Abstract
Intestinal resection resulting in short bowel syndrome (SBS) carries a heavy burden of long-term morbidity, mortality, and cost of care, which can be attenuated with strategies that improve intestinal adaptation. SBS infants fed human milk, compared with formula, have more rapid intestinal adaptation. We tested the hypothesis that the major noncaloric human milk oligosaccharide 2′-fucosyllactose (2′-FL) contributes to the adaptive response after intestinal resection. Using a previously described murine model of intestinal adaptation, we demonstrated increased weight gain from 21 to 56 days (P < 0.001) and crypt depth at 56 days (P < 0.0095) with 2′-FL supplementation after ileocecal resection. Furthermore, 2′-FL increased small bowel luminal content microbial alpha diversity following resection (P < 0.005) and stimulated a bloom in organisms of the genus Parabacteroides (log2-fold = 4.1, P = 0.035). Finally, transcriptional analysis of the intestine revealed enriched ontologies and pathways related to antimicrobial peptides, metabolism, and energy processing. We conclude that 2′-FL supplementation following ileocecal resection increases weight gain, energy availability through microbial community modulation, and histological changes consistent with improved adaptation.
Keywords: short bowel syndrome, intestinal adaptation, human milk oligosaccharide, 2′-fucosyllactose
intestinal failure describes a state of insufficient enteral function in which the intestine cannot support normal fluid balance, electrolyte balance, and growth. When enteral nutrition cannot meet these needs, central venous access is required for daily hydration and nutrition. The presence of a central venous catheter and the use of parenteral nutrition results in significant morbidity, mortality, cost, and lower quality of life (35, 53, 54, 63). The most common cause of intestinal failure in the pediatric population is short bowel syndrome, due to extensive bowel resection (29). Following resection, the remaining intestine undergoes a process of adaptation presumed to compensate for the loss of absorptive surface area and restore full enteral function (12, 43, 64). Thus this process is the primary goal of intestinal rehabilitation as expeditious adaptation results in independence from central venous access and a reduction of its associated risks. Though the specific molecular mechanisms driving the adaptive response are not well understood, the major stimulus appears to be early enteral feeding (25, 60).
Although no consensus exists to suggest whether polymeric or monomeric formula is best for children with short bowel syndrome, a consistent benefit has been observed for human milk (46). Specifically, infants fed human milk achieve enteral autonomy sooner and with less morbidity than those fed formula (2, 32). Many growth factors are exclusively present in human milk and have been studied in the context of bowel resection (15). However, none have solely demonstrated clear and sustained improvement of the adaptive process, prompting investigation into other components of human milk (4). Human milk oligosaccharides are carbohydrate polymers specific to human milk that may be nonnutritive yet able to modulate intestinal epithelial maturation and function by indirect mechanisms including affecting the microbiota (1, 31, 37, 65). Thus they may be responsible for the improved adaptive response observed in infants fed human milk (2).
2′-Fucosyllactose (2′-FL) is the most abundant oligosaccharide found in human milk and is not a component of infant formulas (10, 13, 23, 56). The concentration of 2′-FL is ∼2–3 g/l in milk produced by women with an active FUT2 gene allele, who are known as “secretors” (9, 26, 57, 58). Not only has 2′-FL been shown in vivo to stimulate enterocyte maturation, but it has been shown to act in a prebiotic fashion, encouraging the growth of beneficial bacteria and discouraging the growth of pathogens (3, 31, 37, 65). Though small quantities of 2′-FL may be detected in the blood stream of children receiving secretor milk, it is indigestible and not a caloric source for mammals (14, 28). 2′-FL is attractive for further study to determine direct and indirect effects on the adaptive response to intestinal resection.
We have previously described a mouse model of adaptation following extensive ileocecal resection (ICR) (16). This model demonstrated that murine adaptation is characterized histologically by unsustained increases in villus height and crypt depth. Long-term bowel dilation and lengthening also occur, resulting in a sustained increase in mucosal surface area. Indeed, the improved intestinal function occurring as a result of the adaptive process is observed in a return to preoperative weight following resection, then further gain. In addition to anthropomorphic and histological change, we have also described postoperative changes in the small bowel microbiome (17). We observed a decrease in diversity and shift to a predominance of members of the Firmicutes phylum, specifically the Clostridiaceae family. Thus increased intestinal surface area and microbial community changes characteristic of the adaptive process following intestinal resection occur as weight, a robust marker of intestinal function, increases.
We aimed to examine the effect of 2′-FL supplementation on the adaptive response to ileocecal resection. Specifically, we described the effect of 2′-FL supplementation on a robust measure of adaptation following resection, weight change. We found supplementation with the noncaloric human milk oligosaccharide 2′-FL improved weight gain even before an impact on histological measures of adaptation was observed. Pursuant to mechanistic exploration, we further characterized the fecal microbiome and intestinal transcriptome at the site of resection. We conclude that 2′-FL supplementation augments the long-term adaptive response, not only by increasing mucosal surface area, but by augmenting microbial community shifts, which may improve food energy extraction.
MATERIALS AND METHODS
Male C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) of 8–10 wk of age were weighed and started on an exclusive polymeric formula diet 1 day prior to experiment start (Jevity 1 Cal, Abbott Nutrition, Columbus, OH). All mice were administered one dose of intraperitoneal Zosyn (at ∼100 mg/kg) on experimental day 0. All animals were grouped by operation status and subgrouped into treatment and control arms (Table 1). Liquid diet was refreshed and weights were obtained daily for 7 days, then all mice were transitioned to a standard chow diet with access to water. Animals in both groups were weighed every other day and taken to 21 or 56 days for experiment completion. All animal studies were approved by the Cincinnati Children's Hospital Medical Center Institutional Animal Care and Use Committee.
Table 1.
Guide to experimental group nomenclature
| Nomenclature | Operative Group | Nonoperative Group |
|---|---|---|
| Control group | Control ICR Subgroup | Nonoperative Control Subgroup |
| Treatment group | 2′-FL Supplemented ICR Subgroup | Nonoperative 2′-FL Supplemented Subgroup |
ICR, ileocecal resection; 2′-FL, 2′-fucosyllactose.
Operation status.
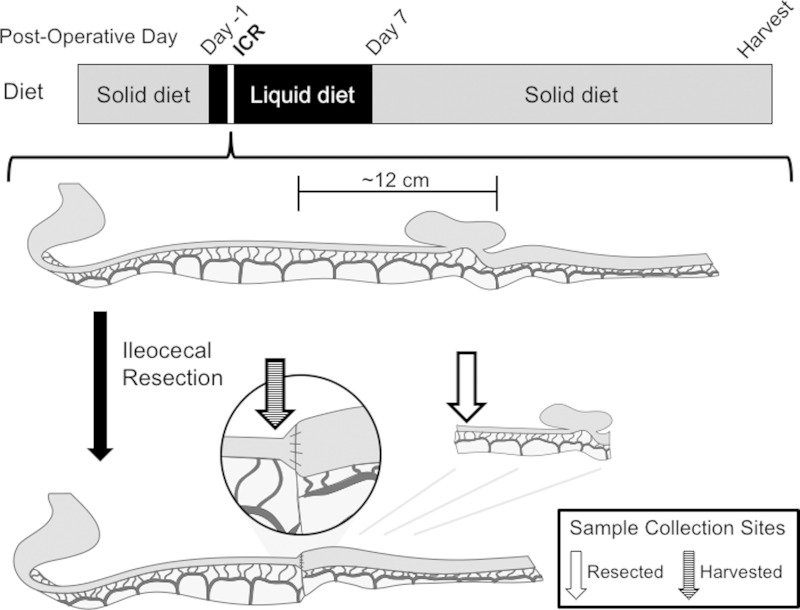
Nonoperated animals were maintained as described above. Operated animals were taken to the operating room on experimental day 0 and underwent ICR as previously described (16). Under the aid of an operating microscope and utilizing sedation with 2% isoflurane, a midline incision was made and the bowel was eviscerated. The ileocecal junction was identified and ∼12 cm of ileum and cecum were resected. Resected small bowel tissue and luminal contents were collected as described below (Fig. 1). Intestinal continuity was restored by end-to-end anastomosis and the abdomen was closed. ICR mice were then assigned to control or treatment subgroups, were administered analgesia with subcutaneous buprenorphine (0.05–0.1 mg/kg), and recovered overnight in a standard neonatal incubator warmed to ∼38°C.
Fig. 1.
All operated male C57Black/6 mice were 8–10 wk of age when placed on a liquid formula diet 1 day prior to undergoing ileocecal resection (ICR). Under sedation, a midline incision was made and the bowel was eviscerated. Approximately 12 cm of ileum and cecum were identified and resected. Bowel continuity was restored by end-to-end anastomosis. Animals were recovered, maintained on liquid formula for 7 days, and then transitioned to chow through harvest occurring on either postoperative day 21 or 56. Sites of resected and harvested tissue collection are indicated by open and hatched arrows, respectively.
Control and treatment subgroups.
Control (nonoperated) animals were maintained as described above. Beginning on experimental day 0, treatment animals were supplemented with 2′-FL (provided courtesy of Glycosyn, Medford, MA). 2′-FL was added to formula, then water to a final concentration of 2.5 g/l. Formula was refreshed daily and water every other day.
Intestinal tissue and small bowel contents preparation.
Only weights were obtained from nonoperated control animals. Resected intestine from operated animals was measured for length then the site of small bowel transection was processed for small bowel contents, histology, and RNA. At the time of experimental completion, small bowel immediately proximal to the site of anastomosis was also harvested for small bowel contents, histology, and RNA from the operated group.
Luminal small bowel contents were expressed into AllProtect Tissue Reagent (Qiagen, Valencia, CA). Bacterial DNA was extracted by using the AllPrep DNA/RNA Mini Kit (Qiagen) according to kit instructions and prior to sequencing (described under 16S sequencing and analysis). Small bowel tissue samples for RNA were placed into RNA Later (Life Technologies, Grand Island, NY). RNA extraction was accomplished by following Qiagen RNeasy Plus Kit instruction (Qiagen). All nucleic acid samples were then stored at −80°C until sequenced (described 16S sequencing and analysis).
Small bowel tissue samples for histology were cut in the longitudinal and transverse section, fixed with 4% paraformaldehyde, mounted in paraffin, and stained with hematoxylin and eosin. Villus height, crypt depth, and bowel circumference were measured in a blinded manner. At least 10 well-oriented villus and crypt domains were assessed for each sample and two samples were averaged per mouse. The serosal circumference was measured twice per sample with two samples per mouse. The average value per mouse was determined. Measurements were performed by use of the Nikon Ti-Eclipse with NIS Element Advanced Research version 4.20 (Nikon, Melville, NY).
Statistical analysis of weight and histological data.
To demonstrate the average trend in the populations measured and to assess significance, animal weight data were analyzed with a generalized estimating equation that incorporates repeated measures. Intestinal length was compared across groups by the Wilcoxon rank-sum test because values were not normally distributed. Histological count data were averaged per mouse across two samples and compared by the Wilcoxon rank-sum test. Weight data were analyzed with Stata version 13.0 (StataCorp, College Station, TX). Remaining morphometric and histological data were analyzed with GraphPad Prism version 5 (GraphPad Software, San Diego, CA).
16S sequencing and analysis.
Following bacterial community DNA extraction from harvested small bowel contents obtained from operated mice taken to 8 wk, samples were quantified using the Qubit ssDNA kit (Life Technologies). The V3 and V4 regions of the 16S gene were then amplified and tagged with region-specific primers (Illumina flowcell compatible sequences), permitting sequencing of up to 576 individual bacterial communities on the same flowcell (24). Two positive and two negative controls were included in each run. FastStart Taq (41) kit (Roche Applied Science, Indianapolis, IN) was used for thermocycling, then equal volumes of each amplicon were pooled and cleaned with the QIAquick PCR cleanup column (Qiagen, MD). The size of library pools was then verified with the Fragment Analyzer CE (AATI, Ames, IA) and quantified with the Qubit high-sensitivity dsDNA kit. Dilution to 1 nM and addition of PhiX V3 library (Illumina) was followed by denaturation and further dilution to 12 pM in Illumina's HT1 buffer. The pool was then loaded to the Illumina MiSeq V2 500 cycle kit cassette, a sample sheet was prepared, and the MiSeq run was initiated for FASTQ generation.
The 16S rRNA amplicon sequences were assembled and processed by use of an integrated, high-throughput analysis pipeline established at Cincinnati Children's Hospital Medical Center. Paired-end reads were assembled and quality filtered by using Pandaseq v2.8 (41). Reads with ambiguous base calls, minimum overlap of 10 nt, or <425 nt were culled. Demultiplexing and removal of barcodes and primers were performed with the FastX-toolkit (47). De novo clustering at 97% sequence similarity and chimera filtering was performed by using UPARSE v7 (21). UCLUST, as implemented in QIIME v1.8, was used for taxonomic classification to the Greengenes v13.8 database (8, 20, 42). PyNast and FastTree were used to align sequence reads and construct a phylogenetic tree (7, 49, 50). Additional integrated analyses included QIIME scripts for the generation of alpha and beta diversity metrics, corresponding visualizations, and summaries and plots of taxonomic composition. Alpha and beta diversity metrics were computed after subsampling to the lowest observed read depth (n = 7,793 reads).
To estimate the treatment effect on alpha diversity metrics adjusted for housing cohort, a generalized ANCOVA model was used. The Chao1, Shannon, Simpson, and Faith's Phylogenetic Diversity indexes were examined. Differences between groups in community composition posttreatment, as measured by the weighted and unweighted UniFrac metrics(39), were tested by permutational ANOVA as implemented by the ADONIS function in the R package vegan (45, 55). Pseudo-F statistics were obtained from sequential sums of squares from 1,000 permutations of the raw data. Differences in the overall abundance of specific OTUs between treatment subgroups at harvest was tested by use of a negative-binomial model as implemented in the R package DESeq2 (38).
RNA sequencing and analysis.
Transcriptional analysis was carried out on resected and harvested small bowel samples obtained from operated mice taken to 8 wk. Murine RNA sequencing libraries were prepared from ∼1.5 μg RNA by using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA) and sequenced on the HiSeq 2000 Sequencing System (Illumina) with single-end 50-bp reads. Following removal of primers and barcodes, sequences were aligned to the mm10 genome by use of reference annotations from University of California, Santa Cruz (51) (n = 36,186 entities). Aligned reads were quantified and used to compute reads per kilobase per million mapped reads; raw counts were then normalized by the DESeq algorithm and each harvested sample was baselined to its own resected sample. We applied a filter to the data, requiring at least three reads in all samples of at least one of the four experimental conditions (n = 14,489 entities). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE72590 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72590) (19).
To characterize the impact of 2′-FL supplementation while accounting for the effect of ileocecal resection, we applied a t-test between resected and harvested samples within control and experimental animals. Significance was set at a P value of ≤0.05 and a fold change of 3, which generated 2,576 differentially regulated entities across two comparisons. Gene set unions and intersections of the control and experimental gene sets were identified through Venn diagram. From the master list, we removed the gene list specific to animals supplemented with 2′-FL to identify genes differentially regulated only by ICR (n = 546 entities); this gene set characterizes the adaptive response. Furthermore, we identified the 2′-FL response (n = 2,030 entities) by removing the adaptive response signature described above from the master list, leaving only genes regulated by 2′-FL supplementation after ICR which were not found in the nonsupplemented adaptive response. Heatmaps were generated by using hierarchical clustering with the Pearson's centered distance metric and the average linkage rule; both entities and samples were clustered. All genomic analyses described above were performed in Strand NGS (Strand Life Sciences, San Francisco, CA).
Gene list functional enrichment was initially discovered by using ToppFun, a member of the ToppGene Suite (11). No statistical correction was selected. Gene lists were further analyzed by using ClueGO, a plugin for Cytoscape designed to decipher functionally grouped gene ontology and pathway annotation networks (5). At the time of data analysis, the annotation files were updated from sources to January 1, 2015. “GO Term Fusion” was enabled to manage ontological term redundancy and network specificity was set to medium. Results were restricted to pathways with P ≤ 0.05 by Benjamini-Hochberg correction and kappa scoring was set to 0.4. Biocyc annotations were assigned a rectangle, gene ontology biological processes were assigned an ellipse, and Wikipathway annotations were assigned a diamond shape. No meaning was assigned to size or color.
RESULTS
The median age (interquartile range) of operated mice at the time of ICR in the 21-day experiment was 93 days (93–94). In the 56-day experiment, mice were operated on at a median age of 76 days (72–77). The age of nonoperated mice in the 21- and 56-day experiments were 66 days and 79 days, respectively. There was no difference in age between control and 2′-FL-supplemented subgroups at each time point.
Weight change.
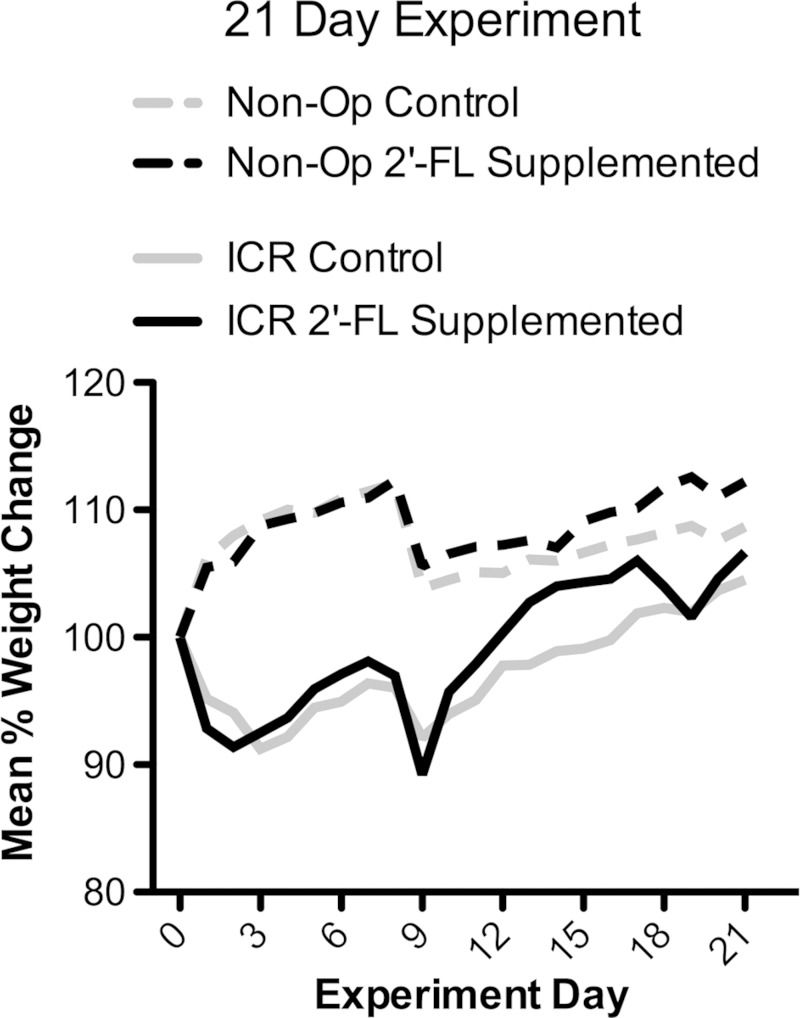
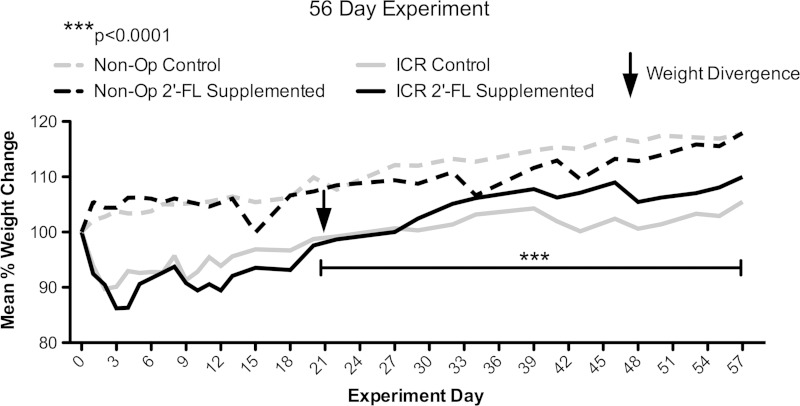
The median weight (interquartile range) of operated mice at the time of ICR in the 21- and 56-day experiments were 23.2 (21.3–25.6) and 25.8 g (24.2–27.3), respectively. As expected, animals in both control and 2′-FL-supplemented ICR subgroups lost ∼10% body weight during the first postoperative week. Both groups returned to their preoperative weight by 3–4 wk after ICR and continued to grow. Animals taken to harvest at postoperative day 21 demonstrated no significant weight difference between control and 2′-FL-supplemented subgroups (Fig. 2). When taken to 56 days, animals supplemented with 2′-FL, compared with controls, demonstrated increased weight on and beyond postoperative day 21 (Fig. 3) (P < 0.001). At 56 days, animals of the control and 2′-FL-supplemented ICR subgroups achieved 105 and 110% of preoperative body weight, respectively.
Fig. 2.
Mean weight change by group and subgroup relative to weight at experiment start. Nonoperative control (Non-Op; n = 4) and 2′-O-fucosyllactose (2′-FL)-supplemented animals (n = 5), and animals subjected to ICR in control (n = 4) and 2′-FL-supplemented subgroups (n = 3) were taken to experimental day 21. No significant difference between subgroups of either operated or nonoperative groups was found. Multiple experiments are shown.
Fig. 3.
Mean weight change by group and subgroup relative to weight at experiment start. Nonoperative control (n = 6) and 2′-FL-supplemented animals (n = 6) and animals subjected to ICR in control (n = 5) and 2′-FL-supplemented subgroups (n = 7) were taken to experimental day 56. Arrow denotes point and bar denotes period of significant weight divergence of ICR subgroups based on the results of a generalized estimating equation indicating that 2′-FL supplementation improves weight following ICR (P < 0.0001). No significant difference between nonoperative subgroups was found. Multiple experiments are shown.
At the time of experiment start, the median weight (interquartile range) of nonoperated mice in the 21- and 56-day groups were 23.7 (21.8–24.0) and 22.6 g (22.4–24.2), respectively. Control and 2′-FL-supplemented nonoperative subgroups both gained an average of 108% body weight at 21 days [not significant (NS)]. When taken to 56 days after ICR, both groups had gained 117% body weight from study start (NS).
Histology.
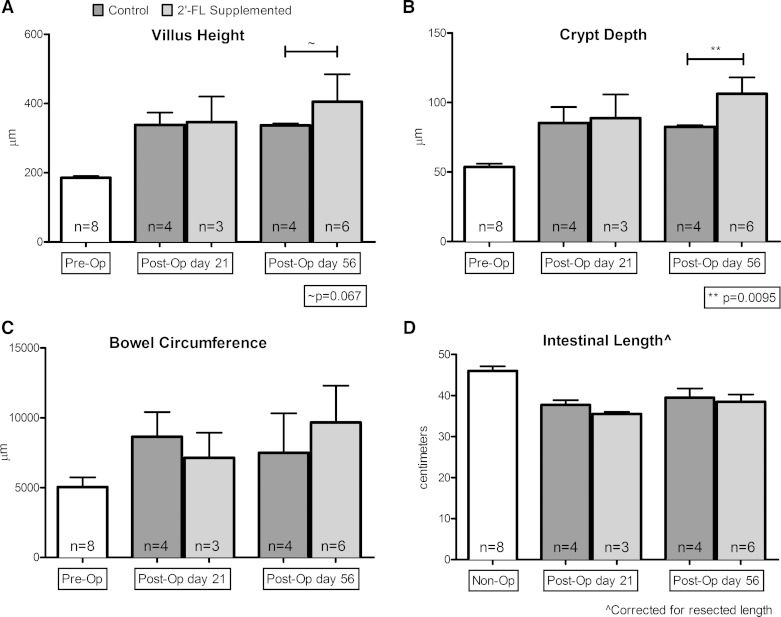
The histological measures of adaptation following ICR were augmented and prolonged with 2′-FL supplementation (Fig. 4). The median baseline crypt depth among all operated animals (interquartile range) was 54 μm (51–56) and increased following ileocecal resection. Crypt depth in control ICR animals was 85 μm (81–97) at 21 days and 82 μm (80–84) at 56 days. Among the 2′-FL-supplemented ICR subgroups, crypts deepened to 89 μm (76–106) at 21 days and further to 106 μm (101–118) on postoperative harvest day 56. There was no difference between control and 2′-FL-supplemented crypt depths on postoperative day 21. On postoperative day 56, the crypts of the 2′-FL-supplemented ICR subgroup were significantly deeper than those of the control group (P = 0.0095).
Fig. 4.
Comparison of histological markers of adaptation following ileocecal resection among control and 2′-FL-supplemented animals. Median and interquartile range of villus height (A), crypt depth (B), bowel circumference (C), and corrected intestinal length (D) from 3 time points are shown. Tissue from preoperative or nonoperated (n = 8) as well as control and 2′-FL-supplemented tissues, respectively, on postoperative days 21 (n = 4, 3) and 56 (n = 4, 6). Comparisons between subgroups per time point by Mann-Whitney test, not significant unless otherwise indicated. Multiple experiments are shown.
The median baseline villus height among all operated animals (interquartile range) was 186 μm (180–190) and also increased following ileocecal resection. Between postoperative days 21 and 56, villus heights among control ICR animals remained similarly elevated over preoperative heights at 338 (323–374) and 337 μm (257–342), respectively. Among the 2′-FL-supplemented ICR subgroups, villus heights increased above baseline to 346 μm (318–420) on postoperative day 21 then 405 μm (340–484) on day 56. There was no difference between control and 2′-FL-supplemented subgroups on postoperative day 21. On postoperative day 56, the villi of those mice supplemented with 2′-FL trended toward a greater height than those of the control subgroup (P = 0.067).
The median baseline distal small bowel circumference was 5,041 μm (4,981–5,739) and increased following ileocecal resection. Between postoperative days 21 and 56, bowel circumferences decreased from 8,643 (7,367–10,413) to 7,491 μm (6,146–10,333), among the respective control ICR subgroups. Among the 2′-FL-supplemented subgroups, small bowel circumferences tended to increase from 7,133 (6,529–8,930) to 9,671 μm (7,851–12,295) from postoperative harvest days 21 to 56. However, no statistical difference in small bowel circumference between control and 2′-FL-supplemented subgroups on postoperative day 21 or 56.
The median baseline small bowel length (interquartile range) was 46 cm (41.38–47.13). Between postoperative days 21 and 56, intestinal lengths (corrected) increased from 37.8 (35.5–38.9) to 39.5 cm (37.3–41.8), respectively, among the control subgroups. Among the 2′-FL-supplemented subgroups, intestinal lengths increased from 35.5 (35–36) to 38.5 cm (37.4–40.3) from postoperative harvest days 21 to 56. There was no difference in intestinal length between control and experimental groups on postoperative day 21, nor was there a difference on postoperative day 56.
Small bowel luminal microbiome.
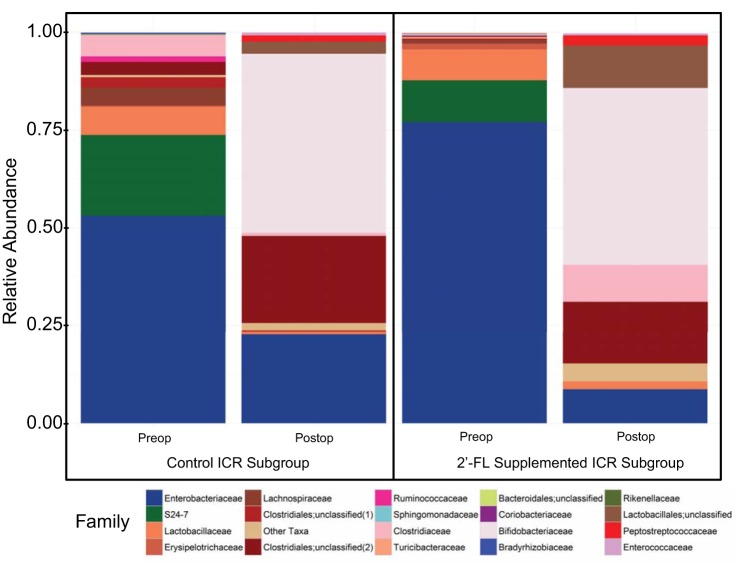
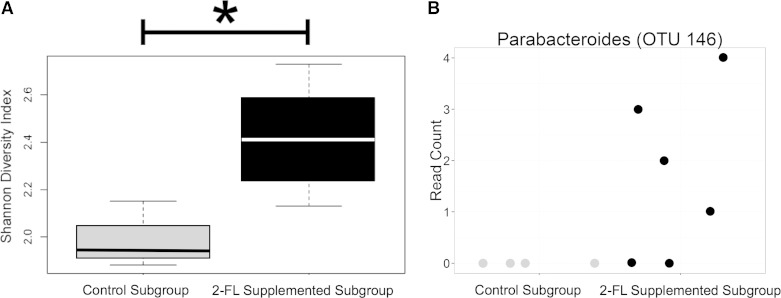
The microbiome of the small bowel luminal contents of operated animal subgroups at 8 wk were analyzed and relative abundance by experimental group displayed (Fig. 5). We tested for differences in alpha diversity between control and 2′-FL-supplemented subgroups after controlling for housing cohort. We found a greater diversity of small bowel bacteria as measured by the Shannon Index among 2′-FL-supplemented animals compared with controls (P < 0.005) (Fig. 6A). No differences were detected for the other alpha-diversity metrics examined. Nor were differences in the weighted or unweighted UniFrac metrics detected between 2′-FL-supplemented animals and controls posttreatment (P = 0.143). Next, we determined the log2-fold change for 2′-FL-supplemented animals compared with controls, adjusted for housing cohort. Sequence reads were enriched among operated, 2′-FL-supplemented animals for a single OTU that could be classified to the genus Parabacteroides: OTU_146 (log2-fold = 4.1, P = 0.035). Parabacteroides was not detected in either study group at baseline, nor was Parabacteroides identified in the controls at follow-up (Fig. 6B).
Fig. 5.
Relative abundance of bacterial families discovered in the luminal contents at the time of (Preop) and following (Postop) ileocecal resection. Families displayed as shades of parent phylum colors including Firmicutes (red and pink), Proteobacteria (blue), Bacteriodetes (green), and Actinobacteria (purple). Enterobacteriaceae were the most abundant taxa among preoperative samples and decreased following resection in both groups though a greater decline was observed among 2′-FL-supplemented animals. A relatively larger bloom in Lachnospiraceae was also observed in this group.
Fig. 6.
Analysis of small bowel luminal contents at 56 days after ileocecal resection among control (n = 4) and 2′-FL-supplemented (n = 6) animals. A: 2′-FL supplementation resulted in greater alpha diversity by Shannon diversity index (*P < 0.005). B: sequence reads classified to the genus Parabacteroides were enriched by 2′-FL supplementation (log2-fold = 4.1, P = 0.035) but not detected in control animals after resection.
Transcriptional analysis of small bowel.
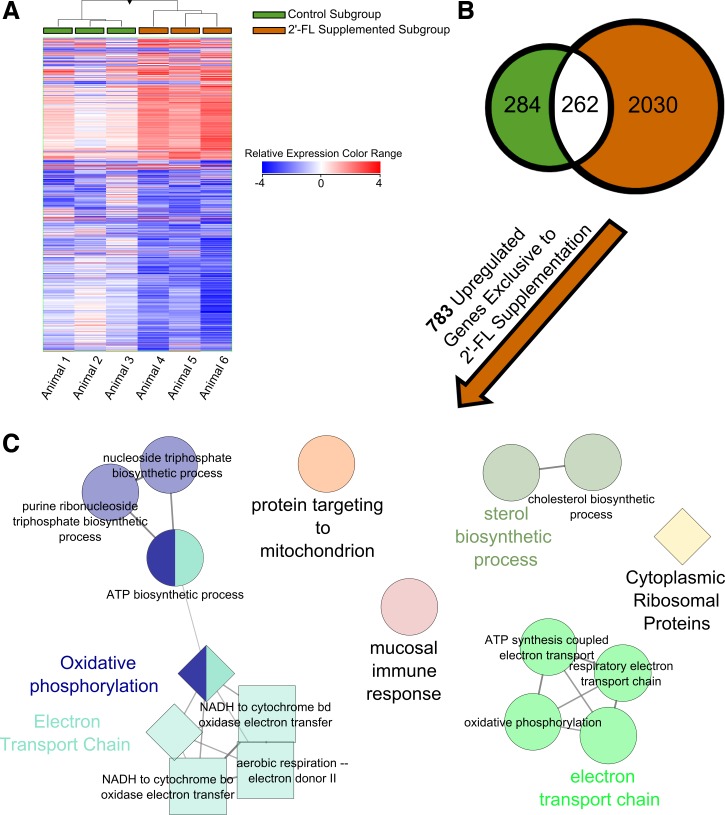
RNA-sequencing analysis was performed in Strand NGS from resected and harvested tissues among those operated subgroups taken to 8 wk. Data were normalized by using the DESeq algorithm and harvested samples were baselined to their respective resected sample. We identified 2,576 differentially regulated entities (P value <0.05 and fold change >2) within control and/or experimental groups. Gene set unions and intersections of these groups were identified through Venn diagram (Fig. 7B). A heatmap of all genes differentially regulated demonstrates an augmentation of the transcriptional directions appreciated among control animals when 2′-FL supplementation is considered (Fig. 7A).
Fig. 7.
Impact of ICR on distal small bowel gene expression among representative control (n = 3) and 2′-FL-supplemented (n = 3) animals. A: hierarchical clustering of 2,567 genes differentially regulated between harvested and resected samples. B: Venn diagram of these genes identifies those exclusively regulated by ICR in the absence of 2′-FL (green), those exclusively regulated by ICR in the presence of 2′-FL (orange), and those shared between both groups (white). C: nonredundant biological functional information was extracted and deciphered from the list of genes exclusively upregulated by ICR in the presence of 2′-FL supplementation. All discovered annotations are present including Biocyc annotations as squares, GO Biological Processes as circles, and Wikipathways as diamonds. Related ontology and pathway groupings by color. No meaning assigned to size.
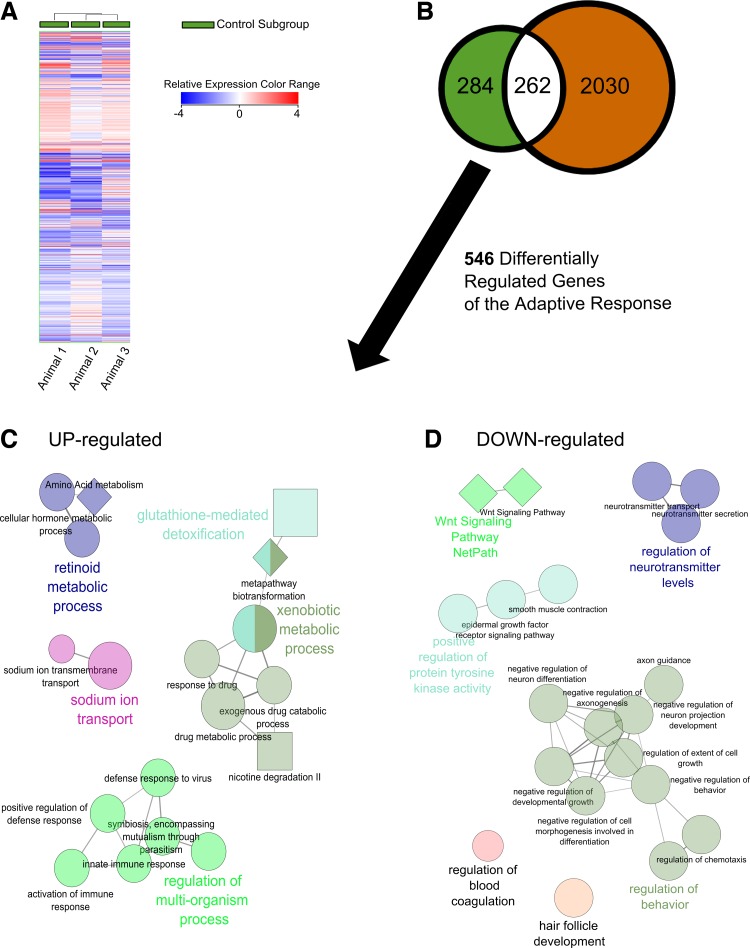
The adaptive response.
The “adaptive response” refers to the genes differentially regulated by harvested to resected comparison in the control subgroup and represents 546 distinct entities (Supplemental Table S1; Supplemental Material for this article is available online at the Journal website). Among the upregulated entities of the adaptive response (n = 154), ontologies pertaining to metabolic processes/metabolism were most salient, including glutathione derivative biosynthetic processes (P = 5.0E−07), organic acid metabolic processes (P = 3.4E−06), cellular modified amino acid metabolic processes (P = 6.9E−06), carboxylic acid metabolic processes (P = 2.7E−05), purine deoxyribonucleoside metabolic processes (P = 1.1E−04), hormone metabolic processes (P = 0.001), metabolism (P = 2.7E−08), fatty acid metabolism (P = 3.0E−07), and glutathione metabolism (P = 5.7E−07), among others. Furthermore, ontologies indicating a response to a shifting microbial community were discovered, including regulation of multiorganism processes (P = 1.4E−04), response to external biotic stimulus (P = 0.001), and response to other organism (P = 0.001). Insulin-like growth factor signaling and purine salvage pathways (P = 0.002 and P = 0.003, respectively) were also discovered. Of the upregulated entities, the most strongly upregulated genes of the adaptive response include Oas1e [fold change (FC) = 55.7], Cyp1a1 (FC = 36.3), Upk3b (FC = 27.1), Ly6g6c (FC = 25.5), and Igfbp6 (FC = 20.4).
Among the downregulated entities (n = 392), ontologies pertaining to the regulation of cellular developmental processes were most salient, including cell development (P = 7.75E−09), regulation of developmental process (P = 2.78E−08), and regulation of cell development (P = 3.54E−07). Other, tissue-specific ontologies related to development include vasculature development (P = 8.05E−06), brain development (P = 4.92E−06), cardiovascular development (P = 1.74E−05), and striated muscle development (P = 1.32E−04). Ontologies related to neural function and generation were also discovered, including neurogenesis (P = 8.70E−09), synaptic transmission (P = 2.77E−07), neurotransmitter secretion (P = 3.73E−06), and axonogenesis (P = 8.62E−06). Finally, MAPK signaling was discovered, including the MAPK cascade (P = 2.16E−06) and regulation of the MAPK cascade (P = 4.53E−06) and may be related to codiscovered, downregulated entities including “positive regulation of epidermal growth factor receptor signaling” (P = 3.55E−05) and “positive regulation of the ERBB signaling pathway” (P = 4.53E−05). Of the downregulated entities, the most strongly downregulated genes of the adaptive response include Dhp (FC = −93.1), Gm129 (FC = −20.1), Bex1 (FC = −13.9), Hlf (FC = −13.0), and Abca1 (FC = −11.9).
Cytoscape's ClueGO application was used to generate nonredundant, functionally grouped gene ontology and pathway annotation networks based on the up- and downregulated gene set of the Adaptive Response (Fig. 8). Importantly, all discovered entities are described. The ClueGO network underscored the importance of multiorganism processes and retinoid metabolic processes in the adaptive response, and further highlighted xenobiotic metabolic processes and sodium ion transport.
Fig. 8.
Impact of ICR on distal small bowel gene expression among representative control animals (n = 3). A: hierarchical clustering of 2,567 genes differentially regulated between harvested and resected samples. B: Venn diagram of these genes identifies those regulated by the adaptive response to ICR (green, white) and those exclusively regulated by 2′-FL supplementation following ICR (orange). Nonredundant biological functional information was extracted and deciphered from the list of genes upregulated (C) and downregulated (D) in the adaptive response to ICR. All discovered annotations are present including Biocyc annotations as squares, GO Biological Processes as circles, and Wikipathways as diamonds. Related ontology and pathway groupings by color. No meaning assigned to size.
2′-FL signature response.
Those genes differentially regulated following ICR in animals supplemented with 2′-FL, less the adaptive response observed in the control group, included 2,030 entities. Among the upregulated entities (n = 783) (Supplemental Table S1), ontologies pertaining to energy presence and processing were most salient, including electron transport chain (P = 1.87E-35), cellular respiration (P = 4.53E-30), mitochondrial ATP synthesis coupled electron transport (P = 2.67E-20), generation of precursor metabolites and energy (P = 3.01E-20), energy derivation by oxidation of organic compounds (P = 4.26E-20), and organic cyclic compound catabolic process (P = 1.10E−08). Also discovered were ontologies suggesting host-microbial interaction, including multiorganism metabolic process (P = 1.59E-22), symbiosis, encompassing mutualism through parasitism (P = 1.47E-13), interspecies interaction between organisms (P = 1.47E-13), multiorganism cellular process (P = 8.20E-13), and mucosal immune response (P = 1.40E−06). Finally, ontologies involving biosynthetic processes were discovered, including sterol biosynthetic process (P = 6.22E−08), cholesterol biosynthetic process (P = 1.76E−07), and various nucleoside biosynthetic processes. Of the upregulated entities, the most strongly upregulated genes of the 2′-FL signature response were Tmem202 (FC = 18.0), Map3k12 (FC = 16.2), Hemt1 (FC = 15.4), Mcpt2 (FC = 13.4), and Fmr1nb (FC = 12.7).
Among the downregulated entities of the 2′-FL signature response (n = 1,247), similar ontologies to those downregulated in the adaptive response were strongly present. These included neurogenesis (P = 7.34E-20), regulation of developmental process (P = 5.83E-18), cardiovascular system development (P = 2.91E-17), circulatory system development (P = 2.91E-17), and axonogenesis (P = 7.60E-16). Furthermore, regulation of development at a cellular level was observed, with ontologies including cell development (P = 2.76E-20), cell morphogenesis involved in differentiation (P = 1.96E-18), epithelial development (P = 2.78E-13), epithelial tube morphogenesis (P = 7.39E-13), cell junction assembly (P = 2.60E−09), and cellular response to growth factor stimulus (P = 4.30E−09). Finally, ontologies relevant to control over cell cycle were discovered, including regulation of Ras protein signal transduction (P = 1.50E-11) and positive regulation of Ras GTPase activity (P = 1.69E-10). Related to this theme were ontologies including positive regulation of cellular biosynthetic process (P = 1.34E-11) and regulation of nucleotide metabolic process (P = 3.41E-11). Of the downregulated entities, the most strongly downregulated genes of the 2′-FL signature response were Fgf15 (FC = −64.6), Irs2 (FC = −43.6), Ttc28 (FC = −24.2), P2ry4 (FC = −22.2), and Rnf150 (FC = −20.4).
To determine all nonredundant, functionally grouped gene ontology and pathway annotation networks based on the gene set of the upregulated 2′-FL signature response, Cytoscape's ClueGO application was used (Fig. 7C). The ClueGO networks discovered underscored the importance of energy processing with ontologies and pathways related to the electron transport chain, oxidative phosphorylation, and protein targeting to the mitochondrion. Furthermore, sterol biosynthesis was discovered. As expected, ontologies involved in the mucosal immune response were also upregulated. When ClueGO was used to generate networks based on the downregulated 2′-FL signature response, ontologies and pathways relating to the IL-7 signaling pathway, positive regulation of Rho GTPase activity, and cell adhesion were discovered (data not shown).
DISCUSSION
2′-Fucosyllactose, the dominant human milk oligosaccharide produced by women who are FUT2 secretors, augments the sustained adaptive response to extensive intestinal resection in mice. We found that operated animals supplemented with 2′-FL gained more weight than control animals, a robust marker of intestinal function. Furthermore, we observed a prolonged but characteristic morphometric adaptive response among supplemented animals though differences were found only after the point of weight divergence, suggesting additional sources of improved growth. 2′-FL buffered microbial changes previously observed after resection, which may have been the stimulus for transcriptional activity most heavily supporting increased energy utilization among supplemented and resected animals. We conclude that supplementation with 2′-FL, an indigestible and noncaloric prebiotic, increases weight gain following ileocecal resection by increasing energy availability through microbial community modulation and directly or indirectly stimulating characteristic histological changes ultimately resulting in improved adaptation. We note this difference among secretor animals capable of decorating their intestinal epithelium with the 2′-FL analog, H-antigen, highlighting the impact of supplementation.
The impact of 2′-FL on weight gain only occurred after ICR, supporting the conclusion of augmented adaptation following intestinal resection over an independent effect on weight. This observation has been supported both when comparing control and 2′-FL-supplemented healthy human infants who did not demonstrate differential weight gain as well as in mechanistic studies demonstrating improved growth following physiological stress among secretor mice (40, 48). Thus 2′-FL seemed to buffer against the stress of intestinal loss while exhibiting little to no effect on weight without insult.
Supplementation with 2′-FL augmented the adaptive increase in absorptive surface area through sustained increases characteristic morphometric markers of gut adaptation. Independently, crypt depth was significantly greater among supplemented animals on postoperative day 56. Furthermore, should the trend observed in villus height represent a true difference, even this modest difference would translate to a significant increase in absorptive surface area in three dimensions. Control and supplemented animals taken to postoperative day 21, the point of weight divergence between both subgroups, were anticipated to experience a greater difference in these measures. This was not so, suggestive of a separate process responsible for the early weight divergence.
Studies of intestinal loss and associated physiological stress reveal a marked decline in alpha diversity, which has been associated with poor adaptation as measured by delayed weaning from parenteral nutrition (22, 34). Our model of adaptation to massive intestinal loss also induces dysbiosis (17). Solutions for buffering or reversing this dysbiosis may improve adaptation and are highly sought in the treatment of short bowel syndrome. One potential solution is 2′-FL supplementation. Secretor status is a key driver of intestinal microbial community composition, where the ability to secrete H antigen or the availability of the H antigen analog, 2′-FL, supports increased community diversity and bolsters microbiota during times of stress (36, 48, 62, 65). We report that 2′-FL supplementation after intestinal resection resulting in massive intestinal loss results in increased gut microbial diversity occurring with improved adaptation.
In this study, we also found a bloom in microbes of the genus Parabacteroides (>4-fold increase among supplemented, resected animals compared with controls). Though the body of literature surrounding this genus is scant, their differential presence may impact both a mucosal inflammatory response to resection and the abundance of available energy to the host. These bacteria have not only been found in higher proportions comparing noninflamed with inflamed enteric samples, but a lysate containing the membranous fraction has been observed to protect from DSS-induced murine colitis (33, 59, 66). A robust and direct interaction between the organism and innate and adaptive immunomodulatory mechanisms occurs, suggesting a rational mechanism of interaction of cellular processes involved in the adaptive response (33). Furthermore, this genus readily ferments indigestible carbohydrates, converting them to beneficial and available organic acids, providing a possible explanation for the growth advantage observed among supplemented animals supporting a bloom in this organism (6).
As expected, the transcriptional analysis of the late adaptive response is characterized by a release of developmental progression pathways and engagement of ontologies involved in diverse metabolic processes. Transcription of the 2′-FL signature response engages many cellular components responsive to energy presence and processing demands, in support of the presumed increase in energy availability among operated animals supplemented with 2′-FL. Furthermore, ontologies relating to energy derivation by oxidation of organic compounds were discovered: the processes by which short-chain fatty acids are converted to energy after undergoing largely passive intracellular diffusion (27). We imply improved energy availability with 2′-FL supplementation among resected animals, likely through short-chain fatty acid production. Indeed, a clear increase in short-chain fatty acid and lactate production, sources of mucosal energy, is observed when 2′-FL is added to in vitro infant fecal samples (65).
The study faces several important limitations, primarily related to the challenges of modeling intestinal resection. Owing to the model complexity, animal numbers were limited, which restricted power in statistical analysis and modeling. Furthermore, animal fragility during the acute postoperative recovery period prevented animal separation and mixing, resulting in subtle differences in resected microbial community composition and an inability to perform feeding efficiency measures. Finally, C57BL/6 animals are all FUT2 positive, hence all animals studied produce H antigen on gut mucosal surfaces. 2′-FL is an analog of the H antigen, and thus differences observed first overcame the effect of physiological H antigen presence among all resected animals. We speculate that individuals lacking H antigen on gut mucosal surfaces may benefit more greatly from 2′-FL supplementation. We also speculate that the effect observed is not specific to 2′-FL but may be seen with other indigestible prebiotic carbohydrates. Notably, carbohydrates historically used as controls, such as inulin, maltodextrin, and galactooligosaccharides, exhibit a prebiotic effect or contribute to overall energy balance (18, 30, 44, 52, 61). Thus no control carbohydrate was provided to reduce the chance of type 1 or type 2 error.
Enhancing the adaptive response is vitally important to improving health and cost outcomes following extensive intestinal resection. We demonstrate augmentation of adaptation with a naturally occurring prebiotic safe for human consumption. Further studies in this model evaluating 2′-FL supplementation and withdrawal, the dose-effect relationship, and outcomes among secretor and nonsecretor animals are needed. Additionally, studies of 2′-FL safety and effectiveness after intestinal resection in humans are needed to support a clinical role. Determining the impact of secretor status on human adaptation may identify a substantial subgroup to benefit most from 2′-FL supplementation. Finally, efforts to understand and modulate gut microbial community changes following intestinal resection promise novel treatment paradigms that may help improve the lives of children suffering from short bowel syndrome.
GRANTS
This work was supported in part by National Institutes of Health (NIH) Grants T32 DK007727 (E. A. Mezoff), P01 HD 13021 (A. L. Morrow), and R01 DK083325-01 (M. A. Helmrath). This work was supported in part by NIH Grant P30 DK078392 of the Digestive Disease Research Core Center in Cincinnati.
DISCLOSURES
A. L. Morrow reports a minority ownership in GlycoSyn, LLC, the company supplying the oligosaccharide studied.
AUTHOR CONTRIBUTIONS
E.A.M., J.A.H., A.L.M., and M.A.H. conception and design of research; E.A.M., J.A.H., and M.A.H. performed experiments; E.A.M., N.J.O., R.K., A.L.M., and M.A.H. analyzed data; E.A.M., J.A.H., N.J.O., R.K., A.L.M., and M.A.H. interpreted results of experiments; E.A.M., N.J.O., and R.K. prepared figures; E.A.M. drafted manuscript; E.A.M., J.A.H., N.J.O., R.K., A.L.M., and M.A.H. edited and revised manuscript; E.A.M., A.L.M., and M.A.H. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Glycosyn, LLC for the provision of 2′-fucosyllactose for the purposes of this study, and David S. Newburg, PhD for consultation and technical assistance on this project.
REFERENCES
- 1.Abrahamsson TR, Sherman PM. Multifaceted effects of human milk oligosaccharides. J Infect Dis 209: 323–324, 2014. [DOI] [PubMed] [Google Scholar]
- 2.Andorsky DJ, Lund DP, Lillehei CW, Jaksic T, Dicanzio J, Richardson DS, Collier SB, Lo C, Duggan C. Nutritional and other postoperative management of neonates with short bowel syndrome correlates with clinical outcomes. J Pediatr 139: 27–33, 2001. [DOI] [PubMed] [Google Scholar]
- 3.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem 286: 34583–34592, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am 60: 49–74, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blatchford P, Bentley-Hewitt KL, Stoklosinski H, McGhie T, Gearry R, Gibson G, Ansell J. In vitro characterisation of the fermentation profile and prebiotic capacity of gold-fleshed kiwifruit. Benef Microbes 6: 829–839, 2015. [DOI] [PubMed] [Google Scholar]
- 7.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castanys-Munoz E, Martin MJ, Prieto PA. 2′-Fucosyllactose: an abundant, genetically determined soluble glycan present in human milk. Nutr Rev 71: 773–789, 2013. [DOI] [PubMed] [Google Scholar]
- 10.Chaturvedi P, Warren CD, Altaye M, Morrow AL, Ruiz-Palacios G, Pickering LK, Newburg DS. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology 11: 365–372, 2001. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–W311, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng SX, Gathungu G, Pashankar D, Jain D, Husain SZ. Jejunal adaptation in a prepubertal boy after total ileal resection and jejunostomy placement: a four-year follow-up. J Clin Gastroenterol 45: 846–849, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coppa GV, Pierani P, Zampini L, Carloni I, Carlucci A, Gabrielli O. Oligosaccharides in human milk during different phases of lactation. Acta Paediatr Suppl 88: 89–94, 1999. [DOI] [PubMed] [Google Scholar]
- 14.Coulet M, Phothirath P, Allais L, Schilter B. Pre-clinical safety evaluation of the synthetic human milk, nature-identical, oligosaccharide 2′-O-fucosyllactose (2′FL). Regul Toxicol Pharmacol 68: 59–69, 2014. [DOI] [PubMed] [Google Scholar]
- 15.Cummins A, Thompson F. Effect of breast milk and weaning on epithelial growth of the small intestine in humans. Gut 51: 748–754, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dekaney CM, Fong JJ, Rigby RJ, Lund PK, Henning SJ, Helmrath MA. Expansion of intestinal stem cells associated with long-term adaptation following ileocecal resection in mice. Am J Physiol Gastrointest Liver Physiol 293: G1013–G1022, 2007. [DOI] [PubMed] [Google Scholar]
- 17.Devine AA, Gonzalez A, Speck KE, Knight R, Helmrath M, Lund PK, Azcarate-Peril MA. Impact of ileocecal resection and concomitant antibiotics on the microbiome of the murine jejunum and colon. PloS One 8: e73140, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dewulf EM, Cani PD, Claus SP, Fuentes S, Puylaert PG, Neyrinck AM, Bindels LB, de Vos WM, Gibson GR, Thissen JP, Delzenne NM. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 62: 1112–1121, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461, 2010. [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998, 2013. [DOI] [PubMed] [Google Scholar]
- 22.Engstrand Lilja H, Wefer H, Nystrom N, Finkel Y, Engstrand L. Intestinal dysbiosis in children with short bowel syndrome is associated with impaired outcome. Microbiome 3: 18, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erney RM, Malone WT, Skelding MB, Marcon AA, Kleman-Leyer KM, O'Ryan ML, Ruiz-Palacios G, Hilty MD, Pickering LK, Prieto PA. Variability of human milk neutral oligosaccharides in a diverse population. J Pediatr Gastroenterol Nutr 30: 181–192, 2000. [DOI] [PubMed] [Google Scholar]
- 24.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2: 6, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feldman EJ, Dowling RH, McNaughton J, Peters TJ. Effects of oral versus intravenous nutrition on intestinal adaptation after small bowel resection in the dog. Gastroenterology 70: 712–719, 1976. [PubMed] [Google Scholar]
- 26.Ferrer-Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, Blancher A, Calafell F, Bertranpetit J, Casals F. A natural history of FUT2 polymorphism in humans. Mol Biol Evol 26: 1993–2003, 2009. [DOI] [PubMed] [Google Scholar]
- 27.Fleming SE, Choi SY, Fitch MD. Absorption of short-chain fatty acids from the rat cecum in vivo. J Nutr 121: 1787–1797, 1991. [DOI] [PubMed] [Google Scholar]
- 28.Goehring KC, Kennedy AD, Prieto PA, Buck RH. Direct evidence for the presence of human milk oligosaccharides in the circulation of breastfed infants. PloS One 9: e101692, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goulet O, Ruemmele F. Causes and management of intestinal failure in children. Gastroenterology 130: S16–S28, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Holscher HD, Bauer LL, Gourineni V, Pelkman CL, Fahey GC Jr, Swanson KS. Agave inulin supplementation affects the fecal microbiota of healthy adults participating in a randomized, double-blind, placebo-controlled, crossover trial. J Nutr 145: 2025–2032, 2015. [DOI] [PubMed] [Google Scholar]
- 31.Holscher HD, Davis SR, Tappenden KA. Human milk oligosaccharides influence maturation of human intestinal Caco-2Bbe and HT-29 cell lines. J Nutr 144: 586–591, 2014. [DOI] [PubMed] [Google Scholar]
- 32.Kohler JA Sr, Perkins AM, Bass WT. Human milk versus formula after gastroschisis repair: effects on time to full feeds and time to discharge. J Perinatol 33: 627–630, 2013. [DOI] [PubMed] [Google Scholar]
- 33.Kverka M, Zakostelska Z, Klimesova K, Sokol D, Hudcovic T, Hrncir T, Rossmann P, Mrazek J, Kopecny J, Verdu EF, Tlaskalova-Hogenova H. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol 163: 250–259, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapthorne S, Pereira-Fantini PM, Fouhy F, Wilson G, Thomas SL, Dellios NL, Scurr M, O'Sullivan O, Ross RP, Stanton C, Fitzgerald GF, Cotter PD, Bines JE. Gut microbial diversity is reduced and is associated with colonic inflammation in a piglet model of short bowel syndrome. Gut Microbes 4: 212–221, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauriti G, Zani A, Aufieri R, Cananzi M, Chiesa PL, Eaton S, Pierro A. Incidence, prevention, and treatment of parenteral nutrition-associated cholestasis and intestinal failure-associated liver disease in infants and children: a systematic review. JPEN J Parenter Enteral Nutr 38: 70–85, 2014. [DOI] [PubMed] [Google Scholar]
- 36.Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, Van Tassell ML, Miller MJ, Jin YS, German JB, Lebrilla CB, Mills DA. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 3: 13, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem 55: 8914–8919, 2007. [DOI] [PubMed] [Google Scholar]
- 38.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marriage BJ, Buck RH, Goehring KC, Oliver JS, Williams JA. Infants fed a lower calorie formula with 2′-fucosyllactose (2′FL) show growth and 2′FL uptake like breast-fed infants. J Pediatr Gastroenterol Nutr 61: 649–658, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13: 31, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDuffie LA, Bucher BT, Erwin CR, Wakeman D, White FV, Warner BW. Intestinal adaptation after small bowel resection in human infants. J Pediatr Surg 46: 1045–1051, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nickerson KP, Homer CR, Kessler SP, Dixon LJ, Kabi A, Gordon IO, Johnson EE, de la Motte CA, McDonald C. The dietary polysaccharide maltodextrin promotes Salmonella survival and mucosal colonization in mice. PloS One 9: e101789, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. Vegan: Community Ecology Package. 2015. https://cran.r-project.org/web/packages/vegan/index.html. [Google Scholar]
- 46.Olieman JF, Penning C, Ijsselstijn H, Escher JC, Joosten KF, Hulst JM, Tibboel D. Enteral nutrition in children with short-bowel syndrome: current evidence and recommendations for the clinician. J Am Diet Assoc 110: 420–426, 2010. [DOI] [PubMed] [Google Scholar]
- 47.Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics 46: 24–36, 1997. [DOI] [PubMed] [Google Scholar]
- 48.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, Chervonsky AV. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514: 638–641, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PloS One 5: e9490, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hickey G, Hinrichs AS, Hubley R, Karolchik D, Learned K, Lee BT, Li CH, Miga KH, Nguyen N, Paten B, Raney BJ, Smit AF, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 43: D670–D681, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salazar N, Dewulf EM, Neyrinck AM, Bindels LB, Cani PD, Mahillon J, de Vos WM, Thissen JP, Gueimonde M, de Los Reyes-Gavilan CG, Delzenne NM. Inulin-type fructans modulate intestinal Bifidobacterium species populations and decrease fecal short-chain fatty acids in obese women. Clin Nutr 34: 501–507, 2015. [DOI] [PubMed] [Google Scholar]
- 53.Spencer AU, Kovacevich D, McKinney-Barnett M, Hair D, Canham J, Maksym C, Teitelbaum DH. Pediatric short-bowel syndrome: the cost of comprehensive care. Am J Clin Nutr 88: 1552–1559, 2008. [DOI] [PubMed] [Google Scholar]
- 54.Squires RH, Duggan C, Teitelbaum DH, Wales PW, Balint J, Venick R, Rhee S, Sudan D, Mercer D, Martinez JA, Carter BA, Soden J, Horslen S, Rudolph JA, Kocoshis S, Superina R, Lawlor S, Haller T, Kurs-Lasky M, Belle SH; Pediatric Intestinal Failure Consortium. Natural history of pediatric intestinal failure: initial report from the Pediatric Intestinal Failure Consortium. J Pediatr 161: 723–728.e2, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, 2015. http://www.R-project.org/. [Google Scholar]
- 56.Thurl S, Muller-Werner B, Sawatzki G. Quantification of individual oligosaccharide compounds from human milk using high-pH anion-exchange chromatography. Anal Biochem 235: 202–206, 1996. [DOI] [PubMed] [Google Scholar]
- 57.Thurl S, Munzert M, Henker J, Boehm G, Muller-Werner B, Jelinek J, Stahl B. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br J Nutr 104: 1261–1271, 2010. [DOI] [PubMed] [Google Scholar]
- 58.Totten SM, Zivkovic AM, Wu S, Ngyuen U, Freeman SL, Ruhaak LR, Darboe MK, German JB, Prentice AM, Lebrilla CB. Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J Proteome Res 11: 6124–6133, 2012. [DOI] [PubMed] [Google Scholar]
- 59.Tyler AD, Knox N, Kabakchiev B, Milgrom R, Kirsch R, Cohen Z, McLeod RS, Guttman DS, Krause DO, Silverberg MS. Characterization of the gut-associated microbiome in inflammatory pouch complications following ileal pouch-anal anastomosis. PloS One 8: e66934, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyson JE, Kennedy KA. Minimal enteral nutrition for promoting feeding tolerance and preventing morbidity in parenterally fed infants. Cochrane Database Syst Rev CD000504, 2000. [DOI] [PubMed] [Google Scholar]
- 61.Vulevic J, Juric A, Walton GE, Claus SP, Tzortzis G, Toward RE, Gibson GR. Influence of galacto-oligosaccharide mixture (B-GOS) on gut microbiota, immune parameters and metabonomics in elderly persons. Br J Nutr 114: 586–595, 2015. [DOI] [PubMed] [Google Scholar]
- 62.Wacklin P, Tuimala J, Nikkila J, Sebastian T, Makivuokko H, Alakulppi N, Laine P, Rajilic-Stojanovic M, Paulin L, de Vos WM, Matto J. Faecal microbiota composition in adults is associated with the FUT2 gene determining the secretor status. PloS One 9: e94863, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wales PW, Kosar C, Carricato M, de Silva N, Lang K, Avitzur Y. Ethanol lock therapy to reduce the incidence of catheter-related bloodstream infections in home parenteral nutrition patients with intestinal failure: preliminary experience. J Pediatr Surg 46: 951–956, 2011. [DOI] [PubMed] [Google Scholar]
- 64.Weser E. Intestinal adaptation to small bowel resection. Am J Clin Nutr 24: 133–135, 1971. [DOI] [PubMed] [Google Scholar]
- 65.Yu ZT, Chen C, Kling DE, Liu B, McCoy JM, Merighi M, Heidtman M, Newburg DS. The principal fucosylated oligosaccharides of human milk exhibit prebiotic properties on cultured infant microbiota. Glycobiology 23: 169–177, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zitomersky NL, Atkinson BJ, Franklin SW, Mitchell PD, Snapper SB, Comstock LE, Bousvaros A. Characterization of adherent bacteroidales from intestinal biopsies of children and young adults with inflammatory bowel disease. PloS One 8: e63686, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.