Abstract
Clinical studies have linked microRNA-155 (miR-155) expression in the tumor microenvironment to poor prognosis. However, whether miR-155 upregulation is predictive of a pro- or antitumorigenic response is unclear, as the limited preclinical data available remain controversial. We examined miR-155 expression in tumor tissue from colon cancer patients. Furthermore, we investigated the role of this microRNA in proliferation and apoptosis, inflammatory processes, immune cell populations, and transforming growth factor-β/SMAD signaling in a chemically induced (azoxymethane-dextran sulfate sodium) mouse model of colitis-associated colon cancer. We found a higher expression of miR-155 in the tumor region than in nontumor colon tissue of patients with colon cancer. Deletion of miR-155 in mice resulted in a greater number of polyps/adenomas, an increased symptom severity score, a higher grade of epithelial dysplasia, and a decrease in survival. Surprisingly, these findings were associated with an increase in apoptosis in the normal mucosa, but there was no change in proliferation. The protumorigenic effects of miR-155 deletion do not appear to be driven solely by dysregulation of inflammation, as both genotypes had relatively similar levels of inflammatory mediators. The enhanced tumorigenic response in miR-155−/− mice was associated with alterations in macrophages and neutrophils, as markers for these populations were decreased and increased, respectively. Furthermore, we demonstrated a greater activation of the transforming growth factor-β/SMAD pathway in miR-155−/− mice, which was correlated with the increased tumorigenesis. Given the multiple targets of miR-155, careful evaluation of its role in tumorigenesis is necessary prior to any consideration of its potential as a biomarker and/or therapeutic target in colon cancer.
Keywords: colon cancer, miR-155, inflammation, immune cell markers, TGFβ/SMAD signaling pathway
noncoding RNAs (microRNAs) control the expression of genes involved in a variety of cellular processes, including inflammation, metabolism, proliferation, and apoptosis (5, 11, 25, 28). One such microRNA, miR-155, has been explored for its potential role in tumorigenesis, given its wide range of targets that are known to impact disease progression. Clinical studies have shown that increased expression of miR-155 correlates with and/or predicts tumor stage in colon tissue (1, 36, 43) and is associated with lymph node metastasis (36). Furthermore, it has been reported that expression levels of miR-155 in serum are increased in colorectal cancer patients, implicating it as a possible tumor biomarker for the diagnosis and assessment of prognosis of colorectal cancer (24). However, whether this microRNA is specifically driving colon tumorigenesis or simply elicited in response to some independent protumorigenic process has yet to be determined.
While the evidence is still relatively sparse, the clinical data supporting a link between miR-155 and colon cancer have prompted research in preclinical models to further define the role of this microRNA in colon tumorigenesis. A number of cell culture studies have used miR-155 manipulation techniques to examine its influence on tumorigenic properties. For example, transfection of SW480 cells with miR-155 enhanced their migration and invasive abilities and altered their morphological appearance but did not affect proliferation (45). Similarly, miR-155 knockdown has been reported to decrease colon cancer cell motility and invasion but, in contrast to the aforementioned study, did affect cell growth (27). To our knowledge, only two animal studies have examined a role for miR-155 in colon tumorigenesis. In 2011, Bakirtzi et al. (1) reported that blocking miR-155 slowed the rate of HCT-166 xenograft tumors following activation with neurotensin. More recently, Chen et al. (4) reported increased multiplicity of colonic neoplasms following chemically induced carcinogenesis in miR-155-deficient (miR-155−/−) mice. Similarly, Chen et al. reported delayed growth of transplantable MC38 colon cancer cells in miR-155−/− mice.
Although limited, the available literature appears to suggest that miR-155 may play a role in promoting colon tumorigenesis. Interestingly, however, this finding has not been consistent across other cancer models. For example, it has been reported that B16–F10 melanoma and Lewis lung carcinoma tumors were larger and heavier in miR-155−/− mice than wild-type (WT) controls (42). Similarly, tumor growth was increased in miR-155−/− compared with WT mice following administration of syngeneic EL4-luc lymphoma, B16–F1, or B16–F10 melanoma cells (14). On the basis of these collective findings, it is possible that manipulation of this microRNA may produce different responses, depending on the cancer model used and, perhaps, even the stage of tumorigenesis at which miR-155 expression has been modulated.
Given the limitations in the available literature, we examined the role of miR-155 in colon cancer. We first sought to determine miR-155 expression in tumor and nontumor colon tissue from colon cancer patients. Subsequently, to investigate the role of miR-155 in inflammation-promoting tumorigenesis, we administered the carcinogen azoxymethane (AOM) in combination with the proinflammatory chemical dextran sulfate sodium (DSS) to miR-155−/− mice. In this AOM-DSS model, animals develop tumors due to several gene mutations that are consistent with those in colon cancer patients (3). Thus this model offers a reliable experimental system, since it mimics a subset of inflammation-associated human cancers.
MATERIALS AND METHODS
miR-155 Expression in Human Colon Tissue
Colon tissues (tumor and nontumor colon tissue) from 15 patients with colon cancer were obtained from the Biorepository Core Facility at the University of South Carolina. An AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) was used to isolate microRNA, and Taq primers (miR-155-5p and RNU48, Applied Biosystems) were used to quantify miR-155 expression in the tumor and nontumor colon tissue.
Animals and AOM-DSS Model
C57BL/6 (WT) and miR-155−/− mice were originally purchased from Jackson Laboratories and subsequently bred at the University of South Carolina. All experimental mice were fed an AIN76A diet (Bio-Serv) with water provided ad libitum for the duration of the experiment.
Experiment 1.
At 10 wk of age, a total of 23 mice were injected with a single 10 mg/kg dose of AOM [Sigma; WT-A/D (n = 12) and miR-155−/− A/D (n = 11)]. DSS (MP Biochemicals; 36,000–50,000 mol wt) was dissolved in filtered drinking water and administered to mice for 7 days at 11 (2% DSS), 14 (1% DSS), and 17 (1% DSS) wk of age. A WT group (n = 11) that was not exposed to AOM or DSS was also included as a noncancer control. At 20 wk of age, mice were euthanized, colons were dissected, polyps were counted using a stereoscope, and a portion of the colon was fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned for hematoxylin-eosin and immunohistochemical staining. Polyps were removed from the remaining colon tissue. The residual colon tissue was divided into two pieces for Western blot and RT-PCR analysis.
Experiment 2.
Another group of 10-wk-old mice [WT-H2O-OG (n = 5), WT-A/D-OG (n = 9), and miR-155−/− A/D-OG (n = 8)] was injected with AOM as described above but received DSS (1/4th of the dose used in experiment 1) via oral gavage (OG). The Institutional Animal Care Committee of the University of South Carolina approved all animal experimentation.
Morphometric Measurements and Food and Water Intake
Body weight and food and water intake were monitored weekly over the duration of the experiment. Body composition was assessed before AOM injection (at 10 wk of age) and after the third cycle of DSS (at 20 wk of age) via dual-energy X-ray absorptiometry (Lunar PIXImus) as previously described (7).
Behavioral and Physiological Measurements
Symptom score was evaluated twice a week beginning at the first cycle of DSS exposure. Briefly, scores of weight loss (WL) (>5% WL = 0, 6–10% WL = 1, and 11–15% WL = 2), fecal consistency (pellet = 0, pasty = 2, and liquid = 4), and blood in stools (assessed with hemoccult kit: negative = 0, positive = 2, and gross bleeding = 4) were combined to monitor animal health (18). Visceral pain was assessed weekly after the first cycle of DSS using von Frey filaments as previously described (6).
Gene Expression
The colon tissue was subjected to quantitative real-time PCR (model 7300 real-time PCR system, Applied Biosciences) after RNA isolation with TRIzol reagent and purification from DSS-treated tissues as described in detail elsewhere (41). The following Taq primers (Applied Biosystems) were used: F4/80, CD11b, CD206, CD11c, MCP-1, TNFα, IL-6, IL-10, IL-17α, and FoxP3.
Western Blot Analysis
The colon was homogenized in Mueller buffer (2), and total protein was determined using the Bradford method. Western blot analysis of 30 μg of colon homogenate was used to determine inflammatory signaling pathways and cell surface markers. Primary antibodies for proliferating cell nuclear antigen (PCNA; Abcam); cleaved caspase-3, NFκB, STAT3, Erk, SOCS1, SOCS3, and SMAD2/3 (Cell Signaling Technology); F4/80 (AbD Serotec); CD11b (Bios); and Lyg6, CD8, and FoxP3 (Abcam) were diluted at 1:1,000. We followed a protocol similar to that described elsewhere (40).
Histology and Immunohistochemistry
Paraffin-block sections of the colonic polyps were stained with hematoxylin-eosin and subsequently evaluated blindly by a pathologist and characterized according to the grade of epithelial dysplasia: no dysplasia, low-grade dysplasia, dysplasia, and high-grade dysplasia. In addition, immunohistochemistry was performed using antibodies for Ki-67 (Abcam), TdT-mediated dUTP nick-end labeling (TUNEL; Millipore), F4/80 (AbD-Serotec), and CD11b (Abcam) according to the manufacturers' instructions.
Statistics
Data were analyzed using Prism 5.0 (GraphPad Software). A one-way ANOVA followed by Newman-Keuls post hoc analysis was used to determine differences between WT, WT-A/D, and miR-155−/− A/D mice. Tumor data were analyzed using an unpaired Student's t-test (WT-A/D and miR-155−/− A/D mice) and Wilcoxon's test (human tumor and normal colon tissue). Any statistical data that did not pass the equal-variance test (Bartlett's test for equal variances) were logarithmically transformed and reanalyzed. Data are presented as means ± SE, and the level of significance was set at P < 0.05.
RESULTS
miR-155 Expression in Tumor and Nontumor Colon Tissues in Patients With Colon Cancer
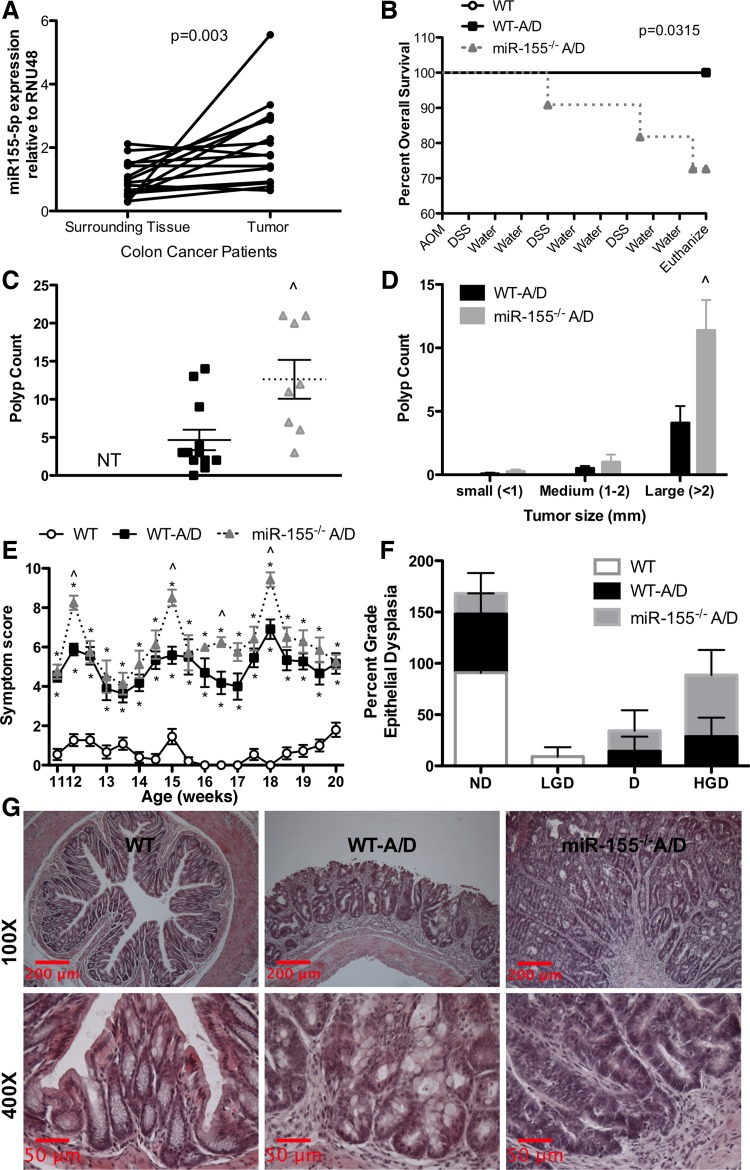
To assess whether miR-155 is dysregulated in colon cancer, we selected tumor and nontumor colon tissue from colon cancer patients (Table 1). Our data indicate higher miR-155 gene expression in the tumor tissue than in the nontumor colon tissue of colon cancer patients (P < 0.05; Fig. 1A).
Table 1.
Patient demographics and clinical characteristics
| n (%) | |
|---|---|
| Age, yr | |
| >60 | 9 (60) |
| 40–60 | 6 (40) |
| Sex | |
| Female | 9 (60) |
| Male | 6 (40) |
| Localization | |
| Jejunum | 1 (6) |
| Cecum | 4 (27) |
| Colon | 4 (27) |
| Sigmoid | 6 (40) |
| Lymph nodule | |
| Positive | 6 (40) |
| Negative | 9 (60) |
| Stage | |
| I | 1 (6.6) |
| II | 12 (80) |
| III | 1 (6.6) |
| IV | 1 (6.6) |
Fig. 1.
Effects of microRNA-155 (miR-155) deficiency on colitis-associated colon cancer. A: RT-PCR assessment of gene expression of miR-155-5p in tumor and normal surrounding tissue of patients with colon cancer. B: impact of azoxymethane (AOM)-dextran sulfate sodium (DSS) treatment on survival of miR-155−/− mice. WT, wild-type mice; WT-A/D, WT mice injected with a single 10 mg/kg dose of AOM; miR-155−/− A/D, miR-155−/− mice injected with a single 10 mg/kg dose of AOM. C and D: polyp count and size after AOM-DSS treatment. NT, no tumor. E: symptom score (body weight loss + blood in stool + diarrhea). F: colonic dysplasia in AOM-DSS-treated mice. ND, no dysplasia; LGD, low-grade dysplasia; D, dysplasia; HGD, high-grade dysplasia. G: representative hematoxylin-eosin-stained sections of colon tissue after AOM-DSS treatment. Values are means ± SE; n = 15 cancer patients, or n = 11–12 mice per group. *P < 0.05 vs. WT. ∧P < 0.05 vs. WT-A/D.
Effects of miR-155 Deficiency on Intestinal Tumorigenesis in the AOM-DSS Model
We next addressed whether knocking out miR-155 would be protective in the AOM-DSS colitis-associated colon cancer model. The percentage of mortality was greater in miR-155−/− A/D than WT-A/D mice (Fig. 1B). In addition, polyp/adenoma multiplicity was higher in miR-155−/− A/D mice (12.6 ± 2.6 vs. 4.7 ± 1.4 tumors per mouse, P < 0.05; Fig. 1C). This difference largely resulted from an increase in the number of large polyps/adenomas in the miR-155−/− A/D group (P < 0.05; Fig. 1D). These results were consistent with other measurements showing worse outcomes in miR-155−/− A/D mice: higher clinical symptom scores (Fig. 1E) and higher grade of epithelial dysplasia (Fig. 1, F and G).
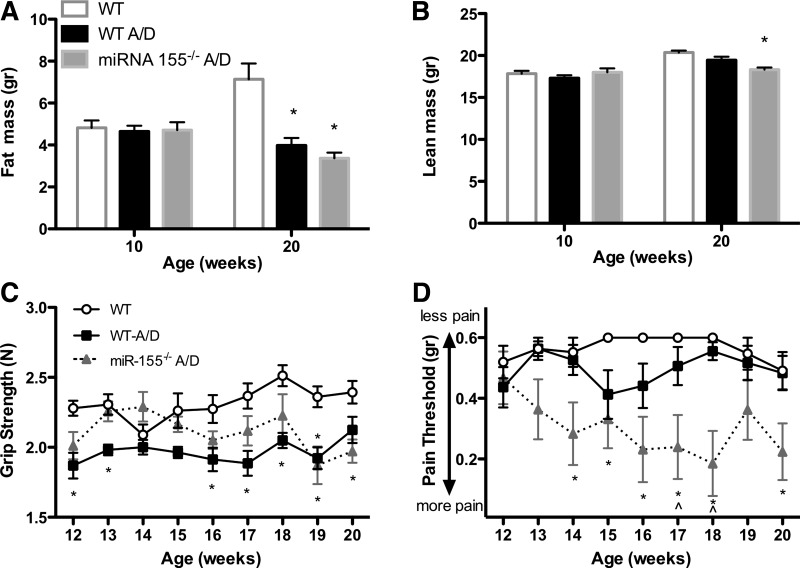
Additionally, we examined body composition changes, complete blood counts, and behavioral measures, given their relevance to cancer prognosis. Both cancer groups (WT-A/D and miR-155−/− A/D mice) showed significant decreases in total fat mass compared with noncancer control WT mice (P < 0.05; Fig. 2A). However, only miR-155−/− A/D mice exhibited a significant decrease in lean mass compared with noncancer control WT mice (P < 0.05; Fig. 2B). A similar degree of splenomegaly and skeletal muscle and visceral fat wasting was observed between WT-A/D and miR-155−/− A/D mice (Table 2). The number of circulating white blood cells was increased in miR-155−/− compared with WT mice (Table 3), but there were no differences between noncancer and cancer groups with respect to red blood cell count and fasting blood glucose (Table 3). Both cancer groups exhibited a decrease in grip strength compared with noncancer control mice (Fig. 2C; P < 0.05). However, only miR-155−/− A/D mice experienced intense visceral pain (Fig. 2D; P < 0.05).
Fig. 2.
Effects of miR-155 deficiency on body composition, grip strength, and referred visceral pain during AOM-DSS treatment. A and B: fat mass and lean mass measured by dual-energy X-ray absorptiometry scan before and after AOM-DSS treatment. C: forelimb and hindlimb force (grip strength) during AOM-DSS treatment. D: visceral pain measured with von Frey filaments during AOM-DSS treatment. Values are means ± SE; n = 8–12 mice per group. *P < 0.05 vs. WT. ∧P < 0.05 vs. WT-A/D.
Table 2.
Effect of miR-155 deletion on lean and fat tissue in the AOM-DSS model of colon cancer
| Group |
|||
|---|---|---|---|
| WT | WT-A/D | miR-155−/− A/D | |
| Spleen, mg | 81 ± 2 | 194 ± 20* | 212 ± 20* |
| Liver, mg | 1,391 ± 70 | 1,507 ± 55 | 1,516 ± 36 |
| Soleus, mg | 8 ± 0.6 | 7±.4 | 7 ± 0.5 |
| Gastrocnemius, mg | 138 ± 2 | 122 ± 5* | 113 ± 2* |
| Fat, mg | |||
| Epididymal | 1,040 ± 129 | 557 ± 102* | 400 ± 40* |
| Kidney | 384 ± 44 | 160 ± 21* | 144 ± 16* |
| Mesenchymal | 633 ± 51 | 343 ± 39* | 266 ± 36* |
Values are means ± SE; n = 8–12 mice/group.
miR-155, microRNA-155; WT, wild-type; A/D, axoxymethane (AOM)-dextran sulfate sodium (DSS).
P < 0.05 vs. WT.
Table 3.
Effect of miR-155 deletion on blood cell count and glucose metabolism in the AOM-DSS model of colon cancer
| Group |
|||
|---|---|---|---|
| WT | WT-A/D | miR-155−/− A/D | |
| RBC, 1012/l | 7.6 ± 0.9 | 8.2 ± 0.9 | 7.0 ± 0.7 |
| Hb, g/dl | 12.0 ± 1.1 | 12.8 ± 1.1 | 11.1 ± 1.2 |
| WBC, 109/l | 7.5 ± 0.8 | 9.1 ± 1.2 | 12.0 ± 1.7* |
| Monocyte, 109 | 0.3 ± 0.07 | 0.3 ± 0.08 | 0.6 ± 0.14† |
| Lymphocyte, 109 | 5.8 ± 0.7 | 7.1 ± 0.9 | 9.3 ± 1.6 |
| Neutrophil, 109 | 1.3 ± 0.3 | 1.7 ± 0.4 | 2.0 ± 0.5 |
| Glucose, mg/dl | 100 ± 8 | 104 ± 9 | 104 ± 10 |
Values are means ± SE; n = 8–12 mice/group.
P < 0.05 vs. WT.
Trend P = 0.069 vs. WT and WT-A/D.
Role of miR-155 in Cell Proliferation and Apoptosis
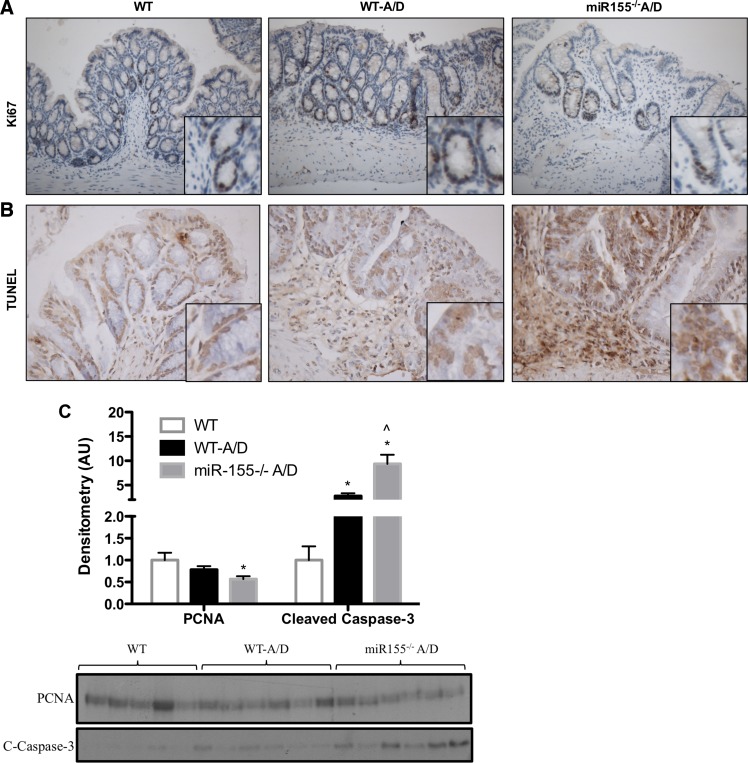
We used Ki-67 and PCNA as markers for cell proliferation and TUNEL and cleaved caspase-3 as indicators of cellular apoptosis to investigate whether an imbalance in cell cycle regulation and programmed cell death could be responsible for the increased adenoma multiplicity in miR-155−/− A/D mice. No significant differences in cell proliferation markers were observed for WT-A/D compared with control noncancer WT mice in the normal mucosal tissue (Fig. 3, A and C). However, miR-155−/− A/D mice showed a small, but significant, reduction in PCNA compared with control noncancer WT mice (Fig. 3C; P < 0.05), but not WT-A/D mice. Examination of apoptotic signal by TUNEL assay and cleaved caspase-3 in the normal mucosa revealed a significant increase in apoptotic cells in WT-A/D compared with control noncancer WT mice (Fig. 3, B and C; P < 0.05) that was further exacerbated in miR-155−/− A/D mice (Fig. 3, B and C; P < 0.05).
Fig. 3.
Effects of miR-155 deficiency on cell proliferation and apoptosis in the AOM-DSS model. A and B: immunohistochemistry for Ki-67 (×100 magnification) and TdT-mediated dUTP nick-end labeling (TUNEL, ×200 magnification) in colon tissue from AOM-DSS-treated mice. C: Western blot densitometry and blots of cell proliferation [proliferating cell nuclear antigen (PCNA)] and apoptosis [cleaved caspase-3 (C-caspase-3)] in colon tissue. Values are means ± SE; n = 2–4 mice per group (immunohistochemistry) and n = 5–6 mice per group (Western blot). *P < 0.05 vs. WT. ∧P < 0.05 vs. WT-A/D.
Examination of Inflammatory Markers and Signaling Pathways in Tissue Surrounding the Colon and Adenomas-Precancerous Lesions of miR-155−/− Mice
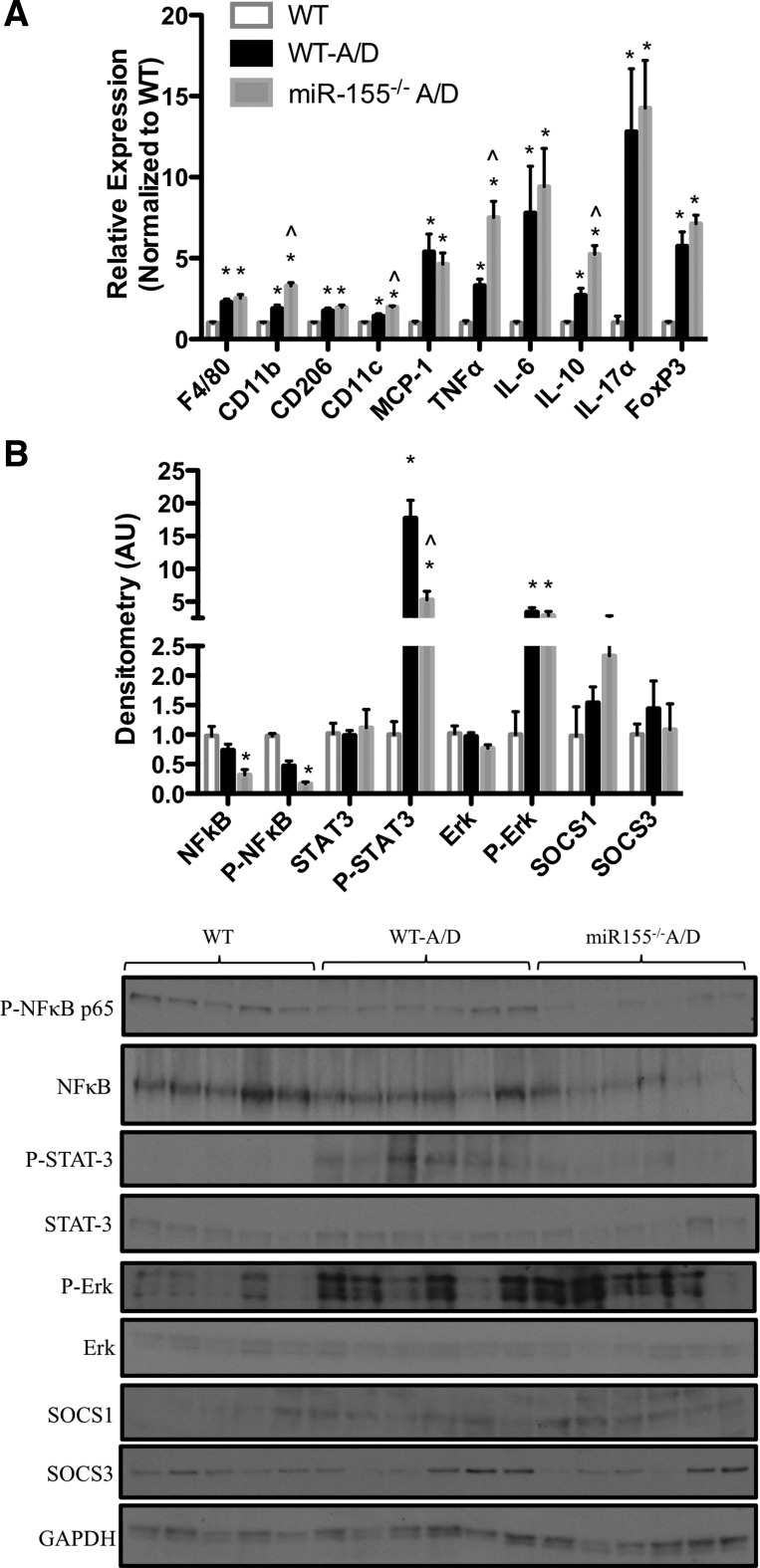
To evaluate whether dysregulation of inflammatory processes by miR-155 was playing a role in tumorigenesis in this model, we next measured gene expression of several inflammatory mediators in the colon tissue (Fig. 4A). As expected, we observed a significant increase in the expression of macrophage markers (F4/80, CD11b, CD206, and CD11c), chemokines (MCP-1 and IL-17α), cytokines (TNFα, IL-6, and IL-10), and T cell-related markers (FoxP3) in AOM-DSS-treated compared with the noncancer control WT mice (P < 0.05). However, minimal changes were observed between genotypes: only CD11b, CD11c, TNFα, and IL-10 were upregulated in the miR-155−/− A/D compared with WT-A/D mice (P < 0.05). We next measured the protein expression of key inflammatory signaling pathways (Fig. 4B). As expected, we observed significantly more activation of STAT3 and Erk (P < 0.05) in AOM-DSS-treated mice than in noncancer control WT mice (P < 0.05). However, activation of STAT3 was significantly suppressed in miR-155−/− A/D compared with WT-A/D mice (P < 0.05). Similarly, NFκB activity was significantly lower in miR-155−/− A/D mice (P < 0.05) than in noncancer control WT, but not WT-A/D, mice. We also measured protein expression of key inflammatory targets of miR-155, including SOCS1 and SOCS3; neither changed with miR-155 deficiency (Fig. 4B).
Fig. 4.
Effects of miR-155 deficiency on inflammation in the AOM-DSS model. A: gene expression of inflammatory markers in colon tissue of mice treated with AOM-DSS. B: Western blot densitometry and blots of inflammatory pathway signaling in tissue surrounding the colon. AU, arbitrary units. Values are means ± SE; n = 8–12 mice per group (RT-PCR), and n = 5–6 mice per group (Western blot). *P < 0.05 vs. WT. ∧P < 0.05 vs. WT-A/D.
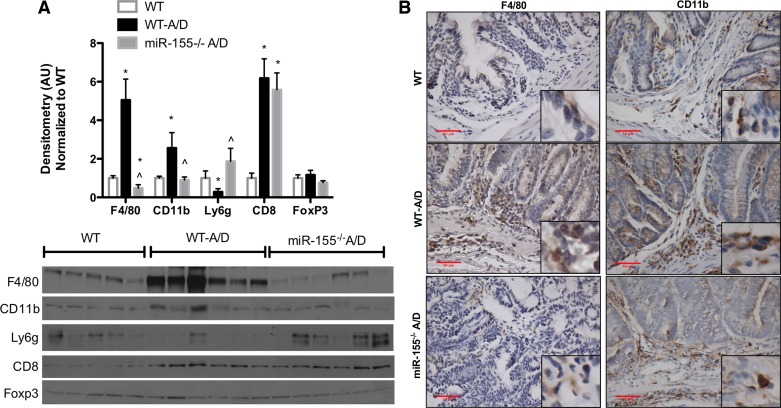
Effects of miR-155 Deficiency on Immune Cell Markers
Since miR-155 is highly expressed in immune cells, we next investigated whether miR-155 affected immune cell markers. We selected nonexclusive conventional immune cell markers, including macrophage (F4-80, CD11b), neutrophil (Ly6g), cytotoxic T cell (CD8), and regulatory T cell (FoxP3). Figure 5A shows significantly elevated protein expression of F4/80, CD11b, and CD8 markers in WT-A/D compared with noncancer control WT mice (P < 0.05). Interestingly, expression of F4/80 and CD11b was significantly reduced in miR-155−/− A/D compared with WT-A/D mice (P < 0.05), and only F4/80 expression was significantly different from noncancer control WT mice (Fig. 5, A and B). In contrast, Ly6g protein expression was significantly decreased in WT-A/D mice but increased in miR-155−/− A/D mice (Fig. 5A; P < 0.05).
Fig. 5.
Effects of miR-155 deficiency on immune cell markers in the AOM-DSS model. A: Western blot densitometry and blots of nonexclusive conventional immune cell markers. B: immunohistochemistry for F4/80 and CD11b in colon tissue of AOM-DSS-treated mice (×400 magnification). Values are means ± SE; n = 8–12 mice per group (RT-PCR), and n = 5 mice per group (Western blot). *P < 0.05 vs. WT. ∧P < 0.05 vs. WT-A/D.
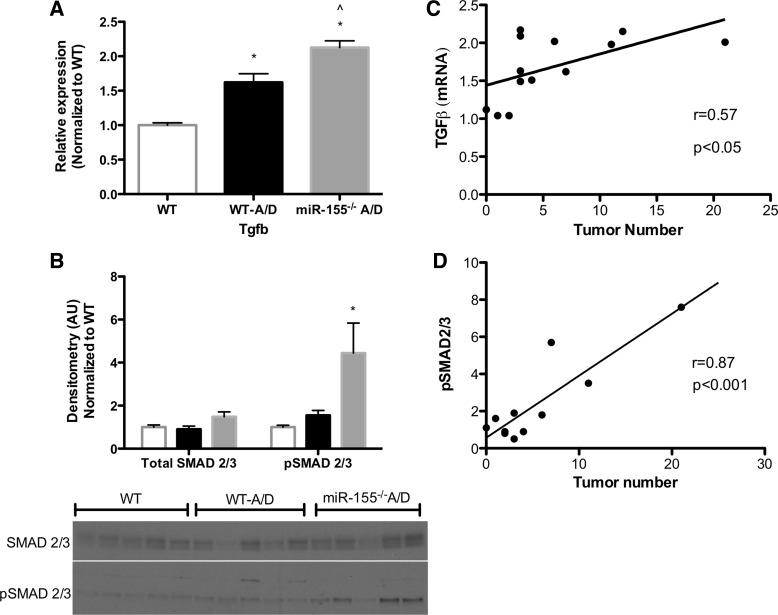
Role of miR-155 in Transforming Growth Factor-β/SMAD Signaling
Previous studies have reported inhibition of the transforming growth factor-β (TGFβ)/SMAD signaling pathway with miR-155 (15, 19). Thus we next analyzed TGFβ gene expression in the colon tissue and found a significant increase in WT-A/D compared with noncancer control WT mice (P < 0.05; Fig. 6A). Interestingly, there was a further increase in miR-155−/− A/D mice (P < 0.05). This result was consistent with an increase in activated SMAD2/3 in tissue surrounding the colon in miR-155−/− A/D mice compared with the noncancer control WT group, which was not different from the WT-A/D group (P < 0.05; Fig. 6B). Even further, tumor number in AOM-DSS-treated groups was highly correlated with TGFβ gene expression and activated SMAD2/3 in tissues surrounding the colon (Fig. 6, C and D).
Fig. 6.
Effects of miR-155 deficiency on transforming growth factor (TGF)-β/SMAD in the AOM-DSS model. A: gene expression of TGFβ (Tgfb) in colon tissue. B: Western blot densitometry and blots of total (SMAD2/3) and activated [phosphorylated (pSMAD2/3)] SMAD2/3 in the colon of mice treated with AOM-DSS. C: correlation between TGFβ gene expression and tumor number in miR-155−/− A/D and WT-A/D mice. D: correlation between SMAD activation in the colon and tumor number in miR-155−/− A/D and WT-A/D mice. Values are means ± SE; n = 8–12 mice per group (RT-PCR), and n = 5 mice per group (Western blot). *P < 0.05 vs. WT. ∧P < 0.05 vs. WT-A/D.
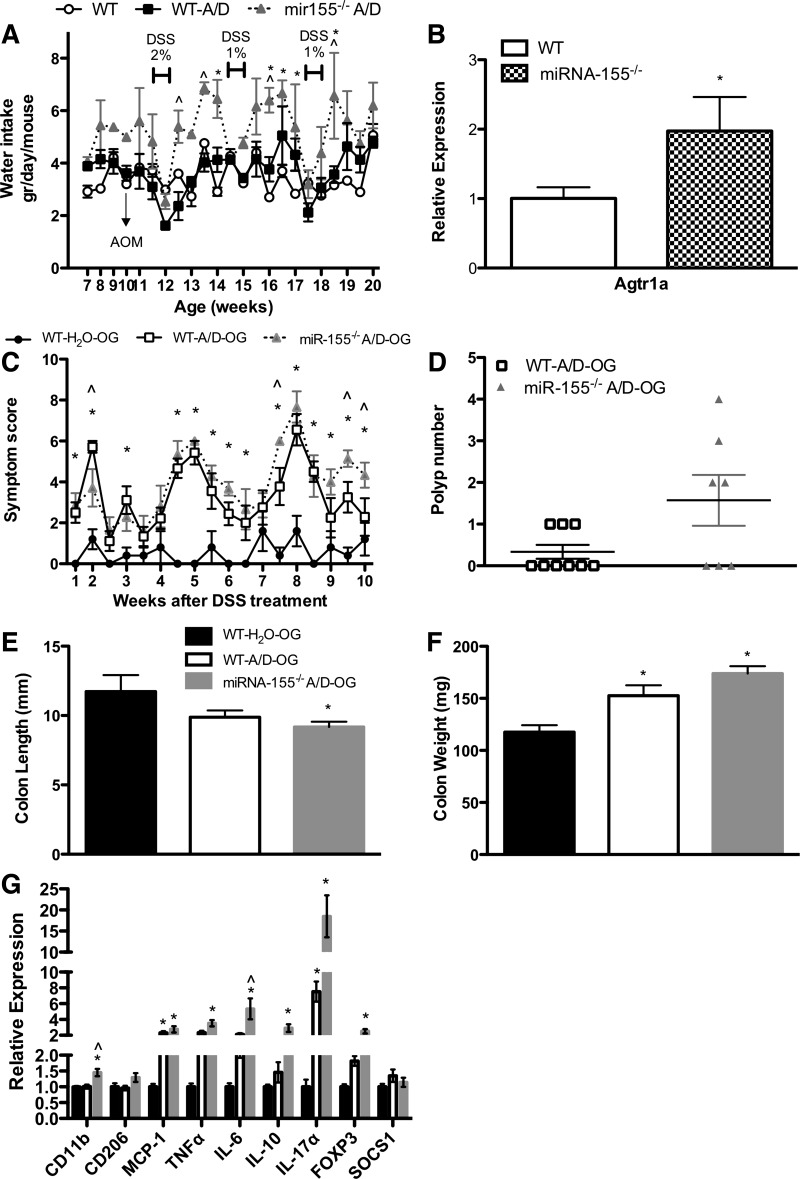
Effects of miR-155 Deficiency on Tumorigenesis Following Controlled Administration of AOM and DSS
An interesting finding during the study (and other studies using miR-155−/− mice in our laboratory) was that miR-155−/− A/D mice, in general, consumed more water than noncancer control WT and WT-A/D mice (P < 0.05; Fig. 7A). Given that DSS, the chemical that drives inflammatory processes and subsequent tumorigenesis in this model, is administered in the drinking water, it was important to evaluate any impact of this increase in water consumption on our findings. We thus evaluated weekly water consumption and found that although water consumption was increased between DSS cycles in the miR-155−/− A/D mice, these mice did not consume more DSS-supplemented water during any of the three DSS cycles (Fig. 7A). Therefore, we do not believe that this phenomenon played a role in the outcomes of this study. Nevertheless, to confirm this, we administered DSS via oral gavage to WT and miR-155−/− mice to standardize the dose of DSS. The dose was calculated according to the DSS-water intake of WT-A/D mice in the initial experiment (mice consumed 0.072 g of DSS in 3.6 ml of drinking water). Administration of DSS via oral gavage (0.072 g of DSS in 0.2 ml of filtered water) was lethal for WT and miR-155−/− mice (data not shown). Therefore, we reduced the DSS concentration to one-fourth of the original administration (0.018 g of DSS in 0.2 ml of filtered water). The reduction in the dose of DSS affected the degree of the disease, but not the overall results. Consistent with our initial findings, miR-155−/− A/D-OG mice showed more clinical symptoms related to colitis: smaller and heavier colons, increased tumor number, and evidence of colitis (Fig. 7, C–G).
Fig. 7.
Effects of miR-155 deficiency on tumorigenesis following controlled administration of DSS. A: daily water consumption of mice treated with DSS in drinking water (experiment 1, n = 8–12 per group). B: gene expression of angiotensin II type 1a receptor (Agt1ra) in the fornix area on the brain of naïve WT and miR-155−/− mice (n = 8 per group). C–G: clinical symptom score (body weight loss, blood in stool, and diarrhea), polyp number, colon length, colon weight, and gene expression of inflammatory markers of mice orally gavaged with water or DSS (experiment 2, n = 5–9 per group). WT-H2O, WT mice orally gavaged with H2O; WT-A/D-OG, WT mice orally gavaged with DSS. Values are means ± SE. *P < 0.05 vs. WT or WT-H2O. ∧P < 0.05 vs. WT-A/D or WT-A/D-OG.
To explore a potential mechanism for the increased water consumption in the miR-155−/− mice, we examined gene expression of angiotensin II type 1 receptor (Agtr1a), an important contributor to water intake, in the fornix (21, 46). Briefly, 10-wk-old naïve mice [WT (n = 8) and miR-155−/− mice (n = 8)] were euthanized, and the fornix was collected for gene expression analysis. We found a significant increase in the gene expression of Agtr1a in the fornix of naïve miR-155−/− compared with naïve WT mice (P < 0.05; Fig. 7B).
DISCUSSION
Recent literature has implied that miR-155 has a role in colorectal cancer. For example, clinical studies have shown that increased expression of miR-155 correlates with and/or predicts tumor stage in colon tissue (1, 36, 43) and is associated with lymph node metastasis (36). Our data corroborate these findings, as we report an increase in miR-155 in colon tumor tissue. However, whether this cancer-associated increase in miR-155 is suggestive of a pro- or antitumor response is not known. Thus we next performed a study using miR-155−/− mice to establish a role for miR-155 in colorectal cancer. Our findings indicate that miR-155−/− mice are more susceptible to tumorigenesis than WT mice in the AOM-DSS model of colon cancer; miR-155−/− mice had a greater number of total polyps/adenomas as well as large polyps/adenomas, increased symptom severity score, higher grade of epithelial dysplasia, and decreased survival. These data indicate that miR-155 protects against the development of colon cancer. Thus the increased miR-155 in human colon tumor tissue in the current study, as well in as in previously reported studies (1, 36, 43), could actually be indicative of an antitumor response by the host.
Interestingly, the increase in tumorigenesis in miR-155−/− mice was not associated with a decrease in apoptosis. In fact, we found an increase in apoptosis in the normal mucosal tissue of miR-155−/− mice. This may not be entirely surprising, as miR-155 has been reported to play a role in apoptosis by targeting antiapoptotic factors such as RPS6KA3, SGK3, RHEB, and KRAS (13). On the other hand, there was no change in proliferation in the normal mucosa with manipulation of miR-155, which was consistent with the finding of Zhang et al. (45) that miR-155 transfection did not affect growth of SW480 cells. However, miR-155 knockdown has been reported to decrease cell growth in a human colon cancer cell line (27). Nonetheless, we do not believe that the protumorigenic effects of miR-155 deficiency in this model can be explained by the findings on apoptosis and proliferation.
In accordance with previous reports in the AOM-DSS model of colon cancer, our data, in general, indicate an increase in inflammation in WT-A/D compared with noncancer control WT mice (16, 26). Of most significance for genotype was the hypothesized decrease in STAT3 activity with miR-155 deficiency; it has been reported that miR-155 can regulate STAT3 signaling (30). However, it is important to point out that STAT3 has also been reported to regulate miR-155 expression (31). Interestingly, the decrease in STAT3 activation in miR-155−/− mice was actually associated with increased tumorigenesis. This may not be completely unexpected, as STAT3 has been reported to play a paradoxical role in tumorigenesis; reductions in STAT3 activation have been linked to tumorigenesis, especially in advanced stages (22). For the rest of the inflammatory mediators measured, only minimal differences in inflammation were observed between miR-155−/− A/D and WT-A/D mice. Thus we do not believe that the increase in adenomas-precancerous lesions in miR-155−/− A/D mice can be explained exclusively by activation of the measured inflammatory signaling pathways alone. Contrary to these findings, our group previously reported that miR-155 deficiency actually protects against DSS-induced colitis, the effects of which are likely mediated through a reduction in inflammation (37). One explanation for the discrepancy between the DSS model of colitis and the AOM-DSS model of colon cancer is that miR-155 may not be directly influencing the inflammatory response in immune cells but, rather, influencing epithelial cell transformation to a dysplastic phenotype. It is also possible that compensatory mechanisms driving inflammation may have occurred in the miR-155−/− mice, given the longer period between initial DSS exposure and measurement of inflammation in the colitis-associated colon cancer model. As such, it would have been informative to examine inflammatory processes during the earlier stages of tumorigenesis to fully evaluate the role of this microRNA in inflammation in this model. It is also important to consider the influence of inflammation on disease progression in each of these models: a reduction in inflammation is vital for treatment of colitis, but once inflammation triggers tumor formation, a reduction in inflammation may not be as important as the modulation of other cellular processes more strongly linked to tumor growth (29). Thus targets of miR-155 independent of inflammatory processes may be more influential in dictating tumorigenesis during the later stages of colon cancer.
In many instances, tumor formation is driven by the activation, recruitment, and accumulation of immune cells in the tumor milieu (12, 34). Thus we next examined nonexclusive conventional immune cell markers to further our understanding of the role of this microRNA in tumorigenesis. Of most interest were the decrease in F4/80 and CD11b and the increase in Ly6g in the miR-155−/− mice, implying a potential role of macrophages and neutrophils in the enhanced tumorigenic response associated with miR-155 deficiency. Macrophages have, in general, been associated with promotion of tumorigenesis. However, they constitute an extremely heterogeneous population that is divided into two main classes (M1 and M2) with documented differential effects on tumorigenesis (38); it is thought that M1 macrophages are cytotoxic against neoplastic cells, whereas M2 macrophages exert protumoral functions (38). Thus, although macrophages are largely associated with poor prognosis, depletion of M1 macrophages, in particular, may actually result in an unfavorable outcome. For instance, a recent study in the AOM-DSS model reported an immediate decrease in both M1 and M2 macrophages as the carcinoma progressed to metastasis (44). Therefore, it is possible that the decrease in F4/80 and CD11b in miR-155−/− mice may lead to immunosuppressive-driven promotion of tumorigenesis in this model. In support of our findings, Ly6g has been associated with increased tumorigenesis in this mouse model, as anti-Ly6g antibody administration reduced tumor number in AOM-DSS-treated mice (35).
Consistent with our findings, others have revealed that deletion of miR-155 results in enhanced immunosuppressive activity of immune cells, whereas miR-155 overexpression may be exerting positive effects, possibly by activating immune cells (14, 42). For example, it has been documented that miR-155−/− mice permit enhanced growth of transplanted EL4-luc lymphoma and B16 melanoma cells compared with WT controls, which was linked to a defect in IFNγ-expressing T cells in tumor-bearing mice (14). Similarly, Wang et al. (42) reported that miR-155 deficiency resulted in accumulation of myeloid-derived suppressor cells in the tumor microenvironment, which was associated with larger and heavier B16–F10 melanoma and Lewis lung carcinoma tumors. Furthermore, it has been reported that miR-155 knockdown promotes mammary tumor growth by impairing classical activation of tumor-associated macrophages, leading to an overall reduction in macrophages, reduced expression of activation markers, and a skewing of tumor immune cells to an M2/Th2 response (47). On the contrary, however, in a recent study using a model of chemically induced colon cancer similar to that used in the current investigation, Chen et al. (4) reported a reduction in accumulation of myeloid-derived suppressor cells, along with a decrease in antitumor CD4+ and CD8+ lymphocytes in miR-155−/− mice, which surprisingly was associated with increased multiplicity of colonic neoplasms following chemically induced carcinogenesis. The reasons for the discrepancies between the current investigation and the report by Chen et al. are unclear, as both studies utilize a similar AOM-DSS protocol, although we used a lower dose of DSS. It is possible that the genetic background, microbiota, and environmental history of the mice used in each study may have contributed to the disparate results (33). The WT and miR-155−/− mice used in our study were bred at the University of South Carolina, whereas the mice used by Chen et al. were purchased from Jackson Laboratories, which may, at least partially, explain the divergent findings. Nonetheless, on the basis of the available literature, the role of miR-155 in the immune response in tumorigenesis appears to be dependent on the model and, most likely, the stage of disease, although microbiota and environmental influences may also be factors in how these findings are interpreted.
Given that miR-155 is known to repress TGFβ/SMAD signaling in immune cell populations (23), a pathway that has been linked to increased tumorigenesis, we next evaluated TGFβ/SMAD signaling as a potential mediator of miR-155 action. Our data confirm that miR-155 deficiency enhances TGFβ expression and SMAD2/3 activation. Furthermore, we show a direct correlation between polyp number and activation of SMAD2/3. Epidemiological studies have documented a positive association between high levels of TGFβ and advanced stage of colon cancer, recurrence, and decreased survival (8, 20). Furthermore, dysregulation of the TGFβ/SMAD signaling pathway is known to produce fibrosis, angiogenesis, immunosuppression, and epithelial-mesenchymal transition (9, 10, 17, 19, 32). Thus it is possible that the activation of this pathway in miR-155−/− mice is contributing to the increased tumorigenesis. A limitation of this analysis is that we were not able to determine whether specific immune cells or tumor cells were triggering the TGFβ/SMAD signaling pathway in the colon. However, a recent report by Suzuki et al. (39) indicated that SMAD2/3-positive cells may actually be stem cells. Furthermore, it has been reported that miR-155 can inhibit TGFβ and SMAD2/3 in macrophages (23), cells that are likely influenced by miR-155 in our model. Thus it is possible that multiple sources may be contributing to the increase in TGFβ/SMAD signaling.
In our study, chemical induction of colon carcinogenesis by AOM injection, as well as ad libitum consumption of DSS in the drinking water or controlled administration of DSS by gavage, convincingly shows an increase in tumorigenesis when miR-155 is deleted. However, it is important to note that these findings may not be translatable to other cancer models as a pro- and antitumorigenic role for this microRNA. This phenomenon is complicated by the existence of numerous targets for this microRNA, many of which play a role in the tumorigenic response. For example, miR-155 has been reported to influence apoptosis, proliferation, inflammation, and immune function (13). Furthermore, the dynamic relationship between the cancer cells and immune cells is likely affected by miR-155, as studies have clearly reported divergent findings on the influence of this microRNA on immunosuppressive activity in the tumor microenvironment and, ultimately, tumorigenesis. As such, future studies using stage-specific models of colon cancer, as well as hematopoietic-specific and/or myeloid-specific miR-155−/− models, are necessary to fully understand the impact of the miR-155 immune response on the tumor microenvironment.
In summary, we found that miR-155 is expressed in the tumor of colon cancer patients. Using a chemical model of colon cancer, we report that this may be a protective response, as miR-155−/− mice have a greater number of total polyps as well as large polyps, an increased symptom severity score, a higher grade of dysplasia, and decreased survival. The mechanisms for this effect appear to be multifactorial in origin, as miR-155−/− mice exhibited alterations in select inflammatory mediators, changes in immune cell markers, and an increase in the TGFβ/SMAD signaling pathway. Given the multiple targets of miR-155, careful evaluation of its role in tumorigenesis is necessary prior to any consideration of its potential as a biomarker and/or therapeutic target in colon cancer.
GRANTS
This work was supported by National Cancer Institute Grants R21 CA-167058 and R21 CA-175636 (to E. A. Murphy), National Center for Complementary and Alternative Medicine Grant K01 AT-007824, and the University of South Carolina [Advanced Support Programs for Innovative Research Excellence (ASPIRE)].
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
K.T.V., R.T.E., J.L.M., T.L.C., U.S., M.N., P.N., D.F., and E.A.M. developed the concept and designed the research; K.T.V., R.T.E., J.L.M., and T.L.C. performed the experiments; K.T.V. and I.C. analyzed the data; K.T.V., R.T.E., J.L.M., T.L.C., I.C., U.S., D.F., and E.A.M. interpreted the results of the experiments; K.T.V. prepared the figures; K.T.V. drafted the manuscript; K.T.V., R.T.E., J.L.M., T.L.C., I.C., U.S., M.N., P.N., D.F., and E.A.M. edited and revised the manuscript; K.T.V., R.T.E., J.L.M., T.L.C., I.C., U.S., M.N., P.N., D.F., and E.A.M. approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank Kei Lam for technical support on RT-PCR, Junfeng Wang for technical support on genotyping mice, the Biorepository Core Facility at the University of South Carolina for the specimens from patients with colon cancer, and NDBbio for assistance with immunohistochemistry.
REFERENCES
- 1.Bakirtzi K, Hatziapostolou M, Karagiannides I, Polytarchou C, Jaeger S, Iliopoulos D, Pothoulakis C. Neurotensin signaling activates microRNAs-21 and -155 and Akt, promotes tumor growth in mice, and is increased in human colon tumors. Gastroenterology 141: 1749–1761, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carson JA, Lee WJ, McClung J, Hand GA. Steroid receptor concentration in aged rat hindlimb muscle: effect of anabolic steroid administration. J Appl Physiol 93: 242–250, 2002. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Huang XF. The signal pathways in azoxymethane-induced colon cancer and preventive implications. Cancer Biol Ther 8: 1313–1317, 2009. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Wang L, Fan J, Ye C, Dominguez D, Zhang Y, Curiel TJ, Fang D, Kuzel TM, Zhang B. Host miR155 promotes tumor growth through a myeloid-derived suppressor cell-dependent mechanism. Cancer Res 75: 519–531, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Siegel F, Kipschull S, Haas B, Frohlich H, Meister G, Pfeifer A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun 4: 1769, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eijkelkamp N, Kavelaars A, Elsenbruch S, Schedlowski M, Holtmann G, Heijnen CJ. Increased visceral sensitivity to capsaicin after DSS-induced colitis in mice: spinal cord c-Fos expression and behavior. Am J Physiol Gastrointest Liver Physiol 293: G749–G757, 2007. [DOI] [PubMed] [Google Scholar]
- 7.Enos RT, Davis JM, Velazquez KT, McClellan JL, Day SD, Carnevale KA, Murphy EA. Influence of dietary saturated fat content on adiposity, macrophage behavior, inflammation, and metabolism: composition matters. J Lipid Res 54: 152–163, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman E, Gold LI, Klimstra D, Zeng ZS, Winawer S, Cohen A. High levels of transforming growth factor-β1 correlate with disease progression in human colon cancer. Cancer Epidemiol Biomarkers Prev 4: 549–554, 1995. [PubMed] [Google Scholar]
- 9.Geissmann F, Revy P, Regnault A, Lepelletier Y, Dy M, Brousse N, Amigorena S, Hermine O, Durandy A. TGF-β1 prevents the noncognate maturation of human dendritic Langerhans cells. J Immunol 162: 4567–4575, 1999. [PubMed] [Google Scholar]
- 10.Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-β signaling in T cells. Nat Med 7: 1118–1122, 2001. [DOI] [PubMed] [Google Scholar]
- 11.He B, Gao SQ, Huang LD, Huang YH, Zhang QY, Zhou MT, Shi HQ, Song QT, Shan YF. MicroRNA-155 promotes the proliferation and invasion abilities of colon cancer cells by targeting quaking. Mol Med Rep 11: 2355–2359, 2015. [DOI] [PubMed] [Google Scholar]
- 12.Heusinkveld M, van der Burg SH. Identification and manipulation of tumor associated macrophages in human cancers. J Transl Med 9: 216, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgs G, Slack F. The multiple roles of microRNA-155 in oncogenesis. J Clin Bioinforma 3: 17, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huffaker TB, Hu R, Runtsch MC, Bake E, Chen X, Zhao J, Round JL, Baltimore D, O'Connell RM. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep 2: 1697–1709, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji H, Li Y, Jiang F, Wang X, Zhang J, Shen J, Yang X. Inhibition of transforming growth factor-β/SMAD signal by miR-155 is involved in arsenic trioxide-induced anti-angiogenesis in prostate cancer. Cancer Sci 105: 1541–1549, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katoh H, Wang D, Daikoku T, Sun H, Dey SK, Dubois RN. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell 24: 631–644, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalil N, Parekh TV, O'Connor R, Antman N, Kepron W, Yehaulaeshet T, Xu YD, Gold LI. Regulation of the effects of TGF-β1 by activation of latent TGF-β1 and differential expression of TGF-β receptors (TβR-I and TβR-II) in idiopathic pulmonary fibrosis. Thorax 56: 907–915, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JJ, Shajib MS, Manocha MM, Khan WI. Investigating intestinal inflammation in DSS-induced model of IBD. J Vis Exp Feb 1: 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor-β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol 28: 6773–6784, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lampropoulos P, Zizi-Sermpetzoglou A, Rizos S, Kostakis A, Nikiteas N, Papavassiliou AG. TGF-β signalling in colon carcinogenesis. Cancer Lett 314: 1–7, 2012. [DOI] [PubMed] [Google Scholar]
- 21.Lazartigues E, Sinnayah P, Augoyard G, Gharib C, Johnson AK, Davisson RL. Enhanced water and salt intake in transgenic mice with brain-restricted overexpression of angiotensin (AT1) receptors. Am J Physiol Regul Integr Comp Physiol 295: R1539–R1545, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Kim JC, Lee SE, Quinley C, Kim H, Herdman S, Corr M, Raz E. Signal transducer and activator of transcription 3 (STAT3) protein suppresses adenoma-to-carcinoma transition in Apcmin/+ mice via regulation of Snail-1 (SNAI) protein stability. J Biol Chem 287: 18182–18189, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-β. J Biol Chem 285: 41328–41336, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv ZC, Fan YS, Chen HB, Zhao DW. Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumour Biol 36: 1619–1625, 2015. [DOI] [PubMed] [Google Scholar]
- 25.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33: 607–619, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onizawa M, Nagaishi T, Kanai T, Nagano K, Oshima S, Nemoto Y, Yoshioka A, Totsuka T, Okamoto R, Nakamura T, Sakamoto N, Tsuchiya K, Aoki K, Ohya K, Yagita H, Watanabe M. Signaling pathway via TNF-α/NF-κB in intestinal epithelial cells may be directly involved in colitis-associated carcinogenesis. Am J Physiol Gastrointest Liver Physiol 296: G850–G859, 2009. [DOI] [PubMed] [Google Scholar]
- 27.Onyeagucha BC, Mercado-Pimentel ME, Hutchison J, Flemington EK, Nelson MA. S100P/RAGE signaling regulates microRNA-155 expression via AP-1 activation in colon cancer. Exp Cell Res 319: 2081–2090, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palma CA, Al Sheikha D, Lim TK, Bryant A, Vu TT, Jayaswal V, Ma DD. MicroRNA-155 as an inducer of apoptosis and cell differentiation in acute myeloid leukaemia. Mol Cancer 13: 79, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pastille E, Bardini K, Fleissner D, Adamczyk A, Frede A, Wadwa M, von Smolinski D, Kasper S, Sparwasser T, Gruber AD, Schuler M, Sakaguchi S, Roers A, Muller W, Hansen W, Buer J, Westendorf AM. Transient ablation of regulatory T cells improves antitumor immunity in colitis-associated colon cancer. Cancer Res 74: 4258–4269, 2014. [DOI] [PubMed] [Google Scholar]
- 30.Rasmussen TK, Andersen T, Bak RO, Yiu G, Sorensen CM, Stengaard-Pedersen K, Mikkelsen JG, Utz PJ, Holm CK, Deleuran B. Overexpression of microRNA-155 increases IL-21 mediated STAT3 signaling and IL-21 production in systemic lupus erythematosus. Arthritis Res Ther 17: 154, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozovski U, Calin GA, Setoyama T, D'Abundo L, Harris DM, Li P, Liu Z, Grgurevic S, Ferrajoli A, Faderl S, Burger JA, O'Brien S, Wierda WG, Keating MJ, Estrov Z. Signal transducer and activator of transcription (STAT)-3 regulates microRNA gene expression in chronic lymphocytic leukemia cells. Mol Cancer 12: 50, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez-Elsner T, Botella LM, Velasco B, Corbi A, Attisano L, Bernabeu C. Synergistic cooperation between hypoxia and transforming growth factor-β pathways on human vascular endothelial growth factor gene expression. J Biol Chem 276: 38527–38535, 2001. [DOI] [PubMed] [Google Scholar]
- 33.Sellers RS, Clifford CB, Treuting PM, Brayton C. Immunological variation between inbred laboratory mouse strains: points to consider in phenotyping genetically immunomodified mice. Vet Pathol 49: 32–43, 2012. [DOI] [PubMed] [Google Scholar]
- 34.Shahar E, Gorodetsky R, Aizenshtein E, Lalush L, Pitcovski L. Modulating the innate immune activity in murine tumor microenvironment by a combination of inducer molecules attached to microparticles. Cancer Immunol Immunother 64: 1137–1149, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang K, Bai YP, Wang C, Wang Z, Gu HY, Du X, Zhou XY, Zheng CL, Chi YY, Mukaida N, Li YY. Crucial involvement of tumor-associated neutrophils in the regulation of chronic colitis-associated carcinogenesis in mice. PLos One 7: e51848, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology 79: 313–320, 2010. [DOI] [PubMed] [Google Scholar]
- 37.Singh UP, Murphy AE, Enos RT, Shamran HA, Singh NP, Guan H, Hegde VL, Fan D, Price RL, Taub DD, Mishra MK, Nagarkatti M, Nagarkatti PS. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology 143: 478–489, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solinas G, Germano G, Mantovani A, Allavena P. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J Leukoc Biol 86: 1065–1073, 2009. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki R, Fukui T, Kishimoto M, Miyamoto S, Takahashi Y, Takeo M, Mitsuyama T, Sakaguchi Y, Uchida K, Nishio A, Okazaki K. Smad2/3 linker phosphorylation is a possible marker of cancer stem cells and correlates with carcinogenesis in a mouse model of colitis-associated colorectal cancer. J Crohns Colitis 9: 565–574, 2015. [DOI] [PubMed] [Google Scholar]
- 40.Velazquez KT, Enos RT, Narsale AA, Puppa MJ, Davis JM, Murphy EA, Carson JA. Quercetin supplementation attenuates the progression of cancer cachexia in ApcMin/+ mice. J Nutr 144: 868–875, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viennois E, Chen F, Laroui H, Baker MT, Merlin D. Dextran sodium sulfate inhibits the activities of both polymerase and reverse transcriptase: lithium chloride purification, a rapid and efficient technique to purify RNA. BMC Res Notes 6: 360, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Yu F, Jia X, Iwanowycz S, Wang Y, Huang S, Ai W, Fan D. MicroRNA-155 deficiency enhances the recruitment and functions of myeloid-derived suppressor cells in tumor microenvironment and promotes solid tumor growth. Int J Cancer 136: E602–E613, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang M, Zhang P, Li Y, Liu G, Zhou B, Zhan L, Zhou Z, Sun X. The quantitative analysis by stem-loop real-time PCR revealed the microRNA-34a, microRNA-155 and microRNA-200c overexpression in human colorectal cancer. Med Oncol 29: 3113–3118, 2012. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, Li X, Zheng D, Zhang D, Peng X, Zhang X, Ai F, Wang X, Ma J, Xiong W, Li G, Zhou Y, Shen S. Dynamic changes and functions of macrophages and M1/M2 subpopulations during ulcerative colitis-associated carcinogenesis in an AOM/DSS mouse model. Mol Med Rep 11: 2397–2406, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS, Zhou T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med 31: 1375–1380, 2013. [DOI] [PubMed] [Google Scholar]
- 46.Zheng L, Xu CC, Chen WD, Shen WL, Ruan CC, Zhu LM, Zhu DL, Gao PJ. MicroRNA-155 regulates angiotensin II type 1 receptor expression and phenotypic differentiation in vascular adventitial fibroblasts. Biochem Biophys Res Commun 400: 483–488, 2010. [DOI] [PubMed] [Google Scholar]
- 47.Zonari E, Pucci F, Saini M, Mazzieri R, Politi LS, Gentner B, Naldini L. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood 122: 243–252, 2013. [DOI] [PubMed] [Google Scholar]