SUMMARY
Trisomy 21, or Down syndrome (DS), is the most common genetic cause of developmental delay and intellectual disability. To gain insight into the underlying molecular and cellular pathogenesis, we conducted a multi-region transcriptome analysis of DS and euploid control brains spanning from mid-fetal development to adulthood. We found genome-wide alterations in the expression of a large number of genes, many of which exhibited temporal and spatial specificity and were associated with distinct biological processes. In particular, we uncovered co-dysregulation of genes associated with oligodendrocyte differentiation and myelination that were validated via cross-species comparison to Ts65Dn trisomy mice. Furthermore, we show that hypomyelination present in Ts65Dn mice is in part due to cell-autonomous effects of trisomy on oligodendrocyte differentiation and results in slower neocortical action potential transmission. Together, these results identify defects in white matter development and function in DS and provide a transcriptional framework for further investigating DS neuropathogenesis.
Keywords: brain development, neurodevelopmental disorders, neocortex, glia, gene expression, genomics, white matter
Graphical Abstract
INTRODUCTION
Trisomy for human chromosome 21 (HSA21) causes Down syndrome (DS) in one out of every 691 live births (Canfield et al., 2006), making it the most common genetic cause of developmental delay and intellectual disability. DS is characterized by a constellation of phenotypes affecting many organ systems, including central nervous system abnormalities such as cognitive and motor impairments, microcephaly, and the early appearance of neuropathological characteristics of Alzheimer's disease (AD; Hartley et al., 2014; Haydar and Reeves, 2012; Letourneau et al., 2014; Lott, 2012).
Prior insights into DS neuropathology have come from studies of human individuals and trisomic mouse models. Morphometric and cellular studies on human brain indicate that trisomy 21 causes complex spatiotemporal disturbances in neural development (Golden and Hyman, 1994; Larsen et al., 2008; Reeves and Haydar, 2011). Mouse models have provided additional insights into the developmental progression of DS, elucidating specific defects in neurogenesis, neuronal differentiation, and impaired synaptic plasticity and learning (Chakrabarti et al., 2007, 2010; Hyde et al., 2001; Kleschevnikov et al., 2004; Siarey et al., 1997, 1999; Tyler and Haydar, 2013; Wang et al., 2013). While it is clear that trisomy of HSA21 is the root cause of DS, which biological processes are affected and which HSA21 and non-HSA21 genes are associated with those processes are not fully understood. Furthermore, the extent to which human postmortem neuropathology is reflected in the mouse models (and vice versa) remains one of the key problems hampering further understanding of DS brain development and function.
Transcriptome profiling has provided insight into the pathophysiological mechanisms underlying neurodevelopmental brain disorders (Mitchell and Mirnics, 2012; State and Geschwind, 2014; Tebbenkamp et al., 2014). While several reports have investigated gene expression in postmortem human DS brains or derived pluripotent stem cells (Letourneau et al., 2014; Lockstone et al., 2007; Mao et al., 2003, 2005), these studies are limited by sparse genome coverage, numbers of samples or developmental time points, and potential variances between in vitro and in vivo gene regulation. We therefore set out to characterize the spatiotemporal dynamics of gene expression in the DS and matched euploid brain and to subsequently decipher the biological processes affected over the course of prenatal and postnatal development.
Genome-wide transcriptional profiling was performed in different regions of male and female postmortem brains spanning from mid-fetal development to adulthood. This approach uncovered a highly dynamic disruption of the DS transcriptome across all chromosomes, with alterations in gene expression in different brain regions over time. Bioinformatic analysis revealed genes and gene networks that were affected throughout the DS brain, revealing key molecular and cellular processes previously implicated in neurological dysfunction in DS as well as many that had not been well characterized in DS brains. Notably, we found transcriptome differences indicating that oligodendrocyte differentiation and myelination are altered in DS. We further identified cell-autonomous deficits in oligodendrocyte differentiation and the production of neocortical myelin, which were accompanied by slower action potential transmission between cerebral hemispheres. Altogether, the data illustrate disruption of distinct gene networks in DS, providing an extensive framework to identify, prioritize, and test gene candidates and biological processes underlying the neuropathophysiology of DS. To facilitate the usefulness and accessibility of the DS transcriptome, we have also generated an internet database ((http://medicine.yale.edu/lab/sestan/resources/).
RESULTS
Genome-Wide Spatiotemporal Alterations of Gene Expression in DS Brains
We used the Affymetrix exon array platform, which features comprehensive genome-wide coverage of the human transcriptome across entire transcripts and each individual exon, to characterize gene expression in different regions of DS brains over the course of development (see Experimental Procedures). Transcriptome profiling was performed using total RNA extracted from eleven regions, including multiple regions of the cerebral neocortex, the hippocampus (HIP), and the cerebellar cortex (CBC) using a standardized protocol from high quality postmortem human brains of DS and clinically and anatomically unremarkable euploid controls (for details on tissue dissections see Extended Experimental Procedures, Figure S1 and Tables S1 and S2). We collected 58 paired tissue samples from 15 euploid control and 15 DS brains ranging in age from 14 post-conception weeks (pcw) to 42 years-old (Table S2) and then allocated them into groups based on a previously described timeline that divides human brain development and adulthood into 15 periods (Kang et al., 2011).
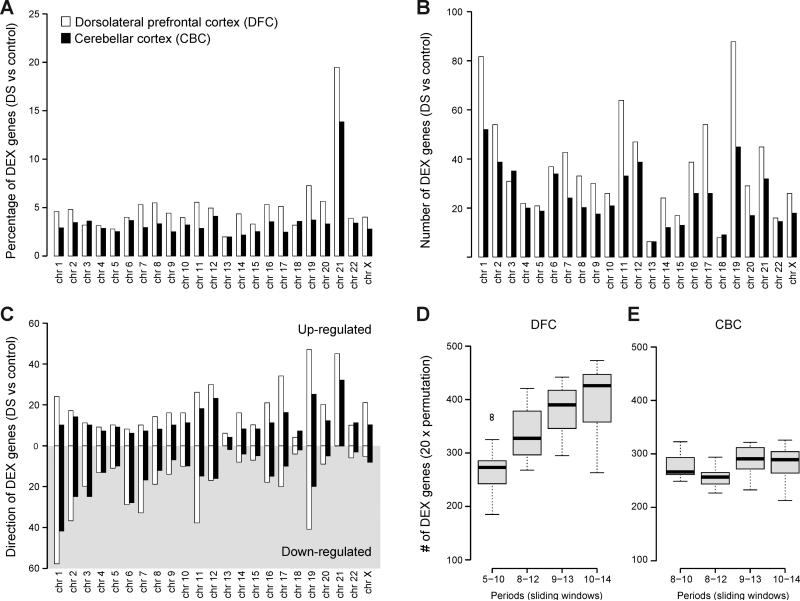
Principal component analysis showed segregation between DS and control samples; however, brain regions and age (developmental periods) contributed more to the global differences in gene expression than disease status (Figure S2), indicating that gene expression in the human brain has a high degree of spatial variability. Table S2). We chose dorsal frontal cortex (DFC) and CBC for our analyses because both regions have been implicated in DS neuropathology (Hartley et al., 2014; Haydar and Reeves, 2012; Letourneau et al., 2014; Lott, 2012) and exhibit diverse transcriptional profiles over the course of development (Kang et al., 2011; Tebbenkamp et al., 2014). Of 17,557 mRNA genes profiled, 842 (4.8%) and 571 (3.25%) genes in DFC and CBC, respectively, were defined as significantly differentially expressed (DEX) between paired DS and euploid samples across all periods (Table S3) (FDR = 0.1; see Extended Experimental Procedures). As expected, the percentage of DEX genes on HSA21 was higher than other chromosomes (Figures 1A and S3). While 19.5% (DFC) and 13.9% (CBC) of the genes localized on HSA21 were DEX genes, a median of 4.4% (DFC) and 3.0% (CBC) DEX genes were found on other chromosomes in DS compared to their matched control. However, the majority of DEX genes were non-HSA21 genes (1,337 genes vs 76 genes on HSA21) (Figure 1B). We also found that while all HSA21 DEX genes were up-regulated in DS samples, only 47.7% (DFC) and 45.2% (CBC) of non-HSA21 DEX genes were up-regulated. Conversely, 52.3% (DFC) and 54.8% (CBC) of non-HSA21 genes were down-regulated (Figure 1C), indicating that the triplication of HSA21 leads to both positive and negative regulation of genes on other chromosomes, (Figure S4). This directionality of HSA21 gene expression profiles suggests that upregulation of genes on HSA21, some of which function as trans factors, is followed by up- and down-regulation of genes distributed throughout the genome. Strikingly, we found that gene expression across all chromosome locations was altered even from 14 pcw, indicating early and widespread changes to the DS transcriptome.
Figure 1. Genes Dysregulated in DS Brains are Globally Distributed and Developmentally Dynamic.
(A) The percentage of differentially expressed genes (DEX) on each chromosome indicating global disruptions in gene expression.
(B) The number of genes on each chromosome that are DEX in Down syndrome (DS). Note that the majority of DEX genes are not located on HSA21.
(C) The number of up-regulated (white shading) and down-regulated (gray shading) DEX genes per chromosome.
(D and E) A permutation analysis of DEX genes in the dorsolateral prefrontal cortex (DFC) (D) and cerebellar cortex (CBC) (E) across 4 sliding windows corresponding to periods from mid-fetal development to adulthood. Periods of human brain development and adulthood are defined as previously described (Kang et al., 2011). Note that the number of DEX genes rises over development for DFC but not CBC. No CBC samples were available for periods 5 to 7.
A combination of sliding-window, paired t-test, and permutation tests was used to investigate temporal dynamics in transcriptional differences between DS and control brains (see Extended Experimental Procedures). This analysis revealed that the number of DEX genes in DFC increased in an age-dependent manner, with significantly more DEX genes emerging over postnatal development and adulthood (periods 8-14) (Figure 1D, and Table S4). A completely different temporal picture emerged in the CBC, where the number of DEX genes remains constant throughout life (Figure 1E, and Table S4). This dichotomy between the DFC and CBC temporal gene expression profiles extended throughout the genome, with more variability across all chromosomes over developmental time in DFC samples (Figure S3). Gene ontology enrichment analysis indicated potential dysregulated biological categories including transmission of nerve impulse (Benjamini-Hochberg-adjusted (BHA) p=2.9×10-8), synaptic transmission (BHA p=1.4×10-5), cell morphogenesis (BHA p=1.4×10-5), cell-cell signaling (BHA p=6.7×10-5), and axon ensheathment (BHA p=4.8×10-2) (Table S5).
Together, these studies demonstrate that many genes throughout the genome are altered in DS, confirming and extending previous findings that non-HSA21 genes may contribute significantly to trisomy 21 phenotypes (Letourneau et al., 2014; Lockstone et al., 2007). Most importantly, our results clearly show that many of these DEX genes display dynamic temporal and spatial differences in expression and highlight specific biological processes potentially disrupted in developing DS brain.
Co-Dysregulation of Genes Associated with Oligodendrocyte Differentiation and Myelination in DS
To integrate the expression differences observed between DS and matched control samples from all 58 paired samples and each brain region into a systems level context, we performed weighted-gene co-expression network analysis (WGCNA) (Zhang and Horvath, 2005) and identified modules of co-expressed genes. We defined 60 gene modules exhibiting altered expression during DS brain development in the CBC, HIP, and multiple neocortical regions (Table S6 and S7). We then carried out gene ontology enrichment analysis of each module to gain insight into the biological significance of these clusters of co-expressed genes (Table S6). This analysis confirmed a number of cellular and molecular processes previously found to be altered in DS or mouse model brains, including cell morphogenesis (Module [M] 24; Figure S5A), RNA processing (M31), gene transcription (M52), immune responses (M54; Figure S5B), neuronal differentiation (M4 and M45; Figure S6A,D), synaptic transmission and regulation (M10 and M34; Figure S6B,C) and electron transport (M14) (Table S6) (Bahn et al., 2002; Lockstone et al., 2007; Sahun et al., 2014; Yoshida et al., 2013; Zampieri et al., 2014).
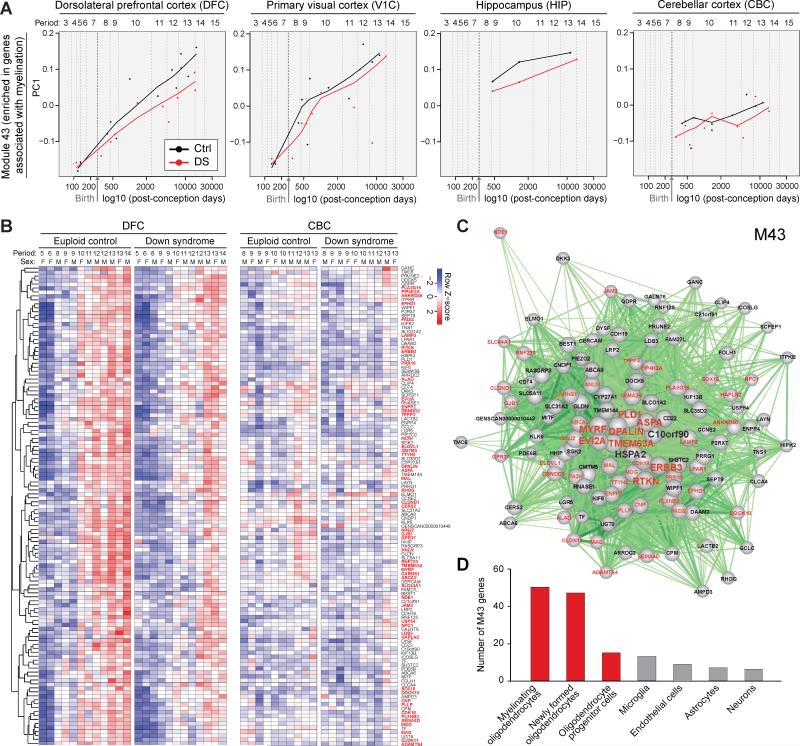
WGCNA also identified co-expression modules enriched for genes associated with biological processes previously not well-characterized in DS. Notably, a module of co-expressed genes, number 43 (M43), was enriched for gene ontology categories related to regulation of action potential (BHA p=1.7×10-2) and axon ensheathment (BHA p=3.7×10-2) and was markedly down-regulated throughout the DS neocortex and in the HIP over development (Figure 2A, B and Table S6). Differences in this module accelerated over the course of DS neocortical development (Figure 2A, B), coinciding with the onset and progression of myelination. Note that the absence of a M43 expression phenotype in the CBC of DS brains (Figure 2A, B) is likely because the most numerous cells in CBC, granule cells, have unmyelinated axons.
Figure 2. A Co-Expression Module Enriched in Genes Associated with Oligodendrocyte Development and Myelination is Decreased over Development in DS.
(A) Gene network analysis identifies distinct modules of co-expressed genes dysregulated in DS, including module (M) 43 that is significantly enriched in genes associated with oligodendrocyte development and myelination. Plots of relative expression of the PC1 of M43 over development in the DFC, primary visual cortex (V1C), hippocampus (HIP), and CBC, indicating higher expression in euploid control versus DS brain that increases in degree over development.
(B) A heat map showing the expression of M43 genes in DFC (left panel) and CBC (right panel), which confirms and details the gene expression trajectories shown in (A). Note that a high proportion of mouse homologs of the M43 genes are expressed in mouse oligodendrocyte lineage cells (gene symbols are highlighted in red).
(C) A network plot of M43 genes and their intramodular connections (cutoff, Pearson correlation > 0.7). 8 of the 10 hub genes (the top ten genes with highest intramodular connectivity; TMEM63A, MYRF, PLD1, RTKN, ASPA, OPALIN, ERBB3, EVI2A) are primarily expressed by mature oligodendrocytes and tightly correlated with other oligodendrocyte-associated M43 genes. Oligodendrocyte enriched genes are shown in red. Note their central position in the network, suggesting high intramodular connectivity.
(D) The number of mouse homologs of M43 genes that are highly expressed in major cell types from mouse cerebral cortex. The majority of the M43 genes are highly expressed specifically in oligodendrocytes. Red bars denote the oligodendrocyte lineage; gray bars denote other cell lineages.
To further characterize M43 genes, we investigated whether they were associated with the developmental program of specific neural cell types (i.e., astrocytes, endothelial cells, microglia, neurons, oligodendrocyte progenitor cells, newly formed oligodendrocytes or myelinating oligodendrocytes). We queried whether M43 genes were highly expressed (FPKM > 20) in different populations of neural cell types purified from mouse cerebral cortex (Zhang et al., 2014) (see Extended Experimental Procedures). We found that of the 121 mouse homologs of genes in M43, 67 met this cutoff for high expression (FPKM > 20) and 82.1% (55/67) of this subset were found to be highly expressed by oligodendrocyte lineage cells (gene names labeled in red in Figure 2B-D). Importantly, 8 of the top 10 genes with highest intramodular connectivity (hub genes) in M43 were primarily expressed by mature oligodendrocytes and tightly correlated with other oligodendrocyte-associated M43 genes (Figure 2C).
To ensure that DEX genes detected were not due to variability in tissue dissections, we analyzed genes highly enriched in astrocytes, neurons, and oligodendrocytes (Zhang et al., 2014) (see Extended Experimental Procedures). We found that intra-individual gene expression for these three cell types was highly correlated in all samples (see Table S8). We further found by pair-wise analysis that gene expression levels for markers of all three cell types were highly correlated for the same brain regions in all control (r > 0.94) or DS individuals (r > 0.95), indicating uniform tissue collection procedures. However, in agreement with our DEX analysis, expression of oligodendrocyte-specific genes were less correlated and significantly different between DS and control DFC (r = 0.89, p=0.018 Wilcoxon signed-rank test; Table S8).
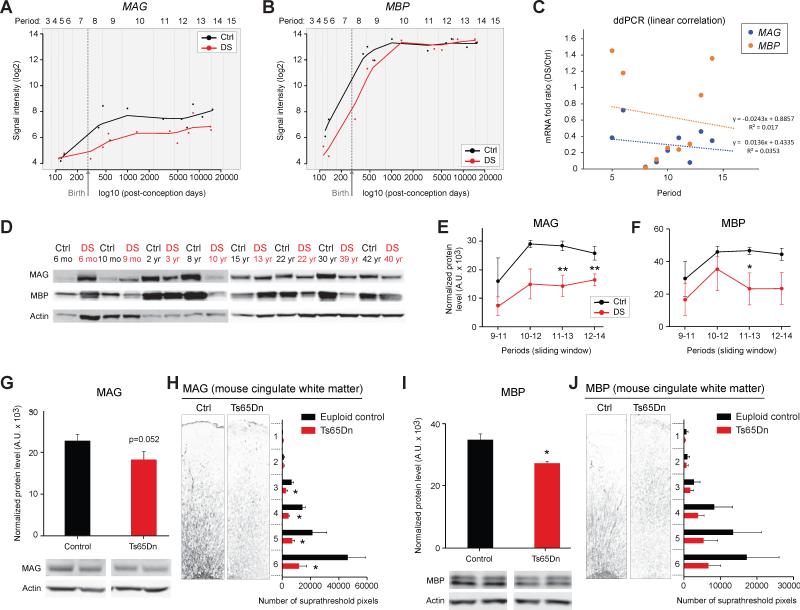
We analyzed individual expression trajectories of several genes within the M43 module known to be expressed in oligodendrocytes and involved in myelination. Lower levels of gene expression spanning from birth to adulthood for myelin associated glycoprotein (MAG; Figure 3A) and from mid-fetal development to early childhood for myelin basic protein (MBP; Figure 3B) were observed. To validate the MAG and MBP exon array data, we assayed the expression of these white matter associated genes and their gene products in the same neocortical samples using digital droplet PCR (ddPCR) (Figure 3C) and immunoblotting (Figure 3D-F). Together, both DEX and WGCNA analyses provide evidence for alterations in neocortical oligodendrocyte differentiation and myelination in developing DS brains at transcript and protein levels.
Figure 3. Expression of the Essential Myelin Components MAG and MBP is Diminished in Developing DS and the Ts65Dn Mouse Brains.
(A) Log2 values of the array signal intensity in human euploid control (Ctrl) and DS DFC show that MAG expression is decreased from birth onwards (periods 8 to 14) in DS. Periods 5-9, p=0.28; periods 8-12, p=0.023; periods 9-13, p=0.025; periods 10-14, p=0.0024; all periods, p=0.014 (paired t-test).
(B) Log2 values of the array signal intensity in human Ctrl and DS DFC show that MBP expression is decreased from mid-fetal development to early childhood (periods 5-9) in DS. Periods 5-9, p=0.078 (one tailed test, p=0.039); periods 8-12, p=0.192; periods 9-13, p=0.52; periods 10-14, p=0.72; all periods, p=0.10 (paired t-test).
(C) ddPCR analysis of MBP and MAG expression in human Ctrl and DS DFC samples confirming decreased expression in developing DS brains.
(D) Representative western blots for MAG and MBP in human Ctrl and DS DFC over development (DS samples in red type).
(E and F) Sliding window analysis of MAG (E) and MBP (F) protein levels identify significant reductions in developing and adult DS brains. *, p≤0.03 (paired t-test).
(G and H) Western blotting of neocortex tissue samples (n=10) (G) and immunostaining of the anterior cingulate cortex (n=9) (H) in postnatal day (P) 30 Ctrl and trisomic Ts65Dn mice identifies a reduction of MAG protein expression in Ts65Dn brains. *, p≤0.05; **, p≤0.001 (paired t-test).
(I and J) Western blotting for MBP in the neocortex (n=3 pairs) (I) and immunostaining for MBP in P30 cingulate cortex (J) of Ctrl and Ts65Dn mice show a trend towards reduced expression of MBP in the white matter of Ts65Dn mice (n=3 pairs). *, p=0.009 (paired t-test).
Given these substantial changes in oligodendrocyte lineage genes and proteins, we sought to confirm previous reports of reduced myelination in the DS forebrain. We imaged tissue sections of the DFC from 5 pairs of human DS and matched control specimens ranging in age from 1-70 years old using the spectral confocal reflectance microscopy (SCoRe) technique (Schain et al., 2014). Consistent with previous reports, we found that the overall density of myelinated fibers is reduced in DS brain across all ages (Figure S6A, B). In addition, we found that myelinated axons in control brains form a grid-like lattice that resembles a rectilinear grid as described previously (Ang et al., 2003; Wedeen et al., 2012) (Figure S6C). However, DS brains did not exhibit evidence of grid-like myelinated fiber orientations, suggesting reduced complexity of fiber pathways in the trisomic human brain.
White Matter Abnormalities in a Trisomic Mouse Model of DS
To determine whether the protein and cellular level consequences of altered oligodendrocyte/myelination associated gene expression in DS are also reflected in mouse models, we examined oligodendrocyte lineage progression, myelination, and neuronal conductivity over development in the Ts65Dn mouse brain. This commonly used DS model is trisomic for ~100 orthologs of the 364 genes on HSA21 and displays many DS-specific phenotypes, including delays in brain development and cognitive defects (Rueda et al., 2012). However, changes in white matter have not been previously described in the Ts65Dn mouse brain. By immunoblotting of neocortex tissue samples (n=10) (G) and immunostaining of the anterior cingulate cortex and underlying corpus callosum (n=9) (H), we found a reduction of MAG (Figure 3G, H) and MBP (Figure 3I, J) protein levels and immunostaining intensity in postnatal day (P) 30 Ts65Dn mice. Reduced immunostaining intensity for 2’,3’-cyclic nucleotide 3’ phosphodiesterase (CNP), an oligodendrocyte and myelin-associated protein, was also observed in the P60 neocortex in Ts65Dn brains (Figure S7). Importantly, these cellular and biochemical changes are first detected during the period of myelin development and refinement and are maintained at later ages (e.g., P60).
Myelin, Axon and Action Potential Conduction Deficits in Trisomic White Matter
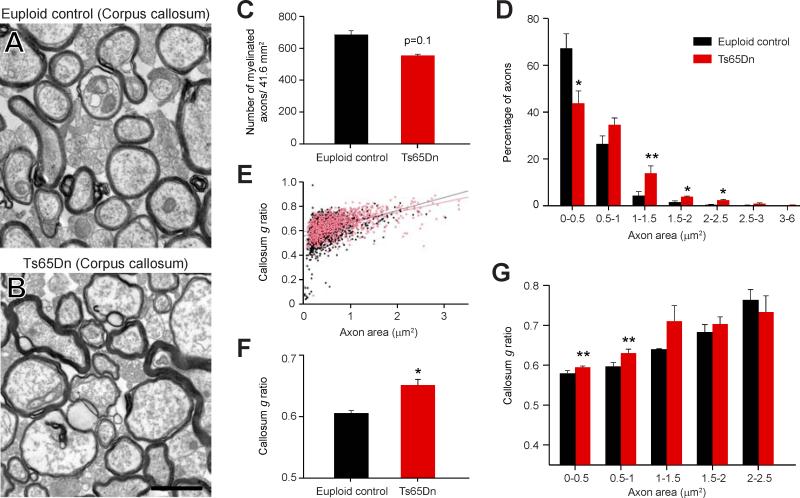
Alterations in white matter associated proteins measured within the trisomic human and mouse brains indicate possible changes in the amount or allocation of myelin. We tested this hypothesis with ultrastructural and immunofluorescent studies in Ts65Dn and euploid mice at P60. Electron microscopy revealed that axon profiles in the Ts65Dn corpus callosum are on average larger than in controls (Figure 4A-B) and there was a trend for fewer myelinated axons in the body of the Ts65Dn corpus callosum (p= 0.1, n=6; Figure 4C). Furthermore, when myelinated axons were binned by axon area, we found a significant decrease in the number of small-bore myelinated axons (0-0.5 μm2; p= 0.046; Figure 4D). We also found that myelinated axons in Ts65Dn brains have significantly thinner myelin sheaths (g-ratio) (p< 0.011; Figure 4E-F). Axons were then binned by size to determine whether a correlation exists between axon diameter and myelin sheath thickness. Specifically, small-bore myelinated Ts65Dn axons within 0-0.5 μm2 and 0.5-1 μm2 have thinner myelin sheaths than corresponding controls (p= 0.084 and p= 0.076, respectively; Figure 4G). Altogether, these results indicate a decrease in the percentage of small-bore myelinated axons and myelin sheath thickness in Ts65Dn white matter.
Figure 4. Diminished Myelin Sheath Thickness in the Ts65Dn Corpus Callosum.
(A and B) Representative electron micrographs of axon cross sections in the P60 euploid control (A) and Ts65Dn (B) corpus callosum. Scale bar = 1 μm.
(C) Graph of the numbers of myelinated axons in euploid versus Ts65Dn revealing a trend towards a decrease in the number of myelinated axons in Ts65Dn mice.
(D) Histogram indicating a decrease in the proportion of small diameter axons and an increase in the proportion of large diameter axons (>1 μm) in Ts65Dn mice. *, p≤0.05; #, p=0.058 (paired t-test).
(E) Plot of g-ratios (y-axis) and the corresponding diameter for all axons assessed. Black dots = euploid control; red dots = Ts65Dn.
(F) Bar graph showing there is a significant increase in the mean g-ratio in Ts65Dn mice indicating thinner myelin sheaths around Ts65Dn axons. *, p=0.011 (paired t-test).
(G) Histogram indicating that the g-ratios of small diameter, but not large diameter axons, are higher in Ts65Dn mice. #, p≤0.08 (paired t-test).
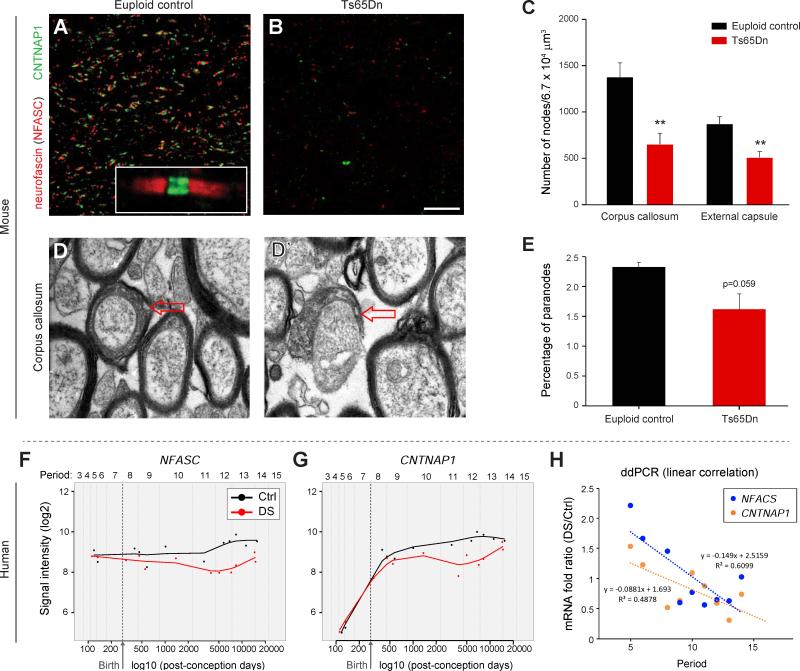
Deficits in myelination and oligodendrocyte maturation can also lead to impaired formation of the nodes of Ranvier (Kaplan et al., 2001; Rasband et al., 1999; Tanaka et al., 2009). Therefore, we immunostained coronal brain sections from Ts65Dn and controls for NF186 (an NFASC isoform) and CNTNAP1 (Thaxton et al., 2010), which mark nodal regions. Quantification of nodal protein profiles in the Ts65Dn brain at P60 revealed a striking decrease in the number of nodes of Ranvier in the corpus callosum and we found similar decreases in the external capsule (Figure 5A-C; p<0.005). Ultrastructural analysis confirmed this result, revealing a trend for fewer paranode cross-sections in Ts65Dn (Figure 5D-E; p=0.059). To determine if these findings implicate changes in node formation as a novel human DS brain phenotype, we evaluated expression levels of both NFASC and CNTNAP1 in the exon array dataset. These genes were significantly down-regulated from birth to adulthood (Figure 5F-G), and subsequent ddPCR analysis confirmed these differences in the DS brain samples (Figure 5H).
Figure 5. Impaired Formation of Nodes of Ranvier in Ts65Dn Mice.
(A) Representative image of nodes of Ranvier in P60 euploid control (Ctrl) corpus callosum identified by immunofluorescent labeling for the paranodal protein NFASC (red) and the internodal protein CNTNAP1 (green) which mark the node of Ranvier.
(B) Representative image of immunostaining for NFASC (red) and CNTNAP1 (green) indicating there are fewer nodes of Ranvier in Ts65Dn mice.
(C) Bar graph demonstrating reduced density of nodes of Ranvier in corpus callosum and external capsule in P60 Ts65Dn brain (n=3 pairs). **, p≤0.002 (paired t-test).
(D and D’) Electron micrographs showing representative images of paranodes (arrows).
(E) Quantification of paranodes reveals that they are reduced in P60 Ts65Dn corpus callosum (n=3 each).
(F) Log2 values of the array signal intensity in human euploid Ctrl and DS DFC show that NFASC expression is decreased from birth onwards (periods 8 to 14) in DS. Periods 5-9, p=0.23; periods 8-12, p=0.016; periods 9-13, p=0.0092; periods 10-14, p=0.0022; all periods, p=0.0028 (paired t-test).
(G) Log2 values of the array signal intensity for DFC expression of CNTNAP1 show that its expression is decreased from birth onwards (periods 8 to 14) in DS. Periods 5-9, p=0.41; periods 8-12, p=0.018; periods 9-13, p=0.017; periods 10-14, p=0.011; all periods, p=0.012 (paired t-test).
(H) ddPCR analysis of human DFC samples confirming decreased expression of NFASC and CNTNAP1 from childhood onwards in DS.
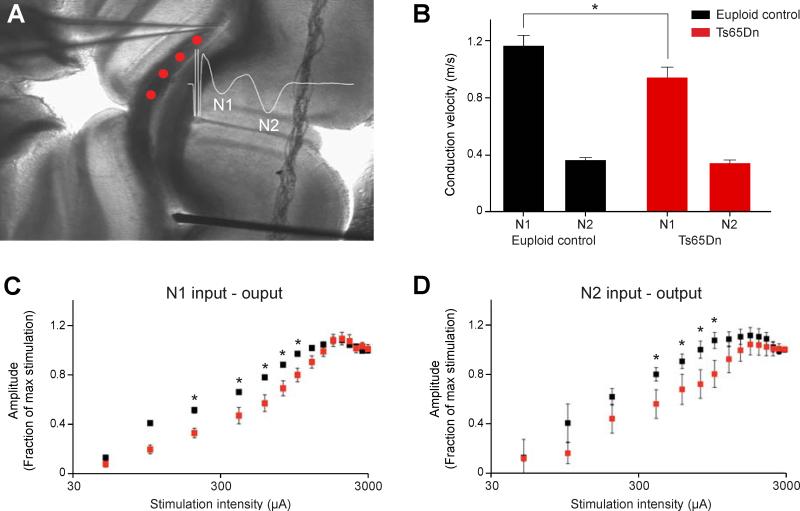
To determine the functional consequences of these white matter defects, we measured compound action potentials (CAP) in the corpus callosum of P30-P55 Ts65Dn forebrain slices (Figure 6A). Our results revealed that action potential transmission is significantly slower in myelinated Ts65Dn axons (N1) compared to euploid controls (Figure 6B; p=0.04, n=10). In contrast, the conduction velocity in unmyelinated axons (N2) is similar between groups (Figure 6B; p=0.4, n=11). Examining the relationship between stimulus intensity and response magnitude (input-output curve) revealed that both types of axons in Ts65Dn are less excitable than those in euploid mice (Figure 6C, D; p<0.001 for N1, p<0.007 for N2). However, the action potential refractory periods were not different between groups, indicating that sodium channel kinetics are less likely to be responsible for the shift in the input-output relationship (data not shown). These data, combined with our cellular and electron microscopy results, indicate that white matter abnormalities found in the trisomic human and Ts65Dn mouse brain contribute to structural and functional defects in neurotransmission.
Figure 6. Slower Axonal Conduction in Ts65Dn Corpus Callosum.
(A) Experimental setup depicting the stimulating electrode (bottom) and recording electrode (top) within P30 corpus callosum of a coronal section. Compound action potentials (CAP) evoked by stimulation were recorded at multiple sites (red dots) to compute conduction velocity. Inset shows a representative example of an evoked CAP depicting the components arising from myelinated fibers (N1) and unmyelinated fibers (N2).
(B) Bar plot showing that conduction velocities for myelinated fibers (N1), but not unmyelinated fibers (N2), were significantly slower in Ts65Dn corpus callosum (*, p=0.04, n=10 for N1; n=11 for N2).
(C) The input-output relationship for myelinated fibers is right-shifted in Ts65Dn corpus callosum suggesting that this fiber type is less excitable (*, p<0.001 for N1 euploid n=5, Ts65Dn n=4).
(D) The input-output relationship for unmyelinated fibers is right-shifted in Ts65Dn corpus callosum suggesting that this fiber type is less excitable (*, p≤0.007 for N2; euploid n=5, Ts65Dn n=4).
Alterations in Oligodendrocyte Lineage Progression in DS and Trisomic Mouse
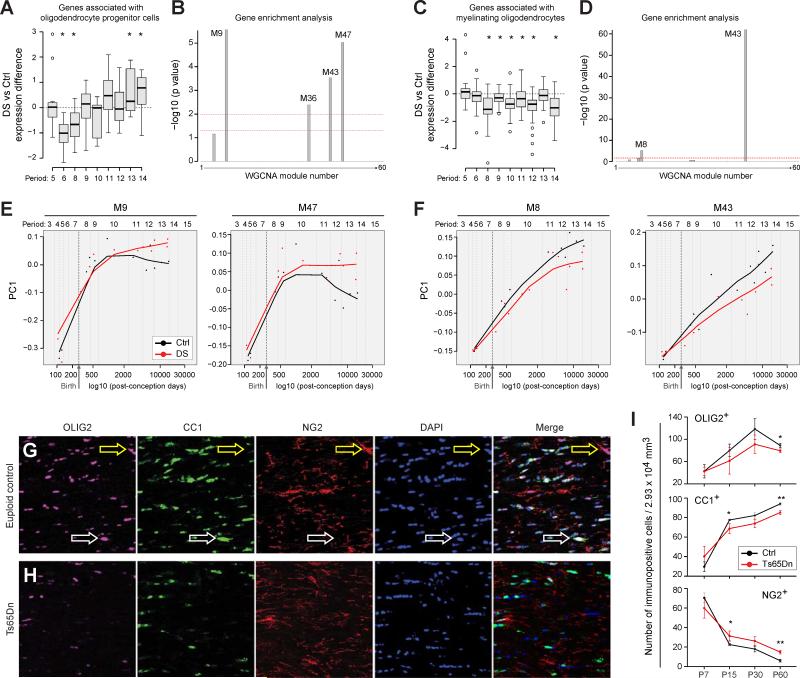
To gain further insight into possible underlying mechanisms for myelin and white matter defects in DS, we examine the expression of genes associated with development of human oligodendrocyte precursor cells and mature oligodendrocytes by generating sets of genes predicted to be selectively enriched in these cell types during human development. These gene sets were generated by overlapping lists of oligodendrocyte lineage candidate genes from previous mouse (Zhang et al., 2014) and human (Kang et al., 2011) studies (for details on the gene selection and analysis see Extended Experimental Procedures and Table S9). We then assessed if there was differential expression of the oligodendrocyte precursor and myelinating oligodendrocyte gene sets in DS versus control DFC. We observed that expression of putative human oligodendrocyte precursor cell-related genes gradually increases in DS compared to controls, persisting until periods 12-14 (Figure 7A). In contrast, putative human myelinating oligodendrocyte genes in the DFC are expressed at lower levels from birth into adulthood (Figure 7C). These data support and extend our DEX and WGCNA analyses (Figure 2, Tables S5-S6-S6) suggesting an overall and long-lasting impairment of oligodendrocyte maturation in DS, which is detectable after birth in the neocortex.
Figure 7. Oligodendrocyte Maturation is Impaired in DS.
(A) Developmental changes in genes associated with oligodendrocyte progenitor cells expressed as ratio of DS vs. Ctrl. *p≤0.05 (paired t-test).
(B) −log10 p-values of expression of oligodendrocyte progenitor cell-enriched genes in weighted gene co-expression network modules reveals significant enrichment in the M9, M36, M43 and M47 modules [y-axis = −log10 (p value)]. Lower dashed line corresponds to p = 0.05; upper dashed line corresponds to p = 0.01.
(C) Developmental changes in genes associated with myelinating oligodendrocytes expressed as ratio of DS vs. Ctrl. *p≤0.05 (paired t-test).
(D) −log10 p-values of enrichment analysis for expression of mature oligodendrocyte enriched genes in gene network co-expression modules demonstrating significant enrichment in modules M8 and M43 [y-axis = −log10 (p value)]. Lower dashed line corresponds to p = 0.05; upper dashed line corresponds to p = 0.01.
(E) PC1 plots of the co-expression modules enriched in oligodendrocyte progenitor cell specific genes (M9 and M47) demonstrating that they are increased in DS and that the differences between Ctrl and DS samples increase over postnatal development.
(F) PC1 plots enriched co-expression modules enriched in specific myelinating oligodendrocyte specific genes (M8 and M43) demonstrating that they are decreased in DS and that the differences between Ctrl and DS samples increase over postnatal development.
(G) Representative immunofluorescent stains of P60 Ctrl corpus callosum for OLIG2 (purple), CC1 (green), NG2 (red), and nuclei (blue). Yellow arrows point to NG2-labeled OPCs and white arrows point to CC1-labeled mature oligodendrocytes.
(H) Representative immunofluorescent stains of P60 Ts65Dn corpus callosum for OLIG2 (purple), CC1 (green), NG2 (red), and nuclei (blue). Fewer CC1-labeled mature oligodendrocytes are apparent.
(I) The numbers of OLIG2 immuno-positive cells in the corpus callosum were counted in image volumes from P7-P60 (n=4 pairs at each age). There was a general trend of fewer OLIG2+ cells in the Ts65Dn white matter which becomes significant at P60. In addition, as a proportion of the total OLIG2+ population, the percentage of NG2+ oligodendrocyte progenitor cells is higher in Ts65Dn corpus callosum from P15-P60. In contrast, the percentage of mature CC1+ oligodendrocytes is reduced in Ts65Dn from P15-P60. *, p≤ 0.05; **, p≤ 0.005.
To characterize these changes at the gene network level, we determined the overlap between WGCNA modules and sets of genes that we predicted to be selectively enriched in oligodendrocyte precursor cells or myelinating oligodendrocytes (Table S9). We found that human oligodendrocyte precursor cell genes were significantly enriched in modules M9, M36, M43, and M47 (Figure 7B); exon array trajectories for the two most significantly enriched modules (i.e., M9 and M47) indicated elevated expression in DS brain samples over the course of development (Figure 7E). Conversely, human mature oligodendrocyte genes were found to be significantly enriched primarily in two modules, M8 and M43, and their trajectories indicate decreased expression in DS brains during development (Figure 7D,F). Thus, the relevant WGCNA modules exhibit similar trajectories when compared with the expression changes of individual oligodendrocyte-lineage genes, demonstrating that these cell type-specific defects in oligodendroglial development are associated with complex gene network disturbances in the DS brain. Altogether, these data indicate that aberrations in the process of oligodendrocyte maturation to the myelinating stage may underlie the white matter abnormalities in DS.
To test whether differences observed through our transcriptomic analyses identify changes in the oligodendrocyte lineage at the cellular level, we performed a developmental study of the Ts65Dn corpus callosum from P7 to P60 (Figure 7G-I). We first immunostained for OLIG2 to mark the oligodendrocyte lineage, and CC1 or NG2 to identify myelinating oligodendrocytes or oligodendrocyte precursor cells, respectively. At P7, around the onset of myelination, we found equivalent numbers of OLIG2+, CC1+ and NG2+ cells in Ts65Dn and control brain white matter. However, during the ensuing periods of active myelination and thereafter (after P7 and up to P60), there were fewer OLIG2+ cells in the Ts65Dn white matter compared to controls, reaching significance at P60; this decline in numbers was not due to higher than normal apoptotic rates (data not shown). More importantly, compared to controls and as predicted by the human transcriptome study, there was a higher percentage of OLIG2+/NG2+ oligodendrocyte precursor cells and a decreased percentage of OLIG2+/CC1+ oligodendrocytes in Ts65Dn white matter from P15 on. These results demonstrate that changes in the ratio of oligodendrocyte precursor cells to mature oligodendrocytes develop during the period of active myelination.
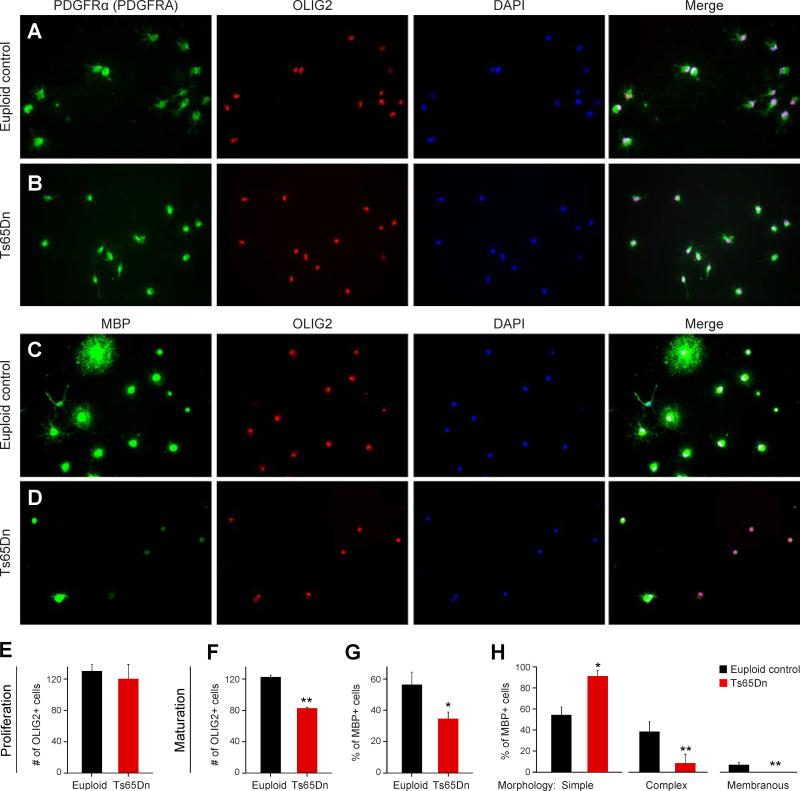
Finally, we sought to determine if there were cell-autonomous effects of Ts65Dn chromosomal triplication on oligodendrocyte development by carrying out proliferation and maturation assays of purified OPCs isolated by immunopanning with an antibody against alpha-type platelet-derived growth factor receptor (PDGRA or PDGFRα) (Figure 8). Effects on OPC proliferation were assayed by plating euploid control and Ts65Dn OPCs at the same density in proliferative conditions for 48 hrs. We found no difference in the number of OLIG2/PDGFRA OPCs in cultures derived from euploid and Ts65Dn mice (Figure 8A, B, & E).
Figure 8. Impaired Maturation and Viability of Immunopurified Oligodendrocyte Progenitor Cells, In Vitro.
(A) Representative micrographs of immunostaining for oligodendrocyte progenitor cells isolated from P7 euploid cortex and proliferated for 48 hours; PDGFRA (green), OLIG2 (red), and nuclei (blue).
(B) Representative micrographs immunostaining for oligodendrocyte progenitor cells isolated from P7 Ts65Dn and proliferated for 48 hours; PDGFRA (green), OLIG2 (red), and nuclei (blue).
(C) Representative micrographs of immunostaining for oligodendrocyte progenitor cells isolated from P7 euploid cortex and cultured in pro-maturation conditions for 72 hours; MBP (green), OLIG2 (red), and nuclei (blue).
(D) Representative micrographs of immunostaining for oligodendrocyte progenitor cells isolated from P7 Ts65Dn cortex and cultured in pro-maturation conditions for 72 hours; MBP (green), OLIG2 (red), and nuclei (blue).
(E) Graph indicating there were no differences between euploid control and Ts65Dn in the number OLIG2+ cells observed after 48 hours in proliferative conditions.
(F) Graph indicating the number OLIG2+ cells was reduced by ~30% after 72 hours in pro-maturation conditions.
(G) Graph indicating the percentage of OLIG2+ cells co-expressing MBP was reduced by ~40% after 72 hours in pro-maturation conditions.
(H) Graph indicating a greater percentage of MBP+ cells from Ts65Dn mice cultured in pro-maturation conditions exhibited a simple morphology (<6 processes) than MBP+ euploid cells, which tended to have a more complex (≥ 6 processes) or membranous morphology. *, p≤ 0.05; **, p≤ 0.01 (unpaired Student's t-test).
To test for effects of trisomy on oligodendrocyte maturation, OPCs were differentiated for 72 hrs. The cells were stained for MBP and OLIG2 (Figure 8C, D). MBP+/OLIG2+ cells were considered mature, whereas MBP- cells were considered immature. We found that the total number of OLIG2 cells was diminished in Ts65Dn cultures after 72 hrs, suggesting that Ts65Dn cell viability is reduced in these conditions (Figure 8C, D, F & G). In addition, the percentage of OLIG2+/MBP+ cells was significantly diminished. To further assess this possibility, we characterized the morphology of MBP+ cells in each culture. We found that a greater proportion of Ts65Dn-derived MBP+ cells displayed a simple morphology compared to euploid cells (Figure 8H). Taken together, these cell culture experiments indicate that Ts65Dn oligodendrocytes exhibit cell-autonomous impairments in oligodendrocyte maturation and viability, but not proliferation.
DISCUSSION
In this study, we provide the most comprehensive developmental analysis of gene expression in postmortem human DS brains and their respective age-matched controls to date, and thereby establish a novel framework for the study of neural development in DS. This effort has elucidated three key aspects of gene expression differences between DS and euploid control brains. First, dysregulated genes are found throughout the genome and are not present solely on HSA21. Second, many of the dysregulated genes exhibited highly specific temporal and regional expression profiles. Third, these dysregulated genes form distinct co-expression networks associated with distinct biological categories, providing novel and unbiased insight into the multiple biological processes affected in developing and adult DS brains. Viewing DS brain development through this new lens, we have uncovered novel and robust abnormalities in the expression of genes associated with oligodendrocyte development and myelination.
Our assessment of Ts65Dn mice provided strong confirmation of the human oligodendrocyte and white matter defects and identified cell autonomous impairments in oligodendrocyte maturation and viability, but not proliferation, as the underlying mechanism. These close similarities allowed us to further assess the cellular and functional consequences of these abnormalities, identifying changes in action potential conductance velocity in the forebrain white matter. These results elucidate a continuum of myelination-associated defects at the molecular, structural and functional levels that likely contribute to developmental delays and life-long intellectual disability in DS.
The chromosomal location of the dysregulated genes identified in our study indicates a scattered, widespread distribution throughout the genome. It has been recently shown by Letourneau et al. (2014) in fibroblasts from monozygotic twins discordant for trisomy 21, that the DS altered gene expression follows a consistent pattern, with increased and decreased gene-expression levels alternating across large chromosomal segments, which they named gene expression dysregulation domains (GEDDs). These GEDDs were not observable when comparing grouped samples due to gene expression differences between individuals, and this is consistent with our results (Figure S4). It therefore remains unclear whether and how these individual-specific GEDDs underlie cardinal features of DS brain development and function. Nevertheless, in the present study, we show that dysregulated genes, despite being present throughout the genome, are organized into multiple gene networks that are robust enough to be measured across multiple samples of unrelated individuals. Most importantly, by elucidating the in vivo impact of the white matter-associated gene modules as an example, we show that these systems level changes impact cellular development and play a functional role in brain maturation.
We uncovered substantial differences between brain regions in the number and identities of altered genes and demonstrated temporal changes in these region-specific expression profiles. When considered together, these data indicate that the neurobiology of DS cannot be characterized by static lists of dysregulated genes or gene networks. Instead, the spatiotemporal nature of the disturbance plays a large role over the course of life. Our transcriptome analyses support a “cascade hypothesis” in which dysregulation of genes on HSA21 is likely the first genome-level disturbance and that gene expression changes on other chromosomes follow quickly thereafter. Indeed, HSA21 genes were already robustly upregulated compared to other chromosomes in the youngest fetal DS samples analyzed in our study, while changes in other regions of the genome increase with age (Figure S3). Moreover, we found that the temporal dynamics of gene module disruptions track the time periods relevant to specific developmental processes. For example, changes in modules 8, 9, 43 and 47 appear during late fetal development and the first several years of postnatal life (Figures 2 and 7), a time in development that coincides with dramatic upregulation of oligodendrocyte/myelination genes (Kang et al., 2011) and rapid expansion of oligodendrocytes in the human brain (Yeung et al., 2014). The record of these expression changes can now be used for the design and application of therapeutics tailored to particular biological process and brain regions at relevant developmental periods.
Myelination is one of the most prolonged neurodevelopmental processes, continuing until the third decade of life (Benes et al., 1994; Fleschig, 1901; Miller et al., 2012; Yakovlev, 1967). Notably, human studies have shown that development and maturation of the white matter correlates with increased motor skills and cognitive functions (Casey et al., 2000; Gibson, 1991; Nagy et al., 2004; Paus et al., 1999; Schmithorst et al., 2005). The present results clearly show a profound deficit in white matter maturation in individuals with DS during infancy and adolescence in DFC, a brain region of late myelination which performs a critical role in the organization of behavioral, linguistic, and cognitive functions (Fuster, 2002; Makinodan et al., 2012). Recent evidence in animal models indicates that ongoing myelin remodeling throughout life may be important for learning, behavior, and cognition throughout adulthood (Liu et al., 2012; McKenzie et al., 2014). This is in agreement both with previous studies showing that DS children manifest learning and memory problems in late infancy which often worsen in adolescence (Koo et al., 1992; Lanfranchi et al., 2010) and with post-mortem studies that show reduced myelin content (Abraham et al., 2012; Wisniewski, 1990; Wisniewski and Schmidt-Sidor, 1989) and fewer oligodendrocytes in DS striatum (Karlsen and Pakkenberg, 2011).
Alterations in white matter or myelin components may also play a role in neuropathology in the aging DS brain with early onset Alzheimer's-like pathology. Nearly all adults with DS develop dementia starting in the third decade of life (Sheehan et al., 2014). While triplication of APP is likely at the root of AD neuropathology in DS, our results raise the intriguing possibility that abnormalities in oligodendrocytes and myelin might contribute to early onset of Alzheimer's-like pathology in DS. Consistent with this possibility, disturbances in myelin have been associated with an increased rate of AD progression and byproducts of homeostatic myelin maintenance can promote the formation of amyloid plaques (Bartzokis, 2007, 2011). Characterizing the axon-oligodendrocyte interactions in DS throughout development and aging will thus be an important topic of future work.
The concordance between the human DS and Ts65Dn mouse samples at the gene, protein and cellular levels clearly demonstrates the value of the Ts65Dn mouse in reflecting the cellular and functional consequences of trisomy 21. Our finding that the Ts65Dn white matter phenotype mirrors that occurring in the human DS brain so robustly suggests that it will be a good model to identify neuronal cues that influence the onset and completion of myelination. Both intrinsic and extrinsic factors have been shown to influence myelination. For example, recent studies have shown that electrical activity drives myelination (Gibson et al., 2014; Ishibashi et al., 2006; Wake et al., 2011) and that synaptogenesis and myelination are linked. However, we also show that myelin abnormalities in DS are at least in part due to a cell-autonomous phenomenon in oligodendrocyte development. Teasing out the relative contributions of cell-autonomous effects and neuron-glial signaling, or other cell extrinsic mechanisms, will be an important goal of future work. These and other queries can now be approached using the developmental expression profiling and multi-species analysis presented here.
In summary, our findings indicate that strategies to enhance myelination may therefore serve as therapeutic targets to attenuate the cognitive and neurological symptoms of DS. Our results also implicate spatiotemporal disturbance in other molecular pathways in DS brain, and provide a powerful data set for further computational and functional analyses. We anticipate that the resources provided by this study will inspire and facilitate future studies of the mechanistic basis for impaired neural development in DS and other cognitive disorders, while also lending insight into the genetic and transcriptional underpinnings of previously described DS phenotypes and the relevance of mouse model studies to the human disease.
EXPERIMENTAL PROCEDURES
Human Tissue
This study was conducted using de-identified postmortem human brain specimens from tissue collections at the Department of Neuroscience, Yale School of Medicine, the University of Maryland Brain and Tissue Bank (Baltimore, MD), Brigham and Women's Hospital Pathology Department, and Boston University Pathology Department. Appropriate informed consent was obtained and all available non-identifying information was recorded for each specimen. The ages of specimens, sex, ethnicity, PMI, RIN, and dissection protocol are detailed Tables S1 and 2 and the extended experimental procedure. Details of the brain regions analyzed and the dissection technique are detailed in Table S1 and S2 and the extended experimental procedures.
Transcriptome Analyses
Full detailed procedures for RNA isolation and cDNA synthesis can be found in Extended Experimental Procedures and were as previously described (Kang et al 2011). Affymetrix Human 1.0 ST arrays and the Affymetrix GeneChip platform were used to obtain transcriptome data, as described in the Extended Experimental Procedures. Data normalization, quality control, and analysis were performed as detailed in the Extended Experimental Procedures. Briefly, the Partek Genomics Suite was used for data normalization and to determine gene level intensities. The expression level of a gene (transcript cluster) was estimated using the median of all exons within the gene.
Principal component analysis was applied to visualize the relatedness of DS and their-matched control samples. A paired t-test was used to identify DEX genes between paired DS and matched control samples during all development periods. FDR-adjusted p-value < 0.1 was used as a cutoff. Signed co-expression networks were built using the WGCNA package in R (Zhang and Horvath, 2005). Modules were generated by hybrid dynamic tree-cutting. For differentially expressed genes and co-expression modules, functional enrichment was assessed using the DAVID Bioinformatics Resource 6.7 (http://david.abcc.ncifcrf.gov/).
For ddPCR, the same human tissue RNA samples used for exon arrays were used for cDNA synthesis.
Mice, Tissue Culture, and Relevant Procedures
Ts65Dn (RRID: MGI_2178111) and euploid control B6EiC3 mice were generated by backcrossing Ts65Dn females to B6EiC3Sn.BLiAF1/J F1 hybrid (B6EiC3) males. Quantitative PCR genotyping was performed on genomic DNA extracted from tail tips (Chakrabarti et al., 2007). All procedures regarding the care and death of these animals was approved by the Institutional Animal Care and Use Committee of Boston University School of Medicine, in accordance with the NIH guide for the care and use of laboratory animal. For procedures relevant to all animal studies, including histology, electron microscopy, image analysis, electrophysiology, immunopanning, oligodendrocyte cultures, and immunoblotting, refer to Extended Experimental Procedures.
Supplementary Material
Highlights.
Genome-wide spatiotemporal dysregulation of gene expression in Down syndrome brains
Transcriptome changes reflects altered oligodendrocyte development and myelination
Oligodendrocyte differentiation and myelination altered in Down syndrome model mice
Speed of action potential propagation is decreased in Ts65Dn neocortical white matter
ACKNOWLEDGEMENTS
We thank Robert Johnson, Andre Sousa and Ronald Zielke for assistance with tissue acquisition and processing, and Manzoor Bhat and William Stallcup for providing NF186 and CASPR, and NG2 antibodies, respectively. We also thank members of the Haydar and Sestan laboratories for their comments. This work is supported by grants NS076503 and MH106934 from the National Institutes of Health and by a grant of the Korea Health Technology R&D Project through KHIDI (HI14C2461). Additional support was provided by the Kavli Foundation, the James S. McDonnell Foundation scholar award, the Ruth Kirschstein NRSA fellowship (NIH), and the China Scholarship Council fellowship. The views expressed in this scientific presentation are those of the author(s) and do not reflect the official policy or position of the U.S. government or the Department of Defense.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization: J.L.O.S., H.J.K., W.A.T., J.C.S., T.F.H., and N.S.; Methodology: J.L.O.S., W.A.T., H.J.K., J.C.S., F.C., Y.Z., T.F.H., and N.S.; Formal Analysis: J.L.O.S., H.J.K., W.A.T., J.C.S., F.C., Y.Z., N.P.C., J.G., T.F.H., and N.S.; Investigation: J.L.O.S., W.A.T., H.J.K., F.C., J.C.S., J.G., C.S., A.P., N.P.C., M.P., L.J-R., and T.F.H.; Resources: A.P., I.D., and J.A.G.; Data Curation: F.C., Y.Z.; Writing – Original Draft: J.L.O.S., H.J.K., J.C.S., W.A.T., T.F.H., N.S.; Writing – Review & Editing: J.L.O.S., W.A.T., J.C.S., F.C., H.J.K., Z.G., T.F.H., N.S.; Visualization: J.L.O.S., H.J.K., W.A.T., J.C.S., F.C., N.P.C., Z.G., J.G., T.F.H., and N.S.; Supervision, Project Administration: T.F.H., and N.S.; Funding Acquisition: H.J.K, Z.G., T.F.H., and N.S.
ACCESSION NUMBER
All transcriptome data reported in this study have been deposited at GEO (http://www.ncbi.nlm.nih.gov/geo/) (GSE59630).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, eight figures, and eight tables and can be found with this article online.
REFERENCES
- Abraham H, Vincze A, Veszpremi B, Kravjak A, Gomori E, Kovacs GG, Seress L. Impaired myelination of the human hippocampal formation in Down syndrome. Int J Dev Neurosci. 2012;30:147–158. doi: 10.1016/j.ijdevneu.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Ang ES, Jr., Haydar TF, Gluncic V, Rakic P. Four-dimensional migratory coordinates of GABAergic interneurons in the developing mouse cortex. J. Neurosci. 2003;23:5805–5815. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn S, Mimmack M, Ryan M, Caldwell MA, Jauniaux E, Starkey M, Svendsen CN, Emson P. Neuronal target genes of the neuron-restrictive silencer factor in neurospheres derived from fetuses with Down's syndrome: a gene expression study. Lancet. 2002;359:310–315. doi: 10.1016/S0140-6736(02)07497-4. [DOI] [PubMed] [Google Scholar]
- Bartzokis G. Acetylcholinesterase inhibitors may improve myelin integrity. Biol. Psychiatry. 2007;62:294–301. doi: 10.1016/j.biopsych.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Bartzokis G. Alzheimer's disease as homeostatic responses to age-related myelin breakdown. Neurobiol. Aging. 2011;32:1341–1371. doi: 10.1016/j.neurobiolaging.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Turtle M, Khan Y, Farol P. Myelination of a key relay zone in the hippocampal formation occurs in the human brain during childhood, adolescence, and adulthood. Arch. Gen. Psych. 1994;51:477–484. doi: 10.1001/archpsyc.1994.03950060041004. [DOI] [PubMed] [Google Scholar]
- Canfield MA, Honein MA, Yuskiv N, Xing J, Mai CT, Collins JS, Devine O, Petrini J, Ramadhani TA, Hobbs CA, et al. National estimates and race/ethnic-specific variation of selected birth defects in the United States, 1999-2001. Birth Defects Res. A Clin. Mol. Teratol. 2006;76:747–756. doi: 10.1002/bdra.20294. [DOI] [PubMed] [Google Scholar]
- Casey BJ, Giedd JN, Thomas KM. Structural and functional brain development and its relation to cognitive development. Biol. Psychol. 2000;54:241–257. doi: 10.1016/s0301-0511(00)00058-2. [DOI] [PubMed] [Google Scholar]
- Chakrabarti L, Best TK, Cramer NP, Carney RS, Isaac JT, Galdzicki Z, Haydar TF. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat. Neurosci. 2010;13:927–934. doi: 10.1038/nn.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L, Galdzicki Z, Haydar TF. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J. Neurosci. 2007;27:11483–11495. doi: 10.1523/JNEUROSCI.3406-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KR. Myelination and behavioral development: A comparative perspective on questions of neoteny, altriciality and intelligence. In: Gibson KR, Petersen A, editors. Brain Maturation and Cognitive Development: Comparative and Cross Cultural Perspectives. Aldine de Gruyter; Hawthorne, N.Y.: 1991. [Google Scholar]
- Golden J, Hyman B. Development of the superior temporal neocortex is anomalous in trisomy 21. J. Neuropathol. Exp. Neurol. 1994;53:513–520. doi: 10.1097/00005072-199409000-00011. [DOI] [PubMed] [Google Scholar]
- Hartley D, Blumenthal T, Carrillo M, DiPaolo G, Esralew L, Gardiner K, Granholm AC, Iqbal K, Krams M, Lemere C, et al. Down syndrome and Alzheimer's disease: Common pathways, common goals. Alzheimers Dement. 2015;11:700–709. doi: 10.1016/j.jalz.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydar TF, Reeves RH. Trisomy 21 and early brain development. Trends. Neurosci. 2012;35:81–91. doi: 10.1016/j.tins.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde LA, Frisone DF, Crnic LS. Ts65Dn mice, a model for Down syndrome, have deficits in context discrimination learning suggesting impaired hippocampal function. Behav. Brain Res. 2001;118:53–60. doi: 10.1016/s0166-4328(00)00313-2. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Dakin KA, Stevens B, Lee PR, Kozlov SV, Stewart CL, Fields RD. Astrocytes promote myelination in response to electrical impulses. Neuron. 2006;49:823–832. doi: 10.1016/j.neuron.2006.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan TL, Bellugi U. Anomalous brain morphology on magnetic resonance images in Williams syndrome and Down syndrome. Arch. Neurol. 1990;47:529–533. doi: 10.1001/archneur.1990.00530050049011. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MR, Cho MH, Ullian EM, Isom LL, Levinson SR, Barres BA. Differential control of clustering of the sodium channels Na(v)1.2 and Na(v)1.6 at developing CNS nodes of Ranvier. Neuron. 2001;30:105–119. doi: 10.1016/s0896-6273(01)00266-5. [DOI] [PubMed] [Google Scholar]
- Karlsen AS, Pakkenberg B. Total numbers of neurons and glial cells in cortex and basal ganglia of aged brains with Down syndrome--a stereological study. Cereb. Cortex. 2011;21:2519–2524. doi: 10.1093/cercor/bhr033. [DOI] [PubMed] [Google Scholar]
- Kleschevnikov AM, Belichenko PV, Villar AJ, Epstein CJ, Malenka RC, Mobley WC. Hippocampal long-term potentiation suppressed by increased inhibition in the Ts65Dn mouse, a genetic model of Down syndrome. J. Neurosci. 2004;24:8153–8160. doi: 10.1523/JNEUROSCI.1766-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, Blaser S, Harwood-Nash D, Becker LE, Murphy EG. Magnetic resonance imaging evaluation of delayed myelination in Down syndrome: a case report and review of the literature. J. Child Neurol. 1992;7:417–421. doi: 10.1177/088307389200700417. [DOI] [PubMed] [Google Scholar]
- Lanfranchi S, Jerman O, Dal Pont E, Alberti A, Vianello R. Executive function in adolescents with Down Syndrome. J. Intellect. Disabil. Res. 2010;54:308–319. doi: 10.1111/j.1365-2788.2010.01262.x. [DOI] [PubMed] [Google Scholar]
- Larsen KB, Laursen H, Graem N, Samuelsen GB, Bogdanovic N, Pakkenberg B. Reduced cell number in the neocortical part of the human fetal brain in Down syndrome. Ann. Anat. 2008;190:421–427. doi: 10.1016/j.aanat.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Letourneau A, Santoni FA, Bonilla X, Sailani MR, Gonzalez D, Kind J, Chevalier C, Thurman R, Sandstrom RS, Hibaoui Y, et al. Domains of genome-wide gene expression dysregulation in Down's syndrome. Nature. 2014;508:345–350. doi: 10.1038/nature13200. [DOI] [PubMed] [Google Scholar]
- Liu J, Dietz K, DeLoyht JM, Pedre X, Kelkar D, Kaur J, Vialou V, Lobo MK, Dietz DM, Nestler EJ, et al. Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat. Neurosci. 2012;15:1621–1623. doi: 10.1038/nn.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockstone HE, Harris LW, Swatton JE, Wayland MT, Holland AJ, Bahn S. Gene expression profiling in the adult Down syndrome brain. Genomics. 2007;90:647–660. doi: 10.1016/j.ygeno.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Lott IT. Neurological phenotypes for Down syndrome across the life span. Prog. Brain Res. 2012;197:101–121. doi: 10.1016/B978-0-444-54299-1.00006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Wang X, Spitznagel EL, Jr., Frelin LP, Ting JC, Ding H, Kim JW, Ruczinski I, Downey TJ, Pevsner J. Primary and secondary transcriptional effects in the developing human Down syndrome brain and heart. Genome Biol. 2005;6:R107. doi: 10.1186/gb-2005-6-13-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Zielke CL, Zielke HR, Pevsner J. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics. 2003;81:457–467. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- McKenzie IA, Ohayon D, Li H, de Faria JP, Emery B, Tohyama K, Richardson WD. Motor skill learning requires active central myelination. Science. 2014;346:318–322. doi: 10.1126/science.1254960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, Fobbs AJ, Sousa AM, Sestan N, Wildman DE, et al. Prolonged myelination in human neocortical evolution. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16480–16485. doi: 10.1073/pnas.1117943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AC, Mirnics K. Gene expression profiling of the brain: pondering facts and fiction. Neurobiol. Dis. 2012;45:3–7. doi: 10.1016/j.nbd.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Westerberg H, Klingberg T. Maturation of white matter is associated with the development of cognitive functions during childhood. J. Cogn. Neurosci. 2004;16:1227–1233. doi: 10.1162/0898929041920441. [DOI] [PubMed] [Google Scholar]
- Paus T, Zijdenbos A, Worsley K, Collins DL, Blumenthal J, Giedd JN, Rapoport JL, Evans AC. Structural maturation of neural pathways in children and adolescents: in vivo study. Science. 1999;283:1908–1911. doi: 10.1126/science.283.5409.1908. [DOI] [PubMed] [Google Scholar]
- Powell D, Caban-Holt A, Jicha G, Robertson W, Davis R, Gold BT, Schmitt FA, Head E. Frontal white matter integrity in adults with Down syndrome with and without dementia. Neurobiol. Aging. 2014;35:1562–1569. doi: 10.1016/j.neurobiolaging.2014.01.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband MN, Peles E, Trimmer JS, Levinson SR, Lux SE, Shrager P. Dependence of nodal sodium channel clustering on paranodal axoglial contact in the developing CNS. J. Neurosci. 1999;19:7516–7528. doi: 10.1523/JNEUROSCI.19-17-07516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda N, Florez J, Martinez-Cue C. Mouse models of Down syndrome as a tool to unravel the causes of mental disabilities. Neural Plast. 2012;2012:584071. doi: 10.1155/2012/584071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahun I, Marechal D, Pereira PL, Nalesso V, Gruart A, Garcia JM, Antonarakis SE, Dierssen M, Herault Y. Cognition and hippocampal plasticity in the mouse is altered by monosomy of a genomic region implicated in Down syndrome. Genetics. 2014;197:899–912. doi: 10.1534/genetics.114.165241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schain AJ, Hill RA, Grutzendler J. Label-free in vivo imaging of myelinated axons in health and disease with spectral confocal reflectance microscopy. Nat. Med. 2014;20:443–449. doi: 10.1038/nm.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmithorst VJ, Wilke M, Dardzinski BJ, Holland SK. Cognitive functions correlate with white matter architecture in a normal pediatric population: a diffusion tensor MRI study. Hum. Brain Map. 2005;26:139–147. doi: 10.1002/hbm.20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan R, Ali A, Hassiotis A. Dementia in intellectual disability. Curr. Opin. Psychiatry. 2014;27:143–148. doi: 10.1097/YCO.0000000000000032. [DOI] [PubMed] [Google Scholar]
- Siarey RJ, Carlson EJ, Epstein CJ, Balbo A, Rapoport SI, Galdzicki Z. Increased synaptic depression in the Ts65Dn mouse, a model for mental retardation in Down syndrome. Neuropharmacology. 1999;38:1917–1920. doi: 10.1016/s0028-3908(99)00083-0. [DOI] [PubMed] [Google Scholar]
- Siarey RJ, Stoll J, Rapoport SI, Galdzicki Z. Altered long-term potentiation in the young and old Ts65Dn mouse, a model for Down Syndrome. Neuropharmacology. 1997;36:1549–1554. doi: 10.1016/s0028-3908(97)00157-3. [DOI] [PubMed] [Google Scholar]
- State MW, Geschwind DH. Leveraging genetics and genomics to define the causes of mental illness. Biol. Psychiatry. 2015;77:3–5. doi: 10.1016/j.biopsych.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ma J, Tanaka KF, Takao K, Komada M, Tanda K, Suzuki A, Ishibashi T, Baba H, Isa T, et al. Mice with altered myelin proteolipid protein gene expression display cognitive deficits accompanied by abnormal neuron-glia interactions and decreased conduction velocities. J. Neurosci. 2009;29:8363–8371. doi: 10.1523/JNEUROSCI.3216-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbenkamp AT, Willsey AJ, State MW, Sestan N. The developmental transcriptome of the human brain: implications for neurodevelopmental disorders. Curr. Opin. Neurol. 2014;27:149–156. doi: 10.1097/WCO.0000000000000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaxton C, Pillai AM, Pribisko AL, Labasque M, Dupree JL, Faivre-Sarrailh C, Bhat MA. In vivo deletion of immunoglobulin domains 5 and 6 in neurofascin (Nfasc) reveals domain-specific requirements in myelinated axons. J. Neurosci. 2010;30:4868–4876. doi: 10.1523/JNEUROSCI.5951-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler WA, Haydar TF. Multiplex genetic fate mapping reveals a novel route of neocortical neurogenesis, which is altered in the Ts65Dn mouse model of Down syndrome. J. Neurosci. 2013;33:5106–5119. doi: 10.1523/JNEUROSCI.5380-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake H, Lee PR, Fields RD. Control of local protein synthesis and initial events in myelination by action potentials. Science. 2011;333:1647–1651. doi: 10.1126/science.1206998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao Y, Zhang X, Badie H, Zhou Y, Mu Y, Loo LS, Cai L, Thompson RC, Yang B, et al. Loss of sorting nexin 27 contributes to excitatory synaptic dysfunction by modulating glutamate receptor recycling in Down's syndrome. Nat. Med. 2013;19:473–480. doi: 10.1038/nm.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeen VJ, Rosene DL, Wang R, Dai G, Mortazavi F, Hagmann P, Kaas JH, Tseng WY. The geometric structure of the brain fiber pathways. Science. 2012;335:1628–1634. doi: 10.1126/science.1215280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski KE. Down syndrome children often have brain with maturation delay, retardation of growth, and cortical dysgenesis. Am. J. Med. Genet. Suppl. 1990;7:274–281. doi: 10.1002/ajmg.1320370755. [DOI] [PubMed] [Google Scholar]
- Wisniewski KE, Schmidt-Sidor B. Postnatal delay of myelin formation in brains from Down syndrome infants and children. Clin. Neuropathol. 1989;8:55–62. [PubMed] [Google Scholar]
- Yakovlev PI, Lecours A. The myelogenetic cycles of regional maturation of the brain. In: Minkowski, editor. Regional Development of the Brain in Early Life. Blackwell Science; Oxford: 1967. [Google Scholar]
- Yeung MS, Zdunek S, Bergmann O, Bernard S, Salehpour M, Alkass K, Perl S, Tisdale J, Possnert G, Brundin L, et al. Dynamics of oligodendrocyte generation and myelination in the human brain. Cell. 2014;159:766–774. doi: 10.1016/j.cell.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, Sanada M, Park MJ, Terui K, Suzuki H, et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat. Gen. 2013;45:1293–1299. doi: 10.1038/ng.2759. [DOI] [PubMed] [Google Scholar]
- Zampieri BL, Biselli-Perico JM, de Souza JE, Burger MC, Silva Junior WA, Goloni-Bertollo EM, Pavarino EC. Altered expression of immune-related genes in children with Down syndrome. PLoS One. 2014;9:e107218. doi: 10.1371/journal.pone.0107218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.