Abstract
Recent studies have demonstrated differences in the intestinal microbiota between patients with irritable bowel syndrome (IBS) and healthy controls (HC), suggesting a role for the intestinal microbiota in the pathogenesis of IBS. Alterations in the microbiota have also been implicated in the pathogenesis of abdominal bloating, a commonly reported symptom in IBS. We investigated the relationship between the intestinal microbiota, abdominal bloating, and altered bowel patterns in a cohort of patients with IBS and HC. The 16S rRNA gene from fresh fecal samples was amplified and pyrosequenced by using Roche-454 Titanium chemistry. A Core Measurable Microbiome (CMM) was generated for Operational Taxonomic Unit (OTU) detected in >75% of all samples and compositional features of CMM were compared between groups by Linear Discriminant Analysis (LDA). IBS differentiated from HC by LDA using continuous variation in the species/OTUs or the CMM genera. When subcategorized based on bloating symptoms and bowel characteristics, the same subjects were also well differentiated from one another and from HC. ANOVA analysis showed quantitative species/OTU differences between the subgroups including IBS with and without bloating, and subtypes based on bowel characteristics. The clear LDA differentiation and the significant microbial taxa differences between the groups imply a significant association of the microbiota with bloating symptoms and bowel characteristics in IBS. These changes in the microbiota may serve as a biomarker for IBS and its clinical subtypes and suggest a role for the intestinal microbiota in the pathogenesis of the main symptoms of the disorder.
Keywords: abdominal bloating, irritable bowel syndrome, intestinal microbiota, 16S rRNA gene, pyrosequencing
alterations in the intestinal microbiota have been implicated in the pathogenesis of functional bowel disorders (24, 27, 29, 33). Recent studies investigating compositional differences of the intestinal microbiota in patients with irritable bowel syndrome (IBS) have demonstrated quantitative and qualitative differences between patients and healthy controls (HCs) and suggested an association between certain bacteria and specific IBS symptoms (3, 5, 7, 22, 28). Most published studies in this area have investigated a mixed group of IBS patients or focused on specific subtypes of IBS based on bowel characteristics (2, 3, 5, 7, 22, 28). However, the association between the intestinal microbiota and abdominal bloating, one of the most common symptoms in IBS that may be specifically related to the intestinal microbiota, has not been systematically investigated. Indeed, population-based epidemiological data show that 82.5% of IBS patients report bloating symptoms and rate abdominal bloating as the second most bothersome symptom after abdominal pain (25). Abdominal distention was originally reported as one of six factors that discriminate IBS from organic conditions and were included in the criteria for the diagnosis of the disorder (26). The presence of abdominal bloating is considered supportive and increases the confidence of the diagnosis of IBS (13).
Several pathophysiological mechanisms have been proposed for the pathogenesis of abdominal bloating including abnormal intraluminal fermentation by colonic bacteria with excessive gas production (17, 19, 30). However, there is a lack of conclusive evidence for these mechanisms and, given the incomplete understanding of the etiology of bloating symptoms, effective treatments for patients with these symptoms are limited. Nevertheless, clinical trials targeting the intestinal microbiota in patients with IBS using antibiotics, probiotics, and prebiotics have demonstrated clinical beneficial effects, specifically on abdominal bloating (8, 20, 26, 31, 32). Collectively, these anecdotal reports point toward a possible role of the intestinal microbiota in the pathogenesis of IBS and bloating symptoms. However, to the best of our knowledge no previous study in IBS patients specifically investigated the intestinal microbiota in relation to symptoms of abdominal bloating.
The goal of the present study was to investigate the relationship between compositional features of the intestinal microbiota and the most common and clinically relevant symptoms of IBS. To achieve this goal we characterized and compared the intestinal microbiota among carefully selected patients with IBS who were clinically categorized based on symptoms of abdominal bloating and bowel characteristics, and HCs, using high throughput pyrosequencing of the 16S rRNA gene and a conservative approach to detect quantitative changes in the relative abundances of taxa.
MATERIALS AND METHODS
Study population.
A total of 80 subjects were examined, 60 of whom met the Rome III criteria for IBS and 20 HCs. Subjects were recruited from the Chapel Hill general population by advertising.
Inclusion criteria consisted of subjects that were 18 years or older and of any gender, race, or ethnicity. Healthy controls had no recurring gastrointestinal (GI) symptoms. IBS subjects had active GI symptoms at mild to moderate severity (IBS-symptom severity scale score 175–300) and unrevealing medical history, physical examination, and routine laboratory tests. Subjects age >50 yr old had a normal colonoscopy within the last 3 years.
Subjects with a history of GI tract surgery (other than appendectomy or cholecystectomy), a history of inflammatory bowel diseases (IBD), celiac disease, lactose malabsorption, or any other diagnosis that could explain their bowel symptoms were excluded. Subjects were also excluded if they used aspirin or other anti-inflammatory medication (e.g., NSAIDs) on a regular basis. An 8-wk washout period was required for subjects who reported use of antibiotics or intentional consumption of probiotics prior to enrollment. All subjects were evaluated by a physician to exclude an alternative diagnosis to IBS. To determine eligibility all subjects were evaluated for their clinical symptoms at baseline using the Rome III IBS module (www.romecriteria.org/pdfs/IBSMode.pdf). In addition, to confirm accurate categorization into the clinically relevant subgroups, all subjects underwent a 2-wk run-in period during which they completed a daily symptom diary (see Supplemental Material; Supplemental Material for this article is available online at the Journal website). All participants were specifically instructed to avoid changing their diets, start new medications, or make significant changes in their life habits during the 2-wk run-in period. The study was approved by the Institutional Review Board and all subjects provided written consent prior to participation in the study.
Sample collection and preparation.
Fresh stool samples were collected on site from all subjects during a study visit. Subjects unable to provide stool samples at the study visit were instructed to collect a specimen at home and return it to study staff the same morning. Each fecal sample was immediately transferred to the laboratory, where it was homogenized, divided into aliquots, and stored at −80°C for future DNA isolation and molecular microbiological analysis.
Isolation of DNA.
Bacterial DNA was isolated from all fecal samples by a phenol/chloroform extraction method combined with physical disruption of bacterial cells and a DNA cleanup kit (Qiagen DNeasy Blood and Tissue extraction kit; Qiagen, Valencia, CA) as previously described (5).
454 Pyrosequencing of 16S rRNA genes.
The V1–V2 region of the 16S ribosomal RNA gene was amplified using bar-coded fusion primers containing Roche-454 A or B Titanium sequencing, followed by a unique 8-base barcode sequence (B) and finally, the 5′ ends of primer A-8FM (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGBBBBBBBBAGAGTTTGATCM-TGGCTCAG) and of primer B-357R (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGBB BBBBBBCTGCTGCCTYCCGTA-3′). Amplicon saturation in the PCR products was confirmed by comparing band intensities of the PCR products after gel electrophoresis with standards using GeneTools software (Syngene). Amplicons from 80 individual samples were pooled in equal amounts, gel-purified, quantified by Pico Green analysis, and used for attachment to individual beads and subsequent emulsion PCR (emPCR). After recovery and enrichment for DNA-containing beads, the emPCR products from the 80-sample pools were sequenced on individual regions of two-region Picotitre plates on a Roche-GS-FLX machine by using Titanium chemistry as previously described (12, 21).
Pyrosequencing data processing pipeline.
16S rRNA raw sequence data generated by the Roche-454 GS-FLX machine was processed through scripts that filter data by the following quality metrics: each sequence contains 1) a complete forward primer sequence and barcode; 2) ≤2 N in a sequence read, where N is equivalent to an interrupted and resumed sequencing signal from sequential flows; 3) a sequence of >200 nucleotides (NT) and <500 NT in length; and 4) an average quality score ≥20 across the entire length of the sequence. After filtering, the remaining reads were trimmed to remove 5′ and 3′ adapter and primer sequences, parsed by barcode into corresponding sample files, automatically associated with a matching .QUAL file containing the quality scores, and uploaded into the in-house-designed CAGE-Microbiome Database. The processed data and the MySQL database tables are stored on a database server and are available to the public at http://cage.unl.edu.
To normalize taxonomic assignment of individual sequence reads, the entire data set was processed through the Multi-CLASSIFIER algorithm, which assigns hierarchical taxonomic status to each sequence read based on a covariance model developed from a training set (36). Sequences were then parsed into “classified” and “unclassified” sets based on meeting threshold limits of 0.8 at the genus level against the Multi-CLASSIFIER model. Classified reads were then assigned species/Operational Taxonomic Unit (OTU)-level status by using a BLAST pipeline that associates the read with taxonomic assignment using a curated database developed from RDP and SILVA 16S ribosomal RNA sequences (21, 36).
Sequences were considered a species match if they achieved 97% identity with a reference sequence over a minimum of 200 bases of contiguous BLAST alignment. Sequence reads that failed to meet the 0.8 scoring threshold at the genus level from the Multi-CLASSIFIER algorithm (unclassified reads) were processed into a separate phylogeny-based, OTU pipeline that binned sequences using the CD-HIT clustering algorithm at a 97% cutoff (12). Reads from the “classified” and “unclassified” portions of the bifurcated pipeline were then combined and quantified relative to the total number of reads for each sample, by calculating the relative proportion for each taxonomic rank according to the following formula:
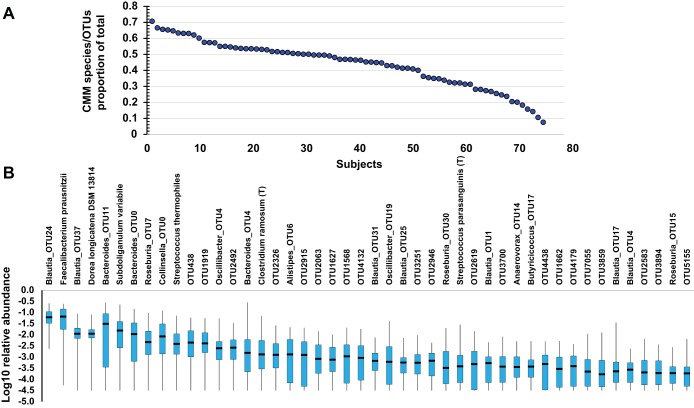
To remove the effects of excessive variation in the data due to taxa that were low abundance and/or sparsely distributed, the taxa were further filtered to remove species and OTUs where sequence reads were not detected across at least 75% of the subjects (1, 10, 11). This resulted in the Core Measurable Microbiome (CMM), which comprised 46 species/OTUs, 21 genera, 14 families, 7 orders, and 4 phyla. The collective CMM species/OTU representation of the total 16S sequencing data across individual subjects ranged from 10 to 70% with a median of 47%. At the genus level, the CMM range from 23 to 76% of the total reads across subjects with a median of 55%.
Statistical analysis.
Statistical analyses were performed on log-transformed data by use of the StatistiXL package. For log-transformation, the relative abundances of the individual taxa were log10-transformed, and samples having no counts for a particular taxon were simply transformed to a value of −4.5 to avoid skewing the data. Linear Discriminant Analysis (LDA) was performed across the 46 species/OTUs comprising the CMM of all 76 subjects (four IBS cases were omitted because of low 16S rRNA sequence reads) in the study. The subjects were categorized with respect to IBS/No IBS, type of IBS, and bloating, and these categories were used as factors in the LDA analyses. To depict the contribution of taxa of interest to the LDA functions, the values for the standardized LDA functions for taxa showing significant differences by ANOVA were plotted in simple bar charts.
ANOVA was used to identify taxa whose variation was significantly associated with the IBS and bloating categories. To visualize the partitioning of taxa between subject groups, taxa showing significant effects IBS or bloating categories (P < 0.05) were plotted with box-and-whisker plots. Descriptive statistics for each taxon showing statistical significance among the groups are listed in Supplemental Tables S1–S3. The post hoc Tukey's test was used to correct for multiple testing where three or more subject groups were concerned and the P value for groups reaching statistical significance are indicted in Supplemental Tables S2 and S3.
RESULTS
Study population.
A total of 80 subjects were investigated of which 76 (56 IBS and 20 HC) had sufficient 16S rRNA sequence data for the final analysis. The study population consisted of 83% women with a mean age of 35 years. Demographics and body mass index (BMI) in all study groups are presented in Table 1. Subjects were subcategorized into the study subgroups of interest based on their reported clinical symptoms. For accuracy, eligibility and clinical category of interest were determined based on consistency of reported symptoms on the daily diary during the 2 wk run-in period. For the primary aim of the study we defined three clinically relevant groups of interest: IBS patients with bloating symptoms (IBS+B), IBS patients without bloating symptoms (IBS−B), and HC. To ensure active symptoms and to avoid overlap in symptoms between the groups, patients were subcategorized and included in the analysis based on their reported symptoms during the run-in period. The IBS+B group (n = 26) included patients who reported abdominal bloating at a severity of 3 or more (on a 0 to 10 Likert scale) at least 3 days per week. Patients who reported less frequent/severe symptoms were not included in the microbiota analysis. The IBS−B (n = 6) and the HC group (n = 16) included subjects who reported no bloating symptoms at screening and did not have bloating symptoms at a severity of >1 (on a 0 to 10 Likert scale) more than 3 days per week during the run-in period. The IBS+B group included 10 patients with symptoms of diarrhea-predominant IBS (D-IBS), 10 patients with symptoms of constipation-predominant IBS (C-IBS), and 6 patients with symptoms of mixed-bowel-pattern IBS (M-IBS). The IBS−B group included 1 patient with D-IBS, 2 patients with C-IBS, and 3 patients with M-IBS.
Table 1.
Characteristics of IBS patients and healthy controls
| Number of subjects | Age (years) mean (range) | % Female | BMI (kg/m2) mean (range) | |
|---|---|---|---|---|
| Total IBS | 56 | 35 (19–61) | 82% | 27 (19–41) |
| Total HC | 20 | 36 (21–60) | 90% | 27 (18–53) |
| IBS + Bloating | 26 | 35 (20–60) | 83% | 26 (19–40) |
| IBS - Bloating | 6 | 37 (19–61) | 88% | 33 (25–41) |
| D-IBS | 21 | 37 (23–60) | 71% | 28 (21–41) |
| C-IBS | 21 | 34 (20–50) | 100% | 27 (19–40) |
| M-IBS | 14 | 34 (19–61) | 78% | 28 (21–39) |
IBS, irritable bowel syndrome; BMI, body mass index; HC, healthy controls; D-IBS, diarrhea-predominant IBS; C-IBS, constipation-predominant IBS; M-IBS, mixed bowel pattern IBS.
The following groups of interest were defined for secondary analysis on the same study samples: IBS (n = 56), D-IBS (n = 21), C-IBS (n = 21), M-IBS (n = 14), and HC (n = 20). Each clinical subcategory included only subjects who provided sufficient data and repeatedly reported GI symptoms that were consistent with the subcategory of interest.
Characterization of the patients and HC fecal microbiota.
A total of 378,693 16S rRNA sequences with acceptable quality were obtained from the V1-2 16S rRNA regions with an average of 8,232 reads per sample (range: 2,939–19,305) (http://www.ncbi.nlm.nih.gov/sra/SRP066323 SRA accession: SRP066323). The number of sequence reads did not significantly differ between the four study groups. To determine the number and abundances of different bacterial groups in each sample we used 3% dissimilarity between 16S rRNA gene sequences as an indicator of a “species level” OTU. Using this procedure, we found a total of 53,191 species and OTUs of which 1,143 could be assigned at least to the genus level. A significant proportion of the species and OTUs are sparse, with 419 species and 12,689 OTUs being singletons occurring only in one individual. Indeed, the vast majority of the total species and OTUs were not detectable in even 50% of the subjects, and among those OTUs any given taxon was on average less than 0.1% of the microbiota. Even in OTUs present in 50–75% of the subjects most were less than 1%. To circumvent the effects of sample error from sparse and low-abundance taxa, we focused our analysis on a core of taxa that are observed in at least 75% of the subjects with at least 5 reads per subject. This resulted in a core of 46 species and OTUs that are log-normally distributed and comprise a shared CMM that varies quantitatively across the study population. Importantly, previous studies have used this method to avoid false discoveries associated with sparse data and have shown that taxa meeting these criteria are least susceptible to sample error (1, 10, 11). Collectively, the species/OTUs of the CMM comprise 10–70% of the sequence reads in any given individual (Fig. 1A) with a median of 47% of the sequence reads from an individual belonging to the CMM. The distribution of the 46 OTUs and species that comprise the CMM are shown in Fig. 1B. Taxonomically, the OUT species of the CMM comprise 20 genera, 14 families, 7 orders, 6 classes, and 4 phyla, representing the major sectors of phylogenetic space occupied by the microbiota.
Fig. 1.
Distributions of the Core Measurable Microbiome (CMM) in subjects of the study. A: dot plot depicts the proportion of the combined CMM species/Operational Taxonomic Unit (OTU)-level abundances out of the total species/OTU-level abundances for each individual in the study. The individuals are arranged in decreasing order of CMM/total from left to right. B: standard box-and-whisker plot depicts the distribution of the abundances of each of the 46 individual species/OTUs comprising the CMM with 50% of the data (box), the range (whiskers), and the median (line in box), plotted on a log10 scale of the relative abundances.
Differentiation of the clinical groups of interest by the intestinal microbiotas.
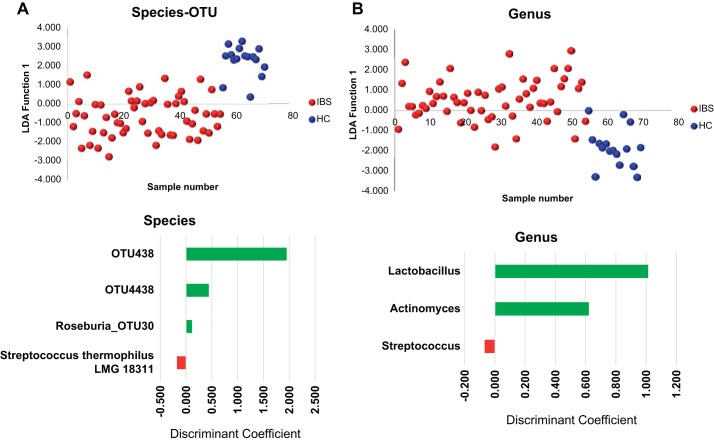
By using continuous variation in the 46 species/OTUs or the 20 genera comprising the CMM as variables, subjects with IBS were differentiated in one dimension by LDA analysis (Fig. 2, A and B). When these same subjects were further categorized by abdominal bloating, both subgroups were differentiated from the HC in the first dimension (x-axis) and the IBS subgroups differentiated from one another in the second (y) dimension, implying that there may be a significant association of the microbiota compositional features with bloating in IBS subjects (Fig. 3A).
Fig. 2.
Linear Discriminant Analysis (LDA) of the fecal microbiota in irritable bowel syndrome (IBS) and healthy controls (HC) by species level (A) and genus levels (B). LDA was performed on log-transformed relative abundances of the CMM with >95% of the variation being captured in a single LDA function when using CMM species or genera represented in the CMM. The values for the standardized LDA functions of taxa showing statistically significant differences between HCs and IBS subjects by ANOVA are plotted in graphs below the LDA plots.
Fig. 3.
LDA of the fecal microbiota in study population by abdominal bloating (A) and by IBS subtypes based on bowel characteristics (B). LDA analysis was performed on the CMM and values for the first 2 LDA functions are plotted for the HC and each of the IBS subtypes. The bar charts below the LDA plots show the standardized LDA functions of taxa showing statistically significant differences between subgroups by ANOVA. CIBS, constipation-predominant IBS; DIBS, diarrhea-predominant IBS; MIBS, mixed-bowel-pattern IBS.
Lastly, when these subjects were categorized based on bowel characteristics, the three IBS bowel subtypes were clearly differentiated from one another, with C-IBS and M-IBS separating in the first dimension and D-IBS separating in the second dimension (Fig. 3B). The HC were largely centered between the C-IBS and M-IBS patients. Thus, based on quantitative differences in the CMM, it appears that the IBS subjects in our study could be categorically differentiated from HCs and they could also be further differentiated from one another based on bloating or IBS bowel subtype.
Identification of bacterial taxa that show partitioning between clinical groups of interest.
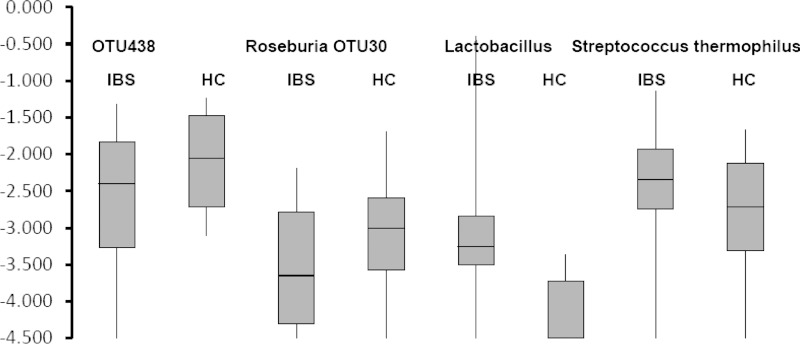
Given the differentiation of the IBS subjects from HC by LDA analysis, we next sought to identify species/OTUs that show significant quantitative differences across IBS categories. ANOVA was used to test significance of variation within and between groups with the Tukey correction to correct for multiple testing where relevant. An OTU belonging to Roseburia and OTUs related to the Lachnospiraceae (OTU438) were significantly decreased in the IBS subjects (Supplemental Table S1, Fig. 4). In contrast, the genus Lactobacillus and a species of Streptococcus that is closely related to S. thermophilus and S. salivarius were elevated in the IBS subjects. In the instance of Lactobacillus, the levels in many of the IBS subjects was close to 0.1% of the fecal population but were near the lower threshold of our CMM criteria in many of the healthy subjects.
Fig. 4.
Box-and-whisker plots showing species/OTUs with significant quantitative differences between IBS (all subgroups) and HCs by ANOVA. The box depicts 50% of the data and whiskers depict the range.
When the IBS subjects were subgrouped by their bloating symptoms, both IBS+B and IBS−B showed statistically significant elevation of the genera Lactobacillus (which comprised L. johnsonii and L. apodemi) and Streptococcus (OTUs most closely related to S. parasanguinis, and S. salivarious/S. thermophilus) vs. HCs (Supplemental Table S2, Fig. 5). The elevation in Lactobacillus and Streptococcus species among the IBS with and without bloating was accompanied by significant decreases in taxa most closely related to the Lachnospiraceae (OTUs 4438 and 2915). Remarkably, the IBS subjects with and without bloating could also be differentiated from one another by their relative abundances of members of the Ruminococcaceae and the Eubacteriaceae. Among the Ruminococcaceae, which were abundant taxa in HC and IBS subjects with bloating, substantial decreases were observed in Subdoligranulum variable members of the genus Anaerotuncus compared with the IBS subjects without bloating. Likewise, Anaerovorax OTU14, belonging to the Clostridiales Family XIII. Incertae Sedis, showed this same pattern of significant decrease in IBS subjects without bloating (Supplemental Table S2, Fig. 5).
Fig. 5.
Box-and-whisker plots showing the distribution of taxa representing the patterns of differences observed between IBS with (IBS+Bloat) and without (IBS−Bloat) bloating and HCs. Taxa depicted are a subset of the taxa from Supplemental Table S2 and represent the different patterns of taxonomic variation that shows statistical significance detected by ANOVA. Significant differences between groups (P < 0.05) were defined by Tukey's post hoc test and are indicated by significant difference between HC and IBS−Bloat (a); significant difference between HC and groups IBS+Bloat and IBS−Bloat (b); and significant difference between IBS +Bloat and groups HC and IBS−Bloat (c).
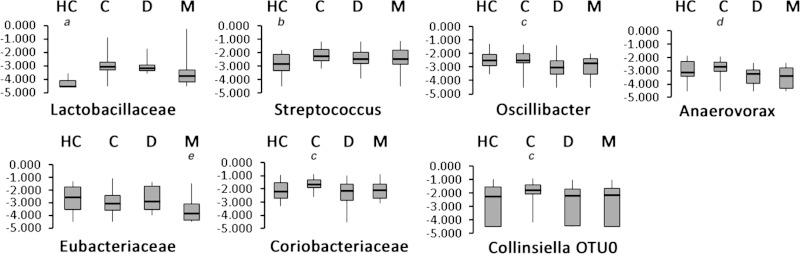
With respect to IBS subtypes, taxa from two different phyla of gram-positive bacteria, the Actinobacteria and the Firmicutes, showed significant differentiation between the subtypes. Within the phylum Actinobacteria, an OTU belonging to the genus Collinsiella (OTU0) was elevated significantly in the C-IBS subgroup and contributed to the statistical significance of the family Coriobacteriaceae (Fig. 6). Although Collinsiella is a significant proportion of the population of Coriobacteriaceae, other unidentified members of this family must also reflect this distribution because the Coriobacteriaceae family showed the same trend (elevation in C-IBS subjects) and statistical significance despite being more abundant overall than the Collinsiella OTU.
Fig. 6.
Box-and-whisker plots illustrating the distribution of taxa that show significant quantitative differences between IBS subgroups (C, constipation predominant; D, diarrhea predominant; M, mixed bowel pattern) and HCs by ANOVA. Significant differences between groups (P < 0.05) were defined using Tukey's post hoc test and are indicated by significant difference between HC and groups C, D, and M (a); significant difference between HC and groups C and D (b); significant difference between C and group D (c); significant difference between C and groups D and M (d); and significant difference between M and groups HC, C, and D (e).
Differentiation between IBS subtypes was observed with several taxa. For example, statistically significant decreases in several members of the Clostridiales, with Oscillibacter, Anaerovorax, and a member of the Incertae sedis XIII group all showing similar patterns, being decreased in subjects with D-IBS and M-IBS but not C-IBS compared with HC (Supplemental Table S3, Fig. 6). The M-IBS subjects also had substantial depletion of the genus Eubacterium and members of the Lachnospiraceae (OTU2063 and OTU438) (Supplemental Table S3) whereas a single species of Roseburia (Roseburia OTU15) was elevated in C-IBS subjects (Supplemental Table S3).
A notable observation in HCs was the lower abundances of members of the genus Lactobacillus, which were nearly at the limits of detection in most HCs but were ∼10-fold higher in subjects with C-IBS and D-IBS (Fig. 6). Despite the magnitude of this difference, no single species of Lactobacillus alone accounted for the difference. L. apodemi and L. johnsonii were the only Lactobacillus species detected but together did not account for total Lactobacillus abundances at the genus level in more than 50% of the individuals. Although a highly diverse genus, Lactobacillus species exhibit relatively low diversity in the 16S genes, and this problem is exacerbated by the relatively short reads of pyrosequencing. Thus many of the Lactobacillus species will fail to resolve below the genus level in our pipeline. Similarly, the genus Streptococcus and the species related to S. thermophilus/salivarius showed this similar pattern with statistically significant differences between HC and all IBS subtypes.
DISCUSSION
IBS patients present a challenging group of GI disorders because of the broad set of symptoms that can manifest in the absence of defined pathophysiological features. The recently discovered associations of IBD and other GI diseases with dysbiosis of the intestinal microbiota has brought study of this complex community to the forefront of investigation of many diseases and is an obvious candidate to test for association with IBS. Our study specifically investigated a cohort of well-defined patients with IBS with a primary focus on identifying relationships between the intestinal microbiota and abdominal bloating. Bloating symptoms are very common in patients with IBS and have a significant impact on the overall symptom severity, quality of life, and healthcare utilization (25). Despite commonality of bloating symptoms among IBS patients, its pathophysiological mechanisms are not well understood. It has been proposed that, at least in some patients, abnormal intraluminal fermentation by the intestinal bacteria could lead to excessive gas production and bloating symptoms (9). Involvement of the intestinal microbiota with abdominal bloating is also supported by the observations that therapeutic interventions targeting the intestinal microbiota, including antibiotics, probiotics, prebiotics, and diet low in fermentable carbohydrates, demonstrate a beneficial effect specifically on bloating symptoms (8, 19, 20, 26, 31, 32, 35).
We used deep pyrosequencing to test shared taxonomic groups of the intestinal microbiota for quantitative differences between HC and IBS phenotypic subgroups categorized based on bloating symptoms and altered bowel patterns. Overall, this approach points toward significant association of the intestinal microbiota with IBS. Although the CMM enables a statistically robust analysis of the data most accurately measured by pyrosequencing, a drawback is that the approach is blind to taxa with between-group differences (e.g., IBS vs. control) that are greater than the dynamic range of the measurements. With typical pyrosequencing depths of ∼10,000 reads per sample, only a 1,000-fold dynamic range can be accurately quantified from a complex ecosystem. With gut microbiota densities ranging in the tens to hundreds of trillions, it is possible that between group differences could range well beyond 1,000-fold dynamic range of typical pyrosequencing depths. Although taxa displaying these essentially binary distributions between groups are indeed of importance (particularly for diagnostic value), their discovery will be accompanied by a high false discovery rate. The vast majority of taxa not making the CMM threshold are relatively low abundance and/or very sparse (Fig. 1B). Such features are highly subject to sample error due to the complexity of the gut microbiota (1, 10, 11) and they will inflate the false discovery when included in an analysis. From a fundamental perspective, reducing the dimensionality of the microbiota to the CMM dramatically lowers the likelihood of false discovery. This same approach (although not necessarily referred to as the CMM) has also been used by others (6, 10, 11, 18, 34). Indeed, observing statistically significant differences among multiple taxa in the CMM between subject groups provides robust support that the microbiota indeed bears significant, measurable biological signatures that are associated with IBS.
Based on quantitative differences in the CMM alone, IBS subjects with or without bloating were differentiated from HCs and from each other. Statistical analysis of individual taxonomic groups identified significant quantitative differences between microbial taxa within the Firmicutes phylum. Specifically, IBS subjects showed significantly higher levels of species of Lactobacillus and Streptococcus with the most significant elevation being observed in IBS subjects without bloating. The Streptococci, in particular, showed dramatic elevation, being present at only 0.1–0.3% of the microbiota in healthy subjects but comprising 0.3–5% of the microbiota in the IBS patients. IBS subjects without bloating constituted the most extreme elevation of Streptococci, ranging from 1–5% of the microbiota, whereas in IBS subjects with bloating the range was 0.3–2% of the microbiota. Lactobacilli, which showed the same trend, were nearly at the limits of detection in healthy subjects (0.01%) but were present at 0.03–0.1% of the microbiota in IBS subjects, with the highest levels being observed in IBS subjects without bloating.
Perhaps one of the most remarkable findings in our study was the tradeoffs observed almost exclusively within the Firmicutes. Concurrent with the substantial elevation of Lactobacilli and Streptococci, all IBS subjects demonstrated significant depletion of species belonging to the Lachnospiraceae (OTU4438 and OTU2915). Additionally, species and OTUs of Subdoligranulum and Anaerovorax (belonging to the families Ruminococcaceae and Eubacteriaceae, respectively) were specifically depleted in the IBS subgroup without bloating (Fig. 5). These quantitative differences observed across significant portions of the Clostridial phylogenetic space illustrate the significance of the association of microbiota composition with IBS as well as specific taxonomic differences that stratify the IBS subgroups. Elevated levels of Lactobacillus and Streptococcus and depletion of members of the Lachnospiraceae in all IBS subjects compared with HCs suggests that these taxa could contribute to the general symptoms of IBS. On the other hand, the extreme elevation of Lactobacillus and Streptococcus coupled with depletion of Ruminococcaceae (particularly Subdoligranulum), may contribute to differentiation of the bloating subgroups of IBS. Whether this pattern is causal or a downstream consequence of IBS itself remains unclear. It is interesting to note that, although portions of this pattern of microbiota composition have been observed in other IBS subjects, the pattern is far from universal. In one study, members of the Ruminococcaceae were negatively associated with IBS symptoms as we observed but, in contrast, these same subjects showed negative correlation between IBS and species of Lactobacillus and Streptococcus (15). Likewise, a study of IBS in children found positive association of Lactobacillus with IBS as we observed, but the depletion of Clostridial species was not observed (23). Lastly, a study of IBS subtypes among a small cohort of subjects found depleted levels of Clostridial species in some IBS subtypes, but in contrast to our observations found elevated levels of Ruminococci (14). Collectively, these data sets suggest that there may be multiple compositional features of the microbiota that can be associated with IBS in different subject populations. It is worth noting that, despite significant differences in the abundances of taxa belonging to the Firmicutes, we did not observe tradeoffs with members of the other dominant phylum of the gut microbiota of humans: the Bacteriodetes. Whether this reflects a unique signature of IBS remains unclear.
In contrast to the bloating, IBS subgroups based on bowel characteristics showed differences associated with taxa from the Actinobacteria and the Firmicutes phyla. The Actinobacteria phyla (in particular the genus Collinsiella) was significantly increased in the C-IBS subgroup compared with all other groups and members of the Firmicutes phyla (Oscillibacter, Anaerovorax, Incertae sedis XIII, Streptococcus, and Eubacteriaceae) were significantly decreased in D-IBS and M-IBS compared with HC and C-IBS.
Previous reports that studied a dysbiotic microbiota in IBS have not specifically investigated the association between the composition of enteric microbes and abdominal bloating symptoms. However, a few intestinal microbiota-based studies in IBS have provided some data on the enteric microbial composition in specific subtypes of this disorder (2, 3, 5). Specifically, using a phylogenetic (HITChip) microarray analysis on fecal samples from 62 patients with IBS (D-IBS n = 25, C-IBS n = 18, M-IBS n = 19) and 46 HC, Rajilic-Stojanovic et al. (22) reported a significant increase in the abundance of Firmicutes (Dorea, Ruminococcus, and Clostridum spp.), Bifidobacterium and Faecalibacterium spp; and a significant decrease the abundance of Bacteroidetes in patients with IBS compared with HC. Additionally, Jeffery at al. (7) have investigated the V4 region of the 16S rRNA gene to characterize the intestinal microbiota in fecal samples collected from 37 patients with IBS (D-IBS n = 15, C-IBS n = 10, M-IBS n = 12) and 20 HC. This study reported a significant increase in the ratio of Firmicutes to Bacteroidetes with an increase in the abundance of Firmicutes and a general decrease in two of the most common genera of Bacteroidetes (Bacteroides and Alistipes). However, no detectable changes in the fecal microbiota were observed in a subgroup of patients with normal Firmicutes-to-Bacteroidetes ratio.
Our study has several unique strengths. We included a well-defined study population that met rigorous inclusion/exclusion criteria, a clinical interview by a physician with expertise in functional GI disorders, and screening laboratory blood and stool tests to exclude possible organic conditions. In addition, we included a 2-wk run-in period to record daily GI symptoms and bowel habits to ensure activity of symptoms at time of sample collection and accurate phenotypic categorization to the clinically relevant subgroups of interest. We used this conservative approach to avoid overlap in symptoms between the groups and to conduct the analysis only on subjects with vs. without significant symptoms. In addition, we carefully screened and excluded subjects who used probiotics or medications that can alter the gut microbiota (e.g., antibiotics and anti-inflammatory drugs). Fecal samples were collected on-site in the majority of the patients or, when not possible, on the same morning of the study visit. All fecal samples were kept at 4°C from time of delivery and were transferred immediately to the laboratory where they were processed and stored until analyzed. We have recently validated our fecal collection protocol and demonstrated a good microbial DNA stability during collection, processing, and sample storage (4). Another strength of our study is the methodology used to extract the DNA, in which we used enzymatic (lysozyme and proteinase K), physical disruption (bead-beating), and chemical (sodium dodecyl sulfate) steps to ensure complete lysis of bacterial cells.
The contribution of the microbiota to the pathophysiology of bloating symptoms is likely to be multifactorial, including its effect on motility, sensation, and fermentation processes. The analysis conducted in this study provides information about the composition but not the function of the intestinal microbiota, and we did not measure fermentation products (e.g., intestinal gas and short-chain fatty acids) in this study. Furthermore, analysis of the microbiota of fecal samples is able to provide only general information about the overall makeup of the intestinal microbiota and does not provide information about localization of microbiota differences (e.g., small vs. large bowel). Such information may be relevant to the pathogenesis of bloating symptoms, particularly in IBS, due to overlap with clinical conditions such as small bowel bacterial overgrowth. It should also be noted that the IBS−B group had a higher BMI than the other groups and therefore may have potential confounding effect. In addition, although we were able to detect differences in microbial community structure between the groups analyzed the sequencing depth generated by the 454 sequencing used in this study (about 1 million reads per run) may not be sufficient to detect additional low-abundance taxa that may differ between groups. Other limitations of our study are the evaluation of the microbiota at a single time point and the lack of information on the effect of diet. The latter is emphasized by the observed higher levels of Lactobacillus found in the IBS subtypes and a similar trend in the levels of species related to Streptococcus thermophilus, raising the possibility that, despite the study-specific instructions and daily diary questions, IBS patients were consuming yogurt whereas the HCs were not. However, we were not able to attribute the effects observed in the Lactobacillus populations to any particular species as the individual species were only sparsely distributed and thus did not meet our distribution criteria (present in 75% of subjects). Given sparse distribution of individual species but a substantial overall effect on the lactobacillus independent of species, we believe the pattern is more consistent with a true disease association rather than a dietary difference. However, as is with other reported studies in this area, these limitations should be addressed in future longitudinal studies that will specifically control for the effects of diet. Another factor that deserves further investigation is the intestinal motility, particularly since the direction of causality of the interaction between the intestinal motility and the intestinal microbiota is not clear.
In conclusion, we demonstrated that IBS patients could be categorically differentiated from HC based on their intestinal microbiota and that they could be further differentiated from HC and from one another when categorized by bloating symptoms or bowel characteristics. We found significant differences in the intestinal microbiota between these clinically relevant IBS subtypes and demonstrated an association between intestinal dysbiosis, symptoms of abdominal bloating, and altered bowel habits. However, although differences in many individual taxa were identified, there was little difference in the relative abundances of major taxonomic groups (e.g., ratio of Bacteriodetes:Firmicutes:Proteobacteria). Taken together the results of our study support the hypothesis that the intestinal microbiota may have an important role in the pathogenesis of IBS and specifically the commonly reported symptoms of bloating in patients with IBS. They provide a rationale and emphasize the need for further research to clarify the role of the altered microbiota in the pathogenesis of IBS and the therapeutic potential of targeting these alterations for prevention and treatment of patients with this disorder.
GRANTS
This research was supported in part by DK084294 and DK075621 grants from the National Institutes of Health awarded to Y. Ringel.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
T.R.-K., A.K.B., and Y.R. conception and design of research; T.R.-K., A.K.B., I.M.C., J.K., R.M.L., and Y.R. performed experiments; T.R.-K., A.K.B., and Y.R. interpreted results of experiments; T.R.-K., A.K.B., and Y.R. drafted manuscript; T.R.-K., A.K.B., I.M.C., and Y.R. edited and revised manuscript; T.R.-K., A.K.B., I.M.C., J.K., R.M.L., and Y.R. approved final version of manuscript; A.K.B., I.M.C., J.K., R.M.L., and Y.R. analyzed data; A.K.B. prepared figures.
Supplementary Material
REFERENCES
- 1.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 107: 18933–18938, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll IM, Chang YH, Park J, Sartor RB, Ringel Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog 2: 19, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll IM, Ringel-Kulka T, Keku TO, Chang YH, Packey CD, Sartor RB, Ringel Y. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 301: G799–G807, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll IM, Ringel-Kulka T, Siddle JP, Klaenhammer TR, Ringel Y. Characterization of the fecal microbiota using highthroughput sequencing reveals a stable microbial community during storage. PLoS One 7: E46953, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterology 24: 521–530, e248, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. The long-term stability of the human gut microbiota. Science 341: 1237439, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeffery IB, O'Toole PW, Öhman L, Claesson MJ, Deane J, Quigley EM, Simrén M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 61: 997–1006, 2012. [DOI] [PubMed] [Google Scholar]
- 8.Kim HJ, Camilleri M, McKinzie S, Lempke MB, Burton DD, Thomforde GM, Zinsmeister AR. A randomized controlled trial of a probiotic, VSL#3, on gut transit and symptoms in diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther 17: 895–904, 2003. [DOI] [PubMed] [Google Scholar]
- 9.King TS, Elia M, Hunter JO. Abnormal colonic fermentation in irritable bowel syndrome. Lancet 352: 1187–1189, 1998. [DOI] [PubMed] [Google Scholar]
- 10.Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM, Huang H, Vangay P, Al-Ghalith GA, Russell C, Sauk J, Knight J, Daly MJ, Huttenhower C, Xavier RJ. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med 6: 107, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leamy LJ, Kelly SA, Nietfeldt J, Legge RM, Ma F, Hua K, Sinha R, Peterson DA, Walter J, Benson AK, Pomp D. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol 15: 552, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Godzik A. CD-HIT: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659, 2006. [DOI] [PubMed] [Google Scholar]
- 13.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology 130: 1480–1491, 2006. [DOI] [PubMed] [Google Scholar]
- 14.Lyra A, Rinttilä T, Nikkilä J, Krogius-Kurikka L, Kajander K, Malinen E, Mättö J, Mäkelä L, Palva A. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol 15: 5936–5945, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malinen E, Krogius-Kurikka L, Lyra A, Nikkilä J, Jääskeläinen A, Rinttilä T, Vilpponen-Salmela T, von Wright AJ, Palva A. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol 16: 4532–4540, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning AP, Thompson WG, Heaton KW, Morris AF. Towards positive diagnosis of the irritable bowel. Br Med J 2: 653–654, 1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maxton DG, Martin DF, Whorwell PJ, Godfrey M. Abdominal distension in female patients with irritable bowel syndrome: exploration of possible mechanisms. Gut 32: 662–664, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKnite AM, Perez-Munoz ME, Lu L, Williams EG, Brewer S, Andreux PA, Bastiaansen JWM, Wang X, Kachman SD, Auwerx J, Williams RW, Benson AK, Peterson DA, Ciobanu DC. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One 7: e39191, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong DK, Mitchell SB, Barrett JS, Shepherd SJ, Irving PM, Biesiekierski JR, Smith S, Gibson PR, Muir JG. Manipulation of dietary short chain carbohydrates alters the pattern of gas production and genesis of symptoms in irritable bowel syndrome. J Gastroenterol Hepatol 25: 1366–1373, 2010. [DOI] [PubMed] [Google Scholar]
- 20.Pimentel M, Lembo A, Chey WD, Zakko S, Ringel Y, Yu J, Mareya SM, Shaw AL, Bortey E, Forbes WP; TARGET Study Group. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med 364: 22–32, 2011. [DOI] [PubMed] [Google Scholar]
- 21.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajilić-Stojanović M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, de Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 141: 1792–1801, 2011. [DOI] [PubMed] [Google Scholar]
- 23.Rigsbee L, Agans R, Shankar V, Kenche H, Khamis HJ, Michail S, Paliy O. Quantitative profiling of gut microbiota of children with diarrhea-predominant irritable bowel syndrome. Am J Gastroenterol 107: 1740–1751, 2012. [DOI] [PubMed] [Google Scholar]
- 24.Ringel Y, Carroll IM. Alterations in the intestinal microbiota and functional bowel symptoms. Gastrointest Endosc Clin N Am 19: 141–150, 2009. [DOI] [PubMed] [Google Scholar]
- 25.Ringel Y, Williams RE, Kalilani L, Cook SF. The prevalence, characteristics and impact of bloating symptoms in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 7: 68–72, 2009. [DOI] [PubMed] [Google Scholar]
- 26.Ringel-Kulka T, Palsson OS, Maier D, Carroll I, Galanko JA, Leyer G, Ringel Y. Clinical trial: Probiotic bacteria lactobacillus acidophilus NCFM and bifidobacterium lactis Bi-07 versus placebo for the symptoms of bloating in patients with functional bowel disorders: a double-blind study. J Clin Gastroenterol 45: 518–525, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salonen A, de Vos WM, Palva A. Gastrointestinal microbiota in irritable bowel syndrome: present state and perspectives. Microbiology 156: 3205–3215, 2010. [DOI] [PubMed] [Google Scholar]
- 28.Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 141: 1782–1791, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saulnier DM, Ringel Y, Heyman MB, Foster JA, Bercik P, Shulman RJ, Versalovic J, Verdu EF, Dinan TG, Hecht G, Guarner F. The intestinal microbiome, probiotics and prebiotics in neurogastroenterology. Gut Microbes 4: 17–27, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serra J, Azpiroz F, Malagelada JR. Mechanisms of intestinal gas retention in humans: impaired propulsion versus obstructed evacuation. Am J Physiol Gastrointest Liver Physiol 281: G138–G143, 2001. [DOI] [PubMed] [Google Scholar]
- 31.Sharara AI, Aoun E, Abdul-Baki H, Mounzer R, Sidani S, El Hajj I. A randomized double-blind placebo-controlled trial of rifaximin in patients with abdominal bloating and flatulence. Am J Gastroenterol 101: 333, 2006. [DOI] [PubMed] [Google Scholar]
- 32.Silk DB, Davis A, Vulevic J, Tzortzis G, Gibson GR. Clinical trial: The effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment Pharmacol Ther 29: 508–518, 2009. [DOI] [PubMed] [Google Scholar]
- 33.Simren M, Barbara G, Flint H, Spiegel BM, Spiller RC, Vanner S, Verdu EF, Whorwell PJ, Zoetendal EG; Rome Foundation Committee. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut 62: 159–176, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivas S, Möller S, Wang J, Künzel S, Zillikens D, Baines JF, Ibrahim SM. Genome-wide mapping of gene-microbiota interactions in susceptibility to autoimmune skin blistering. Nat Commun 4: 2462, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staudacher HM, Whelan K, Irving PM, Lomer MC. Comparison of symptom response following advice for a diet low in fermentable carbohydrates (FODMAPs) versus standard dietary advice in patients with irritable bowel syndrome. J Hum Nutr Diet 24: 487–495, 2011. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, Garrity G, Tiedje J, Cole J. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.