Abstract
Common yet often overlooked, deamidation of peptidyl asparagine (Asn or N) generates aspartic acid (Asp or D) or isoaspartic acid (isoAsp or isoD). Being a spontaneous, non-enzymatic protein post-translational modification, deamidation artifact can be easily introduced during sample preparation, especially proteolysis where higher-order structures are removed. This artifact not only complicates the analysis of bona fide deamidation but also affects a wide range of chemical and enzymatic processes; for instance, the newly generated Asp and isoAsp residues may block or introduce new proteolytic sites, and also convert one Asn peptide into multiple species that affect quantification. While the neutral to mildly basic conditions for common proteolysis favor deamidation, mildly acidic conditions markedly slow down the process. Unlike other commonly used endoproteases, Glu-C remains active under mildly acid conditions. As such, as demonstrated herein, deamidation artifact during proteolysis was effectively eliminated by simply performing Glu-C digestion at pH 4.5 in ammonium acetate, a volatile buffer that is compatible with mass spectrometry. Moreover, nearly identical sequence specificity was observed at both pH’s (8.0 for ammonium bicarbonate), rendering Glu-C as effective at pH 4.5. In summary, this method is generally applicable for protein analysis as it requires minimal sample preparation and uses the readily available Glu-C protease.
Keywords: deamidation, artifact, isoaspartic acid, isomerization, mass spectrometry, calmodulin, exenatide, adrenocorticotropic hormone, Glu-C
INTRODUCTION
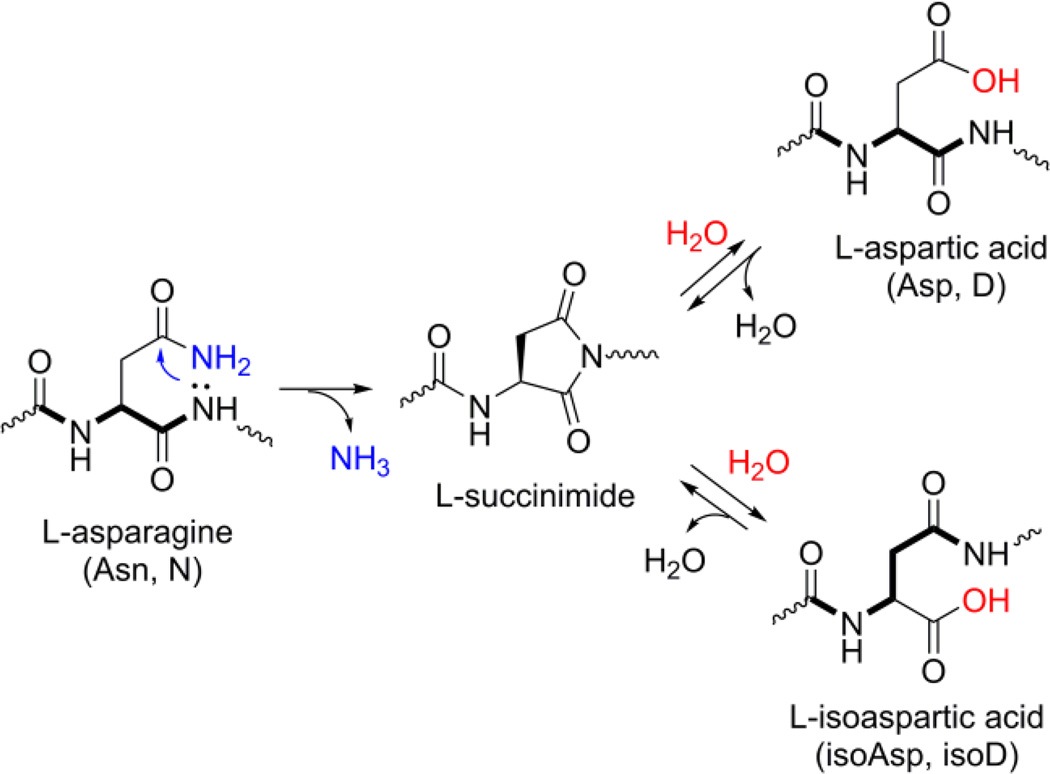
Asparagine deamidation is a common protein post-translational modification (PTM) that arises spontaneously and non-enzymatically. As depicted in Scheme 1, asparagine (Asn or N) is converted into aspartic acid (Asp or D) or isoaspartic acid (isoAsp or isoD) through a succinimide intermediate (Clarke 2003; Reissner and Aswad 2003). As isoAsp has detrimental impacts on biological systems, several repair mechanisms are present for reducing the levels of isoAsp; one prominent pathway is through protein isoaspartate O-methyltransferase (PIMT or PCMT, EC 2.1.1.77). This enzyme catalyzes the transfer of a methyl group from S-adenosyl-L-methionine (AdoMet or SAM) to isoAsp, generating an isoaspartyl methyl ester that can rapidly hydrolyze back to Asp (Alfaro et al. 2008; Biastoff et al. 2006; Mosley et al. 2006; Zang et al. 2009; Zhou et al. 1999). The repair of isoAsp via PIMT methylation is also affected by metabolites in the one-carbon and transsulfuration pathways, such as homocysteine (Gui et al. 2013; Mosley et al. 2006; Perła-Kaján and Jakubowski 2012; Perła-Kaján et al. 2007). It should be noted that deamidation may also occur on glutamine, glycosylated asparagine or other amides, which are negligible under typical proteolysis conditions; in this paper, deamidation refers to asparagine deamidation shown in Fig. 1 unless noted otherwise. Naturally occurring deamidation is ubiquitous and affects both protein structure and function; therefore its analysis is important in its own right (Alfaro et al. 2008; Chen et al. 2013; Lee et al. 2012; O’Connor et al. 2006; Paranandi et al. 1994; Reissner and Aswad 2003; Robinson and Robinson 2001). However, deamidation artifacts during sample preparation are also common, yet often overlooked, and can be highly problematic as detailed next.
Fig. 1.
Deamidation of asparagine (Asn) and the formation of aspartic acid (Asp) and isoaspartic acid (isoAsp) via a succinimide intermediate. Formation of isoAsp confers a beta-peptide linkage in the peptide backbone (highlighted in bold)
The rates of deamidation depend on multiple factors, including the primary sequences and higher-order structures of the proteins, pH, temperature, and components in the solutions (Tyler-Cross and Schirch 1991). For example, most potential deamidation sites are stabilized by higher-order structure, though not always. In comparison, in a linear peptide without defined structure, the residue immediately C-terminal to Asn is the major determinant: Asn-Gly (NG), being most flexible and acidic (backbone amide), is most prone to deamidation with a half-life around 24 h under physiological conditions (pH 7.4, 37 °C); Asn-Ser (NS) and Asn-His (NH), augmented by general acid-catalysis, deamidate faster (half-life about 10 days) than other sequences except NG (Radkiewicz et al. 2001; Robinson et al. 2004). The following mnemonic may help to remember the hotspots for deamidation: Not Good for NG (highly likely to deamidate); Not Sure for NS and Not Happy for NH (likely): and No Problem for NP (not likely). Recent surveys have revealed that at least one NG sequence (also NS) is present in over 50% of the proteins in H. sapiens, M. musculus, E. coli, and S. cerevisiae (Patananan et al. 2014). Therefore, significant and widespread deamidation of peptides or denatured proteins is expected and indeed observed during sample preparation for protein analysis (Li et al. 2008; Wang et al. 2010). For instance, neutral to mildly basic conditions that are typically used for common proteases (e.g., trypsin, Lys-C, Glu-C and chymotrypsin) are also favorable for deamidation. All together, these cumulative factors can easily lead to deamidation artifact during denaturation and proteolysis. In fact, as high as 70%–80% of deamidation artifact have been reported in some proteins after trypsin digestion for 12 h at 37 °C in ammonium bicarbonate (pH 8.0) (Krokhin et al. 2006).
Deamidation artifact has broad, significant and detrimental effects on proteins analysis. First and obviously, these artifacts create misleading results, thus complicating both the identification and quantification of bona fide deamidation. Moreover, deamidation imparts marked changes in the physical, chemical and biochemical properties of the involved proteins and peptides (Chen et al. 2010; Chumsae et al. 2014; Dai et al. 2013; Jiang et al. 2010). For example, the decrease in pI caused by the newly generated carboxylic group in Asp and isoAsp likely alters chromatographic behaviors (Chumsae et al. 2013; Ni et al. 2010; Winter et al. 2009). In addition, deamidation artifact may interfere with common proteolysis (e.g., peptide mapping, amino acid sequencing), as the isoAsp and Asp products may resist enzymatic hydrolysis or introduce new cleavage sites. Asn deamidation also converts one peptide into multiple species that affects quantification. Furthermore, the mass increase of 1 Da imparted by deamidation results in overlay of the isotopic envelope and thus affects analysis based on mass accuracy. Finally, the change in pI and backbone caused by isoaspartic acid can also negatively impact proper protein refolding and induce unexpected precipitation (Noguchi 2010; Orrù et al. 2000).
Conceptually, the best approach to distinguish between bona fide deamidation and artifact is to conduct sample preparation—including proteolysis—in 18O-water, as deamidation is the hydrolysis of an amide (Li et al. 2008; Liu et al. 2013a; Wan et al. 2004; Yao et al. 2001). In practice, however, analysis of 18O-labeled samples can be challenging due to complicated isotopic distributions, as proteolysis also introduces varying degrees of 18O into the newly generated C-termini (Klaene et al. 2014; Liu et al. 2012; Liu et al. 2014). In turn, deamidation artifact in 18O-water also complicates other techniques based on 18O tracing, such as labeling of C-termini and N-linked glycosylation sites using PNGase F (Palmisano et al. 2012). Other ramifications, such as shortening the digestion time and optimizing components of the digestion solution, have been attempted, but with only limited success, as these conditions cannot prevent deamidation.
An obvious and straightforward approach to eliminate deamidation artifact is to process the proteins under mildly acidic conditions (e.g., pH 4.5), as described herein. Deamidation rates follow an inverse bell-shaped curve with the minimum around pH 4 to 5, at which the half-lives for the Asn-Gly peptides are approximately 280 days at 37 °C; in other words, less than 1% deamidation for 24 h (Capasso et al. 1995; Hao et al. 2015; Patel and Borchardt 1990). In fact, preventing deamidation is a primary reason that many protein pharmaceuticals are stored under mildly acidic conditions (Manning et al. 2010). Perhaps not widely known, the commonly used endoprotease Glu-C shows maximal proteolytic activity around both pH 4 and 8 (Drapeau et al. 1972). As detailed below, deamidation artifact is indeed eliminated during Glu-C proteolysis at pH 4.5 in ammonium acetate using bovine calmodulin, exenatide and human adrenocorticotropic hormone (ACTH) peptide, which all contain hotspots for deamidation including the most labile NG sequence (Johnson et al. 1985; Potter et al. 1993). Additionally, since Glu-C is typically used at pH 8, we also evaluated the scope and limitations of proteolysis under mildly acidic conditions and compared the results between pH 4.5 and pH 8. We confirmed that the specificity of Glu-C at both pHs is nearly identical. Altogether, a simple change of buffer and pH renders a practical and general approach for protein digestion without deamidation artifact.
EXPERIMENTAL SECTION
Materials
Recombinant bovine calmodulin (lyophilized powder, C4874) was from Sigma-Aldrich (St. Louis, MO). Exenatide (lyophilized powder, AS-24464, 1HGEGTFTSDLSKQMEEEAVRLFIEWLKNGGPSSGAPPPS39-NH2) and adrenocorticotropic hormone (ACTH) peptide (lyophilized powder, AS-20619, 18RPVKVYPNGAEDESAEAFPLEF39) were from Anaspec (Fremont, CA). The concentrations were determined by UV absorption at 280 nm using extinction coefficients calculated based on amino acid sequence. Sequencing grade Glu-C endoprotease was from Promega (V1651; Madison, WI). Tris(2-carboxyethyl)phosphine (TCEP) hydrochloride was from Hampton Research (Al Jodo, CA). Dithiothreitol (DTT) was from Acros Organics (New Jersey, NJ). 18O water (normalized, 97% – 98% atom percentage) was from Icon Stable Isotopes (Summit, NJ). All aqueous solutions were prepared with MilliQ purified water. All chemicals used were reagent grade or better. The pH of all solutions was determined by EMD Colorphast pH strips with an accuracy of 0.5 unit.
Proteolysis
Calmodulin (final concentration 0.8 mg/mL) was dissolved in 100 mM ammonium acetate (pH 4.5) with 10% acetonitrile or ammonium bicarbonate (pH 8.0) with 10% acetonitrile, reduced by 1 mM DTT and 1 mM TCEP at 37 °C for 20 min; then digested by Glu-C at an enzyme: protein ratio of 1:40 (w/w) at 37 °C. Exenatide (final concentration 1 mg/mL) and ACTH peptide (final concentration 1 mg/mL) were dissolved in 100 mM ammonium acetate (pH 4.5) or ammonium bicarbonate (pH 8.0), then digested with Glu-C using an enzyme: protein ratio of 1:40 (w/w) at 37 °C. Aliquots of the reactions were quenched by adding aqueous trifluoroacetic acid (TFA, 5%) to a final concentration of 0.5%; and the mixture was diluted 10-fold into a solution of 0.1% TFA in 70: 30 water/ acetonitrile (v/v) for mass spectrometry analysis. In parallel, reactions were also carried out in 18O solutions.
Aging of ACTH peptide
ACTH peptide was dissolved in 100 mM ammonium acetate (pH 4.5) and 100 mM ammonium bicarbonate (pH 8.0) to give a final concentration of 1.1 mg/mL (446 µM). The peptide solutions were aged by incubating at 37 °C for 48 hours, and then stored at −80 °C. The aged solutions were diluted 20-fold into a solution of 0.1% TFA in 70: 30 water/ acetonitrile (v/v) for mass spectrometry analysis.
Mass Spectrometry
An Applied Biosystems 5800 MALDI-TOF/TOF analyzer was calibrated using a peptide standard from Anaspec (AS-60882; Calibration mixture 1: Des-Arg1-Bradykinin 904.47 Da, Angiotensin I 1296.68 Da, and Neurotensin 1672.92 Da). The diluted reaction mixtures were mixed 1:1 with 10 mg/mL alpha-cyano-4-hydroxy-cinnamic acid (CHCA) in a solution of 0.1% TFA in 50:50 water/acetonitrile. The mixture (1 uL) was loaded and crystallized on MALDI target plate, and dried at room temperature prior to analysis. MALDI-TOF mass spectra were acquired in both reflectron positive mode and reflectron negative mode, with MS/MS data collected using 2 kV collision energy. Data were processed with Applied Biosystems Data Explorer 4.6 software.
LC-MS data of exenatide was obtained using a H-Class Acquity UPLC system coupled to a Xevo G2-S Q-ToF mass spectrometer (Waters Corp, Milford, MA). Liquid chromatography was performed on a BEH-C18, 2.1 mm × 150 mm column, with pore size of 1.7 µm (Waters Corp, Milford, MA). Mobile phase A consisted of 0.1 % formic acid (v/v) in HPLC grade water and mobile phase B consisted of 0.1 % formic acid (vol/vol) in 100 % HPLC grade acetonitrile (v/v) with a flow rate at 0.2 mL/min. A gradient was applied by starting at 5% mobile phase B for 2 min, increasing to 60% mobile phase B over 40 min, then increasing to 95% mobile phase B over 2 min, holding at 95% mobile phase B for 3 min, and finally decreasing to 5% mobile phase B over 5 min. After liquid chromatography, samples were introduced via an electrospray ion source in-line with the Xevo G2-S Q-ToF. External calibration of m/z scale was performed using sodium cesium iodide. Data were processed manually using Waters UNIFI 1.7.1 software.
RESULTS AND DISCUSSION
As expected and detailed below, deamidation artifact was eliminated during proteolysis at pH 4.5. Additionally, similar sequence specificity was observed for Glu-C digestion at pH 4.5 and 8.0, thus retaining the enzyme’s utility under mildly acidic conditions.
Analysis of Deamidation
Deamidation of Calmodulin
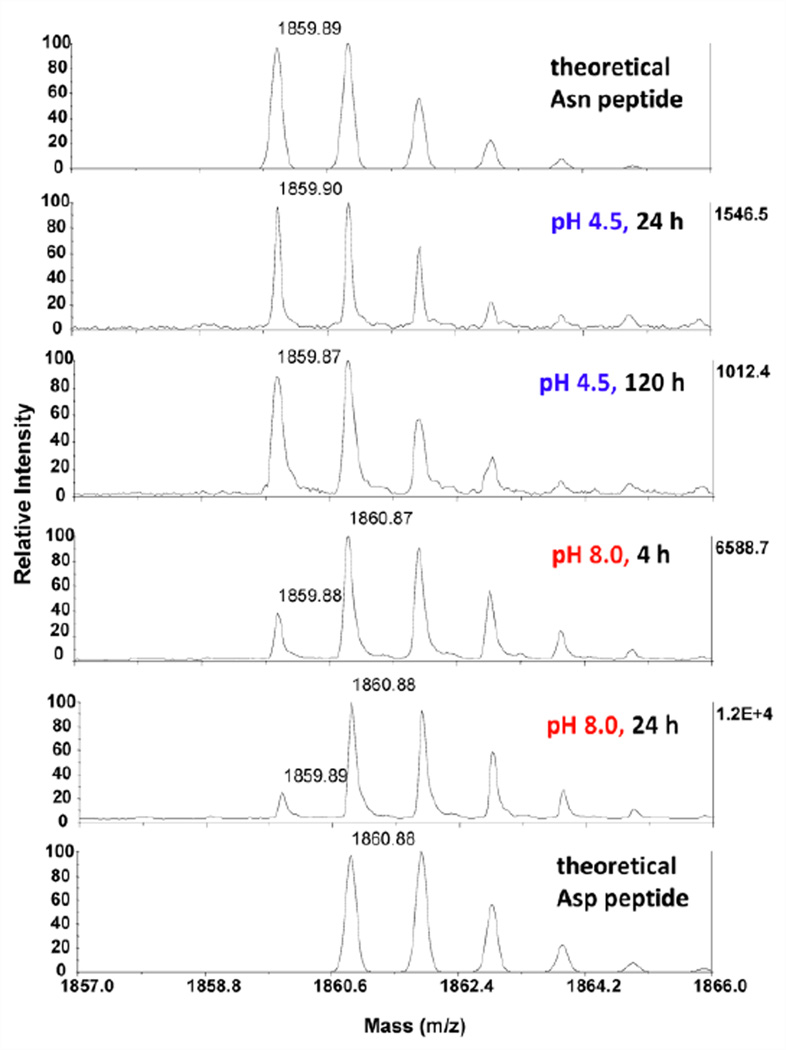
There are six asparagine residues in calmodulin including two Asn-Gly (NG: “Not Good”) sequences that are deamidation hotspots. The MS spectra of one such peptide 88AFRVFDKDGNGYISAAE104 under different proteolytic conditions are shown in Fig. 2. The isotopic envelope of the peptides showed a mixture of deamidated and non-deamidated species. Approximately 50% deamidation of N97 was observed after a 4 h digestion at pH 8, and nearly 80% after 24 h. On the contrary, the same peptide from Glu-C digestion at pH 4.5 for 24 h displayed an isotopic envelope that is nearly identical to the theoretical one; moreover, even after prolonged incubation at pH 4.5 (120 h), only minor deamidation (less than 10%) was observed (see Fig. 2). The corresponding MS/MS spectra can be found in supporting information (Fig. S-1). Altogether, these data showed deamidation was eliminated at pH 4.5 under typical digestion conditions. Since the predominant determinant of deamidation rate in peptides is their primary sequences, our method should be effective for other cases, as demonstrated later.
Fig. 2.
MALDI-TOF MS spectra of calmodulin Glu-C peptide (88AFRVFDKDGNGYISAAE104; theoretical m/z 1859.89) under various conditions; the NG (“Not Good”) tandem (97–98) is prone to deamidation under neutral to basic conditions. At pH 4.5 in ammonium acetate, no deamidation was observed after 24 h digestion, while only minor deamidation (less than 10%) was present after 120 h. At pH 8.0 in ammonium bicarbonate, significant deamidation (~50%) was observed after only 4 h, with nearly complete deamidation after 24 h. Theoretical isotopic envelopes for Asn and Asp isoforms are shown in top and bottom traces respectively
The Glu-C peptide containing the other deamidation hotspot, N60 (NG), was not observed in the positive mode, as the corresponding Glu-C peptide (55VDADGNGTIDFPE67, pI 3.37) may suffer from poor ionization under positive mode. On the other hand, the detection of acidic peptides can sometimes be achieved using negative ionization mode (Dashtiev et al. 2007). Indeed, the second NG-peptide and its deamidation species were detected in negative mode (see Fig. S-2 for spectra). Additionally, as expected, the peptide was fully deamidated at pH 8.0 after 24 h digestion, while digestion at pH 4.5 eliminated deamidation. Aside from N97 and N60, no appreciable levels of deamidation were observed (Fig. S-4 in supporting information) on other asparagine residues of calmodulin: N42 (NP), N53 (NE), N111 (NL), and N137 (NY), which was expected from the sequence dependence on kinetics of deamidation.
Deamidation of Exenatide
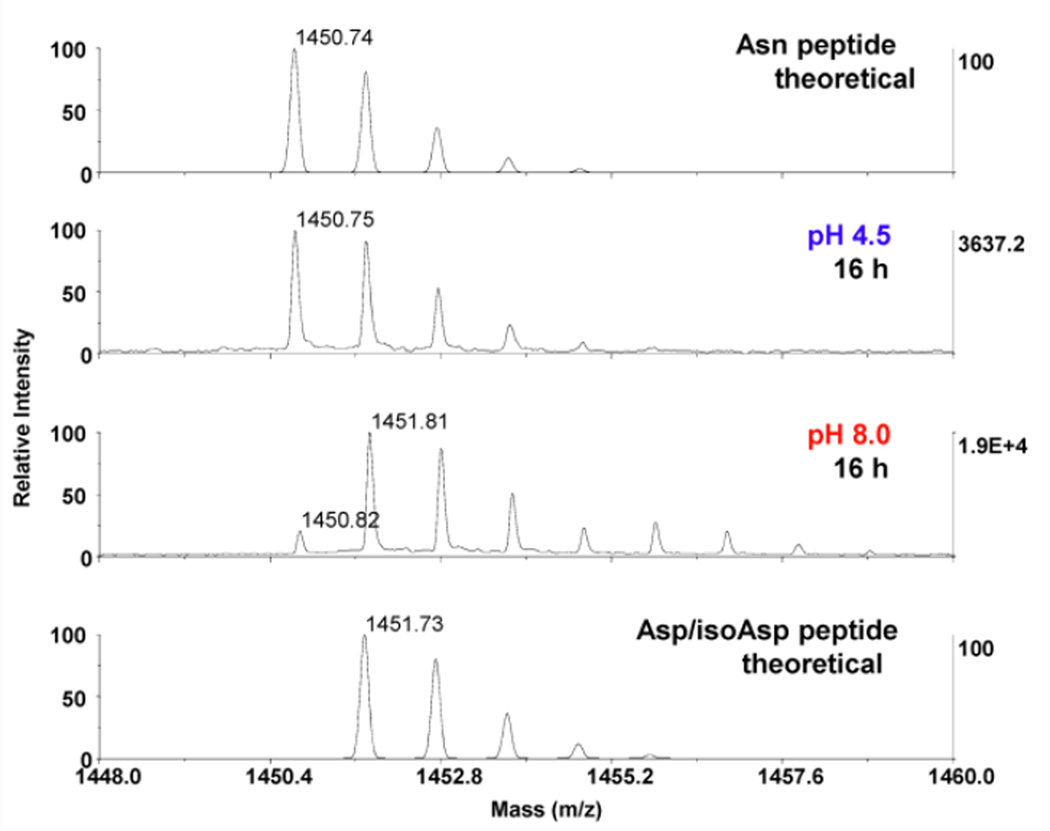
To further evaluate the scope and applicability of our approach, analysis of other deamidation-prone systems was conducted, particularly biotherapeutics, for which deamidation is a major quality attribute. Exenatide (theoretical mass: 4185.01 Da) and adrenocorticotropic hormone peptide (18–39, theoretical mass 2465.20 Da) each contains a labile Asn-Gly sequence (Asn28 and as Asn25 respectively), with the deamidation of ACTH having been previously reported (Gráf et al. 1971; Yu et al. 2013). The MALDI mass spectra of the exenatide Glu-C fragments are shown in Fig. 3. Again, as expected, no deamidation of N28 was detected after overnight (16 h) digestion at pH 4.5 in both 16O and 18O water. In comparison, nearly complete (≥80%) deamidation of N28 occurred after 16 h proteolysis at pH 8.0 in ammonium bicarbonate, and the corresponding 18O mass shift (+3Da) was observed in the peptide digested in 18O water.
Fig. 3.
MALDI-TOF MS spectra of exenatide Glu-C peptide (25WLKNGGPSSGAPPPS39-NH2 (C-terminal amide), theoretical m/z 1450.74 Da) under various conditions. After overnight digestion (16 h), no deamidation was observed at pH 4.5 in ammonium acetate, while nearly complete deamidation occurred at pH 8.0 in ammonium bicarbonate. Theoretical isotopic envelopes for Asn and Asp isoforms are shown in top and bottom traces respectively
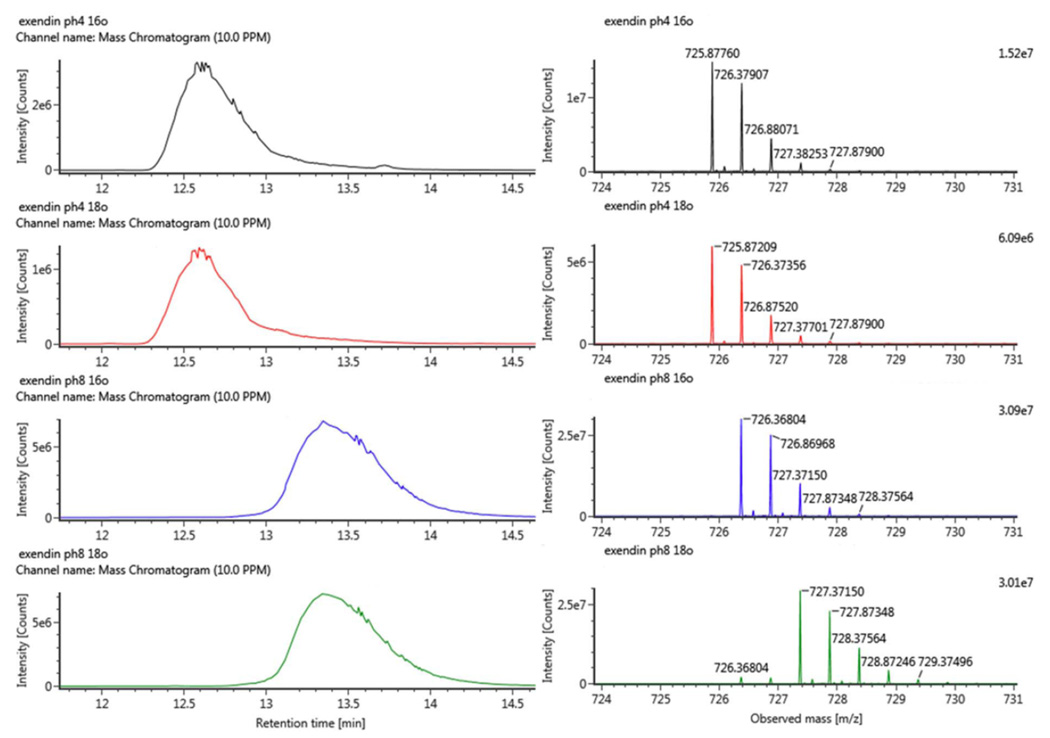
Electrospray ionization (ESI) mass spectrometry is complementary to MALDI, and both are common for protein analysis. Hence, deamidation of exenatide was also investigated by ESI mass spectrometry. As shown in Fig. 4, the deamidated and asparaginyl peptides were resolved by LC and readily distinguished by mass spectrometry, thereby offering even higher sensitivity. Similar to MALDI analysis, complete deamidation was observed after 48 h at pH 8.0 (i.e., no asparaginyl peptide observed). For pH 4.5, with increased sensitivity, about 1% of deamidation was observed after prolonged incubation (48 h). Additionally, 18O labeling confirmed deamidation occurred during sample preparation (Fig. S-3 in supporting information). In practice, deamidation during digestion is negligible under typical digestion conditions (8–24 h).
Fig. 4.
Extracted ion chromatograms (left column) and ESI MS spectra of doubly charged exenatide Glu-C peptide (right column, 25WLKNGGPSSGAPPPS39-NH2 (C-terminal amide), theoretical m/z 725.87 Da) after 48 h proteolysis. The peaks at 12.6 and 13.4 min were for the asparaginyl and deamidated species respectively as confirmed by the mass spectra to the right. Top two traces refer to peptides after proteolysis at pH 4.5 in 16O and 18O water. While bottom two traces refer to peptides after digestion at pH 8.0 in 16O and 18O water; +1 and +3 Da mass shifts that correspond to deamidated species were observed, the small +1 peaks (m/z 726.368) in 18O sample was from the residual 16O (5%) in 18O water
Deamidation of ACTH peptide
Glu-C proteolysis of ACTH peptide also showed similar results, with deamidation artifact being completely eliminated during digestion up to 48 h at pH 4.5. Moreover, deamidation of the intact ACTH peptide was examined as well. The intact ACTH peptides and the Glu-C fragment showed nearly identical deamidation levels under the same conditions; thus indicating that digestion does not affect deamidation and vice versa (Fig. S-4 in supporting information).
No Isomerization at pH 4.5
As the proteolysis conditions change, other potential modifications may occur that should be monitored. Though unlikely, one legitimate concern is isomerization of aspartic acid that generates isoaspartic acid via a succinimide intermediate, analogous to asparagine deamidation, as shown in Scheme 1 (Böhme et al. 2008; Johnson and Aswad 1990). Sequence dependence is similar between isomerization and deamidation, i.e., Asp-Gly is most prone to isomerization. Again, the rates of isomerization are also pH dependent: the half-lives for Asp-Gly in peptides are approximately 50 days at pH 4 and 80 days at pH 8, significantly slower than deamidation and also less sensitive to changes in pH (Oliyai and Borchardt 1993). Therefore, no appreciable isomerization was expected at pH 4.5 in ammonium acetate, but was investigated in calmodulin nonetheless. Once more, 18O-labeling is the best approach to monitor isomerization during sample preparation (Liu et al. 2013b; Yao et al. 2003). Among 17 aspartic acid residues, 12 were detected by mass spectrometry, including five Asp-Gly hotspots. Because 18O was incorporated into the newly formed C-termini, the b ions for the Asp residues can reveal any potential 18O incorporation at that specific residue (Du et al. 2012). As expected, no mass shift, i.e., isomerization, was detected for any of the Asp peptides at both pH 4.5 and 8.0 (Fig. S-5 in supporting information).
Nearly identical proteolytic specificity at pH 4.5 and 8.0
The sequence specificity of Glu-C under mildly acidic conditions has only been reported for a handful of proteins, showing similar specificity as that under neutral conditions. However, many of these studies were prior to the modern mass spectrometry era, so the specificity of Glu-C under both conditions reported herein was compared. It is known that the enzyme specifically cleaves at the C-terminal side of glutamic acid in buffers that include Tris-HCl, bicarbonate and acetate; however, cleavage after both glutamic acid and aspartic acid has been shown to occur in phosphate buffers (Houmard and Drapeau 1972). This was one of the reasons ammonium bicarbonate and ammonium acetate were chosen for this work.
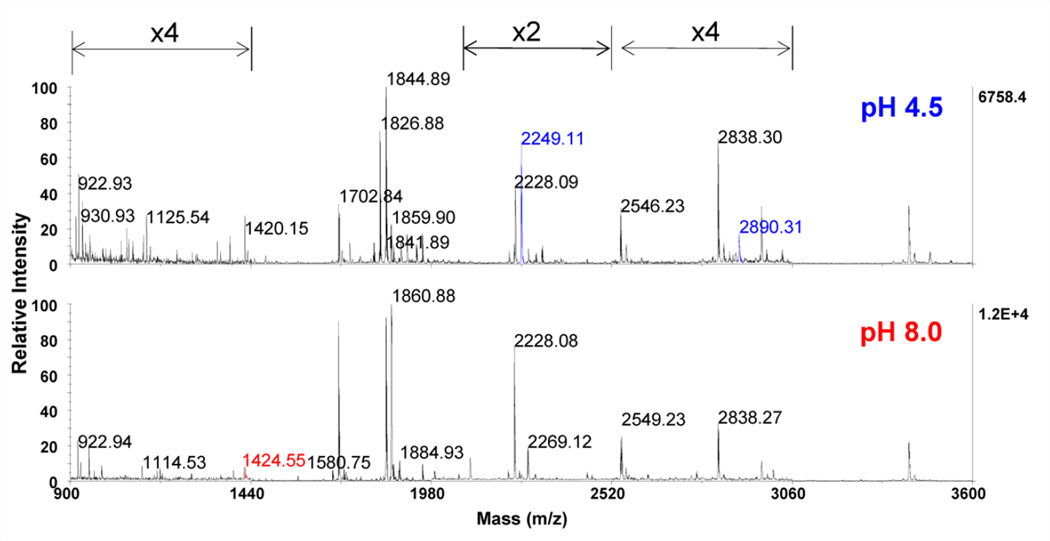
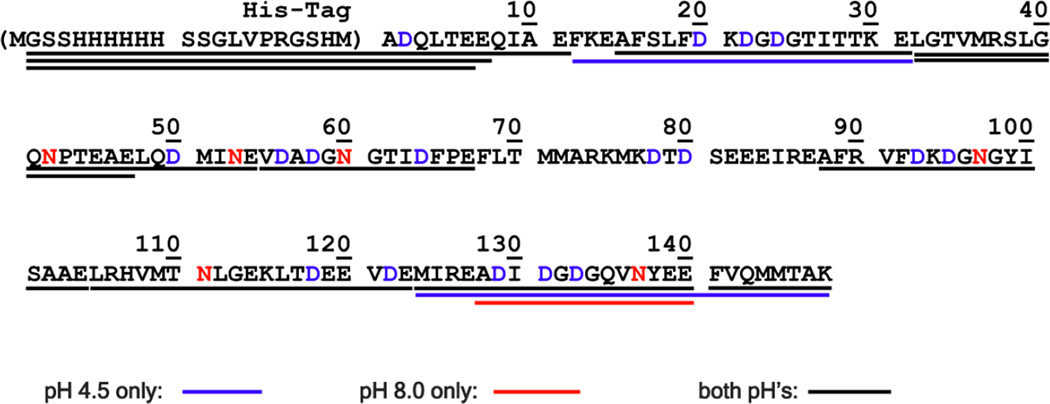
As shown in Fig. 5, the MALDI spectra of the Glu-C digests of calmodulin at pH 4.5 and 8.0 are highly similar (both masses and relative intensity); among 35 major peaks, only three are different (highlighted in red and blue). As summarized in Fig. 6, similar sequence coverage was observed under both pH conditions, with 88% and 87% coverage seen at pH 4.5 and 8.0 respectively. Furthermore, only glutamic acid residues were cleaved under both conditions, with no cleavage at aspartic acid observed after proteolysis at both pH 4.5 and 8.0. While not required, having the same specificity at different pH’s is convenient for the practical application of Glu-C. These observations suggest that the proposed low pH methodology can be a viable alternative to proteolysis at pH 8.
Fig. 5.
MALDI-TOF-MS spectra of calmodulin Glu-C digests at both pH 4.5 (top) and pH 8.0 (bottom). Specific areas of spectra are zoomed in at indicated degree to improve clarity. Glu-C proteolysis at each pH generated similar peptide fragments. Colors are used to indicate peaks only observed at pH 4.5 (blue), only observed at pH 8.0 (red), and at both pH's (black)
Fig. 6.
Sequence coverage of calmodulin by Glu-C proteolysis. Color codes indicate peptides observed at digestion at pH 4.5 only (blue), at pH 8.0 only (red) and at both pH’s (black). The peptides between F68 and E87 were not observed by MALDI. Sequence coverage was found to be 88% and 87% at pH 4.5 and 8.0 respectively
CONCLUSIONS
Deamidation artifact occurs widely and may affect various procedures in protein analysis, but can be eliminated by proteolysis at pH 4.5 in ammonium acetate using the common protease Glu-C, which also retains the same specificity as at pH 8. Since pH is the dominant factor in the rates of deamidation, other proteases that are active under mildly acidic conditions should be equally useful for eliminating deamidation artifact. Judicious control of pH and conditions for sample preparation may also minimize or eliminate other artifacts. For example, proteolysis at low pH has been previously used to minimize disulfide scrambling (Chen et al. 1997; Pompach et al. 2009; Salzano et al. 2011; Tomlinson et al. 1997). Finally, by effectively eliminating deamidation artifact, the bona fide deamidation can be analyzed with greater ease and higher confidence for both biological systems and protein pharmaceuticals alike.
Supplementary Material
Acknowledgments
The work is supported by the National Institutes of Health (Grant GM101396 to Z.S.Z.). We thank Sam Burns, Kalli Catcott, Mike Pablo, Clair Yu, Derek Lin and Wanlu Qu for the helpful discussion and critical reading.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional mass spectra as mentioned in the text.
CONFLICT OF INTEREST: The authors declare that they have no conflict of interest.
REFERENCES
- Alfaro JF, et al. Chemo-Enzymatic Detection of Protein Isoaspartate Using Protein Isoaspartate Methyltransferase and Hydrazine Trapping. Anal Chem. 2008;80:3882–3889. doi: 10.1021/ac800251q. [DOI] [PubMed] [Google Scholar]
- Biastoff S, Teuber M, Zhou ZS, Dräger B. Colorimetric Activity Measurement of a Recombinant Putrescine N-Methyltransferase from Datura stramonium. Planta Med. 2006;72:1136–1141. doi: 10.1055/s-2006-947191. [DOI] [PubMed] [Google Scholar]
- Böhme L, Bär JW, Hoffmann T, Manhart S, Ludwig H-H, Rosche F, Demuth H-U. Isoaspartate residues dramatically influence substrate recognition and turnover by proteases. Biol Chem. 2008;389:1043–1053. doi: 10.1515/BC.2008.123. [DOI] [PubMed] [Google Scholar]
- Capasso S, Kirby AJ, Salvadori S, Sica F, Zagari A. Kinetics and mechanism of the reversible isomerization of aspartic acid residues in tetrapeptides. Journal of the Chemical Society, Perkin Transactions. 1995;2:437–442. [Google Scholar]
- Chen T, et al. Substrates of the Arabidopsis thaliana Protein Isoaspartyl Methyltransferase 1 Identified Using Phage Display and Biopanning. J Biol Chem. 2010;285:37281–37292. doi: 10.1074/jbc.M110.157008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W-Q, Karnaukhova E, Lubec G. The use of native gels for the concomitant determination of protein sequences and modifications by mass spectrometry with subsequent conformational and functional analysis of native proteins following electro-elution. Amino Acids. 2013;44:1381–1389. doi: 10.1007/s00726-013-1477-1. [DOI] [PubMed] [Google Scholar]
- Chen Z-w, Bergman T, Östenson C-G, Efendic S, Mutt V, Jörnvall H. Characterization of Dopuin, a Polypeptide with Special Residue Distributions. Eur J Biochem. 1997;249:518–522. doi: 10.1111/j.1432-1033.1997.t01-2-00518.x. [DOI] [PubMed] [Google Scholar]
- Chumsae C, Gifford K, Lian W, Liu H, Radziejewski CH, Zhou ZS. Arginine modifications by methylglyoxal: discovery in a recombinant monoclonal antibody and contribution to acidic species. Anal Chem. 2013;85:11401–11409. doi: 10.1021/ac402384y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumsae C, et al. Discovery of a chemical modification by citric acid in a recombinant monoclonal antibody. Anal Chem. 2014;86:8932–8936. doi: 10.1021/ac502179m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. Aging as war between chemical and biochemical processes: Protein methylation and the recognition of age-damaged proteins for repair. Ageing Research Reviews. 2003;2:263–285. doi: 10.1016/s1568-1637(03)00011-4. [DOI] [PubMed] [Google Scholar]
- Dai S, Ni W, Patananan AN, Clarke SG, Karger BL, Zhou ZS. Integrated Proteomic Analysis of Major Isoaspartyl-Containing Proteins in the Urine of Wild Type and Protein l-Isoaspartate O-Methyltransferase-Deficient Mice. Anal Chem. 2013;85:2423–2430. doi: 10.1021/ac303428h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashtiev M, Wäfler E, Röhling U, Gorshkov M, Hillenkamp F, Zenobi R. Positive and negative analyte ion yield in matrix-assisted laser desorption/ionization. Int J Mass spectrom. 2007;268:122–130. [Google Scholar]
- Drapeau GR, Boily Y, Houmard J. Purification and Properties of an Extracellular Protease of Staphylococcus aureus. J Biol Chem. 1972;247:6720–6726. [PubMed] [Google Scholar]
- Du Y, Wang F, May K, Xu W, Liu H. Determination of Deamidation Artifacts Introduced by Sample Preparation Using 18O-Labeling and Tandem Mass Spectrometry Analysis. Anal Chem. 2012;84:6355–6360. doi: 10.1021/ac3013362. [DOI] [PubMed] [Google Scholar]
- Gráf L, Bajusz S, Patthy A, Barát E, Cseh G. Revised amide location for porcine and human adrenocorticotropic hormone. Acta Biochim Biophys Acad Sci Hung. 1971;6 [PubMed] [Google Scholar]
- Gui S, Wooderchak-Donahue WL, Zang T, Chen D, Daly MP, Zhou ZS, Hevel JM. Substrate-induced control of product formation by protein arginine methyltransferase 1. Biochemistry. 2013;52:199–209. doi: 10.1021/bi301283t. [DOI] [PubMed] [Google Scholar]
- Hao P, Ren Y, Datta A, Tam JP, Sze SK. Evaluation of the Effect of Trypsin Digestion Buffers on Artificial Deamidation. J Proteome Res. 2015;14:1308–1314. doi: 10.1021/pr500903b. [DOI] [PubMed] [Google Scholar]
- Houmard J, Drapeau GR. Staphylococcal protease: a proteolytic enzyme specific for glutamoyl bonds. Proc Natl Acad Sci U S A. 1972;69:3506–3509. doi: 10.1073/pnas.69.12.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wu S-L, Karger BL, Hancock WS. Characterization of the Glycosylation Occupancy and the Active Site in the Follow-on Protein Therapeutic: TNK-Tissue Plasminogen Activator. Anal Chem. 2010;82:6154–6162. doi: 10.1021/ac100956x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Aswad DW. Fragmentation of isoaspartyl peptides and proteins by carboxypeptidase Y: release of isoaspartyl dipeptides as a result of internal and external cleavage. Biochemistry. 1990;29:4373–4380. doi: 10.1021/bi00470a017. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Freitag NE, Aswad DW. Protein carboxyl methyltransferase selectively modifies an atypical form of calmodulin. Evidence for methylation at deamidated asparagine residues. J Biol Chem. 1985;260:10913–10916. [PubMed] [Google Scholar]
- Klaene JJ, Ni W, Alfaro JF, Zhou ZS. Detection and Quantitation of Succinimide in Intact Protein via Hydrazine Trapping and Chemical Derivatization. J Pharm Sci. 2014;103:3033–3042. doi: 10.1002/jps.24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokhin OV, Antonovici M, Ens W, Wilkins JA, Standing KG. Deamidation of -Asn-Gly- Sequences during Sample Preparation for Proteomics: Consequences for MALDI and HPLC-MALDI Analysis. Anal Chem. 2006;78:6645–6650. doi: 10.1021/ac061017o. [DOI] [PubMed] [Google Scholar]
- Lee J-C, et al. Protein L-isoaspartyl methyltransferase regulates p53 activity. Nat Commun. 2012;3:927. doi: 10.1038/ncomms1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cournoyer JJ, Lin C, O'Connor PB. Use of 18O Labels to Monitor Deamidation during Protein and Peptide Sample Processing. J Am Soc Mass Spectrom. 2008;19:855–864. doi: 10.1016/j.jasms.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang F, Xu W, May K, Richardson D. Quantitation of asparagine deamidation by isotope labeling and liquid chromatography coupled with mass spectrometry analysis. Anal Biochem. 2013a;432:16–22. doi: 10.1016/j.ab.2012.09.024. [DOI] [PubMed] [Google Scholar]
- Liu M, Cheetham J, Cauchon N, Ostovic J, Ni W, Ren D, Zhou ZS. Protein Isoaspartate Methyltransferase-Mediated 18O-Labeling of Isoaspartic Acid for Mass Spectrometry Analysis. Anal Chem. 2012;84:1056–1062. doi: 10.1021/ac202652z. [DOI] [PubMed] [Google Scholar]
- Liu M, Zhang Z, Cheetham J, Ren D, Zhou ZS. Discovery and Characterization of a Photo-Oxidative Histidine-Histidine Cross-Link in IgG1 Antibody Utilizing 18O-Labeling and Mass Spectrometry. Anal Chem. 2014;86:4940–4948. doi: 10.1021/ac500334k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang Z, Zang T, Spahr C, Cheetham J, Ren D, Zhou ZS. Discovery of Undefined Protein Cross-Linking Chemistry: A Comprehensive Methodology Utilizing 18O-Labeling and Mass Spectrometry. Anal Chem. 2013b;85:5900–5908. doi: 10.1021/ac400666p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning M, Chou D, Murphy B, Payne R, Katayama D. Stability of Protein Pharmaceuticals: An Update. Pharm Res. 2010;27:544–575. doi: 10.1007/s11095-009-0045-6. [DOI] [PubMed] [Google Scholar]
- Mosley SL, Bakke BA, Sadler JM, Sunkara NK, Dorgan KM, Zhou ZS, Seley-Radtke KL. Carbocyclic pyrimidine nucleosides as inhibitors of S-adenosylhomocysteine hydrolase. Bioorg Med Chem. 2006;14:7967–7971. doi: 10.1016/j.bmc.2006.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Dai S, Karger BL, Zhou ZS. Analysis of Isoaspartic Acid by Selective Proteolysis with Asp-N and Electron Transfer Dissociation Mass Spectrometry. Anal Chem. 2010;82:7485–7491. doi: 10.1021/ac101806e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi S. Structural changes induced by the deamidation and isomerization of asparagine revealed by the crystal structure of Ustilago sphaerogena ribonuclease U2B. Biopolymers. 2010;93:1003–1010. doi: 10.1002/bip.21514. [DOI] [PubMed] [Google Scholar]
- O’Connor PB, Cournoyer JJ, Pitteri SJ, Chrisman PA, McLuckey SA. Differentiation of Aspartic and Isoaspartic Acids Using Electron Transfer Dissociation. J Am Soc Mass Spectrom. 2006;17:15–19. doi: 10.1016/j.jasms.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Oliyai C, Borchardt R. Chemical Pathways of Peptide Degradation. IV. Pathways, Kinetics, and Mechanism of Degradation of an Aspartyl Residue in a Model Hexapeptide. Pharm Res. 1993;10:95–102. doi: 10.1023/a:1018981231468. [DOI] [PubMed] [Google Scholar]
- Orrù S, Vitagliano L, Esposito L, Mazzarella L, Marino G, Ruoppolo M. For the record: Effect of deamidation on folding of ribonuclease. A Protein Sci. 2000;9:2577–2582. doi: 10.1110/ps.9.12.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmisano G, Melo-Braga MN, Engholm-Keller K, Parker BL, Larsen MR. Chemical Deamidation: A Common Pitfall in Large-Scale N-Linked Glycoproteomic Mass Spectrometry-Based Analyses. J Proteome Res. 2012;11:1949–1957. doi: 10.1021/pr2011268. [DOI] [PubMed] [Google Scholar]
- Paranandi MV, Guzzetta AW, Hancock WS, Aswad DW. Deamidation and isoaspartate formation during in vitro aging of recombinant tissue plasminogen activator. J Biol Chem. 1994;269:243–253. [PubMed] [Google Scholar]
- Patananan AN, Capri J, Whitelegge JP, Clarke SG. Non-repair Pathways for Minimizing Protein Isoaspartyl Damage in the Yeast Saccharomyces cerevisiae. J Biol Chem. 2014;289:16936–16953. doi: 10.1074/jbc.M114.564385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K, Borchardt R. Chemical Pathways of Peptide Degradation. II. Kinetics of Deamidation of an Asparaginyl Residue in a Model Hexapeptide. Pharm Res. 1990;7:703–711. doi: 10.1023/a:1015807303766. [DOI] [PubMed] [Google Scholar]
- Perła-Kaján J, Jakubowski H. Paraoxonase 1 and homocysteine metabolism. Amino Acids. 2012;43:1405–1417. doi: 10.1007/s00726-012-1321-z. [DOI] [PubMed] [Google Scholar]
- Perła-Kaján J, Twardowski T, Jakubowski H. Mechanisms of homocysteine toxicity in humans. Amino Acids. 2007;32:561–572. doi: 10.1007/s00726-006-0432-9. [DOI] [PubMed] [Google Scholar]
- Pompach P, et al. Modified electrophoretic and digestion conditions allow a simplified mass spectrometric evaluation of disulfide bonds. J Mass Spectrom. 2009;44:1571–1578. doi: 10.1002/jms.1609. [DOI] [PubMed] [Google Scholar]
- Potter SM, Henzel WJ, Aswad DW. In vitro aging of calmodulin generates isoaspartate at multiple Asn–Gly and Asp–Gly sites in calcium-binding domains II, III, and IV. Protein Sci. 1993;2:1648–1663. doi: 10.1002/pro.5560021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radkiewicz JL, Zipse H, Clarke S, Houk KN. Neighboring Side Chain Effects on Asparaginyl and Aspartyl Degradation: An Ab Initio Study of the Relationship between Peptide Conformation and Backbone NH Acidity. J Am Chem Soc. 2001;123:3499–3506. doi: 10.1021/ja0026814. [DOI] [PubMed] [Google Scholar]
- Reissner KJ, Aswad DW. Deamidation and isoaspartate formation in proteins: unwanted alterations or surreptitious signals? Cell Mol Life Sci. 2003;60:1281–1295. doi: 10.1007/s00018-003-2287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NE, Robinson AB. Prediction of protein deamidation rates from primary and three-dimensional structure. Proc Natl Acad Sci U S A. 2001;98:4367–4372. doi: 10.1073/pnas.071066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NE, Robinson ZW, Robinson BR, Robinson AL, Robinson JA, Robinson ML, Robinson AB. Structure-dependent nonenzymatic deamidation of glutaminyl and asparaginyl pentapeptides. J Pept Res. 2004;63:426–436. doi: 10.1111/j.1399-3011.2004.00151.x. [DOI] [PubMed] [Google Scholar]
- Salzano AM, Renzone G, Scaloni A, Torreggiani A, Ferreri C, Chatgilialoglu C. Human serum albumin modifications associated with reductive radical stress. Mol BioSyst. 2011;7:889–898. doi: 10.1039/c0mb00223b. [DOI] [PubMed] [Google Scholar]
- Tomlinson AJ, Johnson KL, Lam-Holt J, Mays DC, Lipsky JJ, Naylor S. Inhibition of human mitochondrial aldehyde dehydrogenase by the disulfiram metabolite S-methyl-N,N-diethylthiocarbamoyl sulfoxide: Structural characterization of the enzyme adduct by HPLC-tandem mass spectrometry. Biochem Pharmacol. 1997;54:1253–1260. doi: 10.1016/s0006-2952(97)00359-6. [DOI] [PubMed] [Google Scholar]
- Tyler-Cross R, Schirch V. Effects of amino acid sequence, buffers, and ionic strength on the rate and mechanism of deamidation of asparagine residues in small peptides. J Biol Chem. 1991;266:22549–22556. [PubMed] [Google Scholar]
- Wan W, Zhao G, Al-Saad K, Siems WF, Zhou ZS. Rapid screening for S-adenosylmethionine-dependent methylation products by enzyme-transferred isotope patterns analysis. Rapid Commun Mass Spectrom. 2004;18:319–324. doi: 10.1002/rcm.1335. [DOI] [PubMed] [Google Scholar]
- Wang Z, Rejtar T, Zhou ZS, Karger BL. Desulfurization of Cysteine-Containing Peptides Resulting from Sample Preparation for Protein Characterization by MS. Rapid Commun Mass Spectrom. 2010;24:267–275. doi: 10.1002/rcm.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Pipkorn R, Lehmann WD. Separation of peptide isomers and conformers by ultra performance liquid chromatography. J Sep Sci. 2009;32:1111–1119. doi: 10.1002/jssc.200800691. [DOI] [PubMed] [Google Scholar]
- Yao X, Afonso C, Fenselau C. Dissection of Proteolytic 18O Labeling: Endoprotease-Catalyzed 16O-to-18O Exchange of Truncated Peptide Substrates. J Proteome Res. 2003;2:147–152. doi: 10.1021/pr025572s. [DOI] [PubMed] [Google Scholar]
- Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic 18O Labeling for Comparative Proteomics: Model Studies with Two Serotypes of Adenovirus. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- Yu X, Warme C, Lee D, Zhang J, Zhong W. Characterization of a Low-Level Unknown Isomeric Degradation Product Using an Integrated Online–Offline Top-Down Tandem Mass Spectrometry Platform. Anal Chem. 2013;85:8964–8967. doi: 10.1021/ac401911n. [DOI] [PubMed] [Google Scholar]
- Zang T, Dai S, Chen D, Lee BW, Liu S, Karger BL, Zhou ZS. Chemical methods for the detection of protein N-homocysteinylation via selective reactions with aldehydes. Anal Chem. 2009;81:9065–9071. doi: 10.1021/ac9017132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZS, Peariso K, Penner-Hahn JE, Matthews RG. Identification of the Zinc Ligands in Cobalamin-Independent Methionine Synthase (MetE) from Escherichia coli. Biochemistry. 1999;38:15915–15926. doi: 10.1021/bi992062b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.