesis18. The above results indicate that CLIP captures piRNA biogenesis, complementary retrotransposon targeting and the transient products of Aub slicing activity (Fig. 1g).
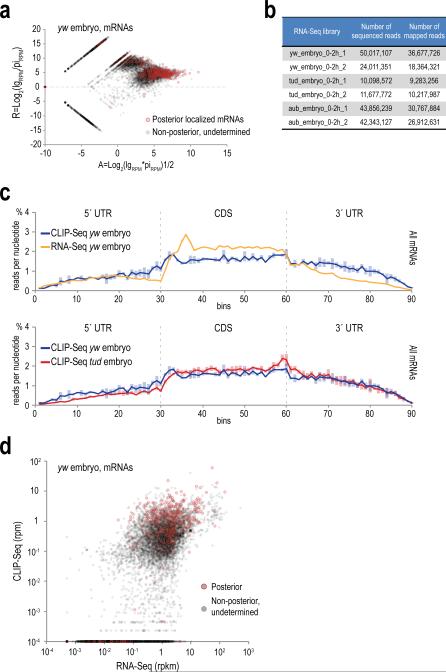
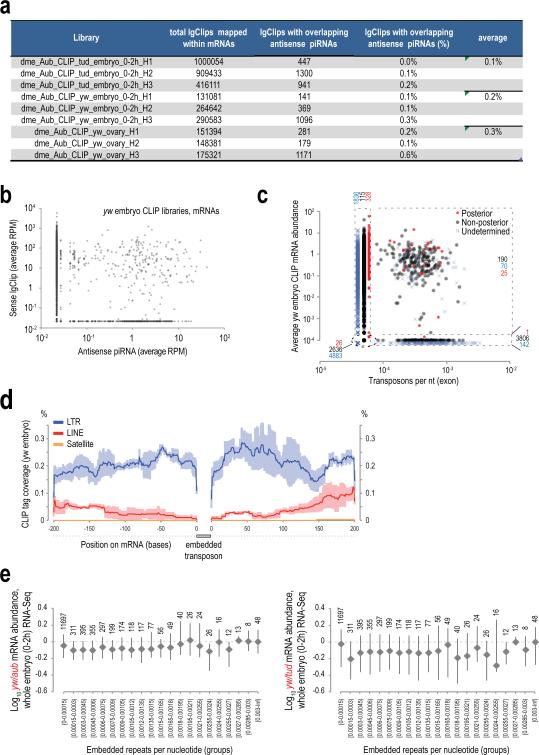
A significant percentage (~50-66%) of lgClips from all CLIP libraries are mRNA-derived (Fig. 1c, Extended Data Fig. 1g). Most Aub-bound mRNAs are not substrates for piRNA processing (Extended Data Fig. 4a). Aub lgClip density is relatively higher within 3′ UTRs compared to RNA-Seq, and overall lgClip abundance is not correlated with mRNA abundance (Extended Data Fig. 4b-d), suggesting specific target mRNA recognition. We cross-indexed Aub-bound mRNAs with the mRNA localization categories (compiled in ref. 19). Strikingly, posterior localization categories are significantly enriched in all three sets of Aub CLIP libraries (embryo: yw and tud, ovary: yw) (Supplementary Table 2). Most importantly, we find 15 posterior and germ cell localization categories significantly depleted, and ubiquitous mRNAs enriched in tud embryo compared to yw embryo CLIP libraries (Supplementary Table 3). Posteriorly localized mRNAs appear marginally upregulated compared to other localization categories in tud versus yw embryo RNA-Seq libraries (two-sided t-test, p=0.01594), ruling out the possibility that the reduced Aub binding is due to reduced posterior mRNA levels in tud embryos. Both Aub (Extended Data Fig. 1a) and germ plasm mRNAs15,20 are uniformly distributed throughout tud embryos; therefore the observed loss of binding specificity towards posterior mRNAs in the absence of Tudor can only be attributed to the disruption of the germ plasm. Thus our experimental approach allows the identification of the mRNAs specifically bound by Aub in the germ plasm, irrespective of the function of Aub in the clearance of maternal mRNAs in the somatic part of the embryo21,22. To identify the primary mRNA targets of Aub within the germ plasm during the formation of germ cells, we calculated the rank product of the normalized lgClip values for mRNAs in the 12 posterior localization categories marked with an asterisk in Supplementary Table 3, from three replicate yw embryo libraries (p-value <0.05). The list contains 220 genes, many of which appear enriched or selectively protected in germ cells10, and with established roles in germ cell specification and development such as cycB, nos, osk, gcl, pgc, hsp83 (Supplementary Table 4). Characterization of Aub RNPs from early embryos provides independent support for the association of germ plasm mRNAs with Aub (Supplemental Results, Extended Data Fig. 5). Four separate analyses provide strong evidence that the extent of the observed Aub binding of mRNAs cannot be explained by piRNA targeting of transposon sequences embedded in mRNAs (Supplemental Results, Extended Data Fig. 6).
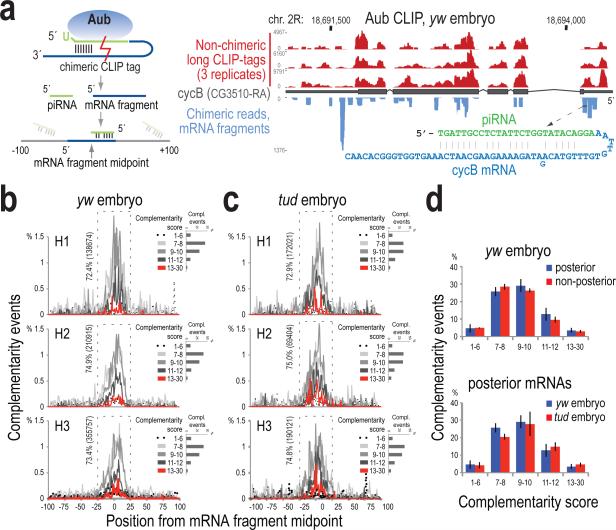
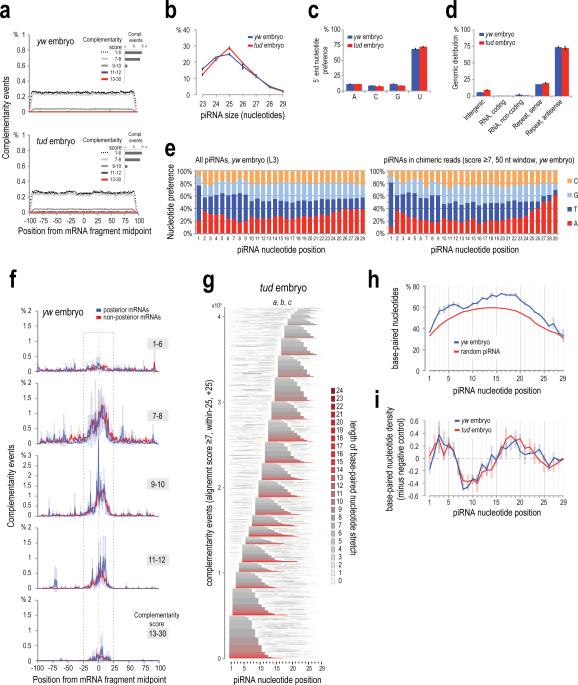
To further investigate the potential of piRNAs to direct Aub to complementary mRNA sequences, we analyzed chimeric lgClips23,24 that each contains an intact piRNA, ligated with a sequence fragment (≥20 nt) that is uniquely aligned on mRNAs (Fig. 2a, Supplementary Table 5). To uncover complementarity patterns we implemented unweighted local alignment between the piRNA (in reverse complement orientation) and the mRNA fragment, scoring matches (+1), mismatches (−1) and indels (−2), and reporting the best alignment for every chimeric read. The search was performed within ±100 bases around the midpoint of the mRNA fragment; this allows the identification of the entire complementary sequence that might be missing from the chimeric fragment, and also provides a reliable estimate of the signal-to-noise ratio. We observed prominent peaks of hundreds of thousands of complementarity events forming around the midpoint and within ±25 nt, in yw and tud embryo CLIP libraries (Fig. 2b-c). Most events score between 7 and 12; therefore, the complementarity is not extensive. The distribution of the complementarity events in the negative control (random piRNA) is completely flat across the search area and has lower scores (Extended Data Fig. 7a), suggesting that the chimeric reads capture genuine sequence-dependent Aub-piRNA:mRNA contacts.
Figure 2. Complementarity analysis between the piRNA and mRNA parts of chimeric CLIP tags.
a. Strategy for chimeric CLIP tag analysis, and genome browser illustrating Aub lgClips on cycB; sequence and base pairing of a chimeric CLIP tag is shown.
b, c. piRNA:mRNA complementarity events (percent) within ±100 bases from the midpoint of the mRNA part of the chimeric read, plotted per alignment score for yw (b) and tud embryo (c) Aub CLIPs (biological triplicates). Percentage and number of total events occurring within ±25 bases (dashed rectangle) are shown. Inset, per sample: barplot of number of complementarity events per score group.
d. Barplots of piRNA:mRNA complementarity events occurring within the ±25 bases window of the midpoint of the search area and with score ≥7, for indicated mRNA localization categories and Aub CLIP libraries. Error bars: ±S.D.; n=3.
piRNAs in chimeric reads are typical Aub piRNAs (Extended Data Fig. 7b-e). piRNA:mRNA complementarities with alignment score ≥7 congregate within a 50-nt window (Fig. 2b-d), so we focused on events that have such scores and locations. piRNA complementarity towards posterior and non-posterior mRNAs is indistinguishable (Fig. 2d, Extended data Fig. 7f), suggesting that the basis of mRNA binding preference by Aub is not sequence specificity. Chimeric reads show substantial overlap (Fig. 2a) and the same enrichment in posterior-localized mRNAs with non-chimeric lgClips (Supplementary Tables 5 and 6), suggesting that they both capture the same RNA binding events.
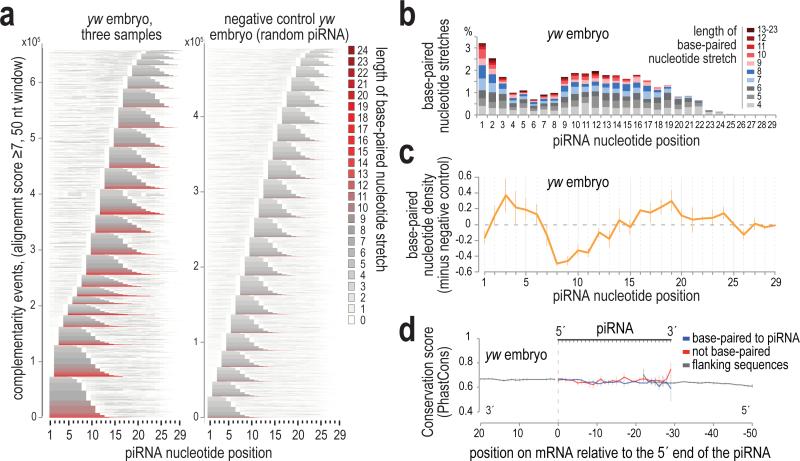
Base-paired nucleotides for every piRNA from three replicate CLIP libraries are summarized in a comprehensive plot (Fig. 3a, Extended Data Fig. 7g), revealing a bimodal distribution of the complementary regions within the piRNA. Many are found at the 5′ end of the piRNA starting at positions 1 and 2 (reminiscent of miRNA seed-type binding); additional base-paired stretches start at positions 9-17 (Fig. 3a, b). This pattern is absent from the negative control (Fig. 3a). Net density of base-paired nucleotides reveals a clear preference for piRNAs to utilize nucleotides at positions 2-6 with additional base pairs in positions 16-24 (Fig. 3c, Extended Data Fig. 7h, i). This profile is strikingly similar in yw and tud libraries, and differs slightly from the miRNA hybridization profile24 in the less frequent base-pairing in the 2-6 region, suggesting that piRNAs do not utilize a conserved seed sequence. The periodicity of the graph in Fig. 3c (Extended Data Fig. 7i) evokes the helical conformation and base-pairing availability of the small RNA in the context of an Ago-miRNA-target RNA tripartite complex25, suggesting that despite the absence of a conserved seed, the mechanics of piRNA complementary binding are analogous to those of microRNAs. Analysis of the evolutionary conservation of paired, unpaired and flanking nucleotides on the mRNA sequence reveals that the piRNA:mRNA contact sites are not preferentially conserved (Fig. 3d).
Figure 3. Characteristics of piRNA base-pairing identified by chimeric CLIP tag analysis.
a. Heat maps showing base-paired nucleotides within the piRNA sequence, for all complementarity events (score ≥7) within ±25 bases window, for yw embryo and negative control. Stacked piRNAs are sorted (bottom to top) by: starting position and length of the longest stretch and total number of base-paired nucleotides. Every nucleotide position is colored according to the length of the stretch of consecutively base-paired nucleotides that runs through that position.
b. Percent of stretches of consecutive base-paired residues per starting position within the piRNA sequence.
c. Base-paired nucleotide density per position minus negative control (random piRNA). Error bars: ±S.D.; n=3.
d. Average mRNA conservation score on and around piRNA:mRNA contact sites. Error bars: ±S.D.; n=3.
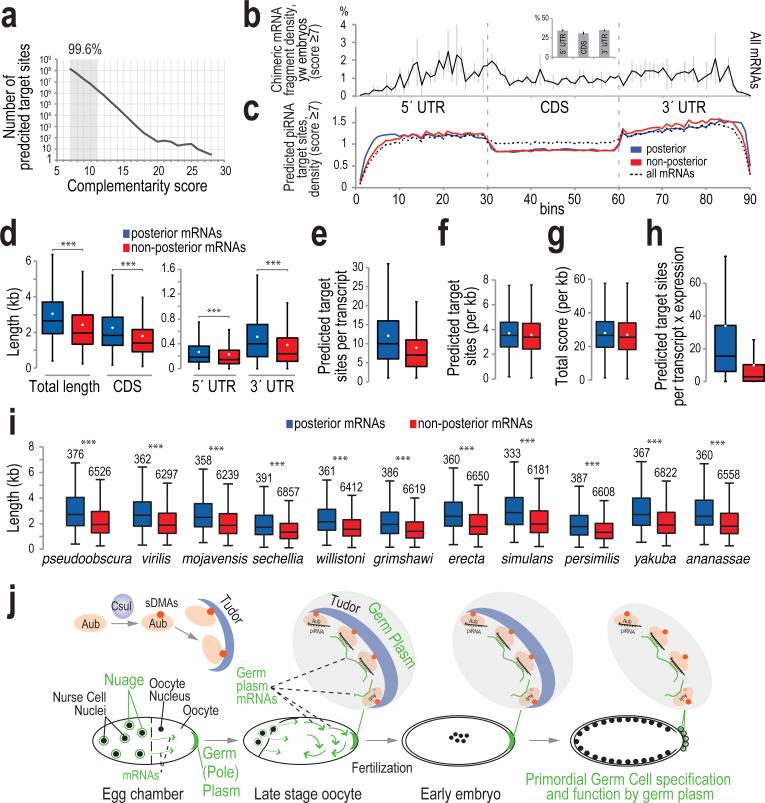
We used the local alignment approach by which we analyzed the chimeric CLIP tags, to identify potential piRNA target sites in the D. melanogaster transcriptome. In 206,400,271 total sites, the vast majority (99.6%) are of scores 7-11 (Fig. 4a). Importantly, the densities of putative piRNA target sites on mRNA regions are essentially identical for mRNAs with or without posterior localization, and very similar to that of the chimeric mRNA fragments (higher densities in the UTRs compared to CDS; Fig. 4b, c, Extended Data Fig. 8).
Figure 4. Transcriptome-wide prediction of piRNA target sites and length differential of posterior-localized mRNAs.
a. Number of predicted piRNA complementary sites on mRNAs, per score.
b, c. Average binned density of: chimeric mRNA fragments (Aub CLIP, yw embryo 0-2 h) along the meta-mRNA (inset: bar plot showing cumulative density in each mRNA region); error bars, ±S.D.; n=3 (b); predicted piRNA complementary sites within all (14058), posterior (380), and non-posterior (6747) localized mRNAs (c).
d – i. Box-and-whisker plots of: lengths of mRNAs expressed in yw embryos (0-2 h); median, black line; mean, white dot, ***: p value <0.005, one-sided t-test (d); number of predicted piRNA complementary sites per mRNA (e); length-normalized number of predicted piRNA complementary sites (f); length-normalized total score of predicted piRNA complementary sites (g); number of predicted piRNA complementary sites per mRNA multiplied with the abundance of each mRNA -RPKM- (h); lengths of orthologous mRNAs in other Drosophila species, ***: p value <10−16, one-sided Wilcoxon exact rank test (i).
j. Aubergine couples piRNA inheritance with germ cell specification in Drosophila. Aub, carrying arginines that are symmetrically dimethylated by Csul, interacts with Tudor, and both are localized in the germ plasm during mid-stage oogenesis. Ooplasmic streaming at later stages promotes diffusion of mRNPs, facilitating random contacts of mRNAs with the germ plasm. AubpiRNAs form an adhesive trap that captures mRNAs forming numerous low complementarity contacts. mRNAs with posterior functions are longer and more abundant than the rest, form more piRNA mediated contacts with the germ plasm, thus their entrapment is enhanced. Tudor-AubpiRNA-mRNA complexes along with other RNA binding proteins form germ granules that contain both piRNAs and mRNAs that induce PGC specification. Aub and its RNA cargo is incorporated in PGCs providing the maternal mRNAs that are necessary for PGC function and the maternal piRNAs that will propagate an RNA immune response against transposons.
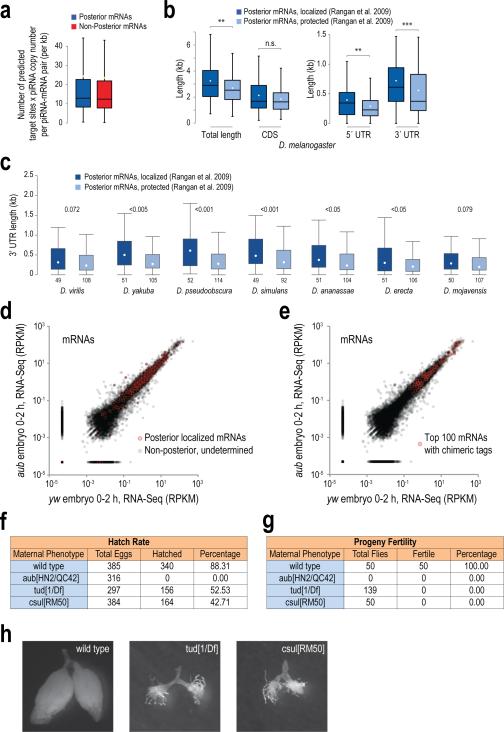
mRNAs in the 12 posterior localization categories are significantly longer than non-posterior localized mRNAs (Fig. 4d)26 and so contain a higher number of piRNA target sites (Fig. 4e); nevertheless, transcript length normalization eliminates this difference (Fig. 4f, g). This holds true when the scores of the predicted sites are accounted for (Fig. 4g), and also when the scores are weighted for the preference of piRNA nucleotides 2-6 and 16-24 to base-pair (not shown). Posterior mRNAs are also more abundant than non-posterior; when factored in, this increases the difference of the target site abundance per transcript for the two localization categories (Fig. 4h). Posterior and non-posterior mRNAs are equally targeted (per kb) by each piRNA even when piRNA copy number is accounted for (Extended Data Fig. 9a). Notably, the size differential (and not the absolute length) of posterior and non-posterior mRNAs is conserved among Drosophilids: the intra-species size differential always favors posterior mRNAs, although non-posterior mRNAs from one species might be longer than the posterior mRNAs of another (Fig. 4i). Therefore, although piRNAs randomly base pair with non-conserved mRNA sequences, this mechanism is biased towards a specific class of mRNAs for germ plasm anchoring. Additionally, from the two categories of posterior localized mRNAs, Localized and Protected10, Localized mRNAs have longer 3′ UTRs than Protected, further supporting the notion that mRNA length positively affects germ plasm enrichment (Extended Data Fig. 9b, c).
The concept of mRNA entrapment at the germ plasm during ooplasmic streaming is well established8,9,27, but the mechanism at the molecular level has been so far elusive. We propose that germ plasm localized Tud-Aub-piRNA complexes play the role of a nondiscriminatory adhesive trap that can form numerous, non-conserved piRNA:mRNA contacts to capture mRNAs and form germ plasm mRNPs (Figure 4j, Supplementary Discussion). This mechanism likely shows preference for posterior mRNAs because they are significantly longer and more abundant26. We believe that the above mechanism acts in addition to specific protein-protein, protein-RNA and RNA-RNA interactions that are necessary for mRNA transfer and anchoring to the posterior, and for translational control10,12,28–30. The multivalence of Aub-Tudor interactions likely contributes to the formation of multimeric germ granule complexes. We propose that germ cell specification and function by maternal mRNAs, and piRNA inheritance converge in Aub. Coupling germ cell specification with piRNA inheritance could be a strategy that increases reproductive fitness by ensuring the propagation of robust transposon silencing mechanisms to germ cells across generations and across the population.
METHODS
Wet-lab methods
Drosophila strains – Tissue collection
The following strains and heteroallelic combinations were used: y1w1118 as the wild-type stock (yw), aub HN2/QC42 (aub), tud1/Df(2R)PurP133 (tud), for aub and tud mutant (loss of function) fly stocks, respectively 31,32,33,15. All stocks were grown at 25 °C with 70% relative humidity on a 12 h light-dark cycle. 2-4 d female flies were crossed to yw males for 2 d in standard cornmeal food supplied with yeast paste before ovary dissection. Embryos harvested at well-defined time-windows were dechorionated in 50% commercial bleach for 2 min, washed extensively in water and collected in PBS or HBSS or fixation solution, depending on downstream applications.
Antibodies
Antibody against Aubergine (Aub-83) was produced by immunizing rabbits with Aub peptide (HKSEGDPRGSVRGRC, where terminal cysteine was used to couple to KLH; Genscript) and selected with peptide-affinity purification of sera. Other antibodies that were used in this study: mouse monoclonal anti-PABP (6E2 clone)34, E7 mouse monoclonal anti-β-tubulin (Developmental Studies Hybridoma Bank) and anti-Tudor mouse monoclonal (gift from M. Siomi).
Immunofluorescence
Fixation and immunohistochemistry of dissected ovaries and embryos was performed according to standard protocols. Primary antibodies against Aub and Tud were used at 1 ng/μL final concentration. Secondary antibodies conjugated to Alexa 488 and 594 (Life technologies) were used at 1:1000 dilution. Ovary and embryo samples were imaged on Leica TCS SPE confocal microscope.
Aub HITS-CLIP
CLIP was performed as previously described for Mili, Miwi and MOV10L117,35,36. The protocol is described in detail in36 and uses stringent buffer conditions to ensure high specificity. 40 mg of Drosophila embryos (0-2 h) or ~80 ovaries from 4-6 d females were collected in ice-cold HBSS and UV-irradiated (3×) at 254 nm (400 mJ/cm2). The tissues were pelleted, washed with PBS and the final tissue pellet was flash-frozen in liquid nitrogen and kept at −80°C. UV light–treated tissues were lysed in 350 μL 1× PMPG [1× PBS (no Mg2+ and no Ca2+), 2% Empigen] with protease inhibitors and RNasin (2 U/μL) and no exogenous ribonucleases; lysates were treated with DNase I (Promega) for 5 min at 37 °C, and then were centrifuged at 100,000 × g for 30 min at 4 °C.
For each IP, approximately 10 μL of our anti-Aub antibody was bound on 150 μL (slurry) of protein A Dynabeads in Ab binding buffer (0.1 M Na-phosphate pH 8 and 0.1% NP-40) at RT for 2 h; Ab-bound beads were washed 3× with 1× PMPG. Antibody beads were incubated with lysates (supernatant of 100,000 × g) for 3 h at 4 °C. Low- and high-salt washes of immunoprecipitation beads were performed with 1× and 5× PMPG (5× PBS, 2% Empigen). RNA linkers (RL3 and RL5), as well as 3′ adaptor labeling and ligation to CIP (calf intestinal phosphatase)-treated RNA CLIP tags were performed as previously described36.
Immunoprecipitation beads were eluted at 70 °C for 12 min using 30 μl of 2× Novex reducing loading buffer. Samples were analyzed by NuPAGE (4%-12% gradient precast gels, run with MOPS buffer). Cross-linked RNA–protein complexes were transferred onto nitrocellulose (Invitrogen LC2001), and the membrane was exposed to film for 1–2 h. Membrane fragments containing the main radioactive signal and fragments up to ~15 kDa higher were excised (Fig. 1a). RNA extraction, 5′ linker ligation, Reverse-transcriptase (RT)-PCR and second PCR step were performed with the DNA primers (DP3 and DP5, DSFP3 and DSFP5) as described previously36. cDNA from two PCR steps was resolved on and extracted from 3% Metaphor 1xTAE gels. Size profiles of cDNA libraries prepared from the main radioactive signal and higher MW were similar (Fig. 1a). DNA was extracted with QIAquick Gel Extraction kit and submitted for deep sequencing. The cDNA libraries were sequenced with Hi-Seq Illumina at 100 cycles.
Solid-support directional (SSD) RNA-Seq
SSD RNA-Seq was performed as previously described17, using total RNA (depleted of ribosomal RNA with Ribozero -EpiCentre-) isolated from 0-2 h embryos of appropriate genotypes.
Nycodenz density gradient ultracentrifugation and subsequent analyses
Nycodenz density gradient separation of RNPs was performed as previously described17 with modifications. A 20%-60% (top to bottom) Nycodenz gradient (4.8 mL) in 1× KMH150 (150 mM KCl, 2 mM MgCl2, 20 mM HEPES pH 7.4, 0.5% NP-40, 0.1 U/μL rRNAsin, and protease inhibitors) was prepared as a step gradient by overlaying 5 equal parts of Nycodenz solutions and was let to diffuse overnight at 4 °C. 0.2 mL of post nuclear yw embryo lysate in 1xKMH was laid over the gradient and centrifuged at 150,000 x g for 20 h. We used embryos of stages 4-6, to avoid earlier stages were mRNAs at the soma form distinct mRNPs than the ones formed in the pole plasm – PGCs. The gradient was collected in 12 equal fractions. Samples from each fraction were used for protein determination by Bradford and RNA extraction with Trizol LS. Right before RNA extraction, 500 ng of in vitro transcript of Renilla Luciferase mRNA was spiked in each fraction for normalization purposes in subsequent steps.
qRT-PCR
Equal volume of RNA extracted from each fraction was reverse transcribed by Supersript III (Invitrogen 18080-051) in the presence of random hexamers. Equal volume of the cDNA was mixed with primers (gcl, osk, hsp83, dhd, cycB: Qiagen QuantiTect Assay; Renilla Luciferase (rLuc), F: 5′-CGCTGAAAGTGTAGTAGATGTG and R: 5′-TCCACGAAGAAGTTATTCTCCA) and Power SYBR Green reaction mix (Applied Biosystems 4367659). The reactions were run on a StepOnePlus™ System (Applied Biosystems) using the default program.
Immunoprecipitation and detection of piRNAs, and preparation of cDNA libraries
Aub immunoprecipitation, 5′ end labeling of piRNAs and cDNA library preparation were carried out as previously described37,38.
Bioinformatic analyses
Code availability
We used CLIPSeqTools39, a bioinformatics suite that we created for analysis of CLIP-Seq datasets (accessible at: https://github.com/mnsmar/clipseqtools and http://mourelatos.med.upenn.edu/clipseqtools/tutorial/) and a Perl programming framework that we developed (M.M., P.A. and Z.M., manuscript in preparation; preprint available at: http://biorxiv.org/content/early/2015/11/03/019265). The latter framework is named GenOO and has been specifically developed for analysis of High Throughput Sequencing data. The source code for GenOO has been deposited in GitHub and can be accessed at https://github.com/genoo/.
Statistics
In statistical analyses, we ensured that the assumptions of each statistical test are met and that the statistical test used is appropriate for the analysis. In all analyses the statistical tests and methods used are clearly stated in relevant sections.
Data
Drosophila (assembly dm3) transcript, exon and repeat genomic locations were downloaded from the UCSC genome browser (downloaded 22 March 2011 from http://genome.ucsc.edu). Repeat consensus sequences were downloaded from Flybase (http://flybase.org/ - transposon_sequence_set v9.42). Localization categories for Drosophila genes were taken from Lécyuer et al., 200719. The localization annotation matrix was downloaded from (http://fly-fish.ccbr.utoronto.ca annotation_matrix.csv). Τransposon categories were as in Malone et al., 200931.
Preprocessing
The 3′ end ligated adaptor (GTGTCAGTCACTTCCAGCGGTCGTATGCCGTCTTCTGCTTG) was removed from the sequences using the cutadapt software and a 0.25 acceptable error rate for the alignment of the adaptor on the read. To eliminate reads in which the adaptor was ligated more than one time, adaptor removal was performed 3 times.
Alignment
Reads for all samples were aligned against the dm3 Drosophila melanogaster genome assembly using the aligner bwa v0.6.2-r126, with the default settings40. Reads were also aligned against the Repeat consensus sequences using the same aligner.
Genomic distribution
All mapped reads were divided in the following genomic categories: repeat, antisense repeat, non-coding RNA, coding RNA. The remaining reads were considered as intergenic reads.
Correlation of replicates
Gene expression was defined as the number of reads that map on each gene and the values were normalized by the upper quartile normalization method41. The log2 gene expression levels of replicates are compared using the Pearson Correlation function in R.
Coincidence with IP
Reads mapping in the same position (same 5′ end mapping) were considered as coinciding. When comparing CLIP with IP libraries, the percentage of piRNA-size CLIP reads that had a coinciding start with any standard IP read were counted as positive.
Significant Localization
For each localization category, the quartile-normalized lgCLIP binding level (“mRNA expression level” in each CLIP library) is compared via two sided t-test between genes that belong to the category vs genes that do not belong to it. To compare two samples, we measure the difference in binding (per gene) between the two conditions (log2(gene.expr.cond1 / gene.expr.cond2)) and then perform a t-test of differences in genes belonging to the category vs genes not belonging in the category.
Early embryo posterior localization categories
The following twelve mRNA localization categories19 were found significantly depleted in tud embryo Aub CLIP libraries compared to yw embryo libraries, and were used in analyses were “posterior localized mRNAs” are mentioned: “1:41:RNA islands”, “1:42:Pole buds”, “1:40:Pole plasm”, “3:265:Perinuclear around pole cell nuclei”, “4:370:Germ cell localization”, “4:403:Germ cell enrichment”, “3:348:Pole cell enrichment”, “2:141:Pole cell localization”, “2:153:Perinuclear around pole cell nuclei”, “2:142:Pole cell enrichment”, “3:347:Pole cell localization”, “1:59:Perinuclear around pole cell nuclei” (http://fly-fish.ccbr.utoronto.ca/). The remaining mRNAs are mentioned as non-posterior localized mRNAs. The following three posterior localization categories were also depleted in tud embryo Aub CLIP libraries compared to yw: “1:39:Posterior localization”, “2:124:Posterior localization”, “3:352:Posterior localization”. Almost all of the mRNAs contained in the above twelve categories are also contained in these three, but these three categories also contain some mRNAs that do not actually localize in the pole plasm or the germ cells (i.e. with apical localization), therefore mRNAs belonging in any of these three localization categories but not in any of the above mentioned twelve posterior categories were not considered for the generation of the Supplementary Table 4. Many mRNAs do not have a designated localization pattern, and they are mentioned as “undetermined localization”. It is worth mentioning that this category contains at least a few mRNAs with clear posterior – pole plasm localization. Through manual searches of the Berkeley Drosophila Genome Project chromogenic ISH database (http://insitu.fruitfly.org/cgi-bin/ex/insitu.pl) we noticed that many Aub bound mRNAs, whose localization is not annotated in the Fly-FISH database, are indeed localized in the germ plasm/cells (such as CG4735/shu, CG7070/PyK, CG4903/MESR4, CG5452/dnk, CG9429/Calr), therefore our analysis is most likely underestimating the true number of Aub bound mRNAs that are important for germline specification and function. Because of this, mRNAs with “undetermined localization” were never mixed with “non-posterior localized” mRNAs in our analyses.
Highly Bound Genes
To identify highly bound genes, we used the rank product method42. Specifically, genes are sorted by expression per sample, and for each gene the product of their ranks is calculated. The probability of this rank product produced by chance is calculated by permutations of all non-zero value genes.
Transcript expression calculation
We calculated the expression for protein-coding transcripts by counting the number of RNA-Seq reads that map within the exons of each transcript. The counts were normalized using RPKM (reads per million divided by the length in kb of the exonic region of the mRNA) and upper quartile normalization, effectively dividing each count by the upper quartile of all counts41. The transcript with the highest RPKM score was used (“best transcript”) unless otherwise noted.
Transcript Aub binding calculation
We calculated the expression for protein-coding transcripts by counting the number of CLIP reads that map within the exons of each transcript in the sense orientation. The counts were normalized using RPM (reads per million) and upper quartile normalization, effectively dividing each count by the upper quartile of all counts41.
RNA-Seq correlation vs CLIP
Upper quartile normalized RPKM for RNA-Seq was compared to similarly normalized CLIP binding levels defined as average number of reads per transcript in CLIP replicates. Correlation was calculated using the Pearson Correlation function in R.
Chimeric CLIP tags
Identification of hybrid reads:
-
1)
Identified lgCLIP size reads (read length >35) that did not align to the genome.
-
2)
Made a set of substrings from both ends of reads from (1) of piRNA size (L=[23,29]).
-
3)
Identified the substring from (2) to full-length piRNAs (L=[23,29]) from corresponding Low samples (table1)
-
4)
The longest aligning piRNAs are retained and coupled with the remainder of the read as piRNA-lgCLIP couples.
-
5)
The piRNA aligning fragment is cut from the read. Very small remainder reads (L=[<,20]) are discarded.
-
6)
The remainders are aligned to the genome (using bwa default settings).
-
7)
Remainders aligned in one single position that is on a known mRNA are retained.
Alignment of piRNAs to regions
-
1)
Regions of 200nt length were cut around the midpoint of the genomic alignment region from step 7 of previous routine. Specifically, if (d=200 the length of the final region we want and L is the length of the read), a genomic region flanking the read on each side of length d/2 was excised from the chromosome sequence. If the alignment was located in the minus strand the sequence was reversed and complemented at this point. This total region has length d+L. We discard an equal number of nucleotides from each side to reach a final length of L (specifically we substring starting from int(L/2) and for d nucleotides. NB: int will always round down). At this point we have a region of length 200nt centered around the alignment region of the fragment.
-
2)
We use a slightly modified Smith-Waterman43 alignment method [weights: match=+1, mismatch=-1, gap=-2] to align piRNAs on the 200-nt long regions from (1).
Differences of our alignment versus Smith-Waterman:
-
a)
No penalties are given to non-matching nucleotides on the edges of the alignment.
-
b)
If there are multiple optimal alignment scores, one is picked randomly.
-
c)
Alignments in which part of one sequence is outside the boundaries of the other sequence are not considered.
-
3)
The midpoint of the alignment (if k nucleotides matched that is the int(k/2) nucleotide) is used for graphs of alignment positioning on regions.
mRNA target prediction for the top 2000 expressed piRNAs
We grouped piRNA sequences into families based on the first 23nt of each piRNA. Using the alignment algorithm described above we aligned one piRNA (the most abundant) for each of the top 2000 families to the longest annotated transcript for each protein-coding gene. These 2000 piRNA families represent ~37% of piRNA reads from Low yw CLIP libraries. To factor in transcript abundance, we multiplied the RNA-Seq (yw embryo 0-2 h) RPKM value for each mRNA with the number of predicted piRNA target sites found within the mRNA. This provides a “targeting potential” of every mRNA species, corrected for its abundance.
We then evaluated the targeting potential of each piRNA-mRNA pair using three different scoring schemes. For the first we sum the alignment score of all putative piRNA binding sites on the mRNA. For the second we calculated a weighted alignment score for each putative piRNA binding site and then we sum all scores similar to the previous scheme. The weighted score for each binding site is calculated based on the following formula ∑ixi * Ai where xi is 1 or 0 based on whether the nucleotide at position i of the piRNA is bound or not and Ai is the weight for nucleotide i. For the third, we multiplied the total number of predicted complementary sites per piRNA, with the piRNA copy number.
Study of the lengths of D. melanogaster orthologous mRNAs in other Drosophila species
Transcript sequences (fasta file) for each species were downloaded from Flybase (ftp://ftp.flybase.net/genomes/ on Sep. 1st 2015, current version used for each genome). For each gene (identified as the “parent” tag in the fasta file header), the longest transcript length was identified. For the analysis of the expressed mRNAs (Fig. 4d), we utilized our yw embryo RNA-Seq data to identify the longest transcript with the highest length normalized abundance. Ortholog gene tables were downloaded from Flybase (gene_orthologs_fb_2015_03.tsv.gz) and were used to identify ortholog genes across species. For each species, all genes that mapped to localized and unlocalized Drosophila melanogaster genes were used in the comparison and were assigned to the corresponding group as their D. melanogaster ortholog. Boxplots were created using the lattice package in R (bwplot) and omitting outliers, p-values were calculated using the Wilcoxon exact rank test (wilcox.test in R) one-sided with the hypothesis that localized genes are longer than nonlocalized.
Extended Data
Extended Data Figure 1. Endogenous Aub localization in genotypes used, sequenced and mapped reads of CLIP-Seq and RNA IP libraries used in this study, and general characteristics of yw ovary and tud embryo (0-2 h) CLIP-Seq libraries.
a. Immunofluorescence of ovary and early embryo of indicated genotypes using antibodies against Aubergine (Aub-83; green) and Tudor (red), and schematic representation of the egg chamber. Aub is localized in the nuage and germ (pole) plasm of WT ovaries, in the germ plasm of early WT embryos (stage 2) and within PGCs as they form in the posterior pole (stage 5) and as they migrate during gastrulation (stage 10). Tud colocalizes with Aub in the germ plasm of early embryos but it is not detected after PGC formation. In Tudor mutant early embryos, Aub is not concentrated in the posterior but it is diffusely present throughout the embryo; PGCs are never specified resulting in agametic adults (see also Extended Data Fig. 9).
b. Sequenced and mapped reads of CLIP-Seq libraries prepared in this study.
c. Sequenced and mapped reads of RNA IP deep sequencing libraries prepared in this study.
d. Size distribution for the three Low and three High yw ovary and tud embryo (0-2 h) Aub CLIP-Seq libraries. The size range of piRNAs (23-29 nt) is indicated with a dashed box.
e. Average 5′ end nucleotide composition for piRNAs (23-29 nt) from three Low yw ovary, tud embryo (0-2 h) and yw embryo (0-2h) Aub CLIP-Seq libraries.
f. Average 5′ end nucleotide composition of CLIP tags from three High yw ovary and tud embryo (0-2 h) Aub CLIP-Seq libraries. piRNAs (23-29 nt) are indicated with a dashed box. Error bars represent one standard deviation (±S.D.; n=3).
g. Genomic distribution of CLIP tags for three High yw ovary and tud embryo (0-2 h) Aub CLIP-Seq libraries. Error bars: ±S.D.; n=3. Overlap of piRNAs from CLIP and IP libraries.
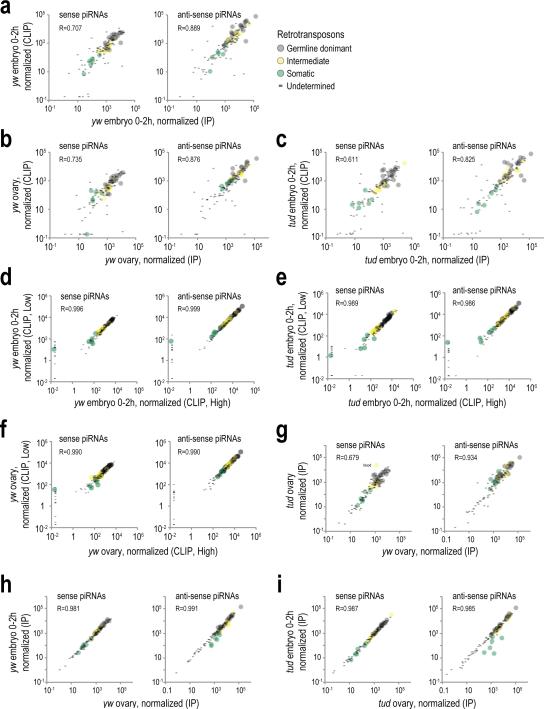
Extended Data Figure 2. Pairwise comparisons of transposon piRNA populations from various libraries.
a, b, c. Scatterplot comparison of normalized abundance of piRNAs mapped on consensus retrotransposon sequences (sense and antisense), from yw embryo (0-2 h) standard Aub IP and Aub CLIP libraries (a); from yw ovary libraries (b); and from tud embryo 0-2 h libraries (c). Pearson correlation is shown for all elements in every plot. Retrotransposon categories are set as in Malone et al., 200931.
d, e, f. Scatterplot comparison of normalized abundance of transposon-derived piRNAs in Aub CLIP libraries prepared from higher MW (Fig. 1a, marked with a light blue line) with the piRNAs found in the libraries prepared from the main radioactive signal (Fig. 1a, marked with a dark blue line) from yw embryo 0-2 h (d); from yw ovary Aub CLIP “High” and “Low” libraries (e); and from tud embryo 0-2 h Aub CLIP “High” and “Low” libraries (f) .These comparisons indicate that the piRNA loads in Low and High CLIP libraries are essentially identical.
g. Scatterplot comparison of normalized abundance of transposon derived piRNAs for yw ovary and tud ovary Aub IP libraries, to evaluate changes of piRNA load in the absence of Tudor. While antisense derived piRNAs are largely unchanged, a few sense-derived piRNAs are changed (blood retrotransposon is indicated).
h, i. Scatterplot comparison of normalized abundance of transposon derived piRNAs for yw ovary and yw embryo 0-2 h Aub IP libraries (h); and for tud ovary and tud embryo 0-2 h libraries (i).
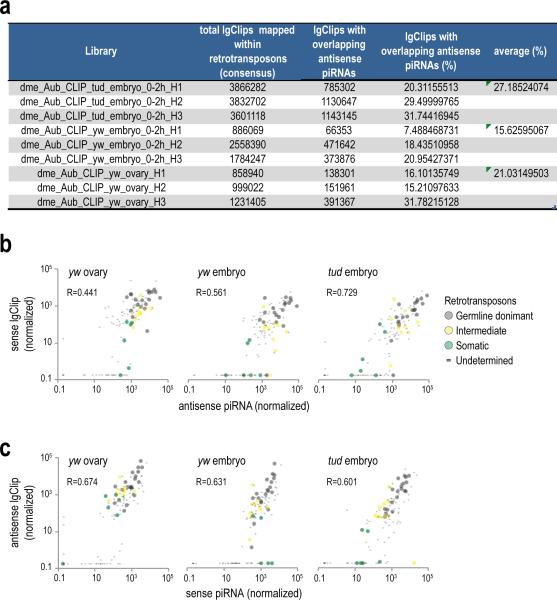
Extended Data Figure 3. Retrotransposon targeting by complementary piRNAs identified by Aub CLIP.
a Overlap of lgClips with complementary piRNAs from CLIP libraries, mapping on retrotransposons.
b, c. Scatterplot of normalized abundance of antisense piRNAs and sense lgClips (b) and for sense piRNAs and antisense lgClips (c) mapped on retrotransposons for the indicated Aub CLIP libraries. Pearson correlation is shown for all elements in every plot. Retrotransposon categories are set as in Malone et al., 200931.
Extended Data Figure 4. CLIP identifies extensive mRNA binding by Aub.
a. Ratio Average (RA) plot of normalized (RPM) Aub CLIP tag (pi, piRNA; lg, lgCLIP) abundance (A value) versus lgClips over piRNA abundance (R value), for all mRNAs. Outlined circles (red) correspond to genes that belong in the 12 posterior localization categories depleted in tud versus yw Aub CLIP libraries. Zero values are substituted with a small (smallest than the minimum) value so that log calculations are possible. This graph strongly suggests that mRNA binding by Aub as captured by CLIP is not for piRNA biogenesis purposes.
b. Sequenced and mapped reads of RNA-Seq libraries prepared in this study.
c. Density of Aub CLIP-Seq tags (yw embryo, and lower panel: tud embryo) and RNA-Seq reads (upper panel: yw embryo) within the untranslated regions and the coding sequence of the meta-mRNA. Each mRNA region is divided in 30 bins and the number of the chimeric mRNA fragments (genomic coordinate of the mRNA fragment midpoint) mapped within each bin is counted. Error bars indicate one S.D. (n=3) for CLIP-Seq; min and max values for the two RNA-Seq replicate libraries.
d. Scatterplot of average normalized mRNA abundance for yw embryo RNA-Seq (rpkm) and Aub CLIP-Seq (rpm). Aub highly bound mRNAs with posterior localizations (Supplementary Table 4) are marked with a red circle. Zero values are substituted with a small (smallest than the minimum) value so that log calculations are possible. CLIP-Seq identifies mRNAs that span the whole expression range of RNA-Seq libraries, indicating that Aub CLIP does not capture transcripts simply based on abundance.
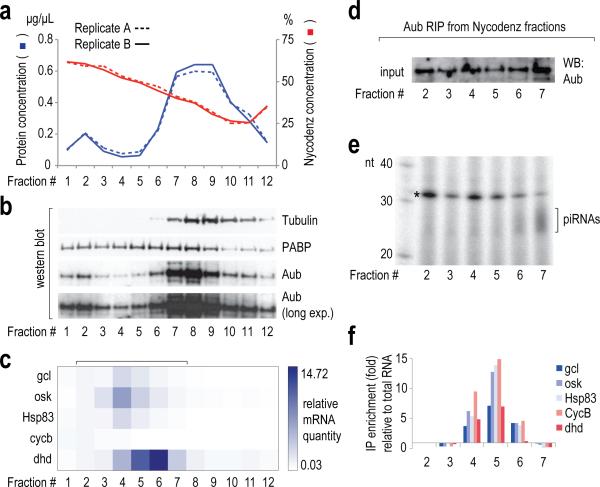
Extended Data Figure 5. Partial purification of Aub RNPs from early embryo supports piRNA independent binding of germ plasm mRNAs by Aub.
a. Fractionation of isopycnic Nycodenz density gradients of post-nuclear yw embryo lysate. Protein and Nycodenz concentration for every fraction is plotted.
b. Western blot detection of indicated proteins in gradient fractions. A short and a long exposure (long exp.) for Aub is shown. Uncropped gels for panels b, d and e can be found in Supplementary Figure 1.
c. Heat map of levels of indicated germ plasm mRNA determined by qRT-PCR, normalized to spiked luciferase RNA, and with fraction 2 as a reference.
d. Western blot detection of Aub in indicated diluted Nycodenz fractions used for Aub RNA IP.
e. Electrophoretic analysis on denaturing polyacrylamide gels of 32P-labeled small RNAs immunoprecipitated with Aub from indicated gradient fractions. A bracket denotes piRNAs, detected primarily in fractions 6 and 7 (asterisk: 2S rRNA).
f. Bar plot showing -fold enrichment (over fraction-extracted total RNA) of indicated germ plasm mRNAs in Aub IPs from gradient fractions, measured by qRT-PCR. Luciferase mRNA was used as a spike.
Extended Data Figure 6. Analysis of Aub CLIP tags mapping to mRNAs with regard to the presence of mRNA embedded transposons.
a. Overlap of lgClips with complementary piRNAs from CLIP libraries, mapping on mRNAs.
b. Scatterplot of yw embryo Aub lgClips mapped in the sense orientation on mRNAs with piRNAs mapped in the antisense orientation. Zero values are substituted with a small (smallest than the minimum) value so that log calculations are possible. Contrary to retrotransposons (Extended Data Fig. 3), there is no correlation, suggesting that extensive piRNA complementarity cannot explain the widespread mRNA binding shown by mRNA lgClips.
c. Scatterplot of yw embryo Aub lgClips mapped in the sense orientation on mRNAs with per base (nt) mRNA embedded retrotransposons (LINE, LTR, Satellite). Posterior, non-posterior and undetermined localizations are marked as indicated. The graph is separated in four quadrants: clockwork from lower left corner: 0 embedded repeats, 0 CLIP tags; 0 embedded repeats, >0 CLIP tags; >0 embedded repeats, >0 CLIP tags, >0 embedded repeats, 0 CLIP tags. The number of genes in the four quadrants is indicated. Zero values are substituted with a small (smallest than the minimum) value (different small value for every localization category was used for clarity) so that log calculations are possible. This graph suggests that there is no correlation between the number of CLIP tags and the number of embedded repeats within the mRNAs.
d. Aub lgClips density surrounding (±200 bases) mRNA embedded retrotransposons (LINE, LTR, Satellite as indicated). This analysis shows that there is no increase in the lgClip density in the areas flanking embedded repeats, suggesting that repeat sequences are not used as target areas for mRNA binding by Aub. Error bars ±S.D.; n=3.
e. Analysis of mRNA expression level in relation to the number of embedded repeats. The number of embedded repeats per nucleotide of exon was plotted with the ratio (log10) of mRNA expression in yw embryo (0-2 h) versus aubQC42/HN2 embryo (0-2 h) (left graph) and yw embryo (0-2 h) versus tud embryo (0-2 h) (right graph). The mRNAs are divided into groups based on the number of embedded repeats. A number above each data point denotes the number of mRNAs in each group. The graphs suggest that there is no proportional or consistent abundance change, decrease or increase, with the number of embedded repeats.
Extended Data Figure 7. Characteristics of piRNA base-pairing with complementary target sites identified from analysis of chimeric CLIP tags.
a. piRNA:mRNA complementarity events for a random piRNA (negative control, average of three yw (upper panel) and tud embryo (lower panel) 0-2 h samples), within ±100 bases from the midpoint of the mRNA part of the chimeric read. Complementarity events are plotted per alignment score group as indicated, for clarity. Inset (per sample): barplot of average complementarity events per score group, error bars ±S.D.; n=3.
b. Size distribution of the piRNAs identified within chimeric CLIP tags, for yw and tud embryo CLIP libraries. Error bars, ±S.D.; n=3. Only the piRNAs implicated in the complementarity events occurring within ±25 nts from the midpoint of the mRNA fragment and with score ≥7 are analyzed in this graph, and the graphs in panels (c, d, e, g, h, i).
c. 5′ end nucleotide preference for the piRNAs identified within chimeric CLIP tags, for yw and tud embryo Aub CLIP libraries. Error bars, ±S.D.; n=3.
d. Genomic distribution for the piRNAs identified within chimeric CLIP tags, for yw and tud embryo Aub CLIP libraries. Error bars, ±S.D.; n=3.
e. Per position nucleotide preference for all piRNAs in Aub yw embryo 0-2 h CLIP library L3 (left), and for the piRNAs identified within chimeric CLIP tags, for yw and tud embryo Aub CLIP libraries.
f. Complementarity events between piRNAs and mRNA fragments of chimeric reads, for posterior and non-posterior localized mRNAs (yw embryo). The plots are separated per score group. Error bars: ±S.D.; n=3.
g. Heatmaps showing base paired nucleotides of piRNAs for all complementarity events identified within chimeric CLIP tags (events occurring within ±25 nts from mRNA fragment midpoint, score ≥7) for tud embryo. Color is according to the length of the consecutive stretch of base paired nucleotides that runs over every position (color code shown on the right). Stacked piRNAs are aligned at their 5′ ends and sorted (bottom to top) following these rules: a) starting position of the longest stretch of consecutive base paired nucleotides, relative to the piRNA end; b) length of longest base-paired stretch; c) total number of base-paired nucleotides.
h. Base-pairing frequency along the piRNA length for yw embryo libraries (blue) and their negative control (red). Error bars: ±S.D.; n=3
i. Net base-pairing frequency along the piRNA length (red) and net density of base paired nucleotides (gray) in mRNAs from chimeric CLIP tags from tud embryo libraries. Error bars: ±S.D.; n=3.
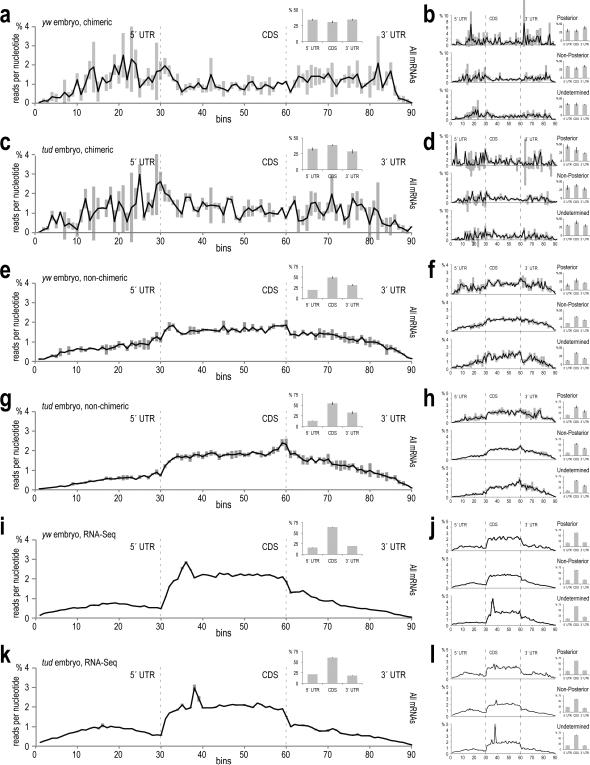
Extended Data Figure 8. Non-chimeric Aub CLIP tag (lgClip), chimeric piRNA-mRNA fragment and RNA-Seq read density along the untranslated and coding sequences of mRNAs.
a. Average density of chimeric mRNA fragments (Aub CLIP, yw embryo 0-2 h) along the three parts of the meta-mRNA. Each mRNA region is divided in 30 bins and the number of the chimeric mRNA fragments (genomic coordinate of the mRNA fragment midpoint) mapped within each bin is counted. Error bars, ±S.D.; n=3. Inset: bar plot showing cumulative density in each mRNA region.
b. Average density of the chimeric mRNA fragments on mRNA regions; mRNAs are separated in three localization groups, posterior localized (12 categories, Supplementary Table 3), non-posterior, and undetermined localization as indicated. Error bars, ±S.D.; n=3. Inset: bar plot showing cumulative density in each mRNA region.
c. Same as (a) for chimeric mRNA fragments from Aub CLIP libraries, tud embryo 0-2 h.
d. Same as (b) for chimeric mRNA fragments from Aub CLIP libraries, tud embryo 0-2 h.
e. Same as (a) for non-chimeric lgClips from Aub CLIP libraries, yw embryo 0-2 h.
f. Same as (b) for non-chimeric lgClips from Aub CLIP libraries, yw embryo 0-2 h.
g. Same as (a) for non-chimeric lgClips from Aub CLIP libraries, tud embryo 0-2 h.
h. Same as (b) for non-chimeric lgClips from Aub CLIP libraries, tud embryo 0-2 h.
i. Same as (a) for RNA-Seq reads, yw embryo 0-2 h.
j. Same as (b) for RNA-Seq reads, yw embryo 0-2 h.
k. Same as (a) for RNA-Seq reads, tud embryo 0-2 h.
l. Same as (b) for RNA-Seq reads, tud embryo 0-2 h.
Extended Data Figure 9. Lengths of posterior localized mRNAs in Drosophila species; characteristics of embryos used in our studies.
a. Box-and-whisker plot of the number of predicted piRNA target sites (per kb of mRNA sequence) for every mRNA-piRNA pair, multiplied by the piRNA copy number. Posterior and Non posterior mRNAs are as indicated. Median, black line. This graph indicates that the “targeting potential” (number of predicted complementary sites multiplied by the piRNA copy number) of every piRNA against each mRNA is the same for the two localization categories, suggesting that the piRNA copy number is not a contributing factor for the observed preference of posterior localized mRNAs for piRNA adhesion.
b. Box-and-whisker plot of the lengths of D. melanogaster mRNAs (and their 5′ UTR, CDS and 3′ UTR parts) that are found in the Enriched and Protected categories, as defined by the Lehmann lab10. Median, black line; mean, white dot; n.s.: p value >0.05; **: p value <0.01; ***:p value <0.001, one-sided Wilcoxon rank sum test.
c. Box-and-whisker plot of the lengths of the 3′ UTRs of mRNAs from the indicated Drosophila species that are orthologous to the D. melanogaster mRNAs found in the Enriched and Protected categories, as defined by the Lehmann lab10. Incomplete annotation did not allow us to perform this analysis for all the species shown on Fig. 4i. Mean, white dot; the p values of the statistical test (one-sided Wilcoxon test) of whether the lengths of the Localized versus Protected mRNAs are different, are displayed for each species.
d, e. RNA-Seq scatterplots from 0-2 h wild-type (yw) and 0-2 h Aub null (aub) embryos. Shown in red are posterior localized mRNAs (d) or the top 100 mRNAs identified from Aub CLIP piRNA-mRNA chimeric reads (e). There is no change in mRNA levels between wild-type and aub mutant 0-2 h embryos.
f, g. Hatch rates (f) and fertility of progeny (g) of embryos from indicated genotypes. Note that, unlike Tud and Csul, the absence of Aub (aub[HN2/QC42]) leads to complete embryo lethality.
h. Gross ovary appearance of wild-type (yw), tudor mutant (tud[1/Df]) and csul mutant (csul[RM50]) adult flies. Note complete absence of germline ovarian tissue in adult flies lacking Tudor or Csul; embryos from these flies develop into agametic adults because PGCs are never specified.
Extended Data Table 1.
Overlap of piRNAs from CLIP and IP libraries Comparisons of piRNA sequences found in CLIP and IP libraries from same tissues.
| a | |||||||
|---|---|---|---|---|---|---|---|
| Library 1 | Library 2 | unique piRNA sequences in library 1 | unique piRNA sequences in library 2 | common | percent1 | percent2 | average percent 2 |
| Aub_IP_yw_embryo_0-2h | Aub_CLIP_yw_embryo_0-2h_H1 | 6913438 | 348812 | 150654 | 2.179147336 | 43.19060124 | 42.22806 |
| Aub_IP_yw_embryo_0-2h | Aub_CLIP_yw_embryo_0-2h_H2 | 6913438 | 838891 | 333876 | 4.829377222 | 39.79968792 | |
| Aub_IP_yw_embryo_0-2h | Aub_CLIP_yw_embryo_0-2h_H3 | 6913438 | 694458 | 284532 | 4.115636822 | 40.97180823 | |
| Aub_IP_yw_ovary | Aub_CLIP_yw_ovary_H1 | 9938639 | 560082 | 286627 | 2.883966306 | 51.17589924 | |
| Aub_IP_yw_ovary | Aub_CLIP_yw_ovary_H3 | 9938639 | 293375 | 156976 | 1.579451673 | 53.50694504 | |
| Aub_IP_yw_ovary | Aub_CLIP_yw_ovary_H2 | 9938639 | 332484 | 176012 | 1.770986953 | 52.93848727 | |
| Aub_IP_tud_embryo_0-2h | Aub_CLIP_tud_embryo_0-2h_L1 | 5147948 | 1257672 | 458182 | 8.900284152 | 36.43096133 | |
| Aub_IP_tud_embryo_0-2h | Aub_CLIP_tud_embryo_0-2h_H2 | 5147948 | 1104187 | 460392 | 8.943213879 | 41.69511143 | |
| Aub_IP_tud_embryo_0-2h | Aub_CLIP_tud_embryo_0-2h_H3 | 5147948 | 2567880 | 948630 | 18.42734231 | 36.94214683 | |
| Aub_IP_tud_embryo_0-2h | Aub_CLIP_tud_embryo_0-2h_H1 | 5147948 | 1040626 | 379030 | 7.362739484 | 36.4232683 | |
| Aub_IP_yw_ovary | Aub_CLIP_yw_ovary_L1 | 9938639 | 1850192 | 874693 | 8.800933407 | 47.27579624 | |
| Aub_IP_yw_ovary | Aub_CLIP_yw_ovary_L2 | 9938639 | 2407082 | 1108175 | 11.15016855 | 46.03810755 | |
| Aub_IP_yw_ovary | Aub_CLIP_yw_ovary_L3 | 9938639 | 3082922 | 1367516 | 13.75959022 | 44.35778784 | |
| Aub_IP_yw_embryo_0-2h | Aub_CLIP_yw_embryo_0-2h_L1 | 6913438 | 2012094 | 722743 | 10.45417634 | 35.91994211 | |
| Aub_IP_yw_embryo_0-2h | Aub_CLIP_yw_embryo_0-2h_L2 | 6913438 | 2161685 | 769241 | 11.12675054 | 35.58524947 | |
| Aub_IP_yw_embryo_0-2h | Aub_CLIP_yw_embryo_0-2h_L3 | 6913438 | 2701578 | 902250 | 13.0506703 | 33.39714789 | |
Supplementary Material
Acknowledgements
Many thanks to former and current lab members for discussions; to M. Siomi (University of Tokyo) for Tudor antibody; to A. Arkov (Murray State University) for tud flies; to G. Dreyfuss (Penn) for PABP antibody; and to J. Schug (Penn) for Illumina sequencing. Supported by a Brody family fellowship to M.M. and NIH Grant GM072777 to Z.M.
Footnotes
Author Contributions
A.V. and Z.M. conceived, and Z.M. supervised, the study. A.V. and N.V. performed the experiments. P.A. performed bioinformatic analyses with contribution by M.M. and A.V. A.V., P.A., N.V., M.M. and Z.M. interpreted data. A.V. wrote the manuscript, with contribution from all authors.
Author Information
Sequences were deposited to Sequence Read Archive (SRA), accession number SRP067739. The authors declare no competing financial interests.
References
- 1.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell. Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 2.Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- 3.Mahowald AP. Assembly of the Drosophila germ plasm. Int Rev Cytol. 2001;203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- 4.Brennecke J, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grentzinger T, et al. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 2012;22:1877–88. doi: 10.1101/gr.136614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khurana JS, et al. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–63. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bucheton A. Non-Mendelian female sterility in Drosophila melanogaster: influence of aging and thermic treatments. III. Cumulative effects induced by these factors. Genetics. 1979;93:131–42. doi: 10.1093/genetics/93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kugler JM, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during drosophila oogenesis. Fly (Austin) 2009;3:15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- 9.Forrest KM, Gavis ER. Live Imaging of Endogenous RNA Reveals a Diffusion and Entrapment Mechanism for nanos mRNA Localization in Drosophila. Curr. Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 10.Rangan P, et al. Temporal and spatial control of germ-plasm RNAs. Curr. Biol. 2009;19:72–7. doi: 10.1016/j.cub.2008.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson T, Liu N, Arkov A, Lehmann R, Lasko P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev. 2008;125:865–873. doi: 10.1016/j.mod.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trcek T, et al. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun. 2015;6:7962. doi: 10.1038/ncomms8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirino Y, et al. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2010;16:70–78. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, et al. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 2010;24:1876–81. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arkov AL, Wang J-YS, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133:4053–62. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- 16.Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 17.Vourekas A, et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol. 2012;19:773–81. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohn F, Handler D, Brennecke J. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348:812–817. doi: 10.1126/science.aaa1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lécuyer E, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–87. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–170. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- 21.Barckmann B, et al. Aubergine iCLIP Reveals piRNA-Dependent Decay of mRNAs Involved in Germ Cell Development in the Early Embryo. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.07.030. doi:10.1016/j.celrep.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouget C, et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–32. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore MJ, et al. miRNA-target chimeras reveal miRNA 3’-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015;6:8864. doi: 10.1038/ncomms9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosswendt S, et al. Unambiguous Identification of miRNA: Target site interactions by different types of ligation reactions. Mol. Cell. 2014;54:1042–1054. doi: 10.1016/j.molcel.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jambor H, et al. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. Elife. 2015;4:e05003. doi: 10.7554/eLife.05003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinsimer KS, Lee JJ, Thiberge SY, Gavis ER. Germ plasm anchoring is a dynamic state that requires persistent trafficking. Cell Rep. 2013;5:1169–77. doi: 10.1016/j.celrep.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Little SC, Sinsimer KS, Lee JJ, Wieschaus EF, Gavis ER. Independent and coordinate trafficking of Drosophila germ plasm mRNAs. Nat. Cell Biol. 2015;17:558–568. doi: 10.1038/ncb3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh S, Marchand V, Gáspár I, Ephrussi A. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat. Struct. Mol. Biol. 2012;19:441–9. doi: 10.1038/nsmb.2257. [DOI] [PubMed] [Google Scholar]
- 30.Gavis ER, Lunsford L, Bergsten SE, Lehmann R. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development. 1996;122:2791–800. doi: 10.1242/dev.122.9.2791. [DOI] [PubMed] [Google Scholar]
- 31.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- 33.Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matunis MJ, Matunis EL, Dreyfuss G. Isolation of hnRNP complexes from Drosophila melanogaster. J. Cell Biol. 1992;116:245–255. doi: 10.1083/jcb.116.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vourekas A, et al. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015;29:617–629. doi: 10.1101/gad.254631.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vourekas A, Mourelatos Z. HITS-CLIP (CLIP-Seq) for Mouse Piwi Proteins. Methods Mol. Biol. 2014;1093:73–95. doi: 10.1007/978-1-62703-694-8_7. [DOI] [PubMed] [Google Scholar]
- 37.Kirino Y, Vourekas A, Khandros E, Mourelatos Z. Immunoprecipitation of piRNPs and Directional, Next Generation Sequencing of piRNAs. Methods Mol. Biol. 2011;725:281–293. doi: 10.1007/978-1-61779-046-1_18. [DOI] [PubMed] [Google Scholar]
- 38.Kirino Y, et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maragkakis M, Alexiou P, Nakaya T, Mourelatos Z. CLIPSeqTools-a novel bioinformatics CLIP-seq analysis suite. RNA. 2015 doi: 10.1261/rna.052167.115. doi:10.1261/rna.052167.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 43.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.