Abstract
The mammalian heart has long been considered to be a postmitotic organ. It was thought that, in the postnatal period, the heart underwent a transition from hyperplasic growth (more cells) to hypertrophic growth (larger cells) due to the conversion of cardiomyocytes from a proliferative state to one of terminal differentiation. This hypothesis was gradually disproven, as data were published showing that the myocardium is a more dynamic tissue in which cardiomyocyte karyokinesis and cytokinesis produce new cells, leading to the hyperplasic regeneration of some of the muscle mass lost in various pathological processes. microRNAs have been shown to be critical regulators of cardiomyocyte differentiation and proliferation and may offer the novel opportunity of regenerative hyperplasic therapy. Here we summarize the relevant processes and recent progress regarding the functions of specific microRNAs in cardiac development and regeneration.
Keywords: microRNA, cardiac regeneration, heart failure, myocardial infarction, heart development
despite undeniable progress in cardiology and cardiac surgery, heart disease remains the leading cause of mortality and morbidity worldwide (80). Advances in our understanding of the molecular processes occurring in failing cardiomyocytes (CMs) in ischemic heart disease and congestive heart failure have indicated that these two diseases result in comparatively large-scale loss of myocardium. Contrary to the theory of the postmitotic heart, the heart has been shown to maintain the capacity for self-renewal; however, analysis of normal unaffected mature CM proliferation indicated a very low renewal rate (7). Technical advances, including the meticulous evaluation of cell cycle events, as well as genetic and metabolic fate mapping, have provided new data about adult CM renewal. Studies performed in the neonatal mouse and adult zebrafish suggest that cell cycle activity during normal growth and after injury leads to functional yet nondividing multinucleated CMs, as well as to new mononucleated CMs that maintain their proliferative capacity (53, 93). These studies also revealed that the majority of regenerated CMs are derived from preexisting CMs through cell division rather than activation of undifferentiated stem or progenitor cells (53, 93). These data point to the possibility of a novel therapeutic strategy: stimulation of the regenerative capacity of the human heart. This regenerative therapy, if made efficient and safe, has the potential to become the primary option to restore function after significant myocardial damage. Cardiac regeneration research is evaluating two major avenues that may be utilized independently or together: 1) exogenous cell transplantation; and 2) stimulation of endogenous regenerative processes (25, 51, 117). This review will focus on the role of microRNAs (miRNAs) in regenerative strategies, including CM proliferation, stem cell differentiation, and cell reprogramming. To aid our review of regeneration, we will begin by discussing the basics of miRNAs, cardiac development, and cardiac regeneration.
OVERVIEW AND BIOGENESIS OF miRNA
miRNAs are a class of natural, endogenously expressed, single-stranded, noncoding RNAs. miRNAs are best known as negative regulators of gene expression at the posttranscriptional level, base pairing in a sequence-dependent manner to target protein-coding mRNAs, often at sites contained in the 3′-untranslated region (UTR) (60). In addition, miRNAs may be sponged up and essentially inactivated by pseudogenes and decoy mRNAs, or they may be targeted to long noncoding RNAs, representing new levels of miRNA regulation and function, as well as important roles of noncoding transcripts that each merit more research (11, 22, 65). miRNAs are important modulators of all cellular pathways, especially cell fate determination, differentiation, proliferation, programmed cell death, and other processes during embryogenesis and in adult life in species ranging from Caenorhabditis elegans and Drosophila melanogaster to humans (58, 62, 112).
In addition to roles in normal physiology, over the past years, the involvement of miRNAs in pathological processes of the heart, such as hypertrophy, cardiomyopathy, fibrosis, apoptosis, and cardiac remodeling, has become better appreciated. This research led to the hope that miRNAs may represent new therapeutic targets. We will begin by discussing miRNA in greater detail.
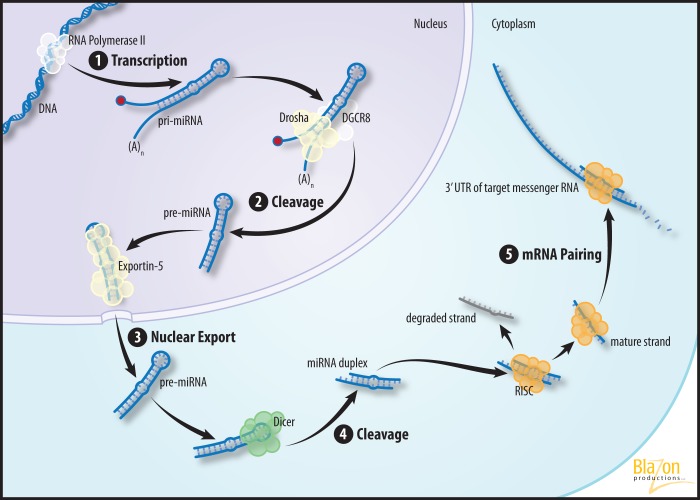
Members of the miRNA family were initially discovered as small transient RNAs that regulate developmental transitions in Caenorhabditis elegans. The linking of the lin-4 and let-7 families, the first described miRNAs, and their target-specific translational inhibition, hinted at a new mechanism of gene regulation during tissue development (98, 119). It was shown that miRNAs are a group of 21–25 nucleotides in length, single-stranded RNAs expressed in invertebrates and vertebrates animals. The biogenesis of miRNA follows the pathway shown in Fig. 1 and briefly described below.
Fig. 1.
Illustration of the process of microRNA (miRNA) biogenesis and function, described in overview and biogenesis of mirna. DGCR8, DiGeorge syndrome critical region gene 8; RISC, RNA-induced silencing complex; pri-miRNA, primary miRNA; UTR, untranslated region.
Transcription of the pri-miRNA
The process of miRNA biogenesis begins when the hairpin primary miRNA (pri-miRNA) is transcribed (with 5′ cap and poly-A tail) by RNA polymerase II, either as an independent transcriptional unit, or as portions of introns of protein-coding RNA polymerase II transcripts (63). Intronic miRNAs are under the regulatory control of response elements of the host gene, and the expression pattern is similar but depends on the miRNA processing machinery. Since miRNA genes are in close proximity to each other in DNA, they could be expressed as a single transcript, if they are present as a single cistron (polycistronic miRNAs), which could be independent or intronic cistrons (99). Recent reports in the literature suggest that some intronic miRNAs might be transcribed and regulated independently as nested genes within their transcription units (95). These findings add to existing knowledge toward understanding the complexity of miRNA biogenesis.
Pri-miRNA cleavage
The relatively large pri-miRNA is processed and cleaved in the nucleus by the RNase III type endonuclease Drosha with the help of the double-stranded RNA binding protein DGCR8 (DiGeorge syndrome critical region gene 8) to produce a hairpin precursor miRNA (pre-miRNA) consisting of ∼70 nucleotides (38). This complex cuts the pri-miRNA transcript within the nucleus into several pre-miRNAs. Drosha is the catalytic subunit of the pri-miRNA processing microprocessor complex, while DGCR8 stabilizes Drosha and recognizes the RNA substrate. Drosha homologs are found only in animals. Drosha protein is capable of orienting itself on pri-miRNA in a way that each RNase III domain is positioned on the correct strand. It was shown that the Drosha RNase III domains and the middle region may recognize the terminal loop or the bottom single-strand RNA region of pri-miRNA. Alternatively, DGCR8 may help Drosha to be correctly positioned on pri-miRNA (61). The two RNase domains of Drosha cleave the 5′ and 3′ arms of the pri-miRNA hairpin, whereas DGCR8 directly and stably interacts with the pri-miRNA and functions as a molecular ruler to determine the precise cleavage site (38).
Export of the pre-miRNA
After cleavage, the pre-miRNA is transported to the cytoplasm via exportin-5 in complex with Ran-GTP. Exportin-5 also protects pre-miRNAs against nuclear digestion. A defined length of the double-stranded stem and the 3′ overhangs are important for successful binding to exportin-5, ensuring the export of only correctly processed pre-miRNAs (55).
miRNA maturation in the cytoplasm
Following export, the RNase III type endonuclease Dicer generates a ∼22-bp double-stranded miRNA duplex. One strand of the duplex is selected to function as a mature miRNA and is incorporated into the RNA-induced silencing complex (RISC), while the other strand is degraded (33). RISC, a ribonucleoprotein complex, is the cytoplasmic effector of the miRNA pathway and contains a single-stranded miRNA guiding it to its target mRNAs. Cytoplasmic miRNA processing is mediated by the RISC loading complex (33). The RISC loading complex is a multiprotein complex composed of the RNase Dicer, the double-stranded RNA-binding domain proteins Tar RNA binding protein and protein activator of PKR, and the core component Argonaute-2 (Ago2). Ago2 proteins participate in miRNA processing and are the RISC effector proteins mediating mRNA degradation, destabilization, or translational inhibition (96). Loss of endogenous Ago2 diminishes the expression and activity of mature miRNA. RISC, after binding with an mRNA target, accumulates in cytoplasmic foci (P-bodies) and stress granules.
Regulation of Target Gene Expression
The final function of miRNAs, regulation of gene expression, is directed by miRNA incomplete base pairing complementarity (in animals, normally sequence specific but imperfect and nonexclusive) to target sequences. These sequences are often located in the 3′-UTR of target mRNAs, but they can also be found in the 5′-UTR or the coding region. Recognition sites may be present in multiple copies in an mRNA sequence to enhance the effect on target gene expression (5).
To date, more than 3,000 miRNAs have been identified in animals, plants, and viruses, including ∼1,000 mature miRNAs in the human genome (34–36, 56, 57). miRNAs can be organized into families based on sequence homology and chromosomal location. The human miRNA genome is predicted to regulate up to 30% of mRNA transcripts (6). Because of this bidirectional lack of exclusivity between miRNA and mRNA, as well as other miRNA targets such as decoy mRNAs and long noncoding RNAs, miRNA-related pathways exhibit significant regulatory complexity.
Together, the above data highlight the essential role of the miRNA biogenesis and suggest that each of these steps serves as a point of regulation and, therefore, provides additional complexity to miRNA-dependent gene regulation. The following sections will discuss the basic knowledge of cardiac development and regeneration and will examine the functions of specific miRNAs in the regulations of these processes.
BASIC DEVELOPMENTAL STAGES OF CARDIOGENESIS
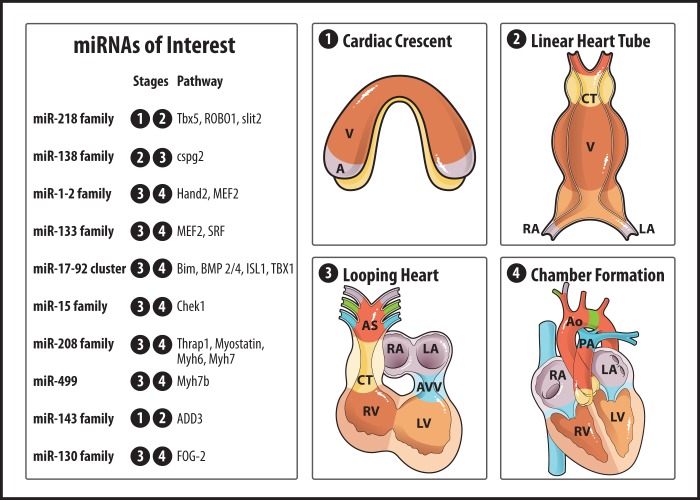
Cardiac development is an intricate multistep process controlled by an evolutionarily conserved complex of genetic pathways and an extended network of transcriptional and posttranscriptional regulatory programs. Beginning mechanical function at day 22 of human embryogenesis to transport the oxygen and nutrients necessary for growth of other tissues and organs; the heart is the first organized and functioning organ in human and vertebrate animals. Therefore, many of the underlying mechanisms are developmentally conserved. In the human during the third week of development, the lateral plate mesoderm splits into two layers: the somatopleuric and splanchnopleuric. The last mesoderm layer contains the myocardial and endocardial cardiogenic precursors in the region of the primary heart fields. The major stages of human cardiac development are depicted in Fig. 2. Briefly, precise signaling-derived changes in cardiac gene expression initiate morphological cardiogenesis, including primordia migration, tube formation, looping, trabeculation, septation, valvulogenesis, and growth, to form the four-chambered heart (26, 78). Despite increasing knowledge of the genes expressed during cardiac development, there are many interactions between genes and mechanical rearrangements during morphogenesis that remain unclear.
Fig. 2.
The four major stages of human cardiac development, along with miRNAs known to play a role. Tbx5, T-box gene family member 5; ROBO1, roundabout axon guidance receptor, homolog 1; slit2, slit homolog 2; cspg2, chondroitin sulfate proteoglycan 2; Hand2, heart and neural crest derivatives-expressed protein 2; MEF2, myocyte enhancer factor-2; SRF, serum response factor; Bim, Bcl-2-like protein 11; BMP 2/4, bone morphogenetic proteins 2 and 4; ISL1, ISL LIM homeobox 1; TBX1, T-box gene family member 1; Chek1, checkpoint kinase 1; Thrap1, thyroid hormone receptor-associated protein 1; Myh6, α-myosin heavy chain; Myh7, β-myosin heavy chain; Myh7b, myosin heavy chain 7b; ADD3, adducin 3; FOG-2, zinc finger protein, friend of GATA family member 2; A, atrium; V, ventricle; CT, Conus truncus; RA, right atrium; LA, left atrium; AS, aortic sac; RV, right ventricle; LV, left ventricle; AVV, atrioventricular valve; Ao, aorta; PA, pulmonary artery.
The heart is derived from multiple cell lineages and must differentiate into distinct regions according to function (26, 78). The major embryonic regions involved in vertebrate heart development include the primary and secondary heart fields, the cardiac neural crest, and the proepicardium. The first heart field gives rise to the left and part of the right ventricles and atria, while the second heart field gives rise to the remaining parts of the right ventricle and atrium and the myocardium surrounding the great vessels and the outflow tract of the mature heart. In the first heart field, the main transcription factors for driving development of mesoderm into myocardium are as follows: Nkx2.5 (NK2 homeobox 5), GATAs (GATA family transcription factors), and tbx5 (T-box gene family member 5). Isl1 (ISL LIM homeobox 1), tbx1 (T-box gene family member 1), FGF-10, Lhx2 (LIM homeobox 2), and others play important roles in outflow tract formation in the second heart field (105). Currently, congenital heart disease (CHD) is present in 7–13 per 1,000 live births and affects ∼1.4 million infants who are born each year worldwide with CHD, presenting a significant clinical problem (100). Molecular and genetic aberrations during embryonic cardiac morphogenesis, including loss of transcription factors, result in most forms of CHD caused by genetic etiologies. So, for example, atrial septal defect formation depends on GATA4, Nkx2.5, Tbx20 (T-box gene family member 20), and tbx5. The discovery of miRNAs has added a new knowledge base of regulatory relations with the described-above transcriptional factors involved in cardiac development. For example, a knockout of Dicer allele using Nkx2.5-Cre ablated the processing of pre-miRNAs into their mature form and led to embryonic lethality (127). Mice with Dicer deletion during epicardial development using GATA5-Cre die immediately after birth with profound coronary vessel defects (103). The detailed roles of miRNAs in the process of cardiac development are investigated next.
miRNAs in Cardiac Development
See Table 1. miRNAs shown to be involved in cardiac development are depicted in Fig. 2, along with relevant genes or proteins and developmental stages. The critical role of miRNAs in cardiac development was first shown through loss of function mutations in the miRNA processing enzymes. Dicer is an ideal target to understand the global role of miRNAs because it is necessary for the processing of all miRNAs into mature products. Gene-targeting studies in mice revealed that Dicer is required for early embryogenesis and embryonic mesoderm formation (8). Loss of Dicer in murine embryonic stem cells (ESCs) leads to a pronounced proliferation defect (82). A knockout of Dicer precluded the processing of pre-miRNAs into their mature form in cardiac progenitors (128) and led to defects in heart development as well as embryonic lethality (127). Additional support for a role of miRNAs in heart development was demonstrated when proliferation defects, similar to those found with Dicer, were observed with knockouts of DGCR8. Mice lacking DGCR8 in muscle tissue die prematurely with signs of heart failure and dilated cardiomyopathy (97). Unlike Dicer, DGCR8-deficient embryonic cells do not fully downregulate pluripotency markers and still express some markers of differentiation (117). Narrowing our discussion from stunting the function of all miRNAs, the role of specific miRNAs in cardiac development is discussed below.
Table 1.
MicroRNA-based cardiac regeneration
| MicroRNA | Model Studied | Mechanism | Targets | Ref. No. |
|---|---|---|---|---|
| Cardiac Development | ||||
| miR-218 | Zebrafish | Overexpression decrease migration of myocardial precursors | TBX5, ROBO1, slit2 | 18, 29 |
| miR-138 | Zebrafish | Required for cardiac maturation and patterning | cspg2, aldh1a2 | 79 |
| miR-1-2 | Mice | Mediates cardiac cell cycle and karyokinesis | HAND2, MEF2 | 127 |
| miR-133 | Mice | Control transcription throughout atrial and ventricular chambers | MEF2, SRF | 69 |
| miR-17/92 | Mice | Regulation of cardiac progenitor genes, repression of fibronectin | Bim, BMP 2/4, ISL1, TBX1 | 102, 116 |
| miR-15 | Mice | Mediates postnatal cardiac myocyte cell cycle arrest | Chek1 | 92 |
| miR-208 | Mice | Negative regulators of muscle growth, necessary for normal cardiac conduction | Thrap1, myostatin, Myh6, Myh7 | 12 |
| miR-499 | Mice | Regulate myosin expression, fiber type gene expression and muscle performance | Myh7b | 21, 111 |
| miR-143 | Zebrafish | Essential for cardiac chamber formation and myocardial cell size and shape | ADD3 | 20 |
| miR-130 | Mice | Myocyte overexpression causes septal defects and wall hypoplasia | FOG-2 | 54 |
| Cardiomyocyte Proliferation | ||||
| miR-133 | Mice, zebrafish | Inhibitor of cardiomyocyte proliferation in vivo | CRF, cyclin D2 | 68, 124 |
| miR-195 | Mice | Mechanism governing cardiomyocyte cell withdrawal and binucleation | Chek1 | 92 |
| miR-1 | Mice | Stimulates regenerative capacity of the heart | Hand2, SRF, myocardin | 127 |
| miR-29 | Rats | Inhibition induces proliferation and upregulates cell cycle gene expression | CDK2, cyclin D2, Akt3 | 14 |
| miR-590 | Mice, rats | Stimulates cardiac regeneration | multiple targets | 27 |
| miR-99/100, Let 7a/c | Mice, zebrafish | Regulate cardiac regeneration | FNTb, SMARCA5 | 1 |
| miR-17/92 | Mice | Stimulates cardiomyocyte proliferation and increase cell number | Bim, BMP 2/4, ISL1, TBX1 | 15 |
| miR-199 | Mice, rats | Stimulates cardiac regeneration | multiple targets | 27 |
| miR-34a | Mice | Induced in aging heart and its silencing reduces cardiomyocyte apoptosis | PNUTS, SRT1 | 10 |
| Stem Cell Differentiation | ||||
| miR-1 | Mice | Represses myocardial differentiation of embryonic stem cells | Hand2, MEF2, Cdk9, Nkx2.5 | 32, 107 |
| miR-133 | Mice | Cardiac progenitor cell protection against apoptosis | Nkx2.5, Bim, Bmf | 44, 107 |
| miR-499 | hCMPC, mice | Reduces cell proliferation and enhances myocyte differentiation | Sox6, ROD1, MYH7b | 41, 105 |
| miR-17/92 | Mice | Regulates proliferative activity of CPC | Bim, BMP 2/4, ISL1, TBX1, Rbl2 | 104 |
| Cell Reprogramming | ||||
| miR-1 | Mice | Promotes mesoderm formation | HAND2, MEF2 | 32, 43, 107 |
| miR-133 | Mice, rats, human | Improves reprogramming from fibroblasts | Nkx2.5, CRF, cyclin D2, Gata4, Mef2c | 43, 44, 107 |
| miR-208 | Mice | Enhances conversion fibroblasts to cardiomyocyte-like cells | Myh6, Myh7 | 45 |
| miR-290 | Mice | Enhances somatic cell reprogramming | Sox2, Klf4, Oct4 | 49 |
| miR-302/367 | Mice, human | Reprograms fibroblasts to iPS cell phenotype | OSKM, Oct4, Sox2 | 2 |
Target genes are as follows:
ADD3, adducin 3; Akt3, protein kinase Bγ; aldh1a2, aldehyde dehydrogenase family 1, subfamily A2; Bim, Bcl-2-like protein 11; Bmf, BCL2 modifying factor; BMP 2/4, bone morphogenetic proteins 2 and 4; CDK2, cyclin-dependent kinase 2; Cdk9, cyclin-dependent kinase 9; Chek1, checkpoint kinase 1; CRF, corticotropin releasing factor; cspg2, chondroitin sulfate proteoglycan 2; cyclin D2; FNTb, farnesyltransferase, CAAX box, β; FOG-2, zinc finger protein, friend of GATA family member 2; Gata4, GATA binding protein 4; Hand2, heart and neural crest derivatives-expressed protein 2; ISL1, ISL LIM homeobox 1; Klf4, Kruppel-like factor 4; MEF2, myocyte enhancer factor-2; Mef2c, myocyte enhancer factor-2c; Myh6, α-myosin heavy chain; Myh7, β-myosin heavy chain; Myh7b, myosin heavy chain 7b; myocardin; myostatin; Nkx2.5, NK2 homeobox 5; Oct4, octamer-binding transcription factor 4; OSKM, Oct4, Sox2, Klf4, Myc; PNUTS, phosphatase nuclear targeting subunit; Rbl2, retinoblastoma-like 2; ROBO1, roundabout axon guidance receptor, homolog 1; ROD1, regulator of differentiation 1; slit2, slit homolog 2; SMARCA5, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 5; Sox2, sex determining region Y-box 2; Sox6, sex determining region Y-box 6; SRF, serum response factor; SRT1, sirtuin 1; TBX1, T-box gene family member 1; Tbx5, T-box gene family member 5; Thrap1, thyroid hormone receptor-associated protein 1. iPS, induced pluripotent; CPC, cardiac progenitor cells.
According to expression profile analysis, 52 target genes of 16 miRNAs show association with congenital heart anomalies (122). Gain and loss-of-function approaches have demonstrated important roles for the miR-15 family in heart development. The miR-15 family consists of six miRNAs, which are clustered on three separate chromosomes (miR-15a, miR-16-1, miR-15b, miR-16-2, miR-195, and miR-497) (28). Inhibition of the miR-15 family was associated with the de-repression of a number of cell cycle genes in the heart and a persistence of CM mitosis beyond the normal developmental window of cell cycle arrest (92). The most highly upregulated member of the miR-15 family is miR-195. Cardiac-specific overexpression of miR-195 during embryogenesis is associated with cardiac malformations, including ventricular septal defect and right ventricular hypoplasia (92). At the molecular level, the miR-15 family targets cell-cycle genes, including Chek1 (checkpoint kinase 1). Chek1 is involved in many functions during DNA repair and mitosis, including chromosome segregation, cytokinesis, and the prevention of genomic instability (90).
A number of myosin genes coexpress miRNAs, named MyomiRs (miR-208a, miR-208b, and miR-499) as introns with vital purposes in cell differentiation, including the direction of cardiac myosin gene expression. Myh6 (α-myosin heavy chain) coexpresses miR-208a, Myh7 (β-myosin heavy chain) coexpresses miR-208b, and Myh7b (myosin heavy chain 7b) coexpresses miR-499. miR-208a was shown to be required for cardiac growth and expression of the β-myosin heavy chain protein, the primary contractile protein of the heart (12). Both miR-208a and miR-208b, members of the miR-208 family, target Thrap1 (thyroid hormone receptor-associated protein 1) and myostatin, important negative regulators of muscle growth. miR-208a and miR-208b are differentially expressed during heart development parallel to the expression of their respective host genes, Myh6 and Myh7. (12). The third miRNA coexpressed by a myosin gene, miR-499, is encoded by intron-19 of the mouse Myh7b gene (73). miR-499 plays a dominant role in the specification of muscle fiber identity by activating slow and repressing fast myofibril gene programs (111). In an example of the complexity of miRNA regulation, cardiac-specific transgene expression was used to compare the consequences of wild-type and a rare mutant miR-499. The mutation was present at the 3′ end of the mature strand, outside of the predicted major mRNA binding region. While both wild-type and mutant miR-499 induce heart failure in mice, the mutation in miR-499 resulted in a less severe phenotype. The mutation negatively impacted target mRNA binding and reduced mRNA silencing, suggesting that miRNA-mRNA binding is reliant on more than the main binding sequence (21).
Muscle-specific miRNAs, especially the miR-1 family and the miR-133 family, have been found to be evolutionarily conserved in most animal species from Drosophila to humans, despite major differences in cardiac anatomy (17). Primary sequences of mature miR-1 or miR-133a families are identical, and both gene families show similar expression patterns. The miR-1 and miR-133 families were demonstrated to be transcribed together (as a single transcript before processing into independent mature miRNAs) in a tissue-specific manner during development. Both of these miRNA families are expressed in the developing heart, and increased expression was found in neonatal mouse hearts (17, 31). miRNA function in cardiac progenitors is necessary for cardiogenesis, and the disruption of just one of two miR-1 family members (miR-1-1 and miR-1-2) has profound consequences for the development and maintenance of the heart. Overexpression of miR-1-1 in Drosophila results in 100% embryonic death at various stages of development due to disrupted myoblast patterning (58). Mice lacking miR-1-2 exhibit a spectrum of structural abnormalities leading to issues in the regulation of morphogenesis and in cardiac conduction (128). miR-1-2 inhibits cardiac development by repressing expression of the Hand2 (heart and neural crest derivatives-expressed protein 2) transcription factor (127). The miR-133 family represses the expression of SRF (serum response factor), an activator of myogenesis (17). The MEF2 (myocyte enhancer factor-2) family of transcription factors was also shown to activate transcription of a bicistronic primary transcript encoding miR-1-2 and miR-133a-1, a miR-133 family member. MEF2 proteins are regulators of the transcriptional and posttranscriptional pathways that control cardiac muscle development. This is mediated by direct and indirect mechanisms, coordinating the regulation of mRNAs and miRNAs in the linear heart tube during embryogenesis and thereafter controlling transcription throughout the atrial and ventricular chambers of the heart (69).
The miR-17/92 cluster was initially reported as a human oncogene, or oncomiR-1, and consists of six miRNAs belonging to four miRNA families (miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a, and miR-92-1) processed from the same gene transcript. miR-17/92 cluster seed sequences are contained within the 3′-UTRs of cardiac progenitor genes, such as Isl1 and tbx1. In miR-17/92 cluster mutant embryos, Isl1 and tbx1 expression failed to be correctly downregulated. Genetic interaction studies provided evidence that the miR-17/92 cluster affects the differentiation of the second heart field (116). Mice deficient in the miR-17/92 cluster die shortly after birth with lung hypoplasia and a ventricular septal defect. Absence of the miR-17/92 cluster miRNAs also leads to increased levels of the pro-apoptotic protein Bim (Bcl-2-like protein 11) and to inhibition of B-cell development at the pro-B to pre-B transition (114). In bone morphogenetic protein mutant embryos, myocardial differentiation is delayed, and multiple miR-17/92 cluster miRNAs are reduced (116). Transgenic mice overexpressing this cluster show overall organ growth retardation, as well as greatly reduced hematopoietic cell lineages. Evidence also supports the notion that overexpression of the miR-17/92 cluster causes cellular defects through repression of fibronectin expression (102).
The evolutionarily conserved miR-138 family (including miR-138-1 and miR-138-2) helps establish the discrete domains of gene expression required for normal cardiac morphogenesis. This family represses cspg2 (chondroitin sulfate proteoglycan core protein 2) and atrioventricular canal-specific transcripts in the developing ventricle via regulation of a network of developmental signals. This contributes to CM maturation and helps to establish the distinct identity of cardiac structures (79).
The miR-143 family is expressed robustly in progenitor cells of the cardiac crescent and highly in myocardial and endocardial cells of the linear heart tube in zebrafish (37). It has been shown that this miRNA family regulates ADD3 (adducin 3), which encodes an F-actin capping protein. This pathway contributes to the correct formation of the ventricular architectures, suggesting that multiple cascades regulate chamber formation in the heart and that the heartbeat-dependent expression of the miR-143 family is necessary for normal development (20). The authors hypothesize that the miR-143 family miRNAs act throughout the heart tube to limit the production of intracellular ADD3 protein and to promote the dissociation of ADD3 from F-actin, thereby allowing the cytoskeleton to be redistributed throughout the cell. In the absence of miR-143 family function, continued ADD3 production might overwhelm other modes of posttranslational regulation, leading to sustained ADD3-actin complex formation (13). Knockdown of the miR-143 family miRNAs in zebrafish produces abnormalities in the outflow tracts and ventricles (75).
The miR-218 family consists of three members (miR-218a-1, miR-218a-2, and miR-218b) and is conserved from humans to zebrafish. The miR-218 family host gene slit2 (slit homolog 2 protein), together with its ligands and Robo (roundabout gene family) receptors, is necessary for proper heart tube formation (29). The miR-218 family is part of a regulatory circuit through which tbx5, a transcription factor mentioned earlier that mediates vertebrate cardiac development, controls heart morphogenesis. Tbx5 overexpression affects heart development in humans and mice, resulting in heart-looping defects and chamber abnormalities (67). Through functional assays in zebrafish, it was demonstrated that there is correlated expression of tbx5 and the miR-218 family member miR-218-1. Furthermore, overexpression of tbx5 or downregulation of the miR-218 family negatively affects early heart morphogenesis. Interestingly, miR-218-1 downregulation rescues the cardiac defects generated by tbx5 overexpression (18).
The miR-130 family (including miR-130a and miR-130b) was shown to be involved in cardiac development via targeting of the transcriptional co-factor FOG-2 (zinc finger protein, friend of GATA family member 2). Analysis of transgenic embryos overexpressing the miR-130 family member miR-130a revealed ventricular wall hypoplasia and ventricular septal defect (54). These articles and more illustrate the vital, yet not fully understood, role of miRNAs in cardiac development.
As stated earlier, cardiac development is a period of high CM proliferation. The following section presents an overview of the major adult process of CM proliferation, cardiac regeneration.
CARDIAC REGENERATION
A number of nonmammalian vertebrate species, including amphibians, reptiles, and zebrafish, can regenerate the heart after myocardial injury throughout life (30, 85). Multiple studies have demonstrated that a zebrafish can fully restore its heart in 30 days after up to a 20% loss of ventricular tissue. This regenerative process is primarily driven by preexisting CMs without scar formation, rather than by progenitor cells, as has been previously suggested (48). Interestingly, after myocardial damage in zebrafish, the epicardium and endocardium respond with proliferation and reexpression of a variety of embryonic genes, such as tbx18 (T-box gene family member 18), wt1 (Wilms tumor 1), and raldh2 (retinaldehyde dehydrogenase 2) (52). CMs in the zebrafish heart are mononucleated and retain proliferative capacity throughout their life (121). By contrast, the mammalian heart grows primarily via proliferation of CMs during all stages of embryogenesis and fetal development. Shortly after birth, many CMs undergo DNA synthesis without cell division; in mice, ∼90% of CMs eventually become binucleated by undergoing mitosis up to, but not including, cytokinesis. Binucleated CMs are thought to be incapable of division, as polyploidy generally indicates terminal differentiation (66). In humans, this proportion (percentage of CMs that are binucleated) is much lower, but the specific value is subject to debate (87).
Due to this, it was long assumed that postnatal cardiac growth in mammals is primarily accomplished through the hypertrophy of CMs (112). Studies in mice indicate that mammals retain regenerative ability after birth, including post-myocardial infarction (MI), and that miRNAs have emerged as important regulators of neonatal mice's heart regenerative capacity (66, 92, 93, 10). Moreover, there is evidence that adult mammalian CMs can slowly self-renew at a rate of 1% of total CMs per year, declining with age. This renewal results in the exchange of ∼50% of adult CMs in a human life span (3, 50). Despite this, moderate to severe infarcts affecting 20–40% of CMs do not fully heal. Interestingly, a thyroid hormone surge can activate the insulin-like growth factor-I/Akt (protein kinase B) pathway during the postnatal period in mice, initiating an intense proliferative burst of predominantly binuclear CMs. This proliferation increases CM number by ∼40%, suggesting a retained capacity for stimulated proliferation after the normal developmental proliferative period (83).
The source of new CMs has been attributed to both division of existing CMs (53) and to progenitors residing within the heart (71), as well as to exogenous niches, such as bone marrow (88). Current evidence indicates that newly generated CMs arise from preexisting CMs through a process of proliferation and maturation (72). These mechanisms are the primary means driving CM replenishment in the mammalian heart following diverse pathological processes (93). However, basic aspects of the mammalian heart's capacity for self-renewal are still unclear (59), and estimates of CM turnover vary widely in different studies (50, 115).
A previous study summarized succinctly the current consensus on baseline cardiac regeneration in mammals (99).
1. CM DNA synthesis decreases with age.
2. New CMs are mostly derived from preexisting CMs during aging.
3. Many CMs undergo DNA replication without completing the cell cycle.
4. Myocardial injury stimulates division of preexisting CMs.
5. Cardiac progenitors do not play a significant role in myocardial homeostasis, and their role after injury is also limited.
The latter conclusion 5 is controversial and premature, as a number of studies have demonstrated the ability to isolate different types of cardiac progenitors from the heart (74) with their clonogenic and multipotent capacities shown in vitro in various animal models (4, 76). While a detailed discussion about cardiac progenitors is beyond the scope of this review, it is likely that CM proliferation is normally a more important mediator of myocardial homeostasis and recovery from injury than progenitor cell differentiation. With this knowledge base in place, we will detail the approaches to enhancing cardiac regeneration.
miRNA and Cardiac Regeneration
See Table 1. The weight of evidence has demonstrated that ESCs, progenitor cells, or reprogrammed cardiac cells can improve heart function in normal and pathological conditions (9, 47, 110). Experience in this field, however, also revealed many challenges regarding the function, homing, survival, and differentiation of these cells (24, 71, 86). Because miRNAs alter gene expression networks, an interesting strategy is to treat cells ex vivo before transplantation to improve the functional capacity of cells.
On the other hand, cardiac developmental growth is largely dependent on CM proliferation, and human CMs are able to proliferate during postnatal life (77). Noncoding RNAs, especially miRNAs, have been shown to participate in cardiac regeneration by CM proliferation. We will begin by discussing the CM proliferation strategy and then move on to stem cells and reprogramming.
miRNA-based CM Proliferation
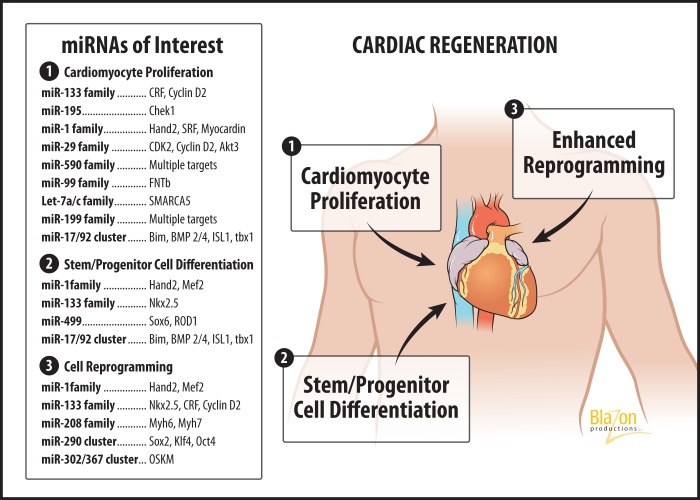
See Table 1. The first of the three major strategies for cardiac regeneration therapy depicted in Fig. 3 is the induction of proliferation in mature CMs. Many early results in various fields suggest the proliferation-regulating effects of miRNAs (19, 64, 84, 120, 125). miRNAs are involved in various aspects of cell cycle control in CMs, including postnatal maturation and terminal differentiation.
Fig. 3.
The three major strategies for hyperplasic cardiac regeneration via miRNAs, along with miRNAs of interest in the field. CRF, corticotropin releasing factor; CDK2, cyclin-dependent kinase 2; Akt3, protein kinase Bγ; FNTb, farnesyltransferase, CAAX box, β; SMARCA5, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 5; Nkx2.5, NK2 homeobox 5; Sox6, sex determining region Y-box 6; ROD1, regulator of differentiation 1; Sox2, sex determining region Y-box 2; Klf4, Kruppel-like factor 4; OSKM: Oct4, Sox2, Klf4, Myc.
In vitro screening identified 204 miRNAs that increase rat neonatal CM proliferation more than twofold compared with CMs in basal conditions, and 331 miRNAs that decrease proliferation without affecting cell viability. The majority of miRNAs that enhance proliferation increase the number of CMs as well (27). Numerous other studies have shown the role of specific miRNAs in CM proliferative capacity in vitro and in vivo.
Several studies revealed the essential roles of the miR-133 family (including miR-133a-1 and miR-133a-2) in CM growth and function. By a combination of gain-of-function and loss-of-function experiments in adult zebrafish, data indicate that the miR-133 family acts as a brake within a circuit regulating regeneration (124). Many phenotypic abnormalities of miR-133a-1/miR-133a-2 null mice can be attributed to the upregulation of two specific mRNA targets encoding SRF and cyclin D2, important in CM growth and proliferation (68, 91). CMs from miR-133a-1/miR-133a-2 double knockout mice show a 2.5-fold increase in proliferation assessed by phospho-histone H3 staining compared with wild type. Furthermore, diminished proliferation was found in response to transgenic overexpression of miR-133a-1 and miR-133a-2, indicating that these miRNAs function as inhibitors of CM proliferation in vivo (68).
miR-195 (a member of the miR-15 family) was one of the most highly upregulated miRNAs examined during the postnatal period, with expression levels almost sixfold higher in ventricles at postnatal day 10 relative to day 1 (92). This miRNA may be part of a vital regulatory mechanism governing CM cell cycle withdrawal and binucleation. miR-195 regulates the expression of a number of cell cycle genes, including Chek1, as mentioned previously (70, 124). It is also implicated in the regulation of mitosis, where it coordinates progression through the G2/M checkpoint and can regulate chromosome segregation and cytokinesis (89). Knockdown of the miR-15 family in neonatal mice with locked nucleic acid-modified anti-miRNAs is associated with depression of Chek1 and an increased number of mitotic CMs. miR-195 transgenic hearts show marked reduction (≈3-fold) in the number of cells undergoing mitosis and an increased proportion of multinucleated CMs (≈3-fold) at the early postnatal period. These data clearly indicate that postnatal upregulation of the miR-15 family in vivo inhibits CM mitotic progression and induces premature cell cycle arrest, contributing to postnatal cell cycle withdrawal (92).
Members of the miR-1 family are critical mediators of cell proliferation and differentiation in cardiac muscle (113, 127). The miR-1 family members miR-1-1 and miR-1-2 were shown to be expressed during cardiogenesis. Both genes are direct targets of SRF and its potent coactivator myocardin, and both are involved in the negative regulation of ventricular CM proliferation through targeting of the Hand2 mRNA (128). It was revealed that the expression of miR-1 is induced by myocardin (cardiac muscle-specific transcription factor) in human smooth muscle cells, and that miR-1 mediates the inhibitory effects of myocardin on smooth muscle cell proliferation (16).
The miR-29 family miRNAs (including miR-29a, miR-29b, miR-29c) are highly conserved among different species. It has been reported that miR-29a is involved in many physiological proliferative processes. Some studies indicate that miR-29a promotes progenitor proliferation through expediting G1 to S/G2 transitions (39). Other studies have found the opposite; transfection of neonatal rat CMs with miR-29a showed that miR-29a overexpression decreases CM proliferation. Quantification of Ki-67-positive CMs (indicative of a proliferative state) revealed a threefold increase at 48 h after miR-29a inhibitor transfection compared with control transfection. The authors found decreased expression of two cell cycle-specific proteins, cyclin D2 and CDK2 (cyclin-dependent kinase 2), in miR-29a-transfected CMs (14). Overexpression of the miR-29 family inhibited mouse C2C12 myoblast cell line proliferation and promoted myotube formation. The miR-29 family specifically targeted Akt3, a member of the serine/threonine protein kinase family responsive to growth factor cell signaling, for posttranscriptional downregulation (118). These findings indicate that the miR-29 family is involved in regulating CM proliferation during postnatal development, although more research is needed to clarify its role.
To test the hypothesis that miRNAs can induce reentry into the cell cycle of fully differentiated cells, CMs from adult rats were transfected with the miR-590 family and miR-199a (of the miR-199 family). These treatments led to an increase in the number of CMs by up to 20% (27). These important and exciting findings demonstrate that these miRNAs are able to induce proliferation of postnatal CMs. AAV9 vectors expressing the miR-590 family or miR-199a were injected intraperitoneally in neonatal mice. At 12 days after injection, the number of mitotic CMs was significantly increased compared with AAV9 control. Next, adult mice (8–12 wk) underwent left anterior descending coronary artery ligation and injection of AAV9 expressing the miRNAs in the peri-infarcted area. At 60 days after delivery, confocal microscopy revealed that most of the CM nuclei belonged to mature CMs, well integrated and with loose myocardial structure, indicative of active proliferation after treatment (27).
Using miR-17/92 knockout and transgenic mice, it was demonstrated that the miR-17/92 cluster is sufficient to induce CM proliferation. The hearts of transgenic mice are enlarged, with increased heart-to-body weight ratio and no histological evidence of CM hypertrophy. It was concluded that the increase in CMs results from an increase in proliferation in embryonic, postnatal, and adult hearts (15). Overexpression of the miR-17/92 cluster was also shown to protect the heart from MI-induced injury. In addition to improvements in echocardiographic functional data, a marked increase in EdU incorporation (indicative of proliferation) in the CMs of the border zone is found with miR-17/92 cluster administration. PTEN (phosphatase and tension homolog), a member of family of protein tyrosine phosphatases, is one of the miR-17/92 cluster functional targets that mediate the function of this cluster to regulate proliferation (15). These recent studies yielded extremely promising results, supporting the hypothesis that miRNAs may be used to replenish lost CMs through CM cell cycle control in animals and possibly humans.
After amputation of ventricular apex of zebrafish, two miRNA families (the miR-99 family and the Let-7a/c family) were found to be significantly downregulated during regeneration. A mechanistic link was established between the identified miRNAs and their targets SMARCA5 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 5) and FNTβ (Farnesyl transferase, β-subunit). Overexpression of either protein (the opposite of the effect of the miRNAs) resulted in mammalian CM de-differentiation and induction of proliferation. Importantly, following MI creation in a murine model, intracardiac delivery of AAV encoding anti-miR-99 or anti-Let-7a/c significantly improved functional heart parameters. These and other data have shown that these two miRNAs are critical regulators of cardiomyogenesis and heart regeneration and explored the therapeutic potential of miRNA (1). The remaining regenerative strategies seek other sources of new CMs.
miRNA-based Stem Cell Differentiation and Cell Reprogramming
In the context of cardiac regeneration, stem cell differentiation seeks to induce various types of stem cells to take on a CM phenotype. Cellular reprogramming involves transforming one adult cell type into another. These two closely related fields comprise the second and third strategies depicted in Fig. 3. We will begin with stem cell differentiation before discussing cellular reprogramming, and, finally, we will discuss induced pluripotent (iPS) cells.
The miR-1 and miR-133 families have been shown to be enriched in ESC-derived CMs and are expressed at the early stages of cardiac mesoderm selection from ESC. Overexpression of either of these miRNA families in ESCs results in enhanced mesoderm gene expression in embryoid bodies, as well as suppressed differentiation into the ectodermal or endodermal lineages. Interestingly, the families had opposing effects on differentiation into cardiac muscle progenitors, with the miR-1 family promoting and the miR-133 family blocking differentiation (43). Controversially, other studies have shown that miR-1 family overexpression reduces the expression of cardiac markers in mouse ESC. In these studies, the levels of these miRNA families were increased during spontaneous myocardial differentiation, but reduced during induced differentiation accomplished via histone deacetylation inhibition (107). Another study found that the miR-1 and miR-133 families are rapidly and progressively upregulated during in vitro differentiation of adult cardiac progenitor cells (CPCs). Overexpression of the miR-1 family enhanced the cardiac differentiation of CPCs, whereas overexpression of the miR-133 family did not modulate this process. On the other hand, the miR-133 family protected CPCs against apoptosis (44). These data represent important subtleties and controversies in miRNAs' role in cardiac stem cell differentiation. The overexpression of miR-1 or miR-133 by lentiviral infection also reduced the expression of a cardiac-specific gene, Nkx2.5, during differentiation of ESCs (107). Despite these niche results showcasing the complexity of miRNA signaling, miR-1 drives CM differentiation in transplanted ESCs, ultimately giving rise to enhanced cardiac repair, regeneration, and function in post-MI hearts (32).
miRNA levels are highly regulated in cultured and differentiated cardiac-derived human CM progenitor cells, with miR-1 family and miR-499 expression being significantly higher in differentiated human CM progenitor cells (105). Expression of miR-499 in human cardiac stem cells represses the target genes SOX6 (sex determining region Y-box 6) and ROD1 (regulator of differentiation 1), enhancing differentiation to CMs in vitro. This miRNA also enhances stem cell differentiation into CMs and potentiates the restoration of myocardial mass and function in the infarcted heart (41).
Pursuing the hypothesis that the miR-17/92 cluster is involved in cardiac differentiation, quantitative PCR analysis demonstrates a more than twofold increase in the expression of all members of the miR-17/92 cluster in neonatal CPCs compared with adult CPCs. Specifically, it was suggested that the reexpression of miR-17/92 in adult CPCs might increase their proliferative potential (104). In addition to stem cells, researchers seek to convert differentiated cells into CMs.
Cellular reprogramming of fibroblasts into specific cell types without passing through a progenitor state offers an alternative approach for cardiac regeneration. Cardiac fibroblasts comprise over 50% of all of the cells in the heart (13). If it were possible to reprogram the resident fibroblasts into beating CMs, these endogenous cardiac fibroblasts would be an exciting potential source of new CMs (42). miRNAs are capable of inducing expression of cardiac markers in fibroblasts and also are efficient at converting the fibroblasts into cells with functional properties characteristic of CMs (such as L-type channel expression, spontaneous calcium oscillations, and contractility) (106).
The miR-1 family alone is sufficient to induce a degree of cardiac reprogramming, but its effects are dramatically enhanced in combination with miR-133a, miR-208a, and miR-499. Notably, this combination of miRNAs did not change the total number of reprogrammed cells found with the miR-1 family alone, but rather enhanced the maturation of converted cells (45). The same combination of miRNAs was later shown to significantly increase the number of reprogramming-related events in vivo, including expression of CM markers, sarcomeric organization, excitation-contraction coupling, and initiation of action potentials characteristic of mature ventricular CMs. Reprogramming is associated with improvement of cardiac function, indicating that correctly applied miRNA combination therapy may promote functional recovery of damaged myocardium (46).
The transformation of differentiated cells to iPS cells added a powerful new tool in stem cell biology, especially for regenerative therapy. This technique represents a fusion of the stem cell differentiation and reprogramming fields; iPS cells are reprogrammed from adult cells into stem cells and then differentiated into CMs as other stem cells would be. miRNAs can be powerful tools for mediating iPS cell reprogramming. A subset of miR-290 cluster (miR-291-3p, miR-294, and miR-295) miRNAs have been shown to enhance iPS reprogramming in the presence of the transcription factors Klf4 (Kruppel-like factor 4), Oct4 (octamer-binding transcription factor 4) and SOX2 (sex-determining region Y-box 2) (49). This study excluded cMyc (Myc proto-oncogene) from the normal OSKM (Oct4, Sox2, Klf4, Myc) reprogramming factor combination, as cMyc binds the promoter of these miRNAs, and they are thought to act as downstream effectors of cMyc. Furthermore, overexpression of the miR-302/367 cluster can directly reprogram mouse and human fibroblasts to a pluripotent stem cell state in the absence of exogenous transcription factors. Reprogramming by the miR-302/367 cluster is up to two orders of magnitude more efficient than with the OSKM factors (2). A greater understanding of the role of miRNA in differentiation and reprogramming could lead to major advances in cardiac regeneration and stem cell research.
CONCLUSION
Our knowledge concerning the heart changes constantly. The heart is a dynamic organ capable of significant remodeling and hypertrophic growth in response to the hemodynamic stress and neuroendocrine signaling associated with changed workloads or chronic cardiovascular disease. The belief that the heart is a postmitotic, non-regenerating organ has changed as well. Regeneration of the human heart is an incredibly important challenge, given the magnitude of heart disease and the absence of effective long-term therapies. The adult mammalian heart cannot efficiently generate new cardiac muscle cells without treatment. This limits the adaptive response to chronic hemodynamic stress. A safe, efficient strategy that increases myocardial mass through CM number rather than cell size would potentially be of immeasurable benefit. There have been many advances in our understanding of the genes and signaling pathways involved in cardiac regeneration, but the overall complexity of these processes suggests that additional regulatory mechanisms remain to be identified (109). The cellular processes and regulatory mechanisms involved in cardiac development during embryogenesis must be compared with mitotic signaling pathways involved in postnatal heart growth and cell cycles.
Two primary sources of new CMs exist in adult mammals: 1) conversion of stem cells or differentiated cells into CMs; and 2) cell cycle reentry and proliferation of mature CMs. It is quite possible that the sufficient restoration of functional heart tissue will necessitate a combination of these approaches. Numerous research studies have demonstrated that miRNAs are deeply involved in the regulation of CM proliferation, in stem cell differentiation and maturation, and in cell reprogramming. In addition, promising recent studies have shown success of regenerative miRNA-based therapy in animal models (15, 27, 41, 46).
Significant challenges to the development of a CM proliferation-based therapy include the low proportion of mononucleated CMs with regenerative potential in the adult mammalian heart, developmental differences between research models and humans, and a lack of data regarding the mechanisms of terminal differentiation. In addition, many issues must be resolved before the resident stem cells of the heart or iPS cells can be clinically induced to differentiate into CMs: doubts regarding the maturity and functional heterogeneity of stem cell-derived CMs, low survival and retention, selection of an appropriate cell source, suboptimal efficiency of cell differentiation toward cardiac lineages, and the potential for oncogenesis (40, 81, 94, 123). For cell reprogramming, similar issues remain: low reprogramming efficiency, low survival, questionable cell maturity (bad contractility-contraction, reproducibility is difficult), and an unknown mechanism (23, 42). Despite these concerns, the results of these fields are extremely promising, especially with miRNAs.
miRNAS have emerged as new therapeutic targets due to their involvement in the pathogenesis of many cardiovascular diseases via cell differentiation, migration, proliferation, and apoptosis. Due to the 5′ end-restricted complementarity to mRNA targets, miRNA-based therapy can regulate cellular processes in multiple signaling pathways through nonexclusive mRNA target recognition. This is a main advantage of miRNA compared with other molecular treatments, as one miRNA can affect multiple target genes in a diverse disease. Moreover, miRNA is present in the blood and can be packaged in microparticles (109) and may function as mediators of cell-to cell communication (126). However, the therapeutic feasibility of miRNA as a target depends on several key issues, including determination of the normal expression level of the miRNA, identification of tissue and cell-specific delivery mechanisms, and avoidance of off-target effects on other miRNAs.
GRANTS
The writing of this paper was supported by National Heart, Lung, and Blood Institute Grant 5R01-HL-083078-08 and grants from the James H. Heineman Foundation.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: M.G.K. and A.P.K. drafted manuscript; M.G.K., A.S.F., A.P.K., R.J.H., and C.R.B. edited and revised manuscript; M.G.K., A.S.F., A.P.K., R.J.H., and C.R.B. approved final version of manuscript.
ACKNOWLEDGMENTS
We would like to acknowledge the Gene Therapy Resource Program (GTRP), as well as Anne Olson for excellent illustrations.
REFERENCES
- 1.Aguirre A, Montserrat N, Zacchigna S, Nivet E, Hishida T, Krause MN, Kurian L, Ocampo A, Vazquez-Ferrer E, Rodriguez-Esteban C, Kumar S, Moresco JJ, Yates JR 3rd, Campistol JM, Sancho-Martinez I, Giacca M, Izpisua Belmonte JC. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell 15: 589–604, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8: 376–388, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anversa P, Leri A, Kajstura J. Cardiac regeneration. J Am Coll Cardiol 47: 1769–1776, 2006. [DOI] [PubMed] [Google Scholar]
- 4.Barile L, Chimenti I, Gaetani R, Forte E, Miraldi F, Frati G, Messina E, Giacomello A. Cardiac stem cells: isolation, expansion and experimental use for myocardial regeneration. Nat Clin Pract Cardiovasc Med 4, Suppl 1: S9–S14, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120: 21–24, 2005. [DOI] [PubMed] [Google Scholar]
- 7.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science 324: 98–102, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet 35: 215–217, 2003. [DOI] [PubMed] [Google Scholar]
- 9.Bolli R, Chugh AR, D'Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Solankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, Anversa P. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet 378: 1847–1857, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Boon RA, Iekushi K, Lechner S, Seeger T, Fisher A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Muller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S. MicroRNA-34a regulates cardiac ageing and function. Nature 495: 107–110, 2013. [DOI] [PubMed] [Google Scholar]
- 11.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 119: 2772–2786, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camelliti P, Borg TK, Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res 65: 40–51, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Cao X, Wang J, Wang Z, Du J, Yuan X, Huang W, Meng J, Gu H, Nie Y, Ji B, Hu S, Zheng Z. MicroRNA profiling during rat ventricular maturation: A role for miR-29a in regulating cardiomyocyte cell cycle re-entry. FEBS Lett 587: 1548–1555, 2013. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, Hu X, Wang G, Lin Z, Wang S, Pu WT, Liao R, Wang DZ. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res 112: 1557–1566, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Yin H, Jiang Y, Radhakrishnan SK, Huang ZP, Li J, Shi Z, Kilsdonk EP, Gui Y, Wang DZ, Zheng XL. Induction of microRNA-1 by myocardin in smooth muscle cells inhibits cell proliferation. Arterioscler Thromb Vasc Biol. 31: 368–375, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38: 228–233, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiavacci E, Dolfi L, Verduci L, Meghini F, Gestri G, Evangelista AM, Wilson SW, Cremisi F, Pitto L. MicroRNA 218 mediates the effects of Tbx5a over-expression on zebrafish heart development. PloS One 7: e50536, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramirez CM, Chamorro-Jorganes A, Wanschel AC, Lasuncion MA, Morales-Ruiz M, Suarez Y, Baldan A, Esplugues E, Fernandez-Hernando C. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle 11: 922–933, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deacon DC, Nevis KR, Cashman TJ, Zhou Y, Zhao L, Washko D, Guner-Ataman B, Burns CG, Burns CE. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development 137: 1887–1896, 2010. [DOI] [PubMed] [Google Scholar]
- 21.Dorn GW 2nd, Matkovich SJ, Eschenbacher WH, Zhang Y. A human 3′ miR-499 mutation alters cardiac mRNA targeting and function. Circ Res 110: 958–967, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4: 721–726, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol 13: 215–222, 2011. [DOI] [PubMed] [Google Scholar]
- 24.Eisenberg CA, Burch JB, Eisenberg LM. Bone marrow cells transdifferentiate to cardiomyocytes when introduced into the embryonic heart. Stem Cells 24: 1236–1245, 2006. [DOI] [PubMed] [Google Scholar]
- 25.Elton TS, Khan M, Terentyev D. MicroRNAs in cardiovascular disease. F1000 Med Rep 3: 10, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Epstein JA. Franklin H. Epstein Lecture. Cardiac development and implications for heart disease. N Engl J Med 363: 1638–1647, 2010. [DOI] [PubMed] [Google Scholar]
- 27.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492: 376–381, 2012. [DOI] [PubMed] [Google Scholar]
- 28.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol 402: 491–509, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fish JE, Wythe JD, Xiao T, Bruneau BG, Stainier DY, Srivastava D, Woo S. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development 138: 1409–1419, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flink IL. Cell cycle reentry of ventricular and atrial cardiomyocytes and cells within the epicardium following amputation of the ventricular apex in the axolotl, Amblystoma mexicanum: confocal microscopic immunofluorescent image analysis of bromodeoxyuridine-labeled nuclei. Anat Embryol (Berl) 205: 235–244, 2002. [DOI] [PubMed] [Google Scholar]
- 31.Fuller AM, Qian L. MiRiad roles for microRNAs in cardiac development and regeneration. Cells 3: 724–750, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glass C, Singla DK. MicroRNA-1 transfected embryonic stem cells enhance cardiac myocyte differentiation and inhibit apoptosis by modulating the PTEN/Akt pathway in the infarcted heart. Am J Physiol Heart Circ Physiol 301: H2038–H2049, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640, 2005. [DOI] [PubMed] [Google Scholar]
- 34.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res 32: D109–D111, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimes AC, Stadt HA, Shepherd IT, Kirby ML. Solving an enigma: arterial pole development in the zebrafish heart. Dev Biol 290: 265–276, 2006. [DOI] [PubMed] [Google Scholar]
- 38.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med 207: 475–489, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hosoda T, Kajstura J, Leri A, Anversa P. Mechanisms of myocardial regeneration. Circ J 74: 13–17, 2010. [DOI] [PubMed] [Google Scholar]
- 41.Hosoda T, Zheng H, Cabral-da-Silva M, Sanada F, Ide-Iwata N, Ogorek B, Ferreira-Martins J, Arranto C, D'Amario D, del Monte F, Urbanek K, D'Alessandro DA, Michler RE, Anversa P, Rota M, Kajstura J, Leri A. Human cardiac stem cell differentiation is regulated by a mircrine mechanism. Circulation 123: 1287–1296, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell stem cell 2: 219–229, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Izarra A, Moscoso I, Levent E, Canon S, Cerrada I, Diez-Juan A, Blanca V, Nunez-Gil IJ, Valiente I, Ruiz-Sauri A, Sepulveda P, Tiburcy M, Zimmermann WH, Bernad A. miR-133a enhances the protective capacity of cardiac progenitors cells after myocardial infarction. Stem Cell Reports 3: 1029–1042, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res 110: 1465–1473, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jayawardena TM, Finch EA, Zhang L, Zhang H, Hodgkinson CP, Pratt RE, Rosenberg PB, Mirotsou M, Dzau VJ. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ Res 116: 418–424, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeevanantham V, Butler M, Saad A, Abdel-Latif A, Zuba-Surma EK, Dawn B. Adult bone marrow cell therapy improves survival and induces long-term improvement in cardiac parameters: a systematic review and meta-analysis. Circulation 126: 551–568, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464: 606–609, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27: 459–461, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kajstura J, Gurusamy N, Ogorek B, Goichberg P, Clavo-Rondon C, Hosoda T, D'Amario D, Bardelli S, Beltrami AP, Cesselli D, Bussani R, del Monte F, Quaini F, Rota M, Beltrami CA, Buchholz BA, Leri A, Anversa P. Myocyte turnover in the aging human heart. Circ Res 107: 1374–1386, 2010. [DOI] [PubMed] [Google Scholar]
- 51.Kartha RV, Subramanian S. MicroRNAs in cardiovascular diseases: biology and potential clinical applications. J Cardiovasc Transl Res 3: 256–270, 2010. [DOI] [PubMed] [Google Scholar]
- 52.Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell 20: 397–404, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464: 601–605, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim GH, Samant SA, Earley JU, Svensson EC. Translational control of FOG-2 expression in cardiomyocytes by microRNA-130a. PloS One 4: e6161, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol 14: 156–159, 2004. [DOI] [PubMed] [Google Scholar]
- 56.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A 102: 18986–18991, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laflamme MA, Murry CE. Heart regeneration. Nature 473: 326–335, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30: 363–364, 2002. [DOI] [PubMed] [Google Scholar]
- 61.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167, 2004. [DOI] [PubMed] [Google Scholar]
- 62.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854, 1993. [DOI] [PubMed] [Google Scholar]
- 63.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem 280: 16635–16641, 2005. [DOI] [PubMed] [Google Scholar]
- 65.Leucci E, Patella F, Waage J, Holmstrom K, Lindow M, Porse B, Kauppinen S, Lund AH. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep 3: 2535, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol 28: 1737–1746, 1996. [DOI] [PubMed] [Google Scholar]
- 67.Liberatore CM, Searcy-Schrick RD, Yutzey KE. Ventricular expression of tbx5 inhibits normal heart chamber development. Dev Biol 223: 169–180, 2000. [DOI] [PubMed] [Google Scholar]
- 68.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 22: 3242–3254, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A 104: 20844–20849, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 36: 5391–5404, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loffredo FS, Steinhauser ML, Gannon J, Lee RT. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell 8: 389–398, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin-Puig S, Fuster V, Torres M. Heart repair: from natural mechanisms of cardiomyocyte production to the design of new cardiac therapies. Ann N Y Acad Sci 1254: 71–81, 2012. [DOI] [PubMed] [Google Scholar]
- 73.McGuigan K, Phillips PC, Postlethwait JH. Evolution of sarcomeric myosin heavy chain genes: evidence from fish. Mol Biol Evol 21: 1042–1056, 2004. [DOI] [PubMed] [Google Scholar]
- 74.Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, Fiordaliso F, Salio M, Battaglia M, Latronico MV, Coletta M, Vivarelli E, Frati L, Cossu G, Giacomello A. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res 95: 911–921, 2004. [DOI] [PubMed] [Google Scholar]
- 75.Miyasaka KY, Kida YS, Banjo T, Ueki Y, Nagayama K, Matsumoto T, Sato M, Ogura T. Heartbeat regulates cardiogenesis by suppressing retinoic acid signaling via expression of miR-143. Mech Dev 128: 18–28, 2011. [DOI] [PubMed] [Google Scholar]
- 76.Mohsin S, Siddiqi S, Collins B, Sussman MA. Empowering adult stem cells for myocardial regeneration. Circ Res 109: 1415–1428, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park SY, Silberstein LE, Dos Remedios CG, Graham D, Colan S, Kuhn B. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci U S A 110: 1446–1451, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moorman AF, Christoffels VM. Cardiac chamber formation: development, genes, and evolution. Physiol Rev 83: 1223–1267, 2003. [DOI] [PubMed] [Google Scholar]
- 79.Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DY, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci U S A 105: 17830–17835, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Després JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MB; American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics–2015 update: a report from the American Heart Association. Circulation 131: e29–e322, 2015. [DOI] [PubMed] [Google Scholar]
- 81.Mummery C. Induced pluripotent stem cells–a cautionary note. N Engl J Med 364: 2160–2162, 2011. [DOI] [PubMed] [Google Scholar]
- 82.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A 102: 12135–12140, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Naqvi N, Li M, Calvert JW, Tejada T, Lambert JP, Wu J, Kesteven SH, Holman SR, Matsuda T, Lovelock JD, Howard WW, Iismaa SE, Chan AY, Crawford BH, Wagner MB, Martin DI, Lefer DJ, Graham RM, Husain A. A proliferative burst during preadolescence establishes the final cardiomyocyte number. Cell 157: 795–807, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA 12: 1161–1167, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. J Exp Zool 187: 249–253, 1974. [DOI] [PubMed] [Google Scholar]
- 86.Oh H, Taffet GE, Youker KA, Entman ML, Overbeek PA, Michael LH, Schneider MD. Telomerase reverse transcriptase promotes cardiac muscle cell proliferation, hypertrophy, and survival. Proc Natl Acad Sci U S A 98: 10308–10313, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olivetti G, Cigola E, Maestri R, Corradi D, Lagrasta C, Gambert SR, Anversa P. Aging, cardiac hypertrophy and ischemic cardiomyopathy do not affect the proportion of mononucleated and multinucleated myocytes in the human heart. J Mol Cell Cardiol 28: 1463–1477, 1996. [DOI] [PubMed] [Google Scholar]
- 88.Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li B, Pickel J, McKay R, Nadal-Ginard B, Bodine DM, Leri A, Anversa P. Bone marrow cells regenerate infarcted myocardium. Nature 410: 701–705, 2001. [DOI] [PubMed] [Google Scholar]
- 89.Peddibhotla S, Lam MH, Gonzalez-Rimbau M, Rosen JM. The DNA-damage effector checkpoint kinase 1 is essential for chromosome segregation and cytokinesis. Proc Natl Acad Sci U S A 106: 5159–5164, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peddibhotla S, Rosen JM. Chking and executing cell division to prevent genomic instability. Cell Cycle 8: 2339–2342, 2009. [DOI] [PubMed] [Google Scholar]
- 91.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev 20: 1545–1556, 2006. [DOI] [PubMed] [Google Scholar]
- 92.Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ, Dorn GW 2nd, van Rooij E, Olson EN. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res 109: 670–679, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science 331: 1078–1080, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qian L, Srivastava D. Direct cardiac reprogramming: from developmental biology to cardiac regeneration. Circ Res 113: 915–921, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramalingam P, Palanichamy JK, Singh A, Das P, Bhagat M, Kassab MA, Sinha S, Chattopadhyay P. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA 20: 76–87, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rand TA, Ginalski K, Grishin NV, Wang X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc Natl Acad Sci U S A 101: 14385–14389, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, Reinhardt F, Liao R, Krieger M, Jaenisch R, Lodish HF, Blelloch R. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 105: 585–594, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906, 2000. [DOI] [PubMed] [Google Scholar]
- 99.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902–1910, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB; American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Executive summary: heart disease and stroke statistics–2012 update: a report from the American Heart Association. Circulation 125: 188–197, 2012. [DOI] [PubMed] [Google Scholar]
- 101.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493: 433–436, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V, Yang BB. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol 11: 1031–1038, 2009. [DOI] [PubMed] [Google Scholar]
- 103.Singh MK, Lu MM, Massera D, Epstein JA. MicroRNA-processing enzyme Dicer is required in epicardium for coronary vasculature development. J Biol Chem 286: 41036–41045, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sirish P, Lopez JE, Li N, Wong A, Timofeyev V, Young JN, Majdi M, Li RA, Chen HS, Chiamvimonvat N. MicroRNA profiling predicts a variance in the proliferative potential of cardiac progenitor cells derived from neonatal and adult murine hearts. J Mol Cell Cardiol 52: 264–272, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sluijter JP, van Mil A, van Vliet P, Metz CH, Liu J, Doevendans PA, Goumans MJ. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol. 30: 859–868, 2010. [DOI] [PubMed] [Google Scholar]
- 106.Srivastava D, Ieda M. Critical factors for cardiac reprogramming. Circ Res 111: 5–8, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takaya T, Ono K, Kawamura T, Takanabe R, Kaichi S, Morimoto T, Wada H, Kita T, Shimatsu A, Hasegawa K. MicroRNA-1 and MicroRNA-133 in spontaneous myocardial differentiation of mouse embryonic stem cells. Circ J 73: 1492–1497, 2009. [DOI] [PubMed] [Google Scholar]
- 108.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459: 708–711, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659, 2007. [DOI] [PubMed] [Google Scholar]
- 110.van der Bogt KE, Sheikh AY, Schrepfer S, Hoyt G, Cao F, Ransohoff KJ, Swijnenburg RJ, Pearl J, Lee A, Fischbein M, Contag CH, Robbins RC, Wu JC. Comparison of different adult stem cell types for treatment of myocardial ischemia. Circulation 118: S121–S129, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 17: 662–673, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 103: 18255–18260, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316: 575–579, 2007. [DOI] [PubMed] [Google Scholar]
- 114.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–886, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Walsh S, Ponten A, Fleischmann BK, Jovinge S. Cardiomyocyte cell cycle control and growth estimation in vivo–an analysis based on cardiomyocyte nuclei. Cardiovasc Res 86: 365–373, 2010. [DOI] [PubMed] [Google Scholar]
- 116.Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, Huang Z, Black BL, Wang F, Martin JF. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell 19: 903–912, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang J, Martin JF. Macro advances in microRNAs and myocardial regeneration. Curr Opin Cardiol 29: 207–213, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wei W, He HB, Zhang WY, Zhang HX, Bai JB, Liu HZ, Cao JH, Chang KC, Li XY, Zhao SH. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis 4: e668, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862, 1993. [DOI] [PubMed] [Google Scholar]
- 120.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326: 1549–1554, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wills AA, Holdway JE, Major RJ, Poss KD. Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development 135: 183–192, 2008. [DOI] [PubMed] [Google Scholar]
- 122.Xing HJ, Li YJ, Ma QM, Wang AM, Wang JL, Sun M, Jian Q, Hu JH, Li D, Wang L. Identification of microRNAs present in congenital heart disease associated copy number variants. Eur Rev Med Pharmacol Sci 17: 2114–2120, 2013. [PubMed] [Google Scholar]
- 123.Yamashita T, Kawai H, Tian F, Ohta Y, Abe K. Tumorigenic development of induced pluripotent stem cells in ischemic mouse brain. Cell Transplant 20: 883–891, 2011. [DOI] [PubMed] [Google Scholar]
- 124.Yin VP, Lepilina A, Smith A, Poss KD. Regulation of zebrafish heart regeneration by miR-133. Dev Biol 365: 319–327, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yin VP, Poss KD. New regulators of vertebrate appendage regeneration. Curr Opin Genet Dev 18: 381–386, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 39: 133–144, 2010. [DOI] [PubMed] [Google Scholar]
- 127.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129: 303–317, 2007. [DOI] [PubMed] [Google Scholar]
- 128.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436: 214–220, 2005. [DOI] [PubMed] [Google Scholar]