Abstract
Chronic liver injury leads to fibrosis and cirrhosis. Cirrhosis, the end stage of chronic liver disease, is a leading cause of death worldwide and increases the risk of developing hepatocellular carcinoma. Currently, there is a lack of effective antifibrotic therapies to treat fibrosis and cirrhosis. Development of antifibrotic therapies requires an in-depth understanding of the cellular and molecular mechanisms involved in inflammation and fibrosis after hepatic injury. Two growth factor signaling pathways that regulate liver fibrosis are transforming growth factor-β (TGFβ) and platelet-derived growth factor (PDGF). However, their specific contributions to fibrogenesis are not well understood. Using a genetic model of liver fibrosis, we investigated whether the canonical TGFβ signaling pathway was necessary for fibrogenesis. PDGF-C transgenic (PDGF-C Tg) mice were intercrossed with mice that lack Smad3, and molecular and histological fibrosis was analyzed. PDGF-C Tg mice that also lacked Smad3 had less fibrosis and improved liver lobule architecture. Loss of Smad3 also reduced expression of collagen genes, which were induced by PDGF-C, but not the expression of genes frequently associated with hepatic stellate cell (HSC) activation. In vitro HSCs isolated from Smad3-null mice proliferated more slowly than cells from wild-type mice. Taken together, these findings indicate that PDGF-C activates TGFβ/Smad3 signaling pathways to regulate HSC proliferation, collagen production and ultimately fibrosis. In summary, these results suggest that inhibition of both PDGF and TGFβ signaling pathways may be required to effectively attenuate fibrogenesis in patients with chronic liver disease.
Keywords: PDGF, TGFβ, Smad, liver fibrosis, hepatic stellate cells
chronic liver injury results in liver fibrosis, which may progress to cirrhosis (31, 42, 45), increasing the risk of developing hepatocellular carcinoma (HCC) (52). Liver fibrosis, a maladaptive form of wound healing, results from the interplay of various intra- and extrahepatic cells activated by a wide variety of cytokine and growth factor mediators (15, 23). Hepatic stellate cells (HSCs) and portal fibroblasts are recognized as the primary cellular mediators of fibrosis as they produce collagen (15, 29, 42, 58). Understanding the molecular pathways involved in coordinated activation and inactivation of HCSs and/or myofibroblasts in fibrogenesis is critical to the development of antifibrotic therapies.
A plethora of cytokines, chemokines, growth factors, and extracellular factors are involved in HSC activation, proliferation, and migration (22, 46, 60). Two families of growth factors appear to be critical regulators of HSCs: transforming growth factor-β (TGFβ) ligands, and platelet-derived growth factor (PDGF) ligands (46). The PDGF ligand family, PDGF-A, B, C, and D, transmit their extracellular signals via tyrosine kinase receptors, PDGF receptor-α (PDGFRα) and -β (PDGFRβ) (1, 5). Stimulation of PDGF receptors induces HSC proliferation in vitro (44, 61), and overexpression of PDGF-A, -B, or -C in the liver (7, 9, 53) results in fibrosis. Hepatic overexpression of PDGF-C results in progressive liver disease, development of tumors, and decreased life span (7), phenotypes not seen when PDGF-A or -B is overexpressed in the liver (9, 53). It is not yet known how the overexpression of these different PDGF ligands results in different liver pathology. Thieringer and Czochra have suggested that the different outcomes may reside with differences in PDGF ligands to stimulate TGFβ ligands (9, 53).
TGFβ is a multifunctional cytokine that regulates cell survival, differentiation, migration, and synthesis of extracellular matrix (ECM) components (12, 25, 59). Elevated levels of TGFβ are seen in organ fibrosis, and Tgfβ overexpression results in liver fibrosis (27, 50). TGFβ stimulates type I collagen production, the hallmark of liver fibrosis, as well as regulating fibrinolysis factors, such as matrix metalloproteinases (MMPs) and their inhibitors (12, 25, 59). TGFβ binds to cell surface receptors to initiate a number of intracellular signal transduction pathways, including activation of Smad proteins, which are transcription factors (12, 25, 59). TGFβ ligand binding stimulates receptor phosphorylation of Smad2 and 3, allowing binding with Smad4-forming complexes that translocate to the nucleus to activate gene expression. Among the genes induced by Smad2/Smad3:Smad4 complexes is the inhibitory Smad, Smad7. Smad7 is regulated by Smad2/3 transcriptional activity and disrupts receptor activation of Smad2/3, which inhibits TGFβ signaling in a negative feedback loop (34, 38, 39).
We have previously reported that hepatic overexpression of PDGF-C results in progressive liver fibrosis, which increases in severity as the mice age, and eventually the development of HCCs (7). Shortly after birth, extensive HSC activation and proliferation is apparent, accompanied by elevated hepatic Tgfβ1 (7, 8). Correlation between collagen deposition, HSC activation, and elevated levels of Tgfβ1 suggests that TGFβ may be a critical regulator in this mouse model of fibrosis. In other hepatic transgenic PDGF mouse strains, increased expression of TGFβ was reported for PDGF-A (53), but not PDGF-B (9). Thus, it is plausible that differences in the amount of TGFβ produced in each of these models (7, 9, 53) may account for the observed differences in the severity of chronic liver disease in these models.
In this study we sought to investigate whether TGFβ regulates PDGF-C-induced liver fibrogenesis by interfering with TGFβ signaling pathways, by deleting the Smad3 allele.1 To test this hypothesis, we generated PDGF-C transgenic (PDGF-C Tg) mice that also lacked Smad3 and found that PDGF-C;Smad3 knockout (KO) mice had less fibrosis compared with PDGF-C Tg mice. Gene expression studies revealed that collagen gene expression was Smad3 dependent, while genes associated with HSC activation were not significantly decreased in PDGF-C;Smad3 KO mice. Isolated Smad3-null HSCs grew more slowly than wild-type (WT) HSCs. In vitro, PDGF ligands stimulated TGFβ protein production and release. In summary, these data indicate that Smad3 is an important mediator of PDGF-C-induced fibrosis and delineate the contributions of TGFβ/Smad3 pathways in this model. PDGF-C regulates both Smad3-dependent and -independent signaling pathways. Thus, combination therapy that targets both TGFβ- and PDGF signaling pathways may provide the most effective long-term therapy for chronic liver disease and prevention of HCC.
MATERIALS AND METHODS
Generation of PDGF-C Tg;Smad3-deficient mice.
PDGF-C transgenic (PTg) mice (7) and Smad3-deleted mice with targeted disruption of exon 8 in the Smad3 gene (Smad3KOex8/ex8) have been previously described (63). Prior to initiation of the experiments described in this study, Smad3KOex8/ex8 were cleared of Helicobacter infection by neonatal rederivation (54), verified by PCR on DNA extracted from fecal matter (35). Additionally, Smad3KOex8/ex8 mice, originally on a mixed 129 background, were backcrossed onto a C57BL6Jax background for six or more generations. To generate experimental mice, wild-type (WT, nontransgenic), heterozygous Smad3 (i.e., PDGF-CWT/WT; Smad3ex8/WT) females were bred with PDGF-C Tg, heterozygous Smad3 gene (i.e., PDGF-CTg/WT; Smad3ex8/WT) male mice. Offspring were genotyped by PCR on DNA extracted from tail tips. Helicobacter-free male and female mice were killed by CO2 inhalation for evaluation of liver histology, liver cell proliferation by bromodeoxyuridine (BrdU) incorporation (7), gene or protein expression, or HSC isolation between the ages of 2 and 6 mo. As previously described, Smad3-null mice were smaller than hemizygous or WT littermates (63). All animal studies were carried out under approved Institutional Animal Care and Use Committee protocols from the University of Washington, which is certified by the Association for Assessment and Accreditation of Laboratory Animal Care International. For the remainder of the article, mice will be referred to as WT, KO (Smad3 KO ex8/ex8), PTg (PDGF-C Tg), and PTg/KO (PDGF-C Tg; Smad3 KOex8/ex8).
Liver histology and immunohistochemistry.
Mouse livers were fixed in 10% neutral-buffered formalin or Methyl Carnoy's solution (60% methanol, 30% chloroform, and 10% acetic acid, vol:vol:vol) overnight, processed to paraffin blocks, sectioned, and stained with hematoxylin/eosin, Masson's trichrome, or Picrosirius Red using standard techniques. Staining was performed as previously described using antibodies specific for mouse anti-BrdU (7) (DAKO, Carpinteria, CA) and rabbit anti-phospho-Smad2/3 (Zymed, now Life Technologies). The Zymed antibody detects both Smad2 and Smad3. Detection of primary antibodies was done using the appropriate biotinylated antibody (Vectastain, Burlingame, CA) and visualized with a peroxidase diaminobenzidine kit (Ventana, Tucson, AZ). The mouse on mouse kit (Vectastain) was used to detect BrdU labeling of both nonparenchymal cells (NPCs) and hepatocytes as a measure of cell proliferation (7, 8). Data are represented as the number of BrdU-positive hepatocyte nuclei or NPCs observed in thirty ×40 fields (i.e., 1.3 mm2; ∼3,000 hepatocytes). Morphometric analysis was performed as described previously (8, 48).
RNA isolation and real-time RT-PCR analysis.
RNA was isolated from whole liver snap frozen at the time of necropsy using TRIzol (Invitrogen, now Life Technologies, Carlsbad, CA) according to the manufacturer's recommendation. For cDNA synthesis, 2 μg RNA were reverse transcribed using the Retroscript kit (Life Technologies), and real-time RT-PCR (qPCR) was performed using FAM-labeled primers (Applied Biosystem, Foster City, CA) as described or primers used with Sybr Green (primer sequences are available on request). Gene expression data were normalized to Gapdh mRNA or 18S rRNA levels using the ΔΔCt method as previously described (62).
Protein extraction and immunoblotting.
Whole liver lysates were prepared using a Triton X-100 lysis buffer with protease inhibitors and quantified using the Bradford method with BSA (8). SDS-PAGE analysis of proteins levels was determined using immunoblotting and the following antibodies: rabbit anti-Smad3 (Zymed, Carlsbad, CA), rabbit anti-Smad2 (Zymed, South San Francisco, CA), mouse anti-Smad4 (Santa Cruz, Santa Cruz, CA), mouse anti-human smooth muscle α-actin (αSMA; DAKO), and rabbit anti-GAPDH conjugated to horseradish peroxidase (HRP) (GeneScript, Piscataway, NJ). Epitope-primary antibody complexes were detected with species-specific secondary antibodies conjugated to HRP followed by ECL (Thermo Fisher Scientific Pierce, Illinois).
Primary HSC isolation and culture conditions.
HSCs were isolated from WT and KO mice (i.e., Smad3ex8/ex8) using pronase/collagenase (36, 57) perfusion and density centrifugation methods using Nycodez. Isolated HSCs were cultured on uncoated tissue culture dishes (1–4 × 104 cells/cm2) in DMEM supplemented with 10% FBS and penicillin in 95% air–5% CO2 humidified atmosphere at 37°C. After 7 days, cultures were placed in serum-free media and incubated with serum-free media or with PDGF-CC in serum-free media for 24 h. Typically, isolated HSCs were combined from two to three mice. Stocks solutions of TGFβ (R&D Systems) were prepared according to the manufacturer's directions.
Cell proliferation in primary HSC cells.
[3H]thymidine incorporation or cell counting was used to measure cell proliferation as previously described (3, 7). Primary HSCs were isolated from 6- to 7-wk-old WT mice or Smad3 KO mice and maintained as described in the previous section. In some experiments, HSC cultures were serum starved overnight prior to ligand treatment and subsequent addition of [3H]thymidine.
Detection of TGFβ1 in HSC media.
Rat CFSC-2G stellate cell cultures were maintained as described by Greenwel and coworkers (18). CFSC-2G cells were serum starved overnight with BSA, treated with the indicated doses of PDGF-CC and -DD [a gift from ZymoGenetics (17)], -AA, -AB, or -BB (R&D Systems), and cell culture media were collected 24 to 72 h later. TGFβ protein levels were determined by ELISA (catalog no. DB100B, Quantikine Human Immunoassay, R&D Systems) after media were treated with HCl to convert latent TGFβ to the active form. This assay recognizes human porcine, mouse, rat, and canine TGFβ1, but not other TGFβ isoforms.
Statistical analysis.
All results are shown as means ± SE. Data were analyzed using nonparametric analyses, including Mann-Whitney test, or Kruskal-Wallis with multiple comparisons with Dunn's post test. P < 0.05 was considered as statistically significant. Statistical analyses were performed with SPSS software for Windows (version 12.0, SPSS, Chicago, IL) or Prism (GraphPad Software, La Jolla, CA).
RESULTS
Elevated TGFβ/Smad signaling in fibrotic liver tissue from PDGF-C Tg mice.
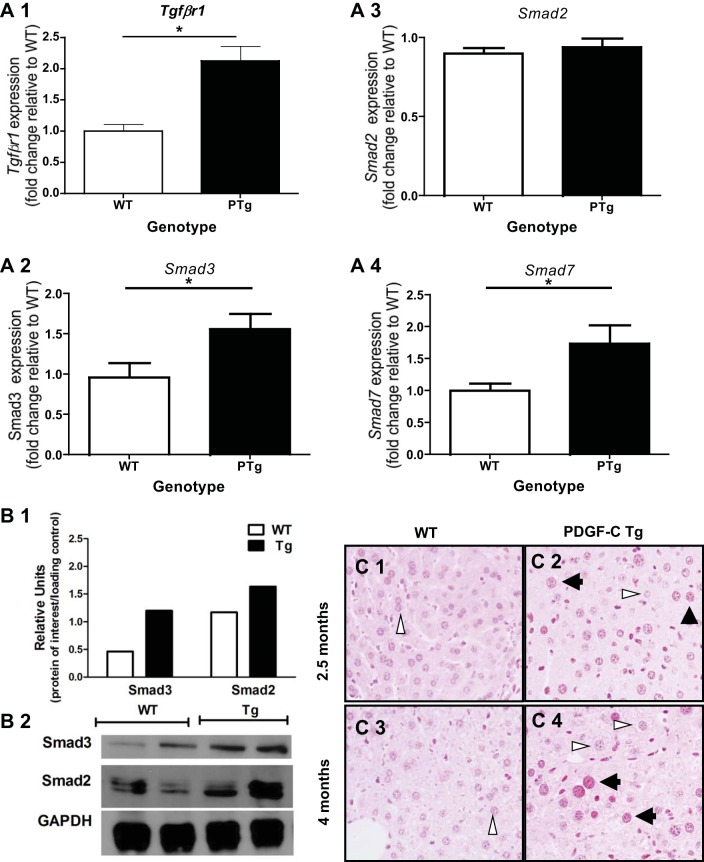
PTg mice develop pericellular and perivenular hepatic fibrosis that progresses to bridging fibrosis as the mice age (7, 8). As TGFβ is a potent profibrotic molecule in many different tissues (13, 16), we hypothesized that TGFβ signaling pathways contributed to the development and progression of fibrosis observed in this model. Previously, we observed that Tgfβ1 mRNA was elevated in PDGF-C PTg mice compared with WT littermates (7). Additionally, cDNA microarray analysis revealed that genes in the TGFβ signaling pathway, including several Smads, were significantly upregulated in liver tissue from PDGF-C PTg relative to WT mice (62) (GEO no. GSE38199). To provide further evidence of TGFβ signaling, we performed real-time PCR, immunoblot and immunohistochemical (IHC) analyses on liver tissue from 8- to 9-wk-old WT and PTg mice (Fig. 1). We observed elevated levels of TGFβ1 receptor 1 (TgfβrI) mRNA (Fig. 1, A1), Smad3 (Fig. 1, A2), but not Smad2 (Fig. 1, A3) in PDGF-C PTg compared with WT mice. In addition, expression of Smad7, a transcriptional target of Smad2/3, was also induced (Fig. 1,A4), demonstrating that components of the TGFβ signaling pathway were present in livers of PDGF-C PTg mice.
Fig. 1.
Evidence of transforming growth factor-β (TGFβ) signaling in livers of PDGF-C transgenic (PTg) mice. Analysis of gene expression (A), protein (B), and phosphorylated Smad2/3 (C) was performed using livers from PTg mice compared with wild-type (WT) mice (8–9 wk of age, n = 6–7). A: TGFβ1 receptor (Tgfβr1) (A1), Smad3 (A2), and Smad7 (A4) gene expression is higher in PTg mice, while Smad2 (A3) expression did not change. Total RNA was prepared from livers of PTg or WT littermates. Real-time PCR was performed to determine mRNA levels of Tgfβr1, Smad2, Smad3, and Smad7 as described in materials and methods. Data are represented as fold change compared with a WT animal. B: immunoblot analysis of Smad2 and Smad3 indicates that Smad3 is elevated in PTg mice liver tissue, while Smad2 levels were similar to WT mice. The bar graph represents the average of densitometry value for two animals after normalization to GAPDH levels. C: phosphorylated Smad2/3 is present in hepatocyte nuclei, and nonparenchymal cells (NPCs) in livers from PTg (C2 and C4), but not WT (C1 and C3) mice. Immunohistochemical analysis was performed on 2-mo-old (C1 and C2), and 4.5-mo-old (C3 and C4) mice as described in materials and methods. Both stained (black arrowheads) and unstained (white arrowheads) hepatocyte nuclei can be seen in the same field. *P < 0.05.
Immunoblot analysis of Smad2 and Smad3 showed increased protein levels of Smad3, but not Smad2 in PTg mice (Fig. 1B). In addition, phosphorylated Smad2/3-positive staining was detected by IHC in PTg mice (Fig. 1, C2 and C4), whereas little nuclear staining was appreciated in liver tissue from WT mice (Fig. 1, C1 and C3). Nonparenchymal cells also appeared to contain phosphorylated Smad2/3, but there was insufficient resolution to determine whether Smad2/3 had translocated into the nucleus. Together, these results suggest that TGFβ signaling pathways were active in livers from PTg mice, and that Smad3 may play a role in mediating TGFβ signaling pathways in this mouse model.
Based on these in vivo observations, we wondered whether PDGF-C could directly stimulate TGFβ production in vitro. It is well known that TGFβ is a potent stimulator of PDGF ligand production in a variety of cell types (5, 41, 46), but less is known about the ability of PDGF-C to stimulate TGFβ production. Rat CFSC-2G stellate cells (18) were exposed to PDGF-CC to determine whether this ligand stimulates TGFβ production. In this established stellate cell line, PDGF-CC treatment resulted in TGFβ1 protein release into the media with PDGF-AB, -BB and -CC ligands being more efficacious than -AA or -DD (data not shown). These results demonstrate that PDGF ligands are capable of stimulating TGFβ release in vitro in an activated myofibroblast-like cell line.
Deletion of Smad3 in PDGF-C Tg mice reverses phenotypic changes induced by PDGF-C.
If PDGF-C-induced liver fibrosis is dependent on TGFβ production and TGFβ regulates collagen production, then disruption of TGFβ signaling pathways should attenuate fibrosis in PTg mice. To test this hypothesis, we intercrossed PTg mice with mice that lack Smad3 (63), a transcription factor that is part of TGFβ canonical signaling. Based on our breeding scheme, six different genotypes were possible, and the following four genotypes were analyzed in detail: wild-type (WT, PDGF-CWT/Smad3WT), Smad3 KO (KO, PDGF-CWT/Smad3KO), PDGF-C Tg (PTg, PDGF-CTg/Smad3WT), and PDGF-C Tg; Smad3KO (PTg/KO, PDGF-CTg/Smad3KO). Over the course of these studies, we observed fewer than expected Smad3KO mice. Mendelian analysis of pups at weaning indicated that mice that lacked the Smad3 allele were born less frequently than expected (Table 1). The higher mortality of Smad3-null weanlings appears to be independent of PDGF-C transgene as PTg/KO mice were born at nearly the same frequency as KO mice.
Table 1.
Genetic analysis of offspring obtained from Smad3 KO;PDGF-C Tg breeding pairs
| Genotype* |
||||||
|---|---|---|---|---|---|---|
| Smad3WT/WT PDGF-CTg/WT | Smad3ex8/WT PDGF-CTg/WT | Smad3ex8/ex8 PDGF-CTg/WT | Smad3WT/WT PDGF-CWT/WT | Smad3ex8/WT PDGF-CWT/WT | Smad3ex8/ex8 PDGF-CWT/WT | |
| Expected number† | 26 | 51 | 26 | 26 | 51 | 26 |
| Actual number | 46 | 53 | 4 | 31 | 66 | 6 |
| Expected percentage† | 12 | 25 | 12 | 12 | 25 | 12 |
| Actual percentage | 22 | 26 | 2 | 15 | 32 | 3 |
χ2 Analysis was performed on genotypes from 206 mice from four different sets or breeders to generate a χ2 value of 15.59. As this number is >11.07 (the value for five degrees of freedom), this analysis indicates that fewer Smad3-null mice are born than expected.
Genotypes were obtained from pups born to breeder pairs consisting of female Smad3ex8/WT;PDGF- CWT/WT and male Smad3ex8/WT;PDGF-CTgWT/WT mice.
These values are the expected numbers of pups of each genotype based on Mendelian ratios. Bold values indicate the lower number of Smad3-null mice that were born.
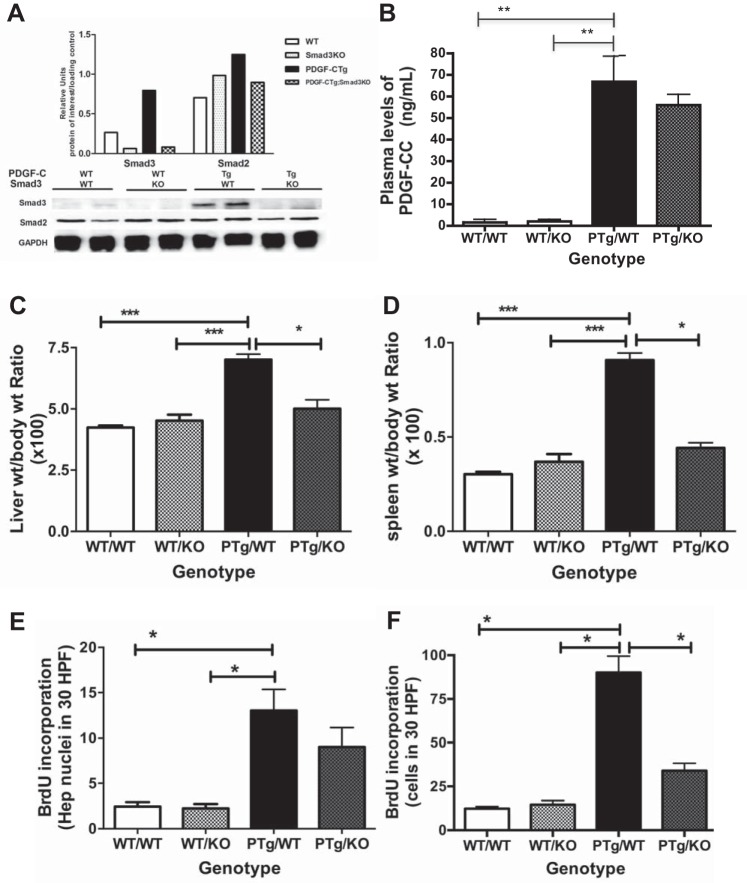
WT, KO, PTg, and PTg;KO mice were killed and analyzed to determine the effects of Smad3 deletion on PDGF-C-induced changes in the liver (Fig. 2). We first examined whether deletion of Smad3 resulted in loss of Smad3 protein. Immunoblot analysis of liver lysates from all four genotypes showed that PTg mice had the highest levels of Smad3 protein, approximately twice the amount seen in WT mice (Fig. 2A). Smad3 protein was not detectable in lysates from KO or PTg/KO mice. Smad2 protein levels did not change with overexpression of PDGF-C or deletion of Smad3 (Fig. 2A), and SMAD4 protein levels were also unchanged (data not shown). These results indicate that PTg/KO mice are deficient for Smad3, but have similar levels of Smad2 and Smad4 as WT, KO, and PTg mice.
Fig. 2.
Deletion of Smad3 attenuates PDGF-C-induced effects in the liver. PTg mice were intercrossed to Smad3 KOex8/ex8, and liver tissue from four groups [wild-type (WT/WT), Smad3 KO (WT/KO), PDGF-C Tg (PTg/WT), and PDGF-C Tg;Smad3 KO (PTg/KO)] was analyzed for Smad2 and Smad3 protein levels (A), circulating levels of human PDGF-CC (B), liver weight-to-body weight ratio (C), spleen weight-to-body weight ratio (D), and hepatocyte (Hep, E) and NPC (F) proliferation (2–5 mo of age, n = 4–5). BrdU, bromodeoxyuridine; HPF, high-power field. *P < 0.05; **P < 0.01; ***P < 0.001 (Mann-Whitney).
PTg mice overexpress the full-length human PDGF-C transgene with an intact CUB domain. For PDGF-C to activate PDGF receptors, the CUB domain must be cleaved from the growth factor domain by extracellular proteases (48, 62). To evaluate circulating levels of human PDGF-CC by ELISA, plasma was collected from mice representing all four genotypes. Only PTg mice had circulating levels of active ligand, and no significant differences were seen between PTg and PTg/KO mice (Fig. 2B). Thus, changes in transgene expression do not appear to account for differences in observed liver phenotypes.
PTg mice have greater liver weight-to-body weight ratios than WT littermates (17), and systemic Smad3 deletion resulted in a statistically significant decrease in liver weight/body weight in PTg mice (Fig. 2C). PDGF-C overexpression also increases the spleen weight relative to body weight, a phenotype that was also partially reversed by deletion of Smad3 (Fig. 2D). Liver cell proliferation is enhanced in PTg mice (7, 8); deletion of Smad3 decreased both hepatocyte (Fig. 2E) and NPC proliferation (Fig. 2F) as measured by BrdU incorporation. No differences in liver cell proliferation were observed between WT and KO mice. These results indicate that TGFβ/Smad3-dependent pathways are in part responsible for prominent cellular changes seen in PTg mice. Moreover, these data indicate that Smad3 signaling is “downstream” of PDGF-C signaling in this model, as loss of Smad3 partially reverses PDGF-C-induced changes in the liver.
Smad3-null HSCs proliferate slowly.
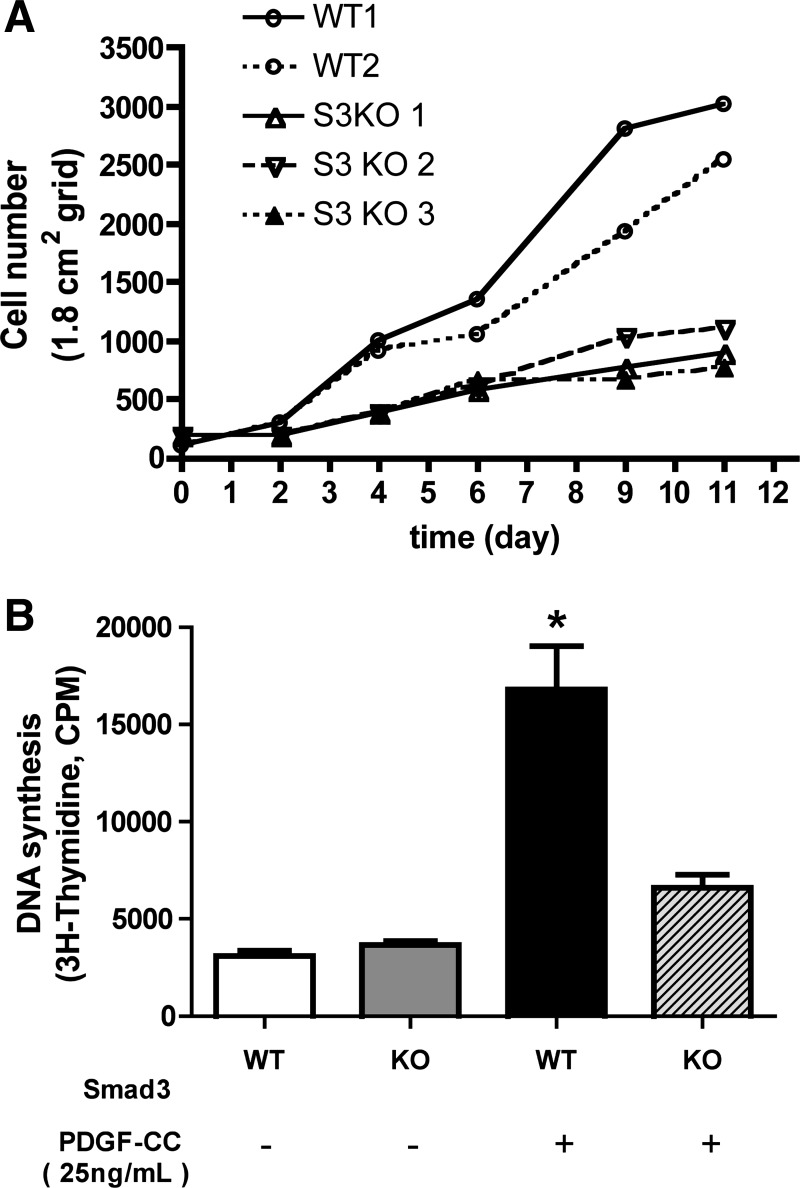
In this study we have shown that NPC proliferation in vivo was significantly decreased in PTg/KO mice compared with PTg mice (Fig. 2F). We thus wondered whether isolated primary HSCs from KO mice would be differentially affected by TGFβ (Fig. 3). In vitro, TGFβ has different effects on cell growth depending on a variety of factors, including the embryologic origin of the cell and its degree of activation or transformation. Mesenchymal cells such as myofibroblasts proliferate or trans-differentiate in response to TGFβ (5, 46, 59). To evaluate the effect of Smad3 gene deletion on HSC proliferation, HSCs were isolated from both WT and KO mice, and cell proliferation was assessed. Primary HSCs isolated from KO mice grew more slowly on plastic dishes than WT HSCs (Fig. 3A). WT cells doubled in approximately 2 days, while Smad3 KO HSCs took 4 days, nearly twice as long to double in number. Nonetheless Smad3-null HSCs were able to proliferate, albeit slowly. To evaluate the effect of Smad3 gene deletion on PDGF-CC-stimulated HSC proliferation, we next plated cultured primary HSCs from WT and Smad3 KO mice on plastic for 7 days, which allowed them to become activated and myofibroblast-like. After removing the serum, the cultures were exposed to PDGF-CC and cell proliferation was measured. PDGF-CC-induced HSC proliferation was significantly attenuated in Smad3-null HSCs (Fig. 3B). Thus Smad3-null HSCs grow more slowly than WT HSCs in the presence of either serum or PDGF-CC in culture. These in vitro results mirror the decreased NPC proliferation observed in vivo in livers of PTg/KO mice (Fig. 2F).
Fig. 3.
Effects of Smad3 deletion on [3H]thymidine incorporation in hepatic satellite cells (HSCs) in vitro. Effect of Smad3 deletion in HSCs was evaluated using primary HSCs isolated from both WT and Smad3 KO (KO) mice in serum-containing media (A) and after PDGF-CC stimulation (B). A: isolated primary Smad3-null HSCs grow more slowly than WT HSCs. Isolated cells were plated in serum-containing media, and cells were counted on days 0, 2, 4, 6, 9, 11. On day 11, WT HSC cultures were confluent; Smad3 KO cultures were not. Each line represents HSCs from individual mice. S3KO, Smad3 knockout. B: PDGF-CC-induced HSC [3H]thymidine incorporation is significantly decreased in Smad3-null HSCs. Isolated HSCs were cultured for 7 days, serum starved, and then either treated with PDGF-CC (25 ng/ml) or left untreated. [3H]thymidine incorporation was used to assess cell proliferation. These data are representative of two independent experiments. *P < 0.05.
Deletion of Smad3 ameliorates PDGF-C-induced liver fibrosis.
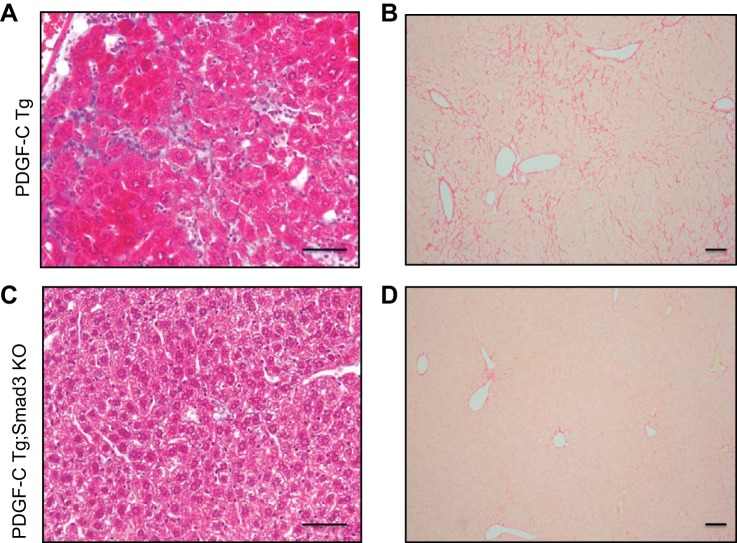
To investigate the role of Smad3 on PDGF-C-induced liver fibrosis, collagen deposition was examined in fixed liver tissues using Masson's trichrome and Picrosirius Red staining (Fig. 4). Architectural derangement of liver and deposition of fibrous filaments was apparent in PTg mouse specimens, but it was diminished in PTg/KO mice (Fig. 4, C and D). Collagen staining was attenuated in 3-mo-old PDGF-C Tg;Smad3 KO mice compared with PDGF-C Tg mice as detected by trichrome (Fig. 4, A and C) or Picrosirius Red (Fig. 4, B and D) staining. Liver specimens from WT and Smad3 KO mice had little collagen deposition, which was less than that seen in PDGF-C Tg;Smad3 KO mice. Differences in collagen deposition were not apparent between WT and Smad3 KO strains (data not shown).
Fig. 4.
PDGF-C Tg;Smad3 KO mice have decreased collagen deposition compared with PTg mice. Liver tissue from PDGF-C Tg and PDGF-C Tg;Smad3 KO mice was stained for collagen deposition using Masson's trichrome (A and C, 4 mo of age) and Picrosirius Red (B and D, 3 mo of age). The extensive collagen deposition seen in PDGF-C Tg mice (A and B) is decreased in PDGF-C Tg;Smad3 KO mice. Scale bar, 50 μm.
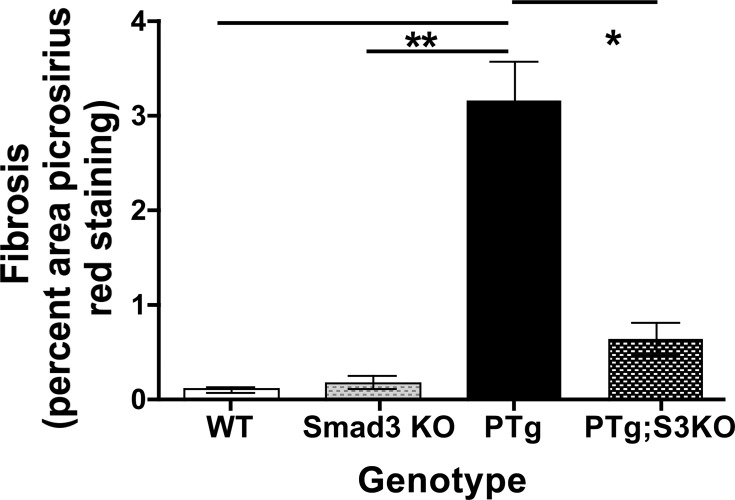
To quantify the differences in collagen deposition, we performed morphometric analysis on Picrosirius Red-stained liver tissue from WT, KO, PTg, and PTg/KO mice (Fig. 5). Liver fibrosis was significantly decreased in PTg/KO mice compared with PDGF-C PTg mice. Loss of Smad3 in PTg mice did not fully reverse the collagen deposition to the levels seen in uninjured WT and KO mice, however. Together, these results indicate that TGFβ/Smad3 signaling pathways partially regulate drive PDGF-C-induced fibrogenesis.
Fig. 5.
Fibrosis is significantly decreased in PDGF-C Tg;Smad3 KO mice. Liver tissue from WT, Smad3 KO, PTg, and PTg/KO mice was stained for collagen deposition using Picrosirius Red staining, and morphometric analysis was done as described in materials and methods (2–5 mo of age, n = 4–5). *P < 0.05; **P < 0.01.
Smad3 regulates hepatic gene expression in PDGF-C Tg mice.
Smad3, a critical component of the canonical TGFβ signaling pathway, transcriptionally regulates expression of genes that contributes to fibrosis. To assess the consequences of Smad3 deletion on PDGF-C-induced changes in gene expression, we performed real-time RT PCR analyses using whole liver RNA from WT, KO, PTg, and PTg/KO mice (Fig. 6). PDGF-C overexpression induces collagen α1(I) (Col1a1), collagen α2(I) (Col2a1), and collagen α1(IV) (Col4a1) expression in the livers of PTg mice, but this induction was blocked in PTg/KO mice (Fig. 6A). Increased smooth muscle α-actin (αSMA, Acta2) is a hallmark of liver fibrosis and is associated with HSC activation. PTg/KO mice have less Acta2 mRNA than PTg mice. Connective tissue growth factor (CTGF, Ctgf) is regulated by TGFβ and is implicated in the regulation of a number of collagen genes (40, 47). PDGF-C overexpression upregulates Ctgf expression, which is attenuated in PTg/KO mice.
Fig. 6.
Loss of Smad3 blunts induction of genes involved in collagen gene expression, but not genes associated with HSC activation. RNA was isolated from liver tissue from WT, Smad3 KO (KO), PDGF-C Tg (Tg), and PDGF-C Tg;Smad3 KO (Tg/KO) mice (2–5 mo of age, n = 4–5). Changes in gene expression were determined using real-time RT PCR normalized to Gapdh as described in materials and methods. Average delta Ct values ± SE are shown for each gene analyzed, and data are displayed for WT (open bars), KO (speckled bars); Tg (black bars), and Tg/SKO (hatched bars) mice. Genes whose expression is decreased by loss of Smad3 (Smad3-dependent genes) (A) and genes that did not show a significant difference when Smad3 is deleted (i.e., Tg compared with Tg/KO mice) (B) are shown. *P < 0.05.
PDGF-C Tg mice induce hepatic genes that are Smad3 independent.
We have previously reported that overexpression of PDGF-C results in the induction of number genes, including Tgfβ1 and the PDGF receptors Pdgfrα and Pdgfrβ (7, 62). In these experiments, we observed a similar induction of these genes in PTg mice, but their expression was not significantly changed when Smad3 was deleted in PTg/KO mice (Fig. 6B). These results suggest that Smad3-independent- or additional signaling pathways regulate cellular retinol binding protein 1 (Crbp1), Tgfβ1, Pdgfrα, and Pdgfrβ gene expression in the PTg liver. Since these genes are often associated with HSC activation and/or proliferation (25, 26, 43, 59), our results suggest that HSC activation may be dependent on PDGF-C-mediated pathways to a greater extent than those mediated by Smad3.
DISCUSSION
TGFβ and PDGF signaling pathways are important regulators of liver fibrosis. However, how these signaling pathways collaborate in liver fibrogenesis is not well understood. The major finding of this study is that PDGF-C-induced hepatic fibrosis is dependent on TGFβ signaling that is regulated by Smad3. Overexpression of PDGF-C results in activation of HSCs, and fibrosis, which increases with severity as Tg mice age. As Tgfβ1 levels are elevated in PDGF-C Tg mice compared with WT mice, we suspected that part of the fibrotic potency of PDGF-C was due to activation of TGFβ signaling pathways. Using a genetic approach, we showed that PTg mice that lack Smad3 have decreased liver and spleen mass, and less collagen deposition compared with PTg mice with intact Smad3. Using in vitro approaches we show that PDGF ligands have the capability to stimulate TGFβ production. These data suggest the following sequence of events in PDGF-C-induced fibrosis. PDGF-C induces Tgfβ expression, which drives the expression and deposition of collagen, resulting in fibrosis. In addition to TGFβ's effects on HSCs, PDGF-C also directly stimulates the proliferation and activation of HSCs.
Our findings are consistent with previous studies in liver fibrosis indicating that Smad3 is a key regulator of collagen deposition. Schnabl and coworkers (51) showed that Col1a1 expression was decreased, while αSMA expression was not after acute carbon tetrachloride (CCl4) injury in Smad3KOexon1 mice. In the present study, Smad3 deletion had a more profound effect on collagen deposition than αSMA expression (Figs. 4 and 5), consistent with reports of less fibrosis in a dimethlynitrosamine model of liver fibrosis using Smad3KOex8/ex8 mice (30). Smad3 KO mice have altered mechano-transduction properties due, in part, to changes in ECM production (2), which are supported by in vitro studies demonstrating that Smad3 regulates cytoskeletal organization (55). Not surprisingly, the role of Smad3 in fibrosis or cirrhosis also depends on the type of fibrotic injury (28), and possibly the underlying etiology in human fibrosis.
It is interesting to note that genetic Smad3 deletion altered the expression of Ctgf, but not Tgfβ in PTg livers. In this model it is surprising that Tgfβ expression was not significantly decreased in PTg/KO mice, because this profibrotic molecule is regulated in an autocrine manner (14, 43), and thus would have been expected to be “downstream” of Smad3. Our studies show that collagen levels decreased despite unchanged levels of Tgfβ gene expression. In our studies, decreased Ctgf expression correlated with decreased collagen gene expression and collagen deposition, suggesting that CTGF is a key fibrogenic regulator that is dependent on Smad3. However, recent studies have demonstrated that Ctgf induction is Smad3 independent, a relationship that may depend on the cell type or injury model that is used (19, 20, 33).
In vivo Smad2 and Smad3 have independent functions as revealed by genetic deletion studies (63). Deletion of Smad2 is embryonic lethal, while Smad3 deletion is permissive during development, regardless of the targeting construct (10, 63, 66). Similarly, Smad3 has separate, nonoverlapping functions from Smad2 in hepatic fibrogenesis and HSC activation. In hepatocytes, Smad2 represses cell growth after injury, while Smad3 blocks apoptosis (26). In the present studies, we did not find evidence that Smad2 compensated for loss of Smad3. Thus, Smad2 and Smad3 regulate different physiological functions despite being 87% similar in amino acid sequence.
Smad3 deletion did not fully reverse effects of PDGF-C overexpression, however. This result was not unexpected as TGFβ stimulates a number of intracellular signaling pathways that are Smad independent (11, 34, 38). Smad3-independent pathways that play key regulatory roles in HSC activation (21, 60) include PDGF receptor-mediated (1, 5), and extracellular fibular collagen and ECM signaling pathways.
Investigation of TGFβ signaling pathways in vitro is complicated by the evolving phenotype of primary HSCs when cultured (32). Isolated HSC become “activated” when disaggregated from the liver and cultured, resulting in changes in Smad2 and Smad3 expression, which impacts their phenotype (32, 55). HSC expression of PDGF ligand and receptor depends on the degree of injury in the liver at the time of isolation, and on the length of time in culture (6). In our study, deletion of Smad3 resulted in decreased proliferation and thymidine incorporation in HSCs. Previously published studies with Smad3 KO cells demonstrated dramatic effects of TGFβ on cell proliferation where myofibroblast proliferation was enhanced after serum or PDGF-BB treatment when Smad3 (exon1) was deleted (51). In contrast, we observed slower cell growth and decreased proliferation after PDGF-CC treatment (Fig. 3). It is important to note that in vitro phenotypes with Smad2- and Smad3-null cells do not recapitulate in vivo phenotypes from mice that lack Smad2 and Smad3 (43, 56, 65). For example, Smad2 compensates for Smad3 in vitro, which is not seen during development.
The major cell types in the liver produce and respond to TGFβ ligands in a manner that is dependent on degree of inflammation and stage of disease. For example, TGFβ functions as a cell cycle inhibitor, consistent with its role as a terminator of liver regeneration, in hepatocytes when inflammation and fibrosis are not apparent. HSCs produce collagen and ECM. In the context of hepatic inflammation and injury, most liver cells produce Tgfβ, including activated HSCs, hepatocytes, and Kupffer cells (24). Cell-specific autocrine and paracrine consequence(s) of myofibroblasts-derived versus Kupffer-derived Tgfβ has not yet been explored. As TGFβ signaling is cell specific and context dependent (37), we do not know whether cell specific deletion of Smad3 would yield the same results as systemic deletion.
An unanswered question from our studies is whether loss of TGFβ/Smad3 signaling would significantly delay hepatocellular carcinogenesis in PTg mice. A long-standing belief is that decreasing fibrosis will delay the onset of tumor development as cirrhotic livers hold the highest risk factor for developing HCCs in humans. As development of HCCs is preceded by severe fibrosis in PTg mice, a logical extension of this study would be to analyze tumorigenesis. However, few Smad3 KO mice survived beyond 5 mo of age, and most died between 3 and 4 mo of age. Smad3 KO mice were smaller than WT or PTg littermates and presented with a range of health issues, including malocclusion, mandible abscesses (64), penile prolapse, and cachexia (data not shown). High morbidity and early mortality has been reported for all Smad3 KO strains, independent of the targeting construct (10, 63, 66). Smad3 KO mice with exon 8 deletion on mixed 129 backgrounds were smaller than their littermates and had higher incidence of inflammatory lesions. In the present study, offspring resulting from Smad3 KOex8/ex8 mice crossed with PDGF-C Tg mice (i.e., PTg/KO mice) had similar mortality as mice that only lacked the Smad3 allele. While it appears that the PDGF-C transgene does not appear to modify the effects of the systemic deletion of Smad3, the high mortality of these mice precluded analysis of hepatocellular carcinogenesis, which develops around 6 to 7 mo of age. Thus, it remains to be determined whether attenuation of fibrosis by blocking TGFβ signaling via Smad3 would result in decreased incidence or prevalence of HCC in this model.
The present study, along with many other studies, provides strong evidence that targeting TGFβ signaling in chronic liver disease would ameliorate liver fibrosis or cirrhosis if selective therapies could be developed. This antifibrotic approach continues to be revised in light of recent studies and previous attempts to block TGFβ signaling in preclinical models (49). The challenge of blocking TGFβ signaling is that this cytokine has cell-specific roles that continually change depending on disease type and stage, which may lead to unanticipated side effects (4). Moreover, our studies suggest that PDGF-C regulates TGFβ signaling pathways and that block of both these signaling pathways may be required for effective long-term therapy for chronic liver disease and prevention of HCC.
GRANTS
This work was supported by the National Institutes of Health (NIH) National Heart, Lung, and Blood Institute, Cardiovascular Pathology Training Grant NIH-HL-007312 (to B. J. Hayes), Herbert Coe Foundation, American Surgical Association Foundation, and the American College of Surgeons (to K. J. Riehle), National Cancer Institute, Mechanisms of PDGF-C induced HCC Grant NIH-CA127228 (to J. S. Campbell).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
J.I.L., J.H.W., K.J.R., and J.S.C. conception and design of research; J.I.L., J.H.W., M.M.J., R.L.B., K.S., S.Y., B.J.H., L.N., and J.S.C. performed experiments; J.I.L., J.H.W., M.M.J., R.L.B., K.S., S.Y., B.J.H., L.N., and J.S.C. analyzed data; J.I.L., J.H.W., L.N., and J.S.C. interpreted results of experiments; J.I.L., R.L.B., and J.S.C. prepared figures; J.I.L. and J.S.C. drafted manuscript; J.I.L., J.H.W., R.L.B., K.S., B.J.H., K.J.R., and J.S.C. edited and revised manuscript; J.I.L., J.H.W., R.L.B., K.S., S.Y., B.J.H., L.N., K.J.R., and J.S.C. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Anita Roberts for providing the Smad3 KO mice, and Drs. Anthony Parks, William Grady, Nelson Fausto, and Deb Gilbertson for insightful discussions and valuable comments on the manuscript.
Present addresses: B. Hayes, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, WA 98109 (e-mail: hayesb2uw@gmail.com); L. Nguyen, Department of Pathology, University of Pittsburgh, 200 Lothrop Street, Pittsburgh, PA 15238 (e-mail: nguyenl@upmc.edu); J. S. Campbell, OncoSec Medical Inc, 5820 Nancy Ridge Rd, San Diego, CA 92121 (e-mail: jcampbell@oncosec.com).
Footnotes
This article is the topic of an Editorial Focus by Wonhyo Seo and Won-Il Jeong (51a).
REFERENCES
- 1.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev 22: 1276–1312, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arany PR, Flanders KC, Kobayashi T, Kuo CK, Stuelten C, Desai KV, Tuan R, Rennard SI, Roberts AB. Smad3 deficiency alters key structural elements of the extracellular matrix and mechanotransduction of wound closure. Proc Natl Acad Sci USA 103: 9250–9255, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argast GM, Campbell JS, Brooling JT, Fausto N. Epidermal growth factor receptor transactivation mediates tumor necrosis factor-induced hepatocyte replication. J Biol Chem 279: 34530–34536, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Biernacka A, Dobaczewski M, Frangogiannis NG. TGF-beta signaling in fibrosis. Growth Factors 29: 196–202, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonner JC. Regulation of PDGF and its receptors in fibrotic diseases. Cytokine Growth Factor Rev 15: 255–273, 2004. [DOI] [PubMed] [Google Scholar]
- 6.Borkham-Kamphorst E, Kovalenko E, van Roeyen CR, Gassler N, Bomble M, Ostendorf T, Floege J, Gressner AM, Weiskirchen R. Platelet-derived growth factor isoform expression in carbon tetrachloride-induced chronic liver injury. Lab Invest 88: 1090–1100, 2008. [DOI] [PubMed] [Google Scholar]
- 7.Campbell JS, Hughes SD, Gilbertson DG, Palmer TE, Holdren MS, Haran AC, Odell MM, Bauer RL, Ren HP, Haugen HS, Yeh MM, Fausto N. Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc Natl Acad Sci USA 102: 3389–3394, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell JS, Johnson MM, Bauer RL, Hudkins KL, Gilbertson DG, Riehle KJ, Yeh MM, Alpers CE, Fausto N. Targeting stromal cells for the treatment of platelet-derived growth factor C-induced hepatocellular carcinogenesis. Differentiation 75: 843–852, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Czochra P, Klopcic B, Meyer E, Herkel J, Garcia-Lazaro JF, Thieringer F, Schirmacher P, Biesterfeld S, Galle PR, Lohse AW, Kanzler S. Liver fibrosis induced by hepatic overexpression of PDGF-B in transgenic mice. J Hepatol 45: 419–428, 2006. [DOI] [PubMed] [Google Scholar]
- 10.Datto MB, Frederick JP, Pan L, Borton AJ, Zhuang Y, Wang XF. Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol Cell Biol 19: 2495–2504, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425: 577–584, 2003. [DOI] [PubMed] [Google Scholar]
- 12.Dooley S, ten Dijke P. TGF-beta in progression of liver disease. Cell Tissue Res 347: 245–256, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffield JS, Lupher M, Thannickal VJ, Wynn TA. Host responses in tissue repair and fibrosis. Annu Rev Pathol 8: 241–276, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flanders KC. Smad3 as a mediator of the fibrotic response. Int J Exp Pathol 85: 47–64, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology 134: 1655–1669, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman SL, Sheppard D, Duffield JS, Violette S. Therapy for fibrotic diseases: nearing the starting line. Sci Transl Med 5: 167sr161, 2013. [DOI] [PubMed] [Google Scholar]
- 17.Gilbertson DG, Duff ME, West JW, Kelly JD, Sheppard PO, Hofstrand PD, Gao Z, Shoemaker K, Bukowski TR, Moore M, Feldhaus AL, Humes JM, Palmer TE, Hart CE. Platelet-derived growth factor C (PDGF-C), a novel growth factor that binds to PDGF alpha and beta receptor. J Biol Chem 276: 27406–27414, 2001. [DOI] [PubMed] [Google Scholar]
- 18.Greenwel P, Rubin J, Schwartz M, Hertzberg EL, Rojkind M. Liver fat-storing cell clones obtained from a CCl4-cirrhotic rat are heterogeneous with regard to proliferation, expression of extracellular matrix components, interleukin-6, and connexin 43. Lab Invest 69: 210–216, 1993. [PubMed] [Google Scholar]
- 19.Gressner OA, Lahme B, Demirci I, Gressner AM, Weiskirchen R. Differential effects of TGF-beta on connective tissue growth factor (CTGF/CCN2) expression in hepatic stellate cells and hepatocytes. J Hepatol 47: 699–710, 2007. [DOI] [PubMed] [Google Scholar]
- 20.Gressner OA, Lahme B, Siluschek M, Rehbein K, Herrmann J, Weiskirchen R, Gressner AM. Activation of TGF-beta within cultured hepatocytes and in liver injury leads to intracrine signaling with expression of connective tissue growth factor. J Cell Mol Med 12: 2717–2730, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson NC, Arnold TD, Katamura Y, Giacomini MM, Rodriguez JD, McCarty JH, Pellicoro A, Raschperger E, Betsholtz C, Ruminski PG, Griggs DW, Prinsen MJ, Maher JJ, Iredale JP, Lacy-Hulbert A, Adams RH, Sheppard D. Targeting of alphav integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med 19: 1617–1624, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson NC, Sheppard D. Integrin-mediated regulation of TGFbeta in fibrosis. Biochim Biophys Acta 1832: 891–896, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol 6: 425–456, 2011. [DOI] [PubMed] [Google Scholar]
- 24.Heymann F, Trautwein C, Tacke F. Monocytes and macrophages as cellular targets in liver fibrosis. Inflamm Allergy Drug Targets 8: 307–318, 2009. [DOI] [PubMed] [Google Scholar]
- 25.Inagaki Y, Okazaki I. Emerging insights into transforming growth factor beta Smad signal in hepatic fibrogenesis. Gut 56: 284–292, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ju W, Ogawa A, Heyer J, Nierhof D, Yu L, Kucherlapati R, Shafritz DA, Bottinger EP. Deletion of Smad2 in mouse liver reveals novel functions in hepatocyte growth and differentiation. Mol Cell Biol 26: 654–667, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanzler S, Lohse AW, Keil A, Henninger J, Dienes HP, Schirmacher P, Rose-John S, zum Buschenfelde KH, Blessing M. TGF-β1 in liver fibrosis: an inducible transgenic mouse model to study liver fibrogenesis. Am J Physiol Gastrointest Liver Physiol 276: G1059–G1068, 1999. [DOI] [PubMed] [Google Scholar]
- 28.Khimji AK, Shao R, Rockey DC. Divergent transforming growth factor-beta signaling in hepatic stellate cells after liver injury: functional effects on ECE-1 regulation. Am J Pathol 173: 716–727, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kisseleva T, Brenner DA. Role of hepatic stellate cells in fibrogenesis and the reversal of fibrosis. J Gastroenterol Hepatol 22, Suppl 1: S73–S78, 2007. [DOI] [PubMed] [Google Scholar]
- 30.Latella G, Vetuschi A, Sferra R, Catitti V, D'Angelo A, Zanninelli G, Flanders KC, Gaudio E. Targeted disruption of Smad3 confers resistance to the development of dimethylnitrosamine-induced hepatic fibrosis in mice. Liver Int 29: 997–1009, 2009. [DOI] [PubMed] [Google Scholar]
- 31.Lim YS, Kim WR. The global impact of hepatic fibrosis and end-stage liver disease. Clin Liver Dis 12: 733–746, vii, 2008. [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Gaca MD, Swenson ES, Vellucci VF, Reiss M, Wells RG. Smads 2 and 3 are differentially activated by transforming growth factor-beta (TGF-beta) in quiescent and activated hepatic stellate cells. Constitutive nuclear localization of Smads in activated cells is TGF-beta-independent. J Biol Chem 278: 11721–11728, 2003. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Liu H, Meyer C, Li J, Nadalin S, Konigsrainer A, Weng H, Dooley S, ten Dijke P. Transforming growth factor-beta (TGF-beta)-mediated connective tissue growth factor (CTGF) expression in hepatic stellate cells requires Stat3 signaling activation. J Biol Chem 288: 30708–30719, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lonn P, Moren A, Raja E, Dahl M, Moustakas A. Regulating the stability of TGFbeta receptors and Smads. Cell Res 19: 21–35, 2009. [DOI] [PubMed] [Google Scholar]
- 35.Maggio-Price L, Treuting P, Zeng W, Tsang M, Bielefeldt-Ohmann H, Iritani BM. Helicobacter infection is required for inflammation and colon cancer in SMAD3-deficient mice. Cancer Res 66: 828–838, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maschmeyer P, Flach M, Winau F. Seven steps to stellate cells. J Vis Exp: 2710. doi: 10.3791/2710, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuzaki K. Smad phospho-isoforms direct context-dependent TGF-beta signaling. Cytokine Growth Factor Rev 24: 385–399, 2013. [DOI] [PubMed] [Google Scholar]
- 38.Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development 136: 3699–3714, 2009. [DOI] [PubMed] [Google Scholar]
- 39.Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci 114: 4359–4369, 2001. [DOI] [PubMed] [Google Scholar]
- 40.Paradis V, Dargere D, Bonvoust F, Vidaud M, Segarini P, Bedossa P. Effects and regulation of connective tissue growth factor on hepatic stellate cells. Lab Invest 82: 767–774, 2002. [DOI] [PubMed] [Google Scholar]
- 41.Parola M, Marra F, Pinzani M. Myofibroblast-like cells and liver fibrogenesis: Emerging concepts in a rapidly moving scenario. Mol Aspects Med 29: 58–66, 2008. [DOI] [PubMed] [Google Scholar]
- 42.Pellicoro A, Ramachandran P, Iredale JP. Reversibility of liver fibrosis. Fibrogenesis Tissue Repair 5, Suppl 1: S26, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piek E, Roberts AB. Suppressor and oncogenic roles of transforming growth factor-beta and its signaling pathways in tumorigenesis. Adv Cancer Res 83: 1–54, 2001. [DOI] [PubMed] [Google Scholar]
- 44.Pinzani M. Platelet-derived growth factor receptor expression in hepatic stellate cells: how too much of a good thing can be bad. Hepatology 22: 997–999, 1995. [DOI] [PubMed] [Google Scholar]
- 45.Pinzani M, Vizzutti F. Fibrosis and cirrhosis reversibility: clinical features and implications. Clin Liver Dis 12: 901–913, 2008. [DOI] [PubMed] [Google Scholar]
- 46.Puche JE, Saiman Y, Friedman SL. Hepatic stellate cells and liver fibrosis. Compr Physiol 3: 1473–1492, 2013. [DOI] [PubMed] [Google Scholar]
- 47.Rachfal AW, Brigstock DR. Connective tissue growth factor (CTGF/CCN2) in hepatic fibrosis. Hepatol Res 26: 1–9, 2003. [DOI] [PubMed] [Google Scholar]
- 48.Riehle KJ, Johnson MM, Johansson F, Bauer RL, Hayes BJ, Gilbertson DG, Haran AC, Fausto N, Campbell JS. Tissue-type plasminogen activator is not necessary for platelet-derived growth factor-c activation. Biochim Biophys Acta 1842: 318–325, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rockey DC. Antifibrotic therapy in chronic liver disease. Clin Gastroenterol Hepatol 3: 95–107, 2005. [DOI] [PubMed] [Google Scholar]
- 50.Sanderson N, Factor V, Nagy P, Kopp J, Kondaiah P, Wakefield L, Roberts AB, Sporn MB, Thorgeirsson SS. Hepatic expression of mature transforming growth factor beta 1 in transgenic mice results in multiple tissue lesions. Proc Natl Acad Sci USA 92: 2572–2576, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schnabl B, Kweon YO, Frederick JP, Wang XF, Rippe RA, Brenner DA. The role of Smad3 in mediating mouse hepatic stellate cell activation. Hepatology 34: 89–100, 2001. [DOI] [PubMed] [Google Scholar]
- 51a.Seo W, Jeong WI. Novel insight into a platelet-derived growth factor-C/Smad3 axis in liver fibrosis. Focus on “Role of Smad3 in platelet-derived growth factor-C-induced liver fibrosis.” Am J Physiol Cell Physiol. 10.1152/ajpcell.00369.2015. [DOI] [PMC free article] [PubMed]
- 52.Severi T, van Malenstein H, Verslype C, van Pelt JF. Tumor initiation and progression in hepatocellular carcinoma: risk factors, classification, and therapeutic targets. Acta Pharmacol Sin 31: 1409–1420, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thieringer F, Maass T, Czochra P, Klopcic B, Conrad I, Friebe D, Schirmacher P, Lohse AW, Blessing M, Galle PR, Teufel A, Kanzler S. Spontaneous hepatic fibrosis in transgenic mice overexpressing PDGF-A. Gene 423: 23–28, 2008. [DOI] [PubMed] [Google Scholar]
- 54.Truett GE, Walker JA, Baker DG. Eradication of infection with Helicobacter spp. by use of neonatal transfer. Comp Med 50: 444–451, 2000. [PubMed] [Google Scholar]
- 55.Uemura M, Swenson ES, Gaca MD, Giordano FJ, Reiss M, Wells RG. Smad2 and Smad3 play different roles in rat hepatic stellate cell function and alpha-smooth muscle actin organization. Mol Biol Cell 16: 4214–4224, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verrecchia F, Chu ML, Mauviel A. Identification of novel TGF-beta/Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J Biol Chem 276: 17058–17062, 2001. [DOI] [PubMed] [Google Scholar]
- 57.Weiskirchen R, Gressner AM. Isolation and culture of hepatic stellate cells. Methods Mol Med 117: 99–113, 2005. [DOI] [PubMed] [Google Scholar]
- 58.Wells RG. Cellular sources of extracellular matrix in hepatic fibrosis. Clin Liver Dis 12: 759–768, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wells Fibrogenesis RG. V. TGF-beta signaling pathways. Am J Physiol Gastrointest Liver Physiol 279: G845–G850, 2000. [DOI] [PubMed] [Google Scholar]
- 60.Wells RG. Tissue mechanics and fibrosis. Biochim Biophys Acta 1832: 884–890, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong L, Yamasaki G, Johnson RJ, Friedman SL. Induction of beta-platelet-derived growth factor receptor in rat hepatic lipocytes during cellular activation in vivo and in culture. J Clin Invest 94: 1563–1569, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wright JH, Johnson MM, Shimizu-Albergine M, Bauer RL, Hayes BJ, Surapisitchat J, Hudkins KL, Riehle KJ, Johnson SC, Yeh MM, Bammler TK, Beyer RP, Gilbertson DG, Alpers CE, Fausto N, Campbell JS. Paracrine activation of hepatic stellate cells in platelet-derived growth factor C transgenic mice: evidence for stromal induction of hepatocellular carcinoma. Int J Cancer 134: 778–788, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, Gu H, Roberts AB, Deng C. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J 18: 1280–1291, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yokozeki M, Afanador E, Nishi M, Kaneko K, Shimokawa H, Yokote K, Deng C, Tsuchida K, Sugino H, Moriyama K. Smad3 is required for enamel biomineralization. Biochem Biophys Res Commun 305: 684–690, 2003. [DOI] [PubMed] [Google Scholar]
- 65.Yoshida K, Matsuzaki K. Differential regulation of TGF-beta/Smad signaling in hepatic stellate cells between acute and chronic liver injuries. Front Physiol 3: 53, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu Y, Richardson JA, Parada LF, Graff JM. Smad3 mutant mice develop metastatic colorectal cancer. Cell 94: 703–714, 1998. [DOI] [PubMed] [Google Scholar]