Abstract
Somatic mutation in neurons is linked to neurologic disease and implicated in cell type diversification. However, the origin, extent and patterns of genomic mutation in neurons remain unknown. We established a nuclear transfer method to clonally amplify the genomes of neurons from adult mice for whole genome sequencing. Comprehensive mutation detection and independent validation revealed that individual neurons harbor ~100 unique mutations from all classes, but lack recurrent rearrangements. Most neurons contain at least one gene disrupting mutation and rare (0-2) mobile element insertions. The frequency and gene bias of neuronal mutations differs from other lineages, potentially due to novel mechanisms governing post-mitotic mutation. Fertile mice were cloned from several neurons, establishing the compatibility of mutated adult neuronal genomes with reprogramming to pluripotency and development.
INTRODUCTION
The genome sequence of a differentiated cell offers a record of the mutational events that occur during its specification, maturation, function and dysfunction. For example, whole genome sequencing of cancer cells has identified distinct mutational signatures that characterize different tumors, raising questions about the extent to which these signatures originate from cell type-specific somatic mutations, differential exposure to mutagens, or impaired genome maintenance due to transformation (Alexandrov et al., 2013; Lawrence et al., 2013; Pleasance et al., 2010a; Pleasance et al., 2010b). In addition, somatic mutations are becoming increasingly recognized as important for human health and disease. In the immune system, developmentally programmed somatic mutations produce cellular diversity for antigen recognition. In other lineages, non-programmed somatic mutations have been shown to cause human developmental and neurologic disorders and may contribute to autism and schizophrenia (Poduri et al., 2013). The accumulation of mutations in different tissues is thought to contribute to the cellular dysfunction that accompanies aging (Kennedy et al., 2012; Vijg, 2014). Somatic mutation also threatens the safety and utility of reprogrammed cells derived from tissues of adults or aged individuals (Lee et al., 2013). Yet, current knowledge of somatic mutation is quite limited and the comprehensive, genome-wide landscape of all classes of mutation has been not been delineated for any somatic cell type.
Among cell types, neurons are of particular interest for genome sequencing. Most neurons are specified during embryonic development, when they exit the cell cycle irreversibly. After this terminal differentiation, neurons maintain their identity without cell division or replacement for the remaining lifetime of the organism. Therefore, many or most somatic mutations in neurons may arise post-mitotically, offering a window into this poorly understood potential source of mutation. During their lifetime, neurons also face unique genomic threats due to their high metabolic rate and their use of epigenetic chromatin remodeling and DNA breaks to alter gene expression in response to neuronal activity (King et al., 2013; Lister et al., 2013; Madabhushi et al., 2015; Suberbielle et al., 2013). Finally, neurons exhibit remarkable cell type diversity and their survival is impacted by genes involved in immune system diversification, leading to suggestions that they may parallel the immune system by employing programmed DNA rearrangements to generate molecular diversity (Chun and Schatz, 1999; Chun et al., 1991; Frank et al., 1998; Gao et al., 1998). These unique features of neurons have raised long-standing questions about the extent to which neurons maintain their genomic integrity and predict that neurons may exhibit unique patterns of mutation relative to other lineages.
Indeed, a series of lower resolution genomic studies indicate that neurons may exhibit unusual levels of aneuploidy or DNA content, and harbor frequent, large-scale DNA copy number variants (CNVs) (Cai et al., 2014; Gole et al., 2013; Kingsbury et al., 2005; Lodato et al., 2015; McConnell et al., 2013; Rehen et al., 2001; Rehen et al., 2005; Westra et al., 2008; Westra et al., 2010). In addition, somatic mobile element insertions (MEIs) have been identified in mouse (Muotri et al., 2005; Muotri et al., 2009), human (Baillie et al., 2011; Coufal et al., 2009; Evrony et al., 2012; Upton et al., 2015), and fly neurons (Li et al., 2013; Perrat et al., 2013), although these studies report widely varying estimates, ranging from 0.07 to 129 per neuron (Coufal et al., 2009; Evrony et al., 2012; Evrony et al., 2014; Perrat et al., 2013; Upton et al., 2015). Understanding whether this variation is due to neuronal subtype diversity or reflects methodological differences is key to discerning the functional impact of MEIs, which have been postulated to contribute to neural diversity (Muotri et al., 2005; Singer et al., 2010; Upton et al., 2015) and/or neurological disorders (Bundo et al., 2014; Coufal et al., 2011; Douville et al., 2011; Jeong et al., 2010; Kaneko et al., 2011; Lathe and Harris, 2009; Li et al., 2012; Muotri et al., 2010; Tan et al., 2012). Together these intriguing first overviews of neuronal genome diversity highlight the importance of applying increasingly sensitive methods to decipher the genome sequences of neurons and survey the complete landscape of neuronal somatic mutations.
One major barrier to such studies is the difficulty in establishing methods to faithfully amplify the genomes of single somatic cells, particularly those that are post-mitotic. In vitro single cell whole genome amplification methods produce stochastic artifacts that are nearly impossible to distinguish from bona fide somatic mutations without access to the original un-amplified genomic material. As such, single cell genomic analyses can only reliably detect large genomic changes such as aneuploidy and copy number variation (CNV), or recurrent somatic mutations that can be distinguished from artifacts by their presence in more than one cell (McConnell et al., 2013). However, as one recent study reported, recurrent SNVs in neurons are rare (1-11 detected per neuron) compared to a higher number of potentially unique SNVs (~1,500) that are difficult to reliably distinguish from errors produced by amplification methods (Lodato et al., 2015). As such, the few reported high-resolution genomic studies of individual non-neuronal cells have relied on clonal expansion and subsequent genome sequencing. Yet, because neurons are post-mitotic and can only be induced to divide via overexpression of multiple mutagenic oncogenes, such approaches are not feasible with this cell type (Ajioka et al., 2007; Behjati et al., 2014; Friedmann-Morvinski et al., 2012; Kim et al., 2011; Young et al., 2012).
Here, we establish a reprogramming based method to produce high-resolution sequences of neuronal genomes and describe and validate the full mutational spectra of six individual post-mitotic neurons. To accomplish this, we reprogrammed adult post-mitotic neurons using somatic cell nuclear transfer (SCNT) of neuronal nuclei into enucleated oocytes. In SCNT, the cytoplasm of the egg faithfully copies the neuronal genome to produce sufficient DNA for whole genome sequencing without in vitro amplification or addition of oncogenes. After SCNT based genome amplification we applied comprehensive bioinformatic detection algorithms to identify all classes of somatic mutation with base-pair resolution, thereby producing the first comprehensive picture of the types, frequency, and patterns of somatic mutation in neuronal genomes. Finally, we generated cloned mice from several neuron-derived embryonic stem cell (ES cell) lines, showing that despite their somatic mutations, genomes of adult neurons can maintain sufficient integrity and plasticity to produce fertile adult mice.
RESULTS
Reprogramming adult post-mitotic neurons by SCNT
We wished to examine the genomes of neurons with an early, well defined birth date that are active and functional throughout the lifetime of an organism. Therefore, we reprogrammed the mitral and tufted neuronal subtype of the olfactory bulb (MT neurons), which are among the earliest born neurons in the brain. The majority of MT neurons exit the cell cycle between embryonic days 9 and 13 (Imamura et al., 2011), and are not produced post-natally (Blanchart et al., 2006; Hinds, 1968a, b). MT neurons are active and functional at birth and exhibit spontaneous and evoked electrical activity throughout the lifetime of an animal (Mair and Gesteland, 1982). In addition, recent single neuron tracing studies predict that MT neurons employ stochastic mechanisms to produce their complex patterns of axonal branching and synaptic connectivity (Ghosh et al., 2011; Miyamichi et al., 2011; Sosulski et al., 2011). These features suggest that the genomes of MT neurons may be expected to harbor genomic signatures of post-mitotic mutation and/or extensive cellular diversification.
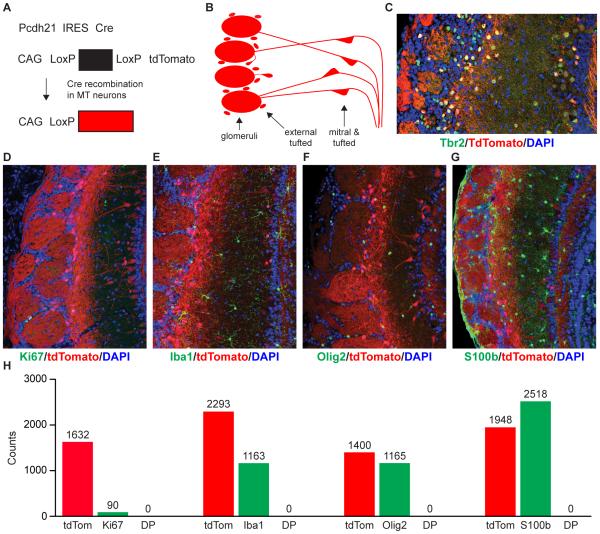
Because SCNT is inherently inefficient (Ogura et al., 2013), we employed a genetic marking strategy to conclusively establish the identity of the donor MT nucleus. We crossed a knock-in Pcdh21/Cre mouse, in which Cre recombinase is co-expressed with the Pcdh21 gene (Boland et al., 2009; Ghosh et al., 2011), with the Ai9 tdTomato-based Cre-reporter mouse (Madisen et al., 2010). In Pcdh21/Cre-Ai9 mice, tdTomato expression is under the control of the Pcdh21 promoter that is active only in MT cells in the olfactory bulb (Figure 1A). As expected, red fluorescence in the olfactory bulbs of these mice is indeed restricted to the MT neuron layers and overlaps with Tbr2, a marker of most MT neurons, but not with markers for astrocytes (S100b), oligodendrocytes (Olig2), microglia (Iba1) or dividing cells (Ki67) (Figure 1B-1H; Extended Experimental Procedures). Rare tdTomato-positive cells were also detected in the granule cell layer of the olfactory bulb (<0.1% of all tdTomato positive cells). However, due to their scarcity, this population is unlikely to be the source of the SCNT-ES cell lines.
Figure 1. Genetic labeling of mitral and tufted (MT) neurons.
(A) Donor animals carry one Pcdh21/Cre allele (top) and one copy of the Ai9 Cre reporter transgene (middle). Cre expression in MT neurons excises the STOP cassette within the Ai9 transgene, resulting in specific tdTomato expression and genetic labeling of MT neurons (bottom). (B) Schematic representation of the MT neuron localization and morphology within the olfactory bulb. MT neurons in the mitral and tufted cell layer, as well as external tufted cells send their dendrites into spherical structures known as glomeruli, where they synapse with olfactory sensory neurons. (C-G) Immunostaining of Pcdh21/Cre-Ai9 mouse olfactory bulb sections for markers of MT neurons, glia and dividing cells. Blue, DAPI nuclear stain; red, endogenous tdTomato fluorescence; green, antibody staining for (C) MT neuron marker Tbr2 (D) dividing cell marker Ki67 (E) microglia marker Iba1 (F) oligodendrocyte marker Olig2 (G) astrocyte and olfactory ensheathing cell marker S100b. (H) Quantification of the absence of co-expression of tdTomato with glial and dividing cell markers. DP: double positive for tdTomato and glial/dividing cell maker. Scale bar in C, 15 µ. Scale bars in D-G 100 µ.
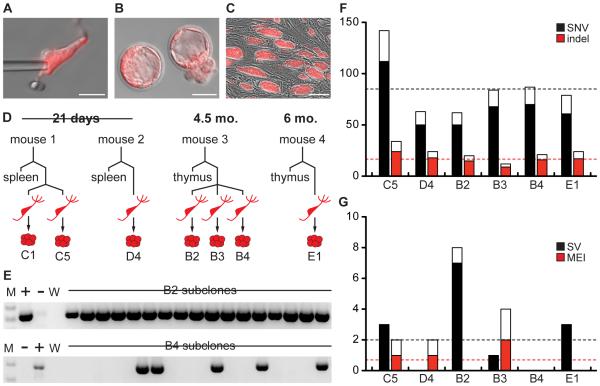
To reprogram MT neurons we performed SCNT on neurons isolated from the olfactory bulbs of Pcdh21/Cre-Ai9 mice (Figure 2A; Extended Experimental Procedures). Previous studies and our pilot experiments indicated that cloning from postnatal or adult CNS neurons is either very inefficient or impossible (Makino et al., 2005; Osada et al., 2002; Wakayama et al., 1998). Therefore, we included the histone deacetylase inhibitor Trichostatin A, which has been reported to improve cloning efficiency for other cell types (Kishigami et al., 2006). Using this method we produced seven SCNT-ES cell lines from four different mice (age range: 3 weeks to 6 months, 1.1% efficiency, Figure 2B and 2C; Table 1; Supplemental Experimental Procedures). These MCNT-ES cell lines each carry the genome of an individual MT neuron and can be expanded indefinitely while maintaining the morphology and gene expression patterns that characterize ES cells (Figure S1A and S1B).
Figure 2. MCNT-ES cells and their whole genome sequences.
(A) Dissociated MT neuron shown with injection pipette. (B) tdTomato positive blastocysts generated from MT neurons. (C) tdTomato positive MCNT-ES cells. (D) Schematic of Pcdh21/Cre-Ai9 donor animals and the MCNT-ES cell lines and control tissues sequenced from each animal. (E) Representative PCR subclone validation for two structural variants (SVs). PCR primers flank the SV breakpoint, and are diagnostic for the presence of the SV mutation. The top SV is somatic, indicated by its presence in all early passage subclones. The bottom SV likely arose during culture or reprogramming, as it is present in only some subclones. Images are cropped to the region of diagnostic band size. M, molecular weight. +, Positive control. –, Negative control. (F) and (G) Observed mutations (black/red bars) and estimated mutational burden based on the false negative rate (FNR; colored plus white bars). For SVs, observed and predicted values for breakpoints are plotted. Scale bar in A, 25 µ. Scale bar in B, 50 µ. Scale bar in C, 100µ. See Figures S1 and S2, and Tables S1 and S2.
Table 1.
Efficiency of SCNT using MT neuron nuclei.
| Donor age |
Oocytes activated |
2-cell embryos (% oocytes activated) |
Morula/blastocysts (% oocytes activated) |
MCNT-ES cell lines (% oocytes activated) |
Independent experiments |
|---|---|---|---|---|---|
| 3 wks | 297 | 137 (46%) | 20 (7%) | 3 (1%) | 7 |
| 4.5-6 mos | 327 | 253 (77%) | 15 (5%) | 4 (2%) | 6 |
Whole genome sequencing and mutation discovery
To produce high-resolution genome sequences of MT neurons, we performed high coverage (32X-59X) whole genome sequencing (WGS) on MCNT-ES cell lines and matched control tissue (thymus or spleen) derived from the same donor animal (Figure 2D; Table S1). We used Novoalign (Hercus) and YAHA (Faust and Hall, 2012) to align reads and developed a custom suite of variant detection pipelines that allowed us to detect single nucleotide variants (SNVs), indels, structural variants (SVs), copy number variants (CNVs) and mobile element insertions (MEIs) (Figure S2A-S2C; Table S2; Supplemental Experimental Procedures). Germline variants were excluded by comparing MCNT-ES cell lines to the matched donor tissues (thymus or spleen) and to a database of mouse strain polymorphisms (Keane et al., 2011). We estimated the sensitivity of each variant detection algorithm by computing the false negative rate at known polymorphisms from the Mouse Genomes Project (Table S3; Supplemental Experimental Procedures) (Keane et al., 2011; Yalcin et al., 2012). To establish false discovery rates we directly validated subsets of calls using PCR and capillary sequencing (Table S4; Supplemental Experimental Procedures).
Our variant calling will detect developmentally acquired neuronal somatic mutations and also identify some mutations that arose during reprogramming or expansion of MCNT-ES cells (culture associated mutations). However, true somatic mutations must have been present on one of the two original chromosomes and therefore these should be heterozygous in all cells in an ES line (Figure S2D). In contrast, culture-associated mutations arising after the first cell division, or even during the short time window of reprogramming prior to division (~24 hours) will be present in only a subset of cells per line (Figure S2F and S2G). The only exception to this would be mutations acquired on both strands of the neuronal genome prior to the first S-phase following nuclear transfer (Figure S2E), however, these are expected to be exceedingly rare (Li et al., 2014; Ma et al., 2014).
A true somatic mutation must be heterozygous yet uniform in a MCNT-ES line. Therefore somatic mutations will exhibit an average variant allele frequency (VAF) of ~50%, while mosaic culture-associated mutations will have lower apparent VAFs (Figure S2D-S2G). Accordingly, for SNVs and indels, we used alignment-based VAF estimates and a VAF cut off of >30% to distinguish high confidence somatic mutations from likely culture associated mutations (Figure S2C). For SVs and MEIs, VAFs are difficult to approximate from sequencing data. We therefore derived subclones of each MCNT-ES cell line at early passages and performed PCR to identify SVs and MEIs that were present in all subclones, while eliminating mosaic variants (Figure 2E).
Genomic variation among MT neurons impacts genes
These analyses revealed that each MT neuron has a unique genome harboring ~100 somatic mutations. The six MT neurons we analyzed contained ~87 (68-139) validated somatic mutations comprising 69 (50-112) SNVs, 17 (9-24) indels, 1.5 (0-3) SVs, 0.7 (0-2) MEIs (Table 2; Tables S5-S8). Applying our false discovery rates, we estimate that MT neurons have a true mutational burden of 112 (89-181) mutations per genome. The notable variation in mutational load per neuron is not dependent on sequence coverage (Figure 2F and 2G). For example, two neurons from 3-week-old mice differed by a factor of ~2 (89 vs. 181), despite having nearly identical sequencing depth and variant detection sensitivity (Table 2; Table S1). These data are consistent with the extreme variability in mutational burden previously observed for large-scale CNVs (>5 mb) using single neuron genome sequencing (Cai et al., 2014; McConnell et al., 2013). However, we did not observe any highly aberrant neuronal genomes marked by multiple large CNVs, and we detected no aneuploidy. One explanation for this may be our sample size. Alternatively, the use of SCNT to reprogram neurons may be less efficient for highly mutated genomes. However, it is important to note that the efficiency of generating MCNT-ES cell lines (Table 1) is similar to those reported for other differentiated cell types, suggesting that the majority of MT neurons are not highly mutated in ways that preclude reprogramming (Hochedlinger and Jaenisch, 2002; Osada et al., 2005; Wakayama et al., 2005).
Table 2.
Somatic mutation discovery statistics.
| C5 | D4 | B2 | B3 | B4 | E1 | Mean | ||
|---|---|---|---|---|---|---|---|---|
| SNVs | Mutation Calls | 112 | 50 | 50 | 68 | 70 | 61 | 68.5 |
| %FDR (n = 69) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -- | |
| %FNR | 21.4 | 20.8 | 19.0 | 18.9 | 19.2 | 22.8 | -- | |
| Estimated Mutations | 142 | 63 | 62 | 84 | 87 | 79 | 86.2 | |
|
| ||||||||
| Indels | Mutation Calls | 25 | 19 | 16 | 9 | 17 | 18 | 17.3 |
| %FDR (n = 23) | 4.3 | 4.3 | 4.3 | 4.3 | 4.3 | 4.3 | -- | |
| %FNR | 28.7 | 25.2 | 24.2 | 24.0 | 24.7 | 28.5 | -- | |
| Estimated Mutations | 34 | 24 | 20 | 12 | 21 | 24 | 22.5 | |
|
| ||||||||
| SVs | Validated Breakpoints | 3 | 0 | 7 | 1 | 0 | 3 | 2.3 |
| Validated Events | 2 | 0 | 3 | 1 | 0 | 3 | 1.5 | |
| %FNR | 13.5 | 12.4 | 13.1 | 8.6 | 8.4 | 13.4 | -- | |
| Estimated Breakpoints | 3 | 0 | 8 | 1 | 0 | 3 | 2.5 | |
|
| ||||||||
| MEIs | Validated Mutations | 1 | 1 | 0 | 2 | 0 | 0 | 0.7 |
| %FNR | 48.2 | 45.8 | 47.6 | 48.3 | 47.7 | 47.0 | -- | |
| Estimated Mutations | 2 | 2 | 0 | 4 | 0 | 0 | 1.3 | |
|
| ||||||||
| Total Estimated Mutations | 181 | 89 | 90 | 101 | 108 | 106 | 112.5 | |
Given that these neuronal genomes harbor ~100 or more new somatic mutations, we asked whether any of these mutations were likely to impact gene function (Supplemental Experimental Procedures). Remarkably, ten of the somatic mutations we identified alter the coding sequence of known genes, at least four of which are expressed in MT neurons (Figure S3). These include missense mutations in Cdc40, Tas2r113, Klf16, Dhx37, and Tekt5, a single codon deletion in Gpr44, an exon deletion in Pkd2l2, an exon duplication in Atp10b, a deletion encompassing the Zic1 and Zic4 genes, and disruption of the Aven gene. In total, five of the six neuronal genomes that we analyzed in depth carry mutated coding sequences, demonstrating that individual neuronal genomes often carry one or more newly mutated genes. However, this did not represent a statistically significant enrichment, perhaps due to our small sample size. In addition, because mutations also fall in intronic and brain-related regulatory regions (Figure S3) these numbers likely underestimate the potential functional consequence of somatic mutation in MT neurons (Wan et al., 2014).
Lack of recurrent mutations in MT neurons
If the majority of somatic mutations in neurons arise early in development, MT neurons derived from the same donor might contain recurrent mutations (Evrony et al., 2014). Here, we analyzed three MCNT-ES lines from one donor mouse (mouse B) and two from another (mouse C), although one MCNT-ES cell line from the latter dataset was excluded from other analyses due to culture-derived loss of one X chromosome that led to a population bottleneck that would generate false positive candidate somatic mutations. None of the mutations we detected were shared. This could indicate that the mutations arose late in development or after neuronal differentiation and mitotic exit. However, these results are also consistent with a diverse embryonic origin for MT neuron precursors as has been reported for human cortical neurons (Lodato et al., 2015).
If neurons employ diversity-generating DNA rearrangements such as those seen in the immune system, we would expect to find identical or highly similar mutations in particular loci in neurons from different animals. However, none of our high confidence somatic mutations exhibited these features, and PCR tests excluded all of the low confidence candidate recurrent SV calls (n=16) (Supplemental Experimental Procedures). Further, the SVs we detected in individual MCNT-ES cell lines did not exhibit hallmarks of programmed rearrangement, such as joining of alternative exons or somatic hypermutation. We also visually inspected the protocadherin gene clusters, which have been proposed as candidate loci for programmed rearrangements in neuronal genomes (Yagi, 2003), and found no evidence of rearrangements. These results strongly argue that DNA rearrangements at a defined locus are not required for neuronal maturation or function. However, we cannot rule out the possibility that rare MT neurons, or neurons from other subtypes employ diversity-generating mutations.
Structural variants and mobile element insertions are rare in MT neurons
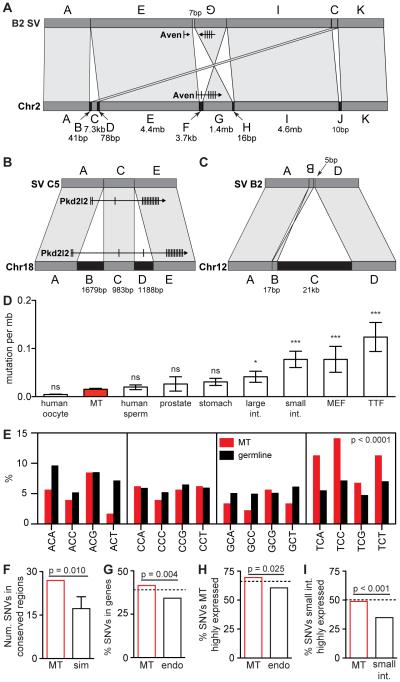
Structural variants (SVs) are of special interest due to their potential to cause large phenotypic effects, and because several lines of evidence suggest that neurons may be especially prone to double-strand breaks (Frank et al., 1998; Gao et al., 1998; Madabhushi et al., 2015; Suberbielle et al., 2013). The six MT neurons harbored nine total SVs, with a range of 0-3 per cell (Figure 2G). Each SV breakpoint had very limited microhomology (0-4 base pairs), and several contained a small number of inserted bases of unknown origin (2-7 base pairs) (Table S7), which is characteristic of repair by non-homologous end joining, the only type of double strand break repair employed by post-mitotic cells. Notably, three of the nine SVs were complex rearrangements (Figure 3A-3C). The most complex variant involved multiple small deletions, a 7.5 kb non-duplicative transposition and a 1.4 mb inversion (disrupting the Aven gene), with none of the breakpoints showing more than 1 bp of microhomology. This variant is best explained by a mechanism similar to chromothripsis involving regional DNA shattering and error-prone NHEJ. Chromothripsis is known to contribute to cancer and rare human disorders (Quinlan and Hall, 2012), but has an unknown role in healthy cell biology (Hatch and Hetzer, 2015). Although we cannot rule out the possibility that any given mutation occurred during reprogramming, our findings suggest that chromothripsis may occur in healthy post-mitotic cells, and that complex rearrangements may play an unanticipated role in neuronal genome diversity.
Figure 3. Mutational features of MT neuron genomes.
(A-C) Complex genomic rearrangements (CGRs) observed in MT neurons. Bottom bar represents wild type, top bar represents mutated configuration. (A) In a chromothripsis like CGR on chromosome 2 in line B2, fragment C is transposed downstream, fragment F is deleted, removing an exon from Aven, and fragment G is inverted, impacting many Aven exons. Small deletions are present at each breakpoint (arrows), and a 7bp insertion is present at the junction between E and G. (B) A CGR on chromosome 18 in line C5 involves two deletions within 3kb. One deletes exon 4 of the Pkd2l2 gene. (C) A 21kb deletion on chromosome 12 in line B2 is comprised of fragment B, a 17bp inversion and a 5bp insertion of unknown origin. (D) Total number of SNVs normalized by the length of the mouse or human diploid or haploid genome. Mean and SEM are plotted. (E) Percent of C→T conversions within each 3 bp context for MT neuron SNVs and germline SNPs. MT neuron SNVs occur significantly more often in the TpCpN context (~44% vs. ~25%, p<0.0001, Fisher’s Exact Test). (F) The number of MT neuron SNVs appearing in evolutionarily conserved regions of the genome is significantly higher than expected by chance (27 actual vs. ~17 simulated, standard deviation shown on graph = ~4, p = 0.010, Monte Carlo). (G) Percent of total MT neuron and endodermal SNVs that fall in genes. SNVs in MT neurons are enriched in genes relative to endodermal SNVs (p = 0.004, Fisher’s Exact Test). The dashed line indicates the percentage of the genome that falls into genes. (H) SNVs found in MT neurons are not depleted in highly expressed genes (top 50%) and are enriched in these genes compared to SNVs found in endodermal cell types (p = 0.025, Fisher’s Exact Test). (I) In contrast, small intestine SNVs are depleted in their own highly expressed genes relative to chance (p = 2.2 × 10−16, Poisson Test). Small intestine SNVs are also depleted in highly expressed genes relative to MT neuron SNVs (p = 7.06 × 10−4, Fisher’s Exact Test). Dotted lines in H and I demonstrate the percent of each transcriptome length that falls within highly expressed genes and represents random chance. MEF, mouse embryonic fibroblast. TTF, tail tip fibroblast. Sim, simulated. Endo, endodermal. See also Figures S3-S4, and Table S9.
The extent to which somatically acquired mobile element insertions (MEIs) contribute to neuronal genome diversity is a major unresolved question in the field. Our application of whole genome sequencing and methods that definitively distinguish true mutations from amplification artifacts and other false positive calls provides a unique opportunity to measure the MEI landscape in single neurons with high sensitivity and accuracy. In total, we predicted five MEIs, four of which were validated by PCR. Individual MT neurons carried 0-2 MEIs (Figure 2G). Our most conservative estimate for MEI detection sensitivity is 52% (Table S3), which predicts an average of at most 1.3 new MEIs per MT neuron genome. Thus, these studies are most consistent with results of two recent single cell sequencing experiments (Evrony et al., 2012; Evrony et al., 2014), and suggest that most MT neurons have a very low MEI burden. However, it is important to note that different neuronal subtypes may exhibit varying degrees of MEI activity, which could have interesting implications for neuronal diversity.
SNVs in MT neurons exhibit unique features, impact genes and may arise post-mitotically
The most common classes of mutations are indels and SNVs. We found that individual MT neurons harbored between 12 and 34 indels and between 62 and 142 SNVs. To determine whether this represented an unusually high or low mutational burden we compared MT neurons with data reported for other dividing and non-dividing cell types (Figure 3D; Supplemental Experimental Procedures). MT neurons carry more SNVs per megabase than reported for human oocytes, which are similar to MT neurons in that they also exit the cell cycle early in development (Kong et al., 2012). However, this is not statistically significant when corrected for multiple comparisons. The average number of SNVs in MT neurons is significantly lower than reported for mouse fibroblasts and intestinal cells and similar to prostate, stomach and sperm cells, all of which are expected to have undergone more cell divisions than MT neurons (Behjati et al., 2014; Kong et al., 2012; Young et al., 2012).
Closer examination of SNVs revealed several intriguing features that may represent unique signatures of neuron-specific and/or post-mitotic mutation. One unique feature emerged from analyses of potential mutational clusters, which identified three clusters of 2-3 SNVs. Two were multiple nucleotide polymorphisms affecting adjacent nucleotides, while the third impacted three nucleotides spanning 249 bp in the Atxn7l1 gene. Given the size of our dataset, these clustered mutations are highly unlikely to have arisen independently and may result from a mutational event similar to kataegis (Nik-Zainal et al., 2012).
MT neurons also exhibit a significant enrichment of C→T conversions within the context of TpCpN trinucleotides: 44% of neuronal SNVs occurred within the TpCpN context, versus ~25% for germline SNPs (p<0.0001, Fisher's Exact) (Figure 3E). To our knowledge, the only mutational process that favors this sequence context is cytosine deamination by Apobec1 (Beale et al., 2004), which has been previously implicated in activity-induced epigenetic modifications in neuronal genomes (Guo et al., 2011). In contrast, the overall MT neuron SNV base conversion spectrum is not obviously distinct from the overall spectrum reported for human germline and does not contain evidence of elevated action of specific mutagens, as has been reported for small intestine cells and some cancers (Figure S4; Supplemental Experimental Procedures) (Behjati et al., 2014; Kong et al., 2012; Pleasance et al., 2010a; Pleasance et al., 2010b). In particular, we did not detect known signatures of oxidative damage such as increased G→T conversions, which one might expect in highly metabolically active cells.
To determine whether SNVs in MT neurons are enriched in particular genomic regions, we compared the distribution of the 395 high confidence autosomal somatic SNVs to various genome annotations (Supplemental Experimental Procedures). Intriguingly, while somatic mutations are distributed randomly with respect to most genomic features (Table S9), they show a significant enrichment in evolutionarily conserved elements, which demarcate functionally relevant features such as genes and regulatory regions (1.6-fold, p = 0.01 by Monte Carlo simulation, Figure 3F).
This result led us to directly investigate whether somatic mutations in MT neurons were preferentially found in genes, compared to simulations that account for gene length and compared to the spectra of somatic mutations found in other cell types (Supplemental Experimental Procedures). The percentage of SNVs found in genes is equivalent to the genic fraction of the genome (Figure 3G). In contrast, neuronal SNVs are 1.2-fold more prevalent in genic regions than SNVs found in endodermal cell types, which are the only other mouse somatic cells sequenced at base-pair resolution at the time of our analyses (p = 0.004, Fisher’s Exact, Figure 3G) (Behjati et al., 2014).
In cancer cells, as well as human white blood cells, SNVs are depleted in expressed genes (Alexandrov et al., 2013; Holstege et al., 2014; Lawrence et al., 2013). To assess whether this was also true of MT neurons, we generated a list of genes that are transcribed in MT neurons using RNA-Seq on flow sorted MT neurons from the Pcdh21-Cre/Ai9 mouse strain (Supplemental Experimental Procedures). MT neuron SNVs are enriched in highly expressed genes (top 50%) compared with SNVs found in endodermal cells (Behjati et al., 2014) (p = 0.025 by Fisher's Exact Test, Figure 3H) and they are not depleted relative to the overall genic content of the genome. For comparison, we performed similar analyses of small intestine SNVs using a recently published RNA-Seq dataset for Lrg5-expressing small intestine stem cells (Sheaffer et al., 2014), which are the same stem cells used to generate the small intestine-derived organoids sequenced by Behjati et al. (Behjati et al., 2014). As predicted by studies of other lineages, mutations from the small intestine are depleted in genes that are highly expressed in small intestine stem cells (p = 2.2 × 10−16 by Poisson Test) and also relative to MT neuron SNVs (p = 7.06 × 10−4 by Fisher's Exact Test, Figure 3I) (Behjati et al., 2014). Therefore MT neurons exhibit a relative bias towards genes and expressed genes that has not been reported for other lineages and is independent of the length of expressed genes.
Overall, SNVs in MT neurons exhibit several unique features. SNVs are found in contexts consistent with Apobec1 action on cytosine bases. In addition, SNVs in MT neurons are biased towards evolutionarily conserved regions and appear to preferentially accumulate in functionally relevant genomic regions. Paired with the prevalence and unique features observed in MT neuron SVs, these studies predict that somatic mutation in MT neurons, as well as other neuronal subtypes, may have functional relevance, particularly if they continue to accumulate throughout the lifetime of the animal.
Developmental potency of MT neuron genomes
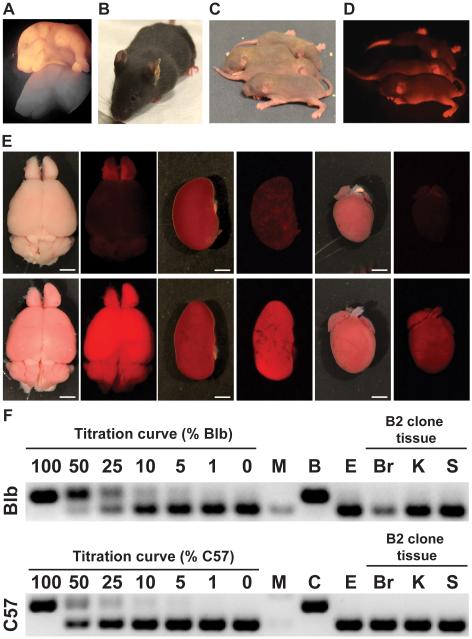
The developmental potency of neurons, and of mutated somatic cell types remains poorly understood, but has relevance for our understanding of neurobiology and for the use of reprogrammed adult cells in regenerative medicine. Having demonstrated that MT neuron genomes harbor many mutations, some of which are large, complex, and impact genes, we next sought to determine whether these genomes maintain sufficient integrity and plasticity to produce fertile adult animals. We therefore tested the developmental potency of six MCNT-ES cell lines using tetraploid embryo complementation (TEC), an assay in which only fully pluripotent cells can produce a viable mouse (Nagy et al., 1990; Nagy et al., 1993). Indeed, three MCNT-ES cell lines maintained full pluripotency based on the production of fertile adult mice, while another line produced full term pups that died shortly after birth (Figure 4A-4D; Table 3). The remaining two lines produced embryos that died in early or mid-gestation, which is consistent with studies of induced pluripotent stem cells and other SCNT-ES cells. The MCNT-mice were derived entirely from MT neurons, based on their ubiquitous expression of tdTomato and lack of detectable microsatellite DNA from the tetraploid host cells (Figure 4E and 4F; Figure S5A-S5E; Supplemental Experimental Procedures) (Boland et al., 2009). Because all tissues in mice generated using TEC are derived entirely from the donor ES cell line, these mice will harbor all of the mutations found in the original neuron in each of their cells. To confirm this we performed PCR on mouse tissues from line B2 and demonstrated the presence of the two expected structural variants (Figure S5F). Mice cloned from these MT neurons were healthy and fertile, with morphologically normal brains and olfactory bulbs (Figure 4E), suggesting that the genomic changes in MT neurons do not necessarily impact their differentiation into or away from their original lineage (Hochedlinger and Jaenisch, 2002). These functional studies are the first demonstration that neuronal nuclei from animals beyond eight weeks of age can maintain pluripotency, showing that that the epigenetic changes that accompany cell cycle exit, terminal differentiation, synaptic refinement, and persistent activity for up to 4.5 months can be accurately reversed by factors in the oocyte cytoplasm.
Figure 4. Mice derived from MT neurons.
(A) Newborn and (B) adult clones generated from MCNT-ES cells. (C) Standard and (D) fluorescence images of offspring of MCNT-mice. Transmission of the tdTomato transgene demonstrates MCNT-ES cells can generate functional germ cells. (E) Alternating standard and fluorescence images of brain, kidney, and heart dissected from Pcdh21/Cre-Ai9 control mice (top row) and MCNT-mice (bottom row). Organs from MCNT-mice exhibit normal morphology and uniform tdTomato expression. (F) Sample microsatellite PCR assay for tetraploid cell contribution to MCNT-mice. Band size distinguishes B2 derived cells from the tetraploid host strains C57 (C57BL/6J-Tyrc-2J) and Blb (Balb/cByJ). DNA titration curve demonstrates 5% detection limit. Analysis of DNA from B2 clone tissues exhibits no detectable tetraploid host DNA. M, molecular weight. E, B2 ES cell DNA. Br, brain. K, kidney. S, spleen. See also Figure S5.
Table 3.
MCNT-ES cell development in the tetraploid embryo complementation assay.
| MCNT- ES cell line |
Age of donor |
Tetraploid blastocysts Injected |
Alive at term (% injected) |
Breathing normally (% injected) |
Perinatal pups (% injected) |
Juvenile animals (% injected) |
Adult animals (% injected) |
|---|---|---|---|---|---|---|---|
| C1 | 3 wks | 152 | 15 (10%) | 10 (7%) | 8 (5%) | 8 (5%) | 8 (5%) |
| C5 | 3 wks | 140 | 8 (6%) | 5 (4%) | 3 (2%) | 2 (1%) | 2 (1%) |
| D4 | 3 wks | 214 | 15 (7%) | 6 (3%) | 1 (0.5%) | 0 | 0 |
| B2 | 4.5 mos | 140 | 32 (23%) | 26 (19%) | 20 (14%) | 19 (14%) | 19 (14%) |
| B3 | 4.5 mos | 140 | 0 | 0 | 0 | 0 | 0 |
| B4 | 4.5 mos | 150 | 0 | 0 | 0 | 0 | 0 |
DISCUSSION
Here, we have established the first high quality base-pair resolution sequence of neuronal genomes. We find that each MT neuron has a unique genome, harboring ~100 somatic mutations of diverse classes, which predicts that an individual brain contains billions of unique genomes. Notably, we did not detect recurrent DNA rearrangements or MEI insertion patterns consistent with the longstanding hypothesis that programmed somatic mutations may be used to generate functional neuronal diversity. However, the majority of neurons carried at least one gene disrupting mutation. These studies also provide evidence that the genomic context and distribution of SNVs in mouse neurons differs from other lineages, providing evidence for variable somatic mutation mechanisms among different cell lineages.
Importantly, these mouse studies establish that several unique features of human neuronal genomes unveiled by single cell sequencing studies can be confirmed by orthogonal methods and therefore are likely to have originated prior to human evolution. Our studies agree with reports that human neurons harbor few MEIs (Evrony et al., 2012). In addition we find that SNVs in mouse neurons exhibit enrichment in evolutionarily conserved regions, as recently reported (Lodato et al., 2015). However, our findings differ from other results based on single cell sequencing of human neurons. First, mouse MT neurons harbor fewer total SNVs than reported for human cortical neurons (~100 vs. ~1,500) (Lodato et al., 2015). Second, we find that the proportion of C to T SNVs in MT neurons is broadly similar to that in other cell types (~35-45% of total, Figure S4), while the human study reports a dramatic increase in C to T mutations (~75-85%) (Lodato et al., 2015). These discrepancies could arise from differences in the fidelity of in vitro and in vivo genome amplification. For example, the known error rate of Φ29 polymerase predicts that one amplification cycle would result in ~500 to 5,000 apparent mutations based on replication errors (Dean et al., 2002), while cell division is reported to produce only ~1 mutation (Behjati et al., 2014; Kong et al., 2012). Similarly, Φ29 based analyses of individual sperm cells report highly elevated frequencies of C to T transitions (Wang et al., 2012), which contrasts with pedigree-based analyses of germline mutation and the results of our study (Kong et al., 2012). Alternatively, it is possible that neuronal mutations vary based on differences between species, neuronal age or neuronal subtype.
SNVs found in mouse and human neurons exhibit unique signatures compared to other cell types. Why might neuronal mutations differ from those in the endodermal lineages, cancer cells, blood cells, and perhaps from other cell types? Mutations are generally assumed to arise during cell division. However, MT neurons exit the cell cycle at early embryonic stages (embryonic days 9-13) after ~14 cell divisions. This would predict a mutation rate of ~4-10 SNVs per division (Supplemental Experimental Procedures), which is higher than estimated for the human male germline or other somatic lineages (0.12 and 1.1 SNVs per cell division, respectively), leading us to consider alternative sources of mutation in MT neurons (Behjati et al., 2014; Kong et al., 2012). One appealing alternative is post-mitotic mutation. While post-mitotic mutation has not been formally described for any cell type, several features of MT neuron mutations are consistent with this mechanism. First, genes that are dynamically regulated by activity in post mitotic neurons are thought to undergo cytosine demethylation, which could explain the Apobec1 signature we observe. Second, we identify an apparent bias towards genes expressed in MT neurons, which is difficult to explain if these mutations arose in precursor cells. However, MT neuron mutations are also enriched in genes in general and we cannot determine whether expressed genes are significantly more vulnerable to mutation with our current sample size. Finally, SVs in MT neurons exhibit signs of NHEJ rather than replication-based mechanisms, consistent with a post-mitotic origin.
If the majority of SNVs in MT neurons arise post-mitotically, it is possible that mutations continue to accumulate throughout the lifetime of an individual. For example, if the ~100 mutations that we observe in a 4.5 month old mouse were to accumulate at that approximate rate (e.g., ~200/year) in humans, neurons from a 50 year old individual might harbor 10,000 somatic SNVs, which is on par with the SNV burden in cancer genomes. However, although our study examined neurons derived from 3 week old and 4.5 month old mice, we did not observe a significant increase of SNVs with mouse age, which is also consistent with another study (Lodato et al., 2015).
It is important to note that our study is specifically designed to provide a "best-case scenario" of genomic mosaicism in neurons, and may underestimate the true mutational burden. SCNT-based cloning may select against the most highly mutated neurons, such as the subset of genomically “aberrant” neurons harboring many CNVs reported previously (McConnell et al., 2013), or neurons harboring deleterious aneuploidies or MEI insertions.
Finally, the design of this study also allows us to functionally test the consequence of neuronal differentiation and maturation on the epigenetic plasticity of neuronal genomes. By generating neonatal and fertile adult mice from genetically labeled MT neurons derived from mice up to 4.5 months old, we demonstrate that despite the difficulty in inducing neurons to re-enter the cell cycle, at least some neurons maintain sufficiently plastic and intact genomes to produce an entire animal. While other studies have reported varying degrees of success (or failure) in efforts to clone from non-embryonic cortical neurons, this study is unique because we conclusively demonstrate the donor cell origin and age, and produce the first clones from adult mice older than 6-8 weeks of age (Kawase et al., 2000; Makino et al., 2005; Mizutani et al., 2015; Osada et al., 2002; Osada et al., 2005; Wakayama et al., 1998; Yamazaki et al., 2001). In addition, because we have deciphered full MT neuronal genome sequences, we show that accumulation of mutations during the development and post-mitotic aging of neurons is compatible with seemingly normal embryonic and postnatal development, which has relevance for the use of adult cells in regenerative medicine.
EXPERIMENTAL PROCEDURES
Somatic cell nuclear transfer and derivation of MCNT-ES cell lines
MT neurons were dissociated and purified as described previously (Brewer and Torricelli, 2007) with modifications (Supplemental Experimental Procedures). Oocytes were harvested from superovulated females and metaphase II spindles were removed and replaced with neuronal nuclei. Embryos were treated with 5 nM Trichostatin A, artificially activated with strontium chloride and cultured to blastocyst stage. Zonae pellucida were removed, embryos were cultured on a MEF feeder layer, and inner cell mass outgrowths were picked and dissociated with trypsin. Resulting ES cell lines were expanded on a MEF feeder layer. See Supplemental Experimental Procedures for further details.
Whole genome sequencing
Early passage MCNT-ES cells were separated from feeders by serial pre-plating. DNA was isolated from MCNT-ES cells and thymus or spleen using standard phenol chloroform extraction, ethanol precipitation and RNaseA digestion. Samples were sequenced by Beijing Genomics Institute using standard library prep for an Illumina Hi-Seq 2000. Reads were aligned with Novoalign against the full mm9 reference. Additional methods are in Supplemental Experimental Procedures.
Identifying somatic mutations
We used the GATK best practices pipeline (DePristo et al., 2011) to call SNPs and indels relative to the reference genome. Putative de novo somatic SNV and indel mutations were identified via a strategy similar to (Kong et al., 2012) (Supplemental Experimental Procedures).
To identify SVs, all unmapped and clipped reads were extracted from the initial alignments, and realigned with YAHA (Faust and Hall, 2012) to find possible split-read mappings spanning SV breakpoints. Discordant read-pairs were used in conjunction with these split-reads as input to LUMPY (Layer et al., 2014) to make SV calls. To identify somatic SVs, we selected calls with evidence in exactly one MCNT-ES cell line, except when searching for shared SVs.
MEIs were identified using a modified version of (Lee et al., 2012) as diagrammed in Figures S2A and S2B. Discordant read-pairs and unmapped and clipped reads were extracted from the initial alignments and realigned against a synthetic reference library formed from known LINE, SINE and LTR sequences (Table S2). Mates of reads that aligned well to the mobile element (ME) library were clustered by their ME type/subtype and their reference coordinates. If two clusters were found with nearly abutting reference genome coordinates and the proper strand orientation, a MEI was called. Evidence for the remaining calls was bolstered by split-read mappings in which one portion of a read was aligned to the ME library and the other to the reference genome within a cluster region. Candidate somatic MEIs were those found to be present in a single MCNT-ES cell line, except when searching for shared MEIs.
Somatic variant validation
To validate candidate de novo mutations, we performed PCR amplification of the genomic region containing the putative mutation on DNA from the MCNT-ES cell line and its control thymus or spleen sample. The products were Sanger sequenced to verify that the mutation was present in the MCNT-ES cell line but not in the control. For SNVs and indels, we tested a random subset of calls. For SVs and MEIs, we tested all calls. Additional experimental details and false discovery rates are in Supplemental Experimental Procedures and Table S4.
Monte Carlo simulation to determine enrichment of SNVs in genomic features
Monte Carlo simulations were carried out separately for each of nine genomic features (Table S9). To simulate our 395 high confidence autosomal SNVs, we randomly distributed 395 simulated SNVs 10,000 times throughout the mm9 autosomes, excluding gap regions and regions in which the total read depth precluded us from making somatic SNV calls. An enrichment p-value was then calculated as the fraction of trials in which the number of randomly distributed SNVs falling within the genomic feature of interest was less than the number of the actual somatic SNVs falling within the feature.
RNA-Seq and analysis
MT neurons were dissociated from Pcdh21/Cre-Ai9 mice as for nuclear transfer and flow sorted to isolate tdTomato positive neurons. RNA was purified and amplified prior to Illumina sequencing. See Supplemental Experimental Procedures for additional details. RNA-Seq data for Lgr5 positive small intestine stem cells was downloaded from the NCBI database (accession ids ERX421326, ERX421327 and ERX421329).
We used TopHat (Trapnell et al., 2009) to measure the expression of genes in each data set, then combined expression estimates by tissue of origin using Cufflinks (Trapnell et al., 2010). Genes exhibiting greater than median expression levels were termed “highly expressed”.
Supplementary Material
HIGHLIGHTS.
Reprogramming neurons by cloning enables high fidelity whole genome sequencing.
Neurons harbor ~100 unique mutations but lack recurrent DNA rearrangements.
Neuronal mutations impact expressed genes and exhibit unique molecular signatures.
Mature adult neurons can generate fertile adult mouse clones.
ACKNOWLEDGMENTS
We would like to thank Kevin Eggan and Chad Cowan for early help with somatic cell nuclear transfer. Michael Duran for assistance in validating mutations, Valentina Lo Sardo for help with cell culture and for feedback on the manuscript, Sohyon Lee for help with the manuscript and Susan Carlson for help with animal husbandry.
This work was supported by funding from the National Institute on Deafness and other Communication Disorders (DC012592 to K.K.B.), the National Institute of Mental Health (MH102698 to K.K.B.), the California Institute for Regenerative Medicine (RB3-02186 to K.K.B.), the Baxter Family, Norris and Del Webb Foundations (K.K.B.), by Las Patronas and the Dorris Neuroscience Center (K.K.B.), the NIH Director’s New Innovator Award (DP2OD006493-01 to I.M.H) and a pre-doctoral fellowship from the California Institute of Regenerative Medicine (J.L.H. & R.K.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.L.H. performed MT cell isolation, characterization of Pcdh21/Ai9 donor mice and mutation validation. SCNT and TEC experiments were performed by A.R. and S.K., with help from J.L.H. and M.B. Derivation of SCNT-ES cell lines was performed by G.M. Microsatellite PCR analyses were performed by W.F and J.L.H. MT neuron FACS was performed by R.T. and J.L.H. RNA-Seq and analyses were performed by R.T., P.C, A.T. G.F and J.L.H. Mutation validations were also performed by S.S. and W.F. Analyses of WGS data were performed by G.F. and I. H. with input from J.L.H. and K.K.B. Experiments were designed and the manuscript was written by J.L.H., G.F., I.H. and K.K.B, with input from the other authors.
The authors declare no potential conflicts of interest.
REFERENCES
- Ajioka I, Martins RAP, Bayazitov IT, Donovan S, Johnson DA, Frase S, Cicero SA, Boyd K, Zakharenko SS, Dyer MA. Differentiated Horizontal Interneurons Clonally Expand to Form Metastatic Retinoblastoma in Mice. Cell. 2007;131:378–390. doi: 10.1016/j.cell.2007.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale A-L, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale RCL, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the Differential Context-dependence of DNA Deamination by APOBEC Enzymes: Correlation with Mutation Spectra in Vivo. Journal of Molecular Biology. 2004;337:585–596. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- Behjati S, Huch M, van Boxtel R, Karthaus W, Wedge DC, Tamuri AU, Martincorena I, Petljak M, Alexandrov LB, Gundem G, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchart A, De Carlos JA, López-Mascaraque L. Time frame of mitral cell development in the mice olfactory bulb. The Journal of Comparative Neurology. 2006;496:529–543. doi: 10.1002/cne.20941. [DOI] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR. Isolation and culture of adult neurons and neurospheres. Nat Protocols. 2007;2:1490–1498. doi: 10.1038/nprot.2007.207. [DOI] [PubMed] [Google Scholar]
- Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, Sunaga F, Toritsuka M, Ikawa D, Kakita A, et al. Increased L1 Retrotransposition in the Neuronal Genome in Schizophrenia. Neuron. 2014;81:306–313. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- Cai X, Evrony Gilad D., Lehmann Hillel S., Elhosary Princess C., Mehta Bhaven K., Poduri A, Walsh Christopher A. Single-Cell, Genome-wide Sequencing Identifies Clonal Somatic Copy-Number Variation in the Human Brain. Cell Reports. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Schatz DG. Rearranging Views on Neurogenesis: Neuronal Death in the Absence of DNA End-Joining Proteins. Neuron. 1999;22:7–10. doi: 10.1016/s0896-6273(00)80671-6. [DOI] [PubMed] [Google Scholar]
- Chun JJM, Schatz DG, Oettinger MA, Jaenisch R, Baltimore D. The recombination activating gene-1 (RAG-1) transcript is present in the murine central nervous system. Cell. 1991;64:189–200. doi: 10.1016/0092-8674(91)90220-s. [DOI] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MCN, Muotri AR, Mu Y, Carson CT, Macia A, Moran JV, Gage FH. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proceedings of the National Academy of Sciences. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, et al. Comprehensive human genome amplification using multiple displacement amplification. Proceedings of the National Academy of Sciences. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville R, Liu J, Rothstein J, Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Annals of Neurology. 2011;69:141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-Neuron Sequencing Analysis of L1 Retrotransposition and Somatic Mutation in the Human Brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony Gilad D., Lee E, Mehta Bhaven K., Benjamini Y, Johnson Robert M., Cai X, Yang L, Haseley P, Lehmann Hillel S., Park Peter J., et al. Cell Lineage Analysis in Human Brain Using Endogenous Retroelements. Neuron. 2014;85:49–59. doi: 10.1016/j.neuron.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust GG, Hall IM. YAHA: fast and flexible long-read alignment with optimal breakpoint detection. Bioinformatics. 2012;28:2417–2424. doi: 10.1093/bioinformatics/bts456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng H-L, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V (D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of Neurons and Astrocytes by Oncogenes Can Induce Gliomas in Mice. Science (New York, NY) 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, et al. A Critical Role for DNA End-Joining Proteins in Both Lymphogenesis and Neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Larson SD, Hefzi H, Marnoy Z, Cutforth T, Dokka K, Baldwin KK. Sensory maps in the olfactory cortex defined by long-range viral tracing of single neurons. Nature. 2011;472:217–220. doi: 10.1038/nature09945. [DOI] [PubMed] [Google Scholar]
- Gole J, Gore A, Richards A, Chiu Y-J, Fung H-L, Bushman D, Chiang H-I, Chun J, Lo Y-H, Zhang K. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat Biotech. 2013;31:1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Junjie U., Su Y, Zhong C, Ming G.-l., Song H. Hydroxylation of 5- Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Hetzer MW. Chromothripsis. Current Biology. 2015;25:R397–R399. doi: 10.1016/j.cub.2015.02.033. [DOI] [PubMed] [Google Scholar]
- Hercus C. Novoalign. ( http://www.novocraft.com)
- Hinds JW. Autoradiographic study of histogenesis in the mouse olfactory bulb I. Time of origin of neurons and neuroglia. The Journal of Comparative Neurology. 1968a;134:287–304. doi: 10.1002/cne.901340304. [DOI] [PubMed] [Google Scholar]
- Hinds JW. Autoradiographic study of histogenesis in the mouse olfactory bulb. II. Cell proliferation and migration. The Journal of Comparative Neurology. 1968b;134:305–321. doi: 10.1002/cne.901340305. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415:1035–1038. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- Holstege H, Pfeiffer W, Sie D, Hulsman M, Nicholas TJ, Lee CC, Ross T, Lin J, Miller MA, Ylstra B, et al. Somatic mutations found in the healthy blood compartment of a 115-yr-old woman demonstrate oligoclonal hematopoiesis. Genome research. 2014 doi: 10.1101/gr.162131.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura F, Ayoub AE, Rakic P, Greer CA. Timing of neurogenesis is a determinant of olfactory circuitry. Nat Neurosci. 2011;14:331–337. doi: 10.1038/nn.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong B-H, Lee Y-J, Carp RI, Kim Y-S. The prevalence of human endogenous retroviruses in cerebrospinal fluids from patients with sporadic Creutzfeldt–Jakob disease. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2010;47:136–142. doi: 10.1016/j.jcv.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase E, Yamazaki Y, Yagi T, Yanagimachi R, Pedersen RA. Mouse embryonic stem (ES) cell lines established from neuronal cell-derived cloned blastocysts. genesis. 2000;28:156–163. [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SR, Loeb LA, Herr AJ. Somatic mutations in aging, cancer and neurodegeneration. Mechanisms of Ageing and Development. 2012;133:118–126. doi: 10.1016/j.mad.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lengner CJ, Kirak O, Hanna J, Cassady JP, Lodato MA, Wu S, Faddah DA, Steine EJ, Gao Q, et al. Reprogramming of Postnatal Neurons into Induced Pluripotent Stem Cells by Defined Factors. Stem Cells. 2011;29:992–1000. doi: 10.1002/stem.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IF, Yandava CN, Mabb AM, Hsiao JS, Huang H-S, Pearson BL, Calabrese JM, Starmer J, Parker JS, Magnuson T, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury MA, Friedman B, McConnell MJ, Rehen SK, Yang AH, Kaushal D, Chun J. Aneuploid neurons are functionally active and integrated into brain circuitry. Proceedings of the National Academy of Sciences. 2005;102:6143–6147. doi: 10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, Bui H-T, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochemical and Biophysical Research Communications. 2006;340:183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Jonasdottir A, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathe R, Harris A. Differential Display Detects Host Nucleic Acid Motifs Altered in Scrapie-Infected Brain. Journal of Molecular Biology. 2009;392:813–822. doi: 10.1016/j.jmb.2009.07.045. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome biology. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Tang C, Rao MS, Weissman IL. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19:998–1004. doi: 10.1038/nm.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Landscape of somatic retrotransposition in human cancers. Science (New York, NY) 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Jin Y, Prazak L, Hammell M, Dubnau J. Transposable Elements in TDP-43-Mediated Neurodegenerative Disorders. PLoS ONE. 2012;7:e44099. doi: 10.1371/journal.pone.0044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Prazak L, Chatterjee N, Gruninger S, Krug L, Theodorou D, Dubnau J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci. 2013;16:529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lu H, Yang W, Yong J, Zhang Z.-n., Zhang K, Deng H, Xu Y. Mouse SCNT ESCs Have Lower Somatic Mutation Load Than Syngeneic iPSCs. Stem Cell Reports. 2014;2:399–405. doi: 10.1016/j.stemcr.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global Epigenomic Reconfiguration During Mammalian Brain Development. Science (New York, NY) 2013:341. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D’Gama AM, Cai X, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science (New York, NY) 2015;350:94–98. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Morey R, O'Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R, Sabatini K, et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Gao F, Pfenning Andreas R., Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao P-C, et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair RG, Gesteland RC. Response properties of mitral cells in the olfactory bulb of the neonatal rat. Neuroscience. 1982;7:3117–3125. doi: 10.1016/0306-4522(82)90234-2. [DOI] [PubMed] [Google Scholar]
- Makino H, Yamazaki Y, Hirabayashi T, Kaneko R, Hamada S, Kawamura Y, Osada T, Yanagimachi R, Yagi T. Mouse embryos and chimera cloned from neural cells in the postnatal cerebral cortex. Cloning Stem Cells. 2005;7:45–61. doi: 10.1089/clo.2005.7.45. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, et al. Mosaic Copy Number Variation in Human Neurons. Science (New York, NY) 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, Taniguchi H, Tasic B, Huang ZJ, He Z, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani E, Oikawa M, Kassai H, Inoue K, Shiura H, Hirasawa R, Kamimura S, Matoba S, Ogonuki N, Nagatomo H, et al. Generation of Cloned Mice from Adult Neurons by Direct Nuclear Transfer. Biology of Reproduction. 2015;92:81. doi: 10.1095/biolreprod.114.123455. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Zhao C, Marchetto MCN, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proceedings of the National Academy of Sciences. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110329. doi: 10.1098/rstb.2011.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada T, Kusakabe H, Akutsu H, Yagi T, Yanagimachi R. Adult murine neurons: their chromatin and chromosome changes and failure to support embryonic development as revealed by nuclear transfer. Cytogenetic and genome research. 2002;97:7–12. doi: 10.1159/000064037. [DOI] [PubMed] [Google Scholar]
- Osada T, Tamamaki N, Song S-Y, Kakazu N, Yamazaki Y, Makino H, Sasaki A, Hirayama T, Hamada S, Nave K-A, et al. Developmental Pluripotency of the Nuclei of Neurons in the Cerebral Cortex of Juvenile Mice. The Journal of Neuroscience. 2005;25:8368–8374. doi: 10.1523/JNEUROSCI.1591-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrat PN, DasGupta S, Wang J, Theurkauf W, Weng Z, Rosbash M, Waddell S. Transposition-Driven Genomic Heterogeneity in the Drosophila Brain. Science (New York, NY) 2013;340:91–95. doi: 10.1126/science.1231965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin M-L, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010a;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin M-L, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010b;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic Mutation, Genomic Variation, and Neurological Disease. Science (New York, NY) 2013:341. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. Characterizing complex structural variation in germline and somatic genomes. Trends in genetics : TIG. 2012;28:43–53. doi: 10.1016/j.tig.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH, Chun J. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proceedings of the National Academy of Sciences. 2001;98:13361–13366. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BSV, Kingsbury MA, Cabral K.t.M.S., McConnell MJ, Anliker B, et al. Constitutional Aneuploidy in the Normal Human Brain. The Journal of Neuroscience. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, Kim R, Aoki R, Elliott EN, Schug J, Burger L, Schübeler D, Kaestner KH. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes & Development. 2014;28:652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, McConnell MJ, Marchetto MCN, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends in Neurosciences. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosulski DL, Lissitsyna Bloom M, Cutforth T, Axel R, Datta SR. Distinct representations of olfactory information in different cortical centres. Nature. 2011;472:213–216. doi: 10.1038/nature09868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suberbielle E, Sanchez PE, Kravitz AV, Wang X, Ho K, Eilertson K, Devidze N, Kreitzer AC, Mucke L. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-[beta] Nat Neurosci. 2013;16:613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Qurashi A, Poidevin M, Nelson DL, Li H, Jin P. Retrotransposon activation contributes to fragile X premutation rCGG-mediated neurodegeneration. Human molecular genetics. 2012;21:57–65. doi: 10.1093/hmg/ddr437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton Kyle R., Gerhardt Daniel J., Jesuadian JS, Richardson Sandra R., Sánchez-Luque Francisco J., Bodea Gabriela O., Ewing Adam D., Salvador-Palomeque C, van der Knaap Marjo S., Brennan Paul M., et al. Ubiquitous L1 Mosaicism in Hippocampal Neurons. Cell. 2015;161:228–239. doi: 10.1016/j.cell.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J. Somatic mutations, genome mosaicism, cancer and aging. Current opinion in genetics & development. 2014;26:141–149. doi: 10.1016/j.gde.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama S, Ohta H, Kishigami S, Van Thuan N, Hikichi T, Mizutani E, Miyake M, Wakayama T. Establishment of Male and Female Nuclear Transfer Embryonic Stem Cell Lines from Different Mouse Strains and Tissues. Biology of Reproduction. 2005;72:932–936. doi: 10.1095/biolreprod.104.035105. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Perry ACF, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake Stephen R. Genome-wide Single-Cell Analysis of Recombination Activity and De Novo Mutation Rates in Human Sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra JW, Peterson SE, Yung YC, Mutoh T, Barral S, Chun J. Aneuploid mosaicism in the developing and adult cerebellar cortex. The Journal of Comparative Neurology. 2008;507:1944–1951. doi: 10.1002/cne.21648. [DOI] [PubMed] [Google Scholar]
- Westra JW, Rivera RR, Bushman DM, Yung YC, Peterson SE, Barral S, Chun J. Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. The Journal of Comparative Neurology. 2010;518:3981–4000. doi: 10.1002/cne.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T. Diversity of the cadherin-related neuronal receptor/protocadherin family and possible DNA rearrangement in the brain. Genes to Cells. 2003;8:1–8. doi: 10.1046/j.1365-2443.2003.00614.x. [DOI] [PubMed] [Google Scholar]
- Yalcin B, Wong K, Bhomra A, Goodson M, Keane T, Adams D, Flint J. The fine-scale architecture of structural variants in 17 mouse genomes. Genome biology. 2012;13:R18. doi: 10.1186/gb-2012-13-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Makino H, Hamaguchi-Hamada K, Hamada S, Sugino H, Kawase E, Miyata T, Ogawa M, Yanagimachi R, Yagi T. Assessment of the developmental totipotency of neural cells in the cerebral cortex of mouse embryo by nuclear transfer. Proceedings of the National Academy of Sciences. 2001;98:14022–14026. doi: 10.1073/pnas.231489398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young Margaret A., Larson David E., Sun C-W, George Daniel R., Ding L, Miller Christopher A., Lin L, Pawlik Kevin M., Chen K, Fan X, et al. Background Mutations in Parental Cells Account for Most of the Genetic Heterogeneity of Induced Pluripotent Stem Cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.