Abstract
The calcineurin B subunit (CnB) is the regulatory subunit of Cn, a Ca2+/calmodulin-dependent serine/threonine protein phosphatase. In this study, we demonstrate that extracellular CnB was effectively internalized through a CD14-independent Toll-like receptor 4 (TLR4) pathway, which led to the phosphorylation of nuclear factor (NF)-kappa-B inhibitor alpha (IκB-α) and upregulation of pro-inflammatory cytokines in human monocytes. CnB-induced IκB-α phosphorylation is completely dependent on TNF receptor-associated factor 3 (TRAF3) but not TRAF6, which is indispensable for IκB-α phosphorylation in response to lipopolysaccharide. The loss-of-function CnB mutants were able to induce IκB-α phosphorylation, further indicating that this novel role of CnB is completely independent of the phosphatase function of Cn. Taken together, these findings demonstrate that CnB is a novel host-derived immunostimulatory factor, having a role as an agonist in monocytes, and specificity in TLR4 signaling through TRAF3 and TRAF6, in response to various agonists.
Host-derived immunostimulators, such as heat-shock proteins, can signal through the same receptors as those used by pathogen-associated molecular patterns (PAMPs), such as lipopolysaccharide (LPS).1 Toll-like receptor 4 (TLR4) is critical for the host's defense against Gram-negative bacteria in the innate immune response.1 On recognition of LPS, TLR4 can initiate two major intracellular signaling pathways, the myeloid differentiation primary response protein-88 (MyD88)-dependent and Toll/IL receptor (TIR) domain-containing adaptor-inducing interferon-β (TRIF)- dependent (MyD88-independent) pathway, through distinct effector functions of TRAF6 and TRAF3, respectively, and induce downstream effects of two transcription factors, nuclear factor-kappa-B (NF-κB) and the interferon regulatory factor 3 (IRF3).2 In both of the different LPS-induced TLR4 signaling pathways, TRAF6, but not TRAF3, is indispensable for NF-κB inhibitor alpha (IκB-α) phosphorylation and NF-κB activation during the pro-inflammatory cytokine response. However, TRAF3 is essential for the activation of IRF3 and the induction of type I interferons and the anti-inflammatory cytokine interleukin-10 (IL-10).2, 3
Calcineurin B (CnB) is a regulatory subunit of Cn in the cytoplasm and functions in signal transduction pathways by upregulating the phosphatase activity of CnA. This results in dephosphorylation and nuclear translocation of NF of activated T cells (NF-AT), which are expressed in most immune cells, thereby indirectly governing the transcription of cytokine genes, such as IL-2.4, 5, 6 Physiological and developmental studies, including those using a targeted/specific deletion of CnB in mice, have suggested a pivotal role for CnB in Cn function and is associated with positive selection of developing T cells in the thymus, skeletal muscle growth, and pancreatic β-cell growth and function.7, 8, 9 Cn has been significantly implicated in the pathogenesis of diseases such as Alzheimer's disease, cervical neoplasia and left ventricular hypertrophy through a reduction of the intracellular Cn enzymatic activity or deletions in the CnB promoter region.10, 11, 12, 13 It is widely recognized that intracellular CnB regulates the activity of the Cn phosphatase and is required for the activation of all cytoplasmic NF-AT proteins.14, 15, 16
The existence of extracellular Cn under physiological conditions has been confirmed and quantitated in the serum, amniotic fluid and cord blood collected from full term pregnant women during normal deliveries.17 The observation of impaired Cn activity in serum has provided new insights into the pathogenesis of many diseases, including idiopathic mental handicap, acute leukemia, type II diabetes mellitus and Duchenne muscular dystrophy.18, 19, 20, 21 Furthermore, the CnA subunit negatively regulates TLR-mediated activation, potentially through interacting with MyD88, TRIF, TLR2 and TLR4 in transfected cells.22 Cn exists in both the cytoplasm and extracellular milieu. We hypothesize that extracellular CnB in vivo may have a novel role as a cytokine and exert potential immunoregulatory effects on various immunocompetent cells. To investigate this possibility and gain insight into the role of extracellular CnB, we identified that CnB elicits its novel function by stimulating cytokine production. We further investigated the molecular mechanism through which CnB functions, which is completely independent of Cn.
Results
CnB-induced endocytosis of TLR4 and upregulation of pro-inflammatory cytokines through a CD14-independent TLR4 pathway
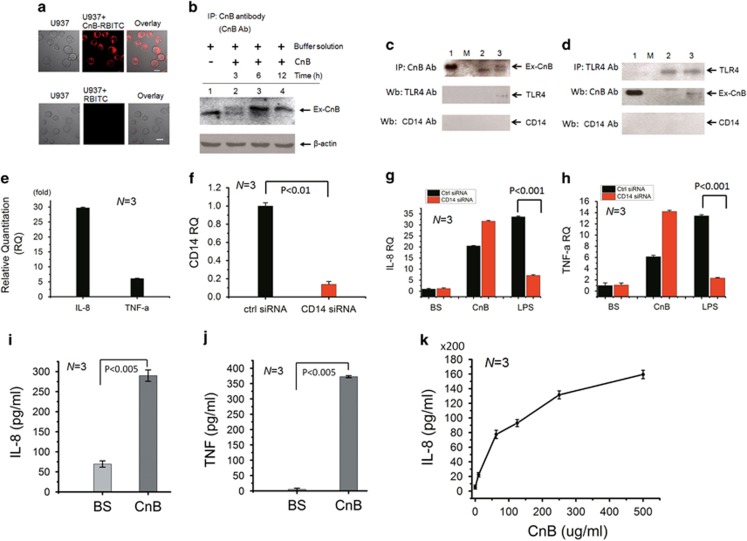
Based on a previous report showing that the CnA subunit interacts with TLRs in transfected cells,22 a CnB protein conjugated to Rhodamine B isothiocyanate (CnB-RBITC) was used as a probe and directly added to the culture medium of human U937 monocytes to investigate whether CnB may target the TLRs on the cell membrane. Confocal imaging showed that the cells were stained by CnB-RBITC protein probe with high efficiency after 10 min and were compared with that of RBITC (Figure 1a). In fact, the internalization occurred rapidly, and, as a result, the cells displayed positive CnB-RBITC staining in <1 min. The internalization of the exogenous CnB was further confirmed by immunoprecipitation (IP) (Figure 1b). To validate whether the endogenous TLR4 is a putative receptor of the exogenous CnB, we performed co-IP experiments. We treated human U937 monocytes with CnB for 30 min and used CnB as the bait protein and TLR4 antibody as a probe, which gave rise to a specific and positive band in the western blot (Figure 1c). CnB–TLR4 association was also demonstrated using endogenous TLR4 as the bait protein in the co-IP and the CnB antibody as the probe (Figure 1d). Although the cluster of differentiation14 (CD14) is a co-receptor for the TLR4 ligand, neither of the two co-IP show results for CnB and TLR4 displayed detectable CD14 signaling in the western blots (Figures 1c and d), indicating a potential CD14-independent TLR4 pathway.
Figure 1.
CnB-induced endocytosis of TLR4 and pro-inflammatory cytokine production are independent of CD14. (a) Confocal microscopy images showing CnB-RBITC and RBITC internalization in U937 cells. (b) Immunoprecipitation analysis of exogenous CnB (Ex-CnB) ingested into U937 cells. Lane 1: U937 cell lysate, the band is the endogenous CnB protein; lanes 2–4: CnB-treated U937 cells at various time points. (c and d) Co-immunoprecipitation (repeated for three times) of exogenous CnB (Ex-CnB) and endogenous TLR4 using exogenous CnB (c) and endogenous TLR4 (d), respectively, as the bait protein, followed by western blotting for CnB, TLR4 and CD14. Lane 1: purified CnB was used as a control added directly to the SDS–PAGE gel; lane M: protein marker; lane 2: buffer solution-treated U937 cells; lane 3: CnB-treated U937 cells. (e) Quantitative PCR of IL-8 and TNF-α, which were normalized to the mRNA transcripts from the buffer solution-treated cells (as a control). (f) Quantitative PCR for CD14 expression in U937 cells transfected with a control siRNA or CD14 siRNA, which were normalized to the β-actin mRNA. CD14 expression was knocked down to 13.9%. (g and h) Quantitative PCR for IL-8 and TNF-α in U937 cells transfected with a CD14 siRNA or control siRNA and treated with CnB, buffer solution (BS) or LPS for 3 h. (i and j) ELISA for IL-8 and TNF-α in U937 cells treated with CnB for 3 h. (k) ELISA for IL-8 in U937 cells treated with different concentrations of CnB (0, 10, 62.5, 125, 250 and 500 μg) for 24 h. The scale bar represents 10 μm in a, and the error bars represent the mean±s.d.
To validate the CD14-independent CnB–TLR4 association in the first stage (within 30 min), we first investigated the mRNA levels of pro-inflammatory cytokines, such as IL-8 and tumor necrosis factor-α (TNF-α), using quantitative PCR. Treatment of human U937 monocytes with CnB resulted in a remarkable upregulation of the expression of IL-8 and TNF-α compared with the buffer solution (BS) (Figure 1e). Next, to validate whether the CnB-induced cytokines are independent of CD14, we knocked down CD14 expression in U937 cells using short interfering RNAs (siRNAs) (Figure 1f) and found that the CnB-induced cytokines, including IL-8 and TNF-α, did not decrease and were completely different from those stimulated by LPS (Figures 1g and h). Furthermore, we tested these cytokines using an Enzyme-linked immunosorbent assay (ELISA) and found that the IL-8 and TNF-α proteins were substantially increased within 3 h (Figures 1i and j). Furthermore, CnB upregulated the expression of these cytokines, including IL-8, in a dose-dependent manner (Figure 1k). Taken together, these findings show that CnB upregulates pro-inflammatory cytokines in human monocytes through a CD14-independent TLR4 pathway.
CnB stimulates cytokine production through IκB-α phosphorylation
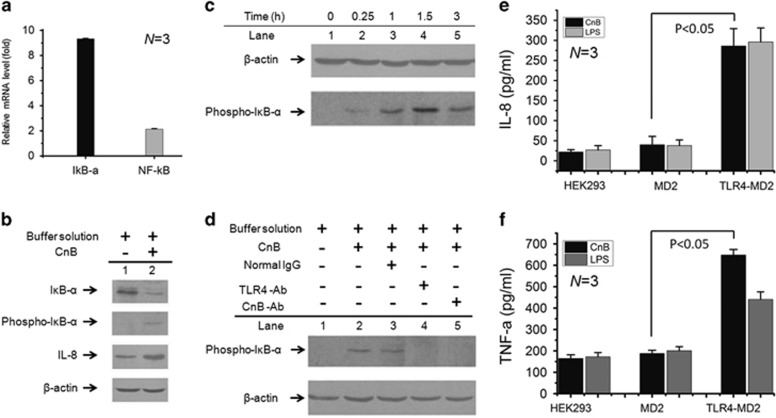
To investigate whether CnB stimulates cytokine production through IκB-α phosphorylation, human U937 monocytes were treated with CnB or its BS alone as a control, resulting in a significant change of gene expression in CnB-induced monocytes compared with the BS-treated monocytes, as revealed by a quantitative PCR analysis (Figures 1e and 2a). The signal transduction cascade activated by exogenous CnB may involve NF-κB, which is complexed with its cytoplasmic inhibitor IκB-α in an inactive state in cytosol and must be activated to upregulate the pro-inflammatory cytokines via the phosphorylation of IκB-α at Ser32 and Ser36.23 We examined phosphorylation of IκB-α by treating the human U937 monocytes with CnB. The amount of IκB-α in the CnB-induced cells (Figure 2b, lane 2) was decreased compared with the BS-induced cells (Figure 2b, lane 1). However, IκB-α phosphorylation was significantly increased in the CnB-treated cells. On phosphorylation of IκB-α, NF-κB is released, migrates to the nucleus and activates the promoters of target genes, resulting in the production of pro-inflammatory cytokines, such as IL-8 and TNF-α (Figure 1).
Figure 2.
The CnB–TLR4 interaction results in cytokine production through IκB-α phosphorylation. (a), Quantitative PCR for NF-κB (2.1-fold) and IκB-α (9.3-fold). P<0.001, N=3. (b) Western blot to detect IκB-α (with or without phosphorylation) and IL-8 in CnB-treated (lane 2) or buffer solution-treated (lane 1) U937 cells; the cells were stimulated with CnB or its buffer solution for 3 h. (c) Western blot to detect IκB-α phosphorylation at various time points in CnB-treated U937 cells. (d) Western blot to detect IκB-α phosphorylation in U937 cells treated with different antibodies. (e and f) ELISA for IL-8 and TNF-α in HEK293 cells transfected with TLR4 and/or MD2 and treated with CnB and LPS for 3 h. The error bars represent the mean±s.d.
The time course of IκB-α phosphorylation was further examined to gain insight into the CnB-induced signal transduction cascade, which may occur through TLR4-mediated NF-κB activation. IκB-α phosphorylation steadily increased and reached the highest level at 1.5 h (Figure 2c). We next investigated whether the signal transduction step in IκB-α phosphorylation results specifically from the CnB–TLR4 interaction. Pre-incubation of human monocytes with an antibody against TLR4, but not normal IgG, completely abrogated CnB-induced IκB-α phosphorylation (Figure 2d, lanes 3 and 4), demonstrating that the signal transduction step in NF-κB activation results from the engagement of TLR4. Furthermore, pre-incubation of cells with an antibody against CnB, but not a control antibody, also sufficiently abrogated IκB-α phosphorylation (Figure 2d, lane 5). Furthermore, to validate the CnB–TLR4 interaction, TLR4 and/or MD2 was transfected into HEK293 cells. Using LPS as a positive control, the ELISA results show that CnB clearly upregulates cytokines, including IL-8 and TNF-α, compared with MD2 (Figures 2e and f). Collectively, these results suggest that the signal, which can be abrogated by antibodies against CnB and TLR4, activates NF-κB and is completely dependent on the CnB–TLR4 interaction. These data demonstrate that CnB is a powerful regulator of both IκB-α phosphorylation and pro-inflammatory cytokine expression in human monocytes, and phosphorylation of the NF-κB/IκB-α complex functions as a potential signaling intermediate in CnB–TLR4 interaction-induced cytokine production.
Effect of CnB on human PBMCs
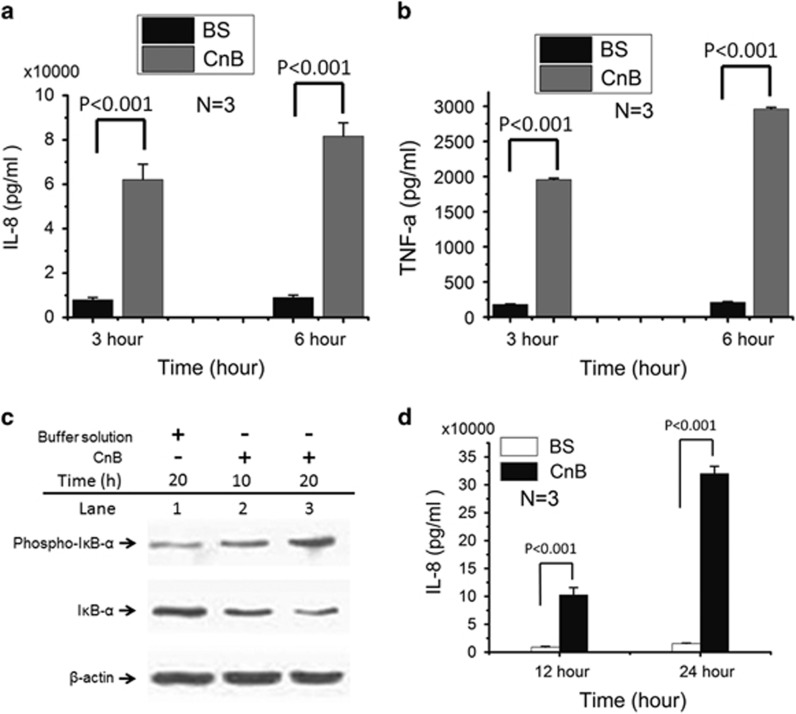
We next examined the effect of CnB on peripheral blood mononuclear cells (PBMCs) under physiological conditions. Treatment of human monocytes from PBMCs with CnB for 3 or 6 h resulted in a remarkable upregulation of cytokines, including IL-8 and TNF-α, compared with the BS (Figures 3a and b). Furthermore, we investigated the time course of IκB-α phosphorylation in PMBCs. After treating the human PBMCs with CnB for 10 or 20 h, we found that while the amount of IκB-α was decreased in the CnB-treated monocytes isolated from PBMCs, IκB-α phosphorylation was considerably increased (Figure 3c), resulting in the transcription and release of pro-inflammatory cytokines, such as IL-8, with an ~10 to 20-fold increase after 12 h (Figure 3d). These results indicate that CnB-induced IκB-α phosphorylation and resulted in downstream effects.
Figure 3.
Effect of CnB on human monocytes from peripheral blood mononuclear cells (PBMCs). (a and b) ELISA for IL-8 and TNF-α in human monocytes from PBMCs that were treated with CnB or its buffer solution (BS) for 3 or 6 h. (c) Western blot analysis of IκB-α and its phosphorylation in human monocytes treated with CnB or its buffer solution (BS). (d) Human monocytes were incubated with CnB or its buffer solution alone as negative control for 12 or 24 h, and analyzed using a human IL-8 ELISA kit. IL-8 was translated and released in a time-dependent manner and showed a 10-fold increase after 12 h and a 20-fold increase after 24 h. The error bars represent the mean±s.d.
A new role for CnB independent of CnA, but dependent on TRAF3
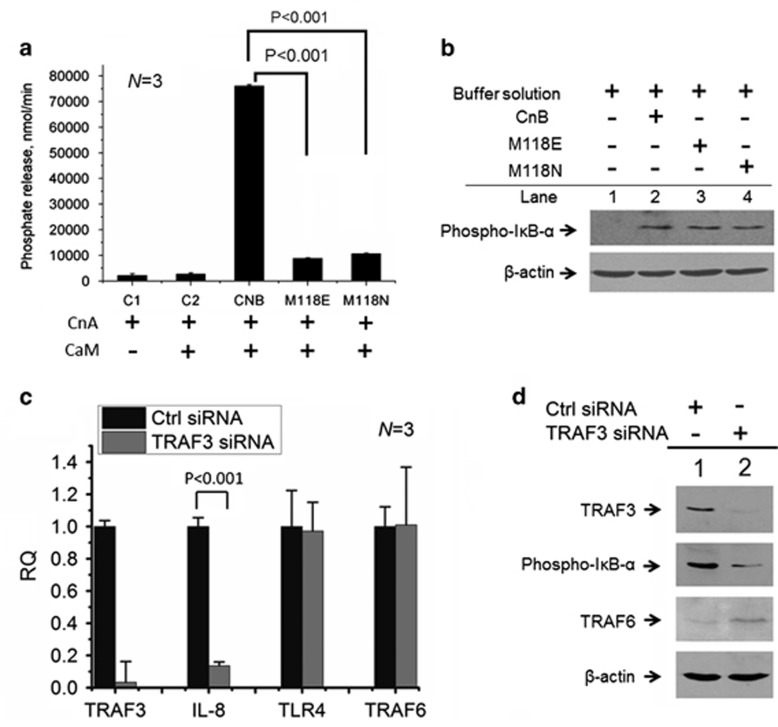
CnB Met118 mutants are detrimental to Cn's phosphatase activity,24, 25 and these loss-of-function CnB mutants have been checked using [32P] RII, a 32P-labeled peptide substrate for Cn14 (Figure 4a). Furthermore, we determined whether these mutants are also unable to phosphorylate IκB-α. Unexpectedly, in contrast to wild-type CnB, these mutants still robustly induced IκB-α phosphorylation (Figure 4b), thus displaying a role for CnB independent of Cn's dephosphorylation activity.
Figure 4.
The role of exogenous CnB as an agonist independent of Cn activity, but dependent on TRAF3. (a) Dephosphorylation activity of CnA (CnA subunit) toward [32P] RII regulated by wild-type CnB and its mutants. Controls 1 and 2 (C1 and C2) did not include CnB or its mutants. CaM, calmodulin; N=3. (b) Western blot analysis of IκB-α phosphorylation induced by the buffer solution, CnB or its mutants in U937 cells; the cells were stimulated with buffer solution, CnB and its mutants for 3 h. (c) Quantitative PCR for TRAF3 IL-8, TLR4 and TRAF6 in U937 cells transfected with TRAF3 siRNA or its control siRNA, respectively, and then induced by CnB. (d) Western blot analysis to detect the expression of TRAF3 and TRAF6 and IκB-α phosphorylation; lane 1: control siRNA and lane 2: TRAF3 siRNA. The error bars represent the mean±s.d.
TNF receptor-associated factor family members (TRAFs) are adaptor molecules linking upstream receptor signals to downstream gene activation.3 We next investigated whether TRAFs have a potential effect on CnB-induced IκB-α phosphorylation using small interfering RNAs. Using a small interfering RNA targeting TRAF3 in human U937 monocytes (Figure 4c), we showed that TRAF3-knockdown cells downregulated the expression of pro-inflammatory cytokines, including IL-8, but did not exhibit changes in TRAF6 and TLR4 expression (Figure 4c). We further validated the phosphorylation of IκB-α using western blotting and demonstrated the TRAF3-dependent phosphorylation of IκB-α (Figure 4d).
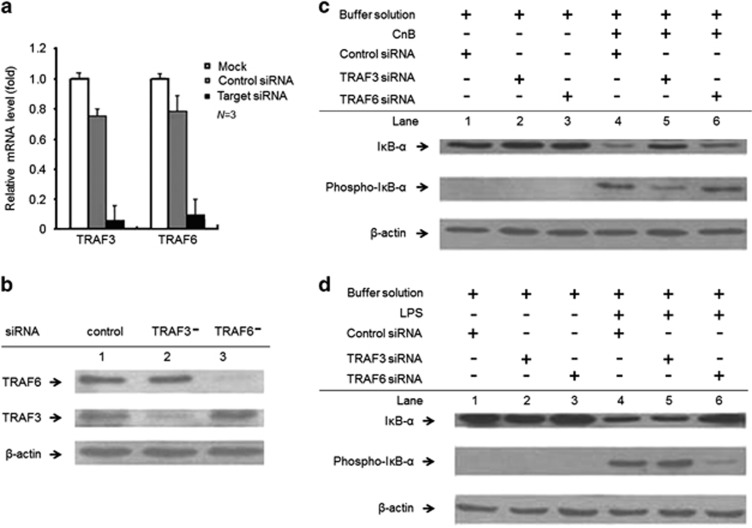
Specificity in TLR4 signaling, through TRAF3 and TRAF6, in response to different agonists
TRAF3-deficient cells overproduce PAMP-induced pro-inflammatory cytokines.3 Thus, the dual effector functions of TRAF3 in TLR4 signaling pathway have been emerging based on TRAF3-dependent NF-κB activation (Figures 4c and d). We further knocked down both TRAF3 and TRAF6 expression in human U937 cells (TRAF3−cells and TRAF6−cells) at the mRNA (Figure 5a) and protein levels (Figure 5b). In contrast to the results obtained in TRAF6−cells (Figure 5c, lane 6) and normal cells (Figure 5c, lane 4), CnB-induced IκB-α phosphorylation was obviously decreased in the TRAF3−cells (Figure 5c, lane 5), suggesting that CnB-mediated NF-κB activation is dependent on TRAF3 rather than TRAF6. On the other hand, TRAF6, but not TRAF3, is critical for LPS-induced IκB-α phosphorylation,3 which was validated in the normal (Figure 5d, lane 4), TRAF3−(Figure 5d, lane 5) and TRAF6−cells (Figure 5d, lane 6). Thus, these results reveal an essential difference in the TRAFs that activate NF-κB in response to CnB and LPS, and further eliminate the possibility of contamination of purified CnB with LPS. However, most importantly, these findings demonstrated TRAF3's specificity in TLR4 signaling, with dual effector functions in response to endogenous immunostimulators and PAMPs. Furthermore, these findings demonstrated specificity in TLR4 signaling in response to host-derived immunostimulators and PAMPs, which were dependent on different TRAFs to activate the downstream effects of NF-κB.
Figure 5.
TLR4 signaling through TRAF3 and TRAF6 in response to different agonists. (a and b) Quantitative PCR (a) and western blot (b) analyses of TRAF3 and TRAF6 expression. The TRAF3 and TRAF6 mRNA levels were decreased by ~6% and ~9% (shown by qRT–PCR), respectively, compared with the levels in the mock transfected cells (without siRNA), or by ~7 and ~12%, respectively, compared with the levels in the cells transfected with the negative control siRNA. P<0.001. (c and d) Western blot analysis of CnB-induced or LPS-induced IκB-α phosphorylation in the siRNA-transfected cells; the cells were stimulated with LPS, CnB or its buffer solution for 3 h. The LPS-treated, TRAF6-knockdown cells remarkably decreased the phosphorylation of IκB-α compared with the negative control and TRAF3-knockdown cells. Furthermore, the TRAF3-knockdown cells slightly increased LPS-induced IκB-α phosphorylation, but considerably impaired CnB-induced IκB-α phosphorylation. The experiments were repeated three times. The error bars represent the mean±s.d.
Discussion
We have identified a mechanism by which extracellular CnB triggers a potent pro-inflammatory immune response in human monocytes. Extracellular CnB elicits its novel function in a CD14-independent TLR4 signaling pathway, resulting in rapid and effective receptor-mediated internalization. To gain insight into the emerging function of extracellular CnB, a widely known substrate of the Cn phosphatase in the cytosol for the dephosphorylation of NF-AT, we have found that TLR4-mediated CnB signaling profoundly stimulated monocytes and resulted in IκB-α phosphorylation and upregulation of pro-inflammatory cytokines in a potential TRAF3-dependent pathway.
CD14, as co-receptor for the TLR4 ligand, is critical for the host's defense against Gram-negative bacteria through two major intracellular signaling pathways, that is, the MyD88-dependent and TRIF-dependent pathways, but is not required for the CnB-induced endocytosis of TLR4 according to our co-IP results. Interestingly, CnB upregulates the expression of pro-inflammatory cytokines, which cannot be abrogated by CD14 siRNA and are completely different from those upregulated by LPS, indicating that the CnB–TLR4 association is not dependent on CD14. Thus, CD14 is not a receptor for CnB to promote TLR4 endocytosis and its downstream effects. We further tested CnB-induced pro-inflammatory cytokines such as IL-8 and TNF-α using HEK293 cells and human monocytes isolated from PBMCs and found that the CnB-induced pro-inflammatory cytokines are correlated with the expression of TLR4, suggesting a CD14-independent TLR4 signaling pathway. Furthermore, we investigated whether TRAFs have a potential effect on the CnB-induced pro-inflammatory cytokines using small interfering RNAs and found that TRAF3, but not TRAF6, is important for the upregulation of the CnB-induced pro-inflammatory cytokines, suggesting that TRAF3 acts as an adaptor molecule and functions in a CnB-induced, CD14-independent TLR4 signaling pathway.
Previous work has shown that the functions of TRAF3 and TRAF6 in TLR4 pathway are distinct,3 largely in the transcription factors they activate and their downstream effects. TRAF6, but not TRAF3, is critical for IκB-α phosphorylation and the subsequent activation of nuclear factor NF-κB. In response to LPS, TRAF3-deficient cells overproduced pro-inflammatory cytokines due to defects in the anti-inflammatory cytokine IL-10;2, 3 thus, the TRAF3-knockdown cells displayed stronger and detectable IκB-α phosphorylation. Until now, TRAF3 was dispensable for NF-κB activation and the production of pro-inflammatory cytokines in the LPS-induced TLR4 signaling pathway. Interestingly, in contrast, our TRAF3 results demonstrated a pivotal role of TRAF3 in CnB-induced NF-κB activation. According to Matzinger's danger model, host-derived immunostimulators can signal through the same receptors as those used by PAMPs to induce the innate immune response.1 Our evidence in TLR4-mediated CnB signaling suggested that while the host-derived immunostimulators and TLR4-mediated PAMPs converge at NF-κB activation, they ultimately depend on different TRAFs. Furthermore, TRAF3 exhibited a dual effector function in response to PAMPs and endogenous immunostimulators such as CnB.
By definition, cytokines are proteins that are secreted by cells and have regulatory effects on other cells. Thus, a protein with regulatory effects that co-exists in the cytoplasm and extracellular milieu will be a potential cytokine. We postulate that in vivo, CnB may act as a cytokine and be secreted by cells, based on the large-scale identification of secreted and membrane-associated gene products and ascertainment of the extracellular Cn concentrations under physiological conditions in both serum and amniotic fluid.17, 26 Furthermore, our observation that an antibody against CnB abrogated its regulatory effects on human monocytes implicates a new physiological role for CnB as a cytokine, which, unexpectedly, was independent of Cn's dephosphorylation function in human monocytes, as evidenced by CnB mutants that have an impaired regulatory role for CnA enzymatic activity but facilitate IκB-α phosphorylation. Taken together, in serum, CnB acts as a powerful cytokine that targets TLR4 and regulates the function of immunocompetent cells through a previously unknown TRAF3-dependent pathway. The discovery of this novel function of CnB raises the possibility that CnB may be a potent inducer of antitumor immunity and a potential adjuvant for therapeutic treatments.
Methods
Preparation and fluorescent labeling of the proteins
The constructions of CnB and its mutants, as well as their expression and purification have been described in detail elsewhere.24, 27 After routine removal of pyrogens with the Detoxi-Gel Endotoxin Removing Gel using immobilized polymixin B (Pierce Biotechnology, 20344, Rockford, IL, USA), the endotoxin content of CnB, and the M118E and M118N mutants was further measured by the Limulus amebocyte lysate assay kit (Zhanjiang Bokang Marine Biological co. Ltd, Zhanjiang, China), with a sensitivity of 0.25EU ml−1, according to the manufacturer's instructions. RBITC and CnB-RBITC were used (Beijing Biosynthesis Biotechnology, Beijing, China) for protein tracing. LPS (30 μg ml−1) was used as a control for the subsequent experiments.
Confocal microscopy
Human U937 monocytes (ATCC Number: CRL-1593.2, Manassas, VA USA) were maintained in RPMI 1640 medium (GIBCO, Grand Island, NY, USA), containing 10% (v/v) heat-inactivated fetal calf serum. For the confocal experiment, all cells were cultured in a 35-mm petri dish (MatTek, Ashland, MA, USA), without any additional intervention. The images of the live cells were captured by confocal microscopy (Olympus FV300, Tokyo, Japan) from 1 to 10 min. The protein concentration was determined using the optical density at 280 and 260 nm (quantified by a GBC cintra 10e, Dandenong, VIC, Australia) and calculated using the following equation: protein concentration (mg ml−1)=1.45 × OD280−0.74 × OD260. CnB (0.25 μM) was used for confocal experiment.
Small interference RNA and quantitative RT–PCR
Each siRNA duplex was a pool of three 20–25 nt target-specific siRNAs provided by Ribobio (Guangzhou, China). The siRNAs (100 nM of each siRNA) were co-transfected into U937 cells (5 × 106 cells, suspended in 0.4 ml of OptiMEM I reduced serum medium, Invitrogen, Carlsbad, CA, USA) by electroporation using a Bio-Rad Genepulser Xcell system (BIO-RAD, Hercules, CA, USA) set at 240 V and 975 μF. To maximize the efficiency of the siRNAs, the transfection was repeated at ~24 h after the first transfection. At ~30 h after the second transfection, the cells were collected for both RNA extraction and to prepare protein lysates. The efficiency of target gene suppression was monitored by quantitative reverse transcription (qRT–PCR) and western blotting. Cells transfected with an unrelated siRNA, that is, a control siRNA, were used as a negative control. Furthermore, the U937 cells were treated with 250 μg ml−1 CnB and BS alone for 3 h, and then the total RNAs were prepared using the RNApure High-purity Total RNA Rapid Extraction Kit (BioTeke, Beijing, China). The cDNAs were synthesized from 2 μg of the total RNA in a 20 μl reaction system, including reverse transcriptase M-MLV, oligo-d(T)18 primers, ribonuclease inhibitor and dNTP mixture (TaKaRa, Dalian, China).
For qRT–PCR, all reactions designed to yield ~200 bp sequences were performed using the LightCycler 1.2 RealTime PCR System (Roche, Rotkreuz, Switzerland) and the LightCycler-FastStart DNA Master SYBR Green I Kit (Roche). Each 20 μl PCR reaction was prepared as follows: 13.8 μl of water, 1.6 μl of MgCl2 (3 mM), 0.8 μl of each primer (10 mM), 2 μl of Fast Start DNA Master SYBR Green I, 1 μl RT product and water up to 20 μl. The qRT–PCR reactions began with incubation for 10 min at 95 °C and then 40 cycles of amplification of 15 s at 95 °C, 5 s at ‘Ta' °C and 15 s at 72 °C. The plate was read for fluorescence data collection at 76 °C. Here, ‘Ta' represents the annealing temperature. To check the specificity of the amplified product, melting curve analyses were performed from 75 to 95 °C and the products were analyzed by 1.5% agarose gel electrophoresis. The comparative threshold cycle (CT) method was used to calculate the fold amplification. The expression level of each gene was normalized to β-actin. The quantitative RT–PCR primers (sequence 5′ to 3′) are shown below—forward primer (FP); reverse prime (RP).
IL-8—FP: 5′-AGGAATTGAATGGGTTTGCTAG-3′ RP: 5′-CACTGTGAGGTAAGATGGTGGC-3′: TNF-α—FP: 5′-CGAAGTGGTGGTCTTGTTGCT-3′ RP: 5′-GACCTTTTCAACTTGGCTTCC-3′: NF-κB—FP: 5′-GACCTTTTCAACTTGGCTTCC-3′; RP: 5′-TGGCGACCGTGATACCTTTA-3′: IκB-α—FP: 5′-CGTCTGACGTTATGAGTGCAAAG-3′ RP: 5′-CACAAAAGCAACAAAATGAGGG-3′: β-Actin—FP: 5′-CATGTACGTTGCTATCCAGGC-3′ RP: 5′-CTCCTTAATGTCACGCACGAT-3′: TLR4—FP: 5′-CCTGTCCCTGAACCCTATGAACTT-3′ RP: 5′-GGACTTCTAAACCAGCCAGACCTT-3′: TRAF3—FP: 5′-GCAGAAACACGAAGACACCGACT-3′ RP: 5′-TGCTGGGGGCATTGACACAC-3′: TRAF6—FP: 5′-CCAGCGACCCACAATCCC-3′ RP: 5′-TCCCTCCGAAGGCTACCC-3′: CD14—FP: 5′-AAGCCACAGGACTTGCACTTT-3′ RP: 5′-CCCAGCGAACGACAGATTG-3′.
Immunoprecipitation
To validate the receptor-mediated internalization by IP (Figure 1), human U937 monocytes (5 × 106 cells) were treated with 250 μg ml−1 CnB at various time points (Figure 1b) or for 0.5 h (Figures 1c and d), and maintained in RPMI 1640 medium containing 10% (v/v) heat-inactivated fetal calf serum at 37 °C. The cells were then washed twice with PBS and lysed in CLWI buffer. The lysates were precleared for 1 h with protein A sepharose (Beyotime Biotechnology, Shanghai, China) and normal IgG and then incubated for ~3–5 h with 2 μg ml−1 of the appropriate antibody. The immunocomplexes were collected with protein A sepharose for ~1–2 h before washing in lysis buffer according to the manufacturer's instructions. The immune complexes were eluted at 95 °C for ~5–10 min in 2 × SDS sample buffer supplemented with 50 mM DTT and then detected by western blotting.
Extract preparation and western blotting
U937 monocytes (2 × 106 cells) were treated with 250 μg ml−1 CnB, its mutants or BS alone (Figures 2b, c, and 4b). Alternatively, the cells were pre-treated with small interfering RNAs (Figures 4d, 5c and d), BS alone, 5 μg ml−1 of a control antibody, 5 μg ml−1 of a TLR4 antibody, or 5 μg ml−1 of a CnB antibody (Figure 2d) at 37 °C in humidified air containing 5% CO2 for 1 h and then treated with CnB for an additional 1.5 h. After washing twice with PBS, the cells were lysed in ~150 μl of CLWI buffer (cell lysis buffer for western blotting and IP: P0013, Beyotime Biotech. The whole cell lysates were collected by centrifugation at 4 °C for 5 min at 12 000 g and then incubated 95 °C for 5 min in 1 × SDS sample buffer.
After separation by SDS–PAGE, the target proteins were transferred to nitrocellulose membranes (Millipore, Billerica, MA, USA), and probed with the appropriate antibody at the concentration indicated by the manufacturer, including PP2B-B1/2 (FL-170): sc-33166, TLR4 (H-80): sc-10741, CD14 (M-305): sc-9150, IL-8 (H-60): sc-7922 (Santa Cruz Biotechnology, Dallas, TX, USA); IκB-α antibody: #9242, phospho-IκB-α (Ser32/36) (5A5) mouse mAb: #9246, TRAF6 antibody: #4743, TRAF3 antibody: #4729 (Cell Signaling Technology, Danvers, MA, USA); β-actin: bs-0061R (Biosynthesis biotechnology, Beijing, China); peroxidase-conjugated affinipure goat anti-rabbit IgG (H+L), peroxidase-conjugated affinipure goat anti-mouse IgG (H+L) (Jackson, West Grove, PA, USA). The membranes were then developed using a SuperEnhanced chemiluminescence detection kit (Applygen Technologies, Inc., Beijing, China). The protein bands were visualized after exposing the membranes to Kodak X-ray film (Kodak, Rochester, New York, NY, USA).
Cn phosphatase activity assay
The phosphatase activity assay was performed using two separate mixtures: (1) 10 μl of the enzyme solution, including 0.2 μM of each Cn subunit; and (2) 10 μl of the assay solution, including 40 μM of the 32P-labeled RII peptide, 0.6 μM calmodulin and 0.2 mM CaCl2, in a uniform buffer system (50 mM Tris-HCl, 0.1 mg ml−1 BSA, 0.5 mM DTT and 0.5 mM Mn2+, pH 7.4). After mixing these two solutions and then incubating them at 30 °C for 10 min, the reaction was instantly terminated by the addition of 0.18 ml of 83.3 mM H3PO4. The free 32Pi released from the RII peptide was isolated by AG50W-X8 chromatography (AGXC, BIO-RAD) and quantified by scintillation counting (TRI-CARB 2900TR, Packard Bioscience, Arvada, CO, USA), as previously described.14 Each reaction was performed in triplicate, with variations <10%, and blank assays lacking the enzyme were routinely conducted through the AGXC step.
ELISA
A recombinant plasmid containing TLR4 and/or MD2 (Addgene, Cambridge, MA, USA) was transfected into HEK293s using the Lipofectamine 3000 Transfection Reagent (Invitrogen), and then selected with neomycin for 6 days. The HEK293 cell line was maintained in DMEM medium (Sigma, Beijing, China) containing 10% (v/v) heat-inactivated fetal calf serum (Sigma). Furthermore, the transfected and untransfected U937 and HEK293 cells were treated with 250 μg ml−1 CnB or 30 μg ml−1 LPS for 3 h. The supernatants were collected for IL-8 and TNF-α ELISAs (RayBio, Norcross, GA, USA). The assay procedure is summarized as follows. (1) Prepare all reagents, samples and standards. (2) Add 100 μl of the standard or sample to each well and incubate for 2.5 h at room temperature. (3) Add 100 μl of the prepared biotin antibody to each well and incubate for 1 h at room temperature. (4) Add 100 μl of the prepared streptavidin solution and incubate for 45 min at room temperature. (5) Add 100 μl of the TMB one-step substrate reagent (RayBio) to each well and incubate for 30 min at room temperature. (6) Add 50 μl of the stop solution to each well and record the absorbance at 450 nm.
PBMCs and monocyte isolation
Human PBMCs were isolated from the PB of healthy donors by density gradient centrifugation with lymphocyte separation medium (density=1.077; HaoYang, Shandong, China). The monocytes were then isolated through the MACS sort system (Miltenyi Biotec, Bergisch Gladbach, Germany) using positive selection with immunomagnetic beads specific for CD14 (Miltenyi Biotec), according to the manufacturer's protocol. The supernatants were collected from the cell cultures at 3, 6, 12 or 24 h after stimulation with 250 μg ml−1 CnB. Then, IL-8 and TNF-α were quantified by ELISA according to the manufacturers' protocols (NeoBioscience, Beijing, China and RayBio). The collected cells were also used for western blotting.
Acknowledgments
This work was partially supported by the National Natural Science Foundation of China, the International Cooperation Project, the National Important Basic Research Project and the National Important Novel Medicine Research Project. Zongchao Jia is supported by the Canadian Institute of Health Research and the Natural Science and Engineering Research Council of Canada and is a Canada Research Chair.
The authors declare no conflict of interest.
References
- Seong SY, Matzinger P. Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nat Rev Immunol 2004; 4: 469–478. [DOI] [PubMed] [Google Scholar]
- Zhong B, Tien P, Shu HB. Innate immune responses: crosstalk of signaling and regulation of gene transcription. Virology 2006; 352: 14–21. [DOI] [PubMed] [Google Scholar]
- Häcker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 2006; 439: 204–207. [DOI] [PubMed] [Google Scholar]
- Griffith JP, Kim JL, Kim EE, Sintchak MD, Thomson JA, Fitzgibbon MJ et al. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell 1995; 82: 507–522. [DOI] [PubMed] [Google Scholar]
- Milan D, Griffith J, Su M, Price ER, McKeon F. The latch region of calcineurin B is involved in both immunosuppressant-immunophilin complex docking and phosphatase activation. Cell 1994; 79: 437–447. [DOI] [PubMed] [Google Scholar]
- Parry RV, June CH. Calcium-independent calcineurin regulation. Nat Immunol 2003; 4: 821–823. [DOI] [PubMed] [Google Scholar]
- Dadley-Moore D. Thymocyte development: positive about calcineurin. Nat Rev Immunol 2004; 4: 318. [Google Scholar]
- Dunn SE, Simard AR, Prud'homme RA, Michel RN. Calcineurin and skeletal muscle growth. Nat Cell Biol 2002; 4: E46. [DOI] [PubMed] [Google Scholar]
- Heit JJ, Apelqvist AA, Gu X, Winslow MM, Neilson JR, Crabtree GR et al. Calcineurin/NFAT signalling regulates pancreatic beta-cell growth and function. Nature 2006; 443: 345–349. [DOI] [PubMed] [Google Scholar]
- Ladner CJ, Czech J, Maurice J, Lorens SA, Lee JM. Reduction of calcineurin enzymatic activity in Alzheimer's disease: correlation with neuropathologic changes. J Neuropathol Exp Neurol 1996; 55: 924–931. [DOI] [PubMed] [Google Scholar]
- Padma S, Sowjanya AP, Poli UR, Jain M, Rao B, Ramakrishna G. Downregulation of calcineurin activity in cervical carcinoma. Cancer Cell Int 2005; 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Arnett DK, Devereux RB, Panagiotou D, Province MA, Miller MB et al. Identification of a novel 5-base pair deletion in calcineurin B (PPP3R1) promoter region and its association with left ventricular hypertrophy. Am Heart J 2005; 150: 845–851. [DOI] [PubMed] [Google Scholar]
- Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol 2006; 7: 589–600. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Klee CB, Bierer BE, Burakoff SJ. Calcineurin phosphatase activity in T lymphocytes is inhibited by FK 506 and cyclosporin A. Proc Natl Acad Sci USA 1992; 89: 3686–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef IA, Chen F, Chen L, Kuo A, Crabtree GR. Signals transduced by Ca(2+)/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell 2001; 105: 863–875. [DOI] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell 2002; 109: S67–S79. [DOI] [PubMed] [Google Scholar]
- Padma S, Subramanyam C. Extracellular calcineurin: identification and quantitation in serum and amniotic fluid. Clin Biochem 1999; 32: 491–494. [DOI] [PubMed] [Google Scholar]
- Bindu LH, Rani PU, Reddy PP. Calcineurin activity in children with Mental handicap. Indian J Clin Biochem 2007; 22: 32–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padma S, Subramanyam C. Clinical significance of serum calcineurin in acute leukemia. Clin Chim Acta 2002; 321: 17–21. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan TK, Sethi BK, Subramanyam C. Serum calcineurin activity in relation to oxidative stress and glycemic control in type II diabetes mellitus. Clin Biochem 2005; 38: 218–222. [DOI] [PubMed] [Google Scholar]
- Sundaram JS, Rao VM, Meena AK, Anandaraj MP. Decreased calcineurin activity in circulation of Duchenne muscular dystrophy. Clin Biochem 2007; 40: 443–446. [DOI] [PubMed] [Google Scholar]
- Kang YJ, Kusler B, Otsuka M, Hughes M, Suzuki N, Suzuki S et al. Calcineurin negatively regulates TLR-mediated activation pathways. J Immunol 2007; 179: 4598–4607. [DOI] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 1998; 16: 225–260. [DOI] [PubMed] [Google Scholar]
- Wu W, Jia Z, Liu P, Xie Z, Wei Q. A novel PCR strategy for high-efficiency, automated site-directed mutagenesis. Nucleic Acids Res 2005; 33: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Wu W, Li J, Wei Q. The polarity of the amino acid residue 118 of calcineurin B is closely linked to calcineurin enzyme activity. IUBMB Life 2010; 62: 561–567. [DOI] [PubMed] [Google Scholar]
- Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat Genet 2000; 25: 58–62. [DOI] [PubMed] [Google Scholar]
- Wei Q, Lian ML, Jing FZ, Zhang N, Yan MS, Chen Y et al. Studies of calcineurin B subunit from genetic engineering for use in medicine. Drug Dev Res 2002; 56: 40–43. [Google Scholar]