Abstract
HMG domain transcription factor, Sox2, is a critical gene for the development of cochlear hair cells, the receptor cells for hearing, but this has been ascribed to expansion of the progenitors that become hair cells. Here, we show that Sox2 activated Atoh1, a transcription factor important for hair cell differentiation, through an interaction with the 3′ enhancer of Atoh1. Binding to consensus sequences in the Atoh1 enhancer was dependent on the level of Sox2, and the extent of enhancer binding correlated to the extent of activation. Atoh1 activation by Sox2 was required for embryonic hair cell development: deletion of Sox2 in an inducible mutant, even after progenitor cells were fully established, halted development of hair cells, and silencing also inhibited postnatal differentiation of hair cells induced by inhibition of γ-secretase. Sox2 is thus required in the cochlea to both expand the progenitor cells and initiate their differentiation to hair cells.
Sox2, an HMG domain transcription factor, plays multiple roles, most prominently in cellular reprogramming and stem cell pluripotency. It accomplishes this by inhibiting transcription of differentiation factors for specific cell types, acting as a transcriptional repressor at a large number of genes and their regulatory regions1. The effector activity of Sox2 is exerted through DNA binding and is dependent on level2,3. It is widely expressed in the embryo and in progenitor cells of various organs as well as the developing and mature nervous system. In the otic placodes from which the ear develops, Sox2 is prominent4,5, and it is a marker for the prosensory domain in the developing cochlea from which the cochlear and vestibular epithelia develop5,6. It is extinguished shortly after birth in cochlear hair cells but continues to be expressed in type 2 vestibular hair cells and in supporting cells of vestibular and cochlear epithelium6. Evidence from a number of mouse mutants suggested a role for Sox2 in development of the sensory epithelium of the inner ear4,5. In particular its expression was required for establishment of the progenitor cells5, and, indeed, the lack of hair cells in Sox2 hypomorphic mice was attributed to the failure to establish a prosensory domain5. Later studies suggested that Sox2 was antagonistic to bHLH transcription factor, Atoh14, similar to what has been shown for proneural transcription factors in other systems7,8,9, but other recent studies showed an upregulation of Atoh1 by Sox210,11,12,13. Sox2 was also shown to be important for auditory neuron differentiation14. We sought to determine its overall role in the development of hair cells, the receptor cells for hearing.
Hair cells are present in limited number and although spontaneous regeneration of hair cells has been noted in early postnatal ears15,16,17, this capacity is lost in the adult ear18. As a result, loss of hair cells due to environmental toxins, noise, and aging can lead to permanent deafness. A better understanding of mechanisms for the differentiation of hair cells could contribute to new therapies to regenerate hair cells. Supporting cells surround hair cells in the sensory epithelium; they include a subset expressing leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5) that can act as Wnt-responsive progenitors for hair cells19. Notch signaling inhibits bHLH transcription factor, Atoh1, a key gene for hair cell development20,21,22,23. Notch inhibition increases Atoh1 expression when Wnt signaling is active24. Notch signaling influences cochlear progenitor cell fate by its effect on bHLH transcription factor expression, and the effects of Notch on cochlear progenitors could be mediated by Sox2, which is regulated by Notch. We show here that Sox2 expression is required for the late stages of progenitor cell differentiation to hair cells and stimulates Atoh1 expression through a concentration dependent molecular interaction with the 3′ enhancer of Atoh1.
Results
Interaction between Sox2 and the Atoh1 3′ Enhancer
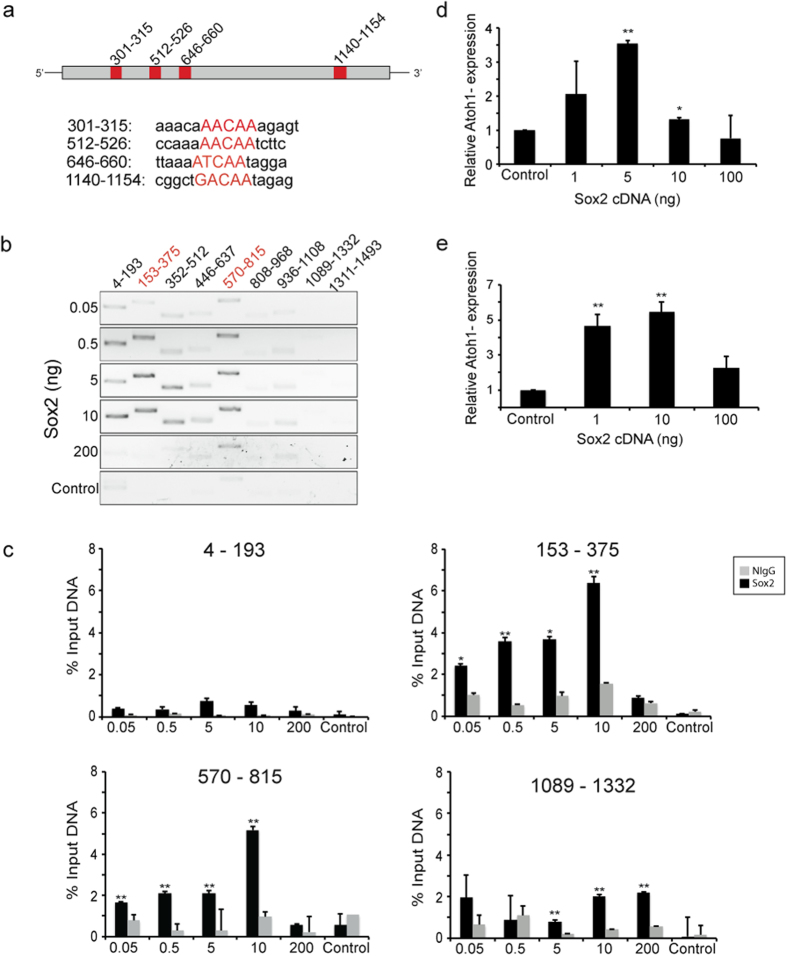
Sox2 is necessary for normal cochlear development, because of its function in the establishment of the prosensory epithelium5,25,26. We and others have shown that Sox2 upregulates bHLH transcription factors, Atoh1 and Neurog1, during determination of cochlear progenitor cell fate10,12,27, as it does in neural progenitor and Merkel cells11,13,28. We found that the level of Sox2 was an important determinant for progenitor cell differentiation or proliferation27, as it is for the determination of ES cell fate29. To examine the role of Sox2 in Atoh1 regulation, we assessed Sox2 binding to the Atoh1 enhancer. Sox2 is an HMG domain transcription factor; its interaction with consensus binding sequences has been characterized30. We attempted to determine whether DNA binding to Sox2 was level-dependent and whether alterations in binding of Sox2 to the Atoh1 enhancer at increasing concentrations correlated to changes in Atoh1 expression. We assessed binding to Sox2 sites (Fig. 1a shows sites at 301–315, 512–526, 646–660, and 1140–1154 in the Atoh1 enhancer) by chromatin immunoprecipitation (ChIP) in OC-1 cells, an inner ear cell line that endogenously expresses Atoh131. Precipitation of the DNA with an HA antibody after transfection of OC-1 cells with Sox2-HA indicated active binding sites in the Atoh1 enhancer. These sites could be detected by PCR using 9 consecutive primer sets to span the entire enhancer (Fig. 1b; Table S1). The ChIP indicated 2 active binding sites at 301–315 and 646–660 (contained in the primer sets shown in red in Fig. 1a), and usage of the sites was sensitive to the concentration of Sox2. Quantitative ChIP at these sites showed that the strength of binding changed with the level of Sox2. At 301–315, where the highest level of binding was seen, a plateau of DNA binding was reached at 10 ng (Fig. 1c). At this site, higher concentrations decreased the percent of input recovered in the ChIP. Low signals or signals that did not increase monotonically with Sox2 concentration were attributed to incomplete shearing of DNA and these sequences, which included areas with (1089–1332) and without (4–193) canonical sites (Fig. 1c), were assumed to lack active binding sites. Binding to the site at 1089–1332, however, could not be ruled out by the results of the ChIP analysis.
Figure 1. Activation of the Atoh1 Enhancer by Sox2 Binding.
(a) The Sox2 binding sequences in the Atoh1 3′- enhancer are shown (red boxes) with the sequences identified by MatInspector (Genomatix). (b) ChIP was performed in OC-1 cells after transfection of Sox2-HA and the Atoh1 enhancer. DNA was precipitated with HA antibody (Sox2), followed by RT-PCR with overlapping primer sets covering the entire enhancer. The control represents transfection with empty vector. Positive primer sets, 153–375 and 570–815 are shown in red and encompass the binding sites at 301–315 and 646–660, respectively. The bands in adjacent primer sets were ascribed to incomplete shearing of DNA. (c) Quantitative ChIP after transfection of OC-1 cells with Sox2-HA showed increased binding with Sox2 concentration up to 5–10 ng/40,000 cells and decreased binding at the higher concentration. Binding at 153–375 and 570–815 sites showed a monotonic increase with Sox2 concentration. The low signal at 4–193 was thought to represent overlap with the site amplified by 153–375. As in (b), concentration-dependent binding was not observed at 1089–1332. The control represents transfection with empty vector. ChIP for Sox2 was compared to ChIP for NIgG (*P < 0.05; **P < 0.01). (d) Increasing concentrations of Sox2 cDNA led to maximum upregulation of Atoh1 at 5 ng, as measured by quantitative RT-PCR in OC-1 cells. (e) Upregulation of Atoh1 was greatest at 10 ng of Sox2 cDNA, as measured by quantitative RT-PCR in differentiating neurospheres.
Further testing of Sox2 binding to the Atoh1 enhancer was performed in HEK cells, and binding was compared to OC-1 cells to determine whether the result was generalizable outside the cochlea. Whereas the overall pattern was similar, differences in the distribution of Sox2 suggested that tissue-specific expression of DNA-binding components or protein interacting partners may fine-tune the binding to specific sites (Fig. S1a). Additional quantitative ChIP was carried out through the use of a construct with a 4X repeat of the Sox2 binding site at 301–315. Sox2 binding to this construct was also dose-dependent up to a maximum, with a decrease at higher concentrations (Fig. S1b).
Atoh1 mRNA levels were measured in vitro in OC-1 cells and inner ear neurospheres by quantitative RT-PCR to assess the effect of Sox2. In OC-1 cells, Atoh1 expression increased with Sox2 expression to a maximum and decreased upon elevation of Sox2 to higher levels (Fig. 1d). The results were similar in inner ear neurospheres (Fig. 1e).
We assessed the activation of the Atoh1 enhancer and the effect of different levels of Sox2 using a construct that drove luciferase with the Atoh1 enhancer to further understand the role of Sox2 in its expression. Due to low levels of transfection in OC-1 cells, these experiments were performed in Atoh1-expressing intestinal cell line, IEC-6. The level of enhancer activity declined when Sox2 concentrations were increased further after reaching a maximum at 0.5 ng/well i.e. at the high levels, Sox2 had decreasing effects on Atoh1 (Fig. S1c). Additional experiments conducted with a luciferase vector made with the 4X repeat of the Sox2 binding site at position 301–315 demonstrated a similar Sox2 level dependence, although reaching a maximum at lower concentrations (Fig. S1d). Together, these experiments demonstrate a concentration-dependent binding and enhancer activation of Atoh1 by Sox2 that was seen in both cochlear cells and cell lines derived from other tissues.
Sox2 Expression in Embryonic Hair Cell Progenitors
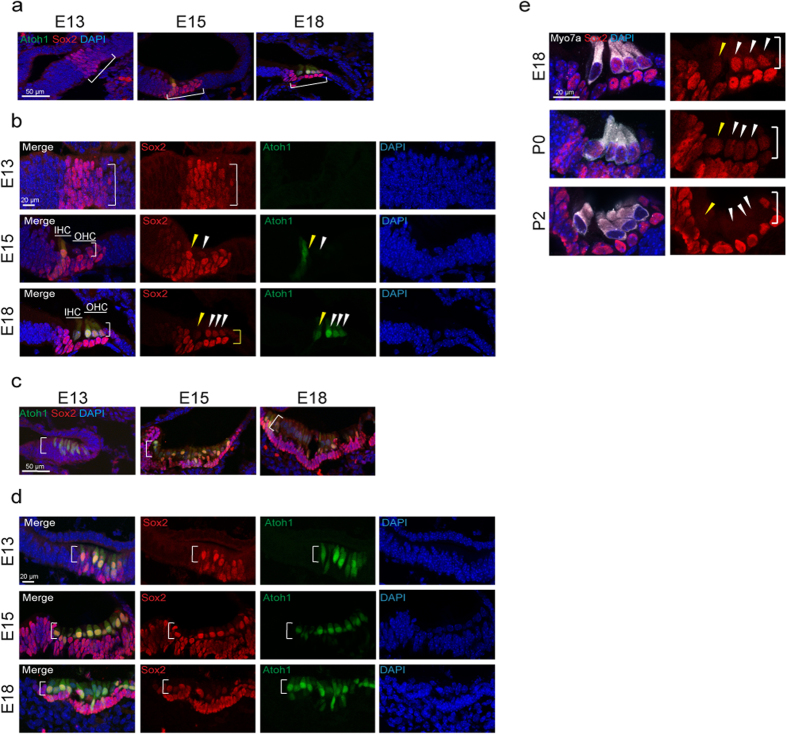
We next analyzed the expression of Sox2 during development of the mouse cochlea. Expression of Sox2 was observed in the prosensory cells of the cochlea at E13 (Fig. 2a,b), and initiation of Atoh1 expression was observed between E13 and E15 (Fig. 2b).
Figure 2. Sox2 Expression in the Developing Cochlea and Vestibular System.
(a) Sox2 (red) was expressed in the prosensory cells of the cochlea at E13 (bracket). Sox2 was expressed in the developing sensory epithelium at E15 and E18. (b) Atoh1-nGFP-positive cells (green) were first found in the sensory epithelium at E15 (bracket and arrowheads). Inner hair cells (IHC, yellow arrowhead), which develop before outer hair cells (OHC, white arrowheads), expressed Sox2 when newly formed at E15, and the newly formed inner and outer hair cells continued to express Sox2 at E18. Sox2 expression in hair cells had begun to decrease at that time (arrowheads) but was strong in supporting cells (yellow bracket). The mid-basal region is shown. (c) Atoh1-nGFP positive cells could be seen in the developing vestibular system as early as E13 (white bracket) and the cells expressed Sox2. (d) Atoh1-nGFP positive cells continued to express Sox2 at E15 (white bracket). Atoh1-nGFP-positive hair cells showed reduced Sox2 expression at E18 (white bracket). (e) Cochlear hair cells expressed myosin VIIa (Myo7a, white) at E18, P0 and P2. Sox2 was still expressed in hair cells in the cochlea until P0, but was undetectable in these cells at P2. It continued to be expressed in supporting cells at P2 (bracket and arrowheads).
The area of the developing otocyst marked by Sox2 expression corresponds to the prosensory epithelium (Fig. 2b). At E15, expression of Sox2 was apparent in the ventromedial region (Fig. 2b) and was accompanied by GFP expression from the Atoh1 enhancer. These newly differentiating hair cells that co-expressed Sox2 and Atoh1 can be seen in the upper layer of the sensory epithelium (Fig. 2b). Sox2 expression was seen in cells destined to become both hair cells and supporting cells. Developing hair cells, identified by their expression of Atoh1 as well as myosin VIIa, still expressed Sox2 at E18 (Fig. 2b,e) and loss of Sox2 in hair cells in a gradient from base to apex occurred between P0 (Fig. 2e, P0) and P2 (Fig. 2e, P2).
Sox2 was broadly expressed in the developing vestibular epithelia at E13 (Fig. 2c). Atoh1-expressing cells could be detected at E13 in the vestibular organs, which differentiate earlier than the cochlea (Fig. 2c,d). Co-expression of Sox2 continued in the newly developing, Atoh1-positive, cochlear hair cells (Fig. 2d). Hair cells at E18 expressed myosin VIIa and Sox2 and had not completely lost Sox2 expression in the early postnatal period (Fig. S2). The results in both balance and hearing organs of the developing inner ear thus suggest that expression of Sox2 is maintained throughout the differentiation of hair cells, before becoming undetectable at P2 in cochlear hair cells.
Sox2 is Necessary for Hair Cell Differentiation
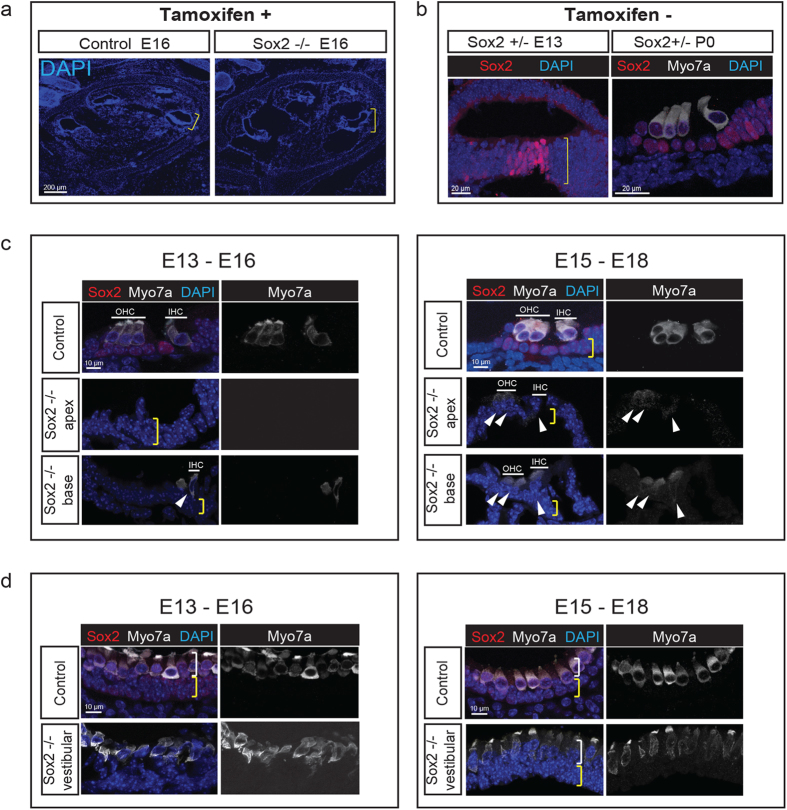
Sensory progenitor cells in the developing sensory epithelium are established by E1332. After tamoxifen administration to delete the floxed Sox2 gene at E13, we found an overall normal gross morphology at E16 (Fig. 3a). To test whether progenitor cells were present in Sox2-Cre-ER; Sox2flox/+ mice at this time point, we assessed expression by immunolabeling for Sox2. The prosensory domain contained the normal complement of cells in both surface views (Fig. S3) and sections (Fig. 3b, E13) and generated hair cells normally (Fig. 3b, P0) in controls that were not treated with tamoxifen. Examination of the cochlea at E16 after administration of tamoxifen at E13, showed that hair cells had differentiated partially at the base (where hair cell differentiation occurs first), but that deletion of Sox2 had halted the further differentiation of hair cells. No hair cells were found in the middle and apical regions (Fig. 3c). Few hair cells developed when the gene was deleted at E15 and examined at E18 (Fig. 3c).
Figure 3. Sox2 is Required for Differentiation of Cochlear and Vestibular Hair Cells.
(a) The structure of the cochlea was intact when Sox2 was deleted by tamoxifen administration at E13 and analyzed at E16 (a Cre-negative Sox2flox/flox control was compared to a Sox2-Cre-ER; Sox2flox/+ knockout, Sox2 −/−). (b) The prosensory progenitors were present at E13, and hair cells had formed normally at P0 in an undeleted Sox2-Cre-ER; Sox2flox/+ mouse (Sox2 +/−). (c) After deletion of Sox2 at E13, the cochlea showed a nearly complete absence of hair cells at E16 (no hair cells were seen in the mid and apical regions and one row of dysmorphic inner hair cells was observed at the base). The position of myosin VIIa-positive (white) inner and outer hair cells are indicated by lines labeled IHC and OHC, and the position of the supporting cell layer is indicated by the yellow bracket (note the absence of Sox2 in the tamoxifen-treated animal). One inner and two outer hair cell rows at the base and a small number of dysmorphic hair cells in the apical region (arrowheads) were apparent at E18 when Sox2 was deleted 2 days later at E15. (d) After deletion of Sox2 at E13, the vestibular organs had weakly myosin VIIa-positive (white), dysmorphic hair cells at E16 in a layer (white bracket) above the supporting cell layer (yellow bracket; compare to the Sox2-positive (red) supporting cells and myosin VIIa/Sox2 double-positive hair cells in control). A greater number of hair cells were observed (white bracket) at E18 after deletion of Sox2 at E15. Nuclei were stained with DAPI (blue).
Deletion of Sox2 at E13 (Fig. 3d) did not affect initial appearance of hair cells in the vestibular sensory epithelia, consistent with earlier development of hair cells in these organs, although the hair cells were short and irregularly shaped, with low myosin VIIa expression, and pyknotic nuclei. Again, further development was seen when the deletion was performed at E15. The abrogation of hair cell differentiation by deletion of Sox2 in ears after the formation of sensory progenitor cells indicates that Sox2 is necessary not only for establishing the sensory progenitors5, but for the differentiation of hair cells from the sensory progenitors.
Expression of Sox2 in Progenitor Cells Differentiating to Hair Cells
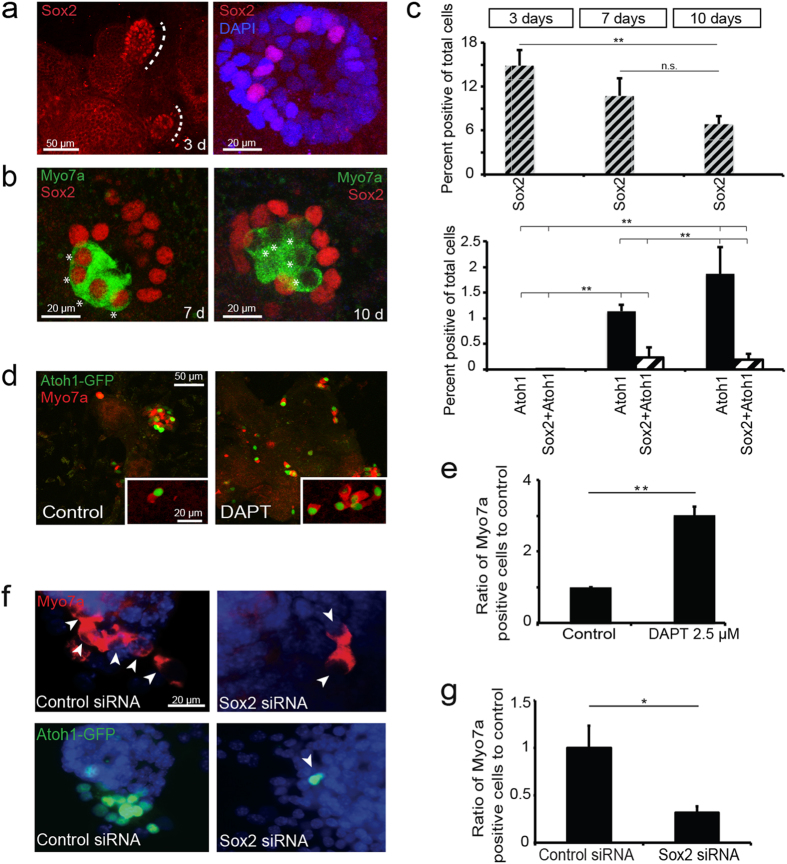
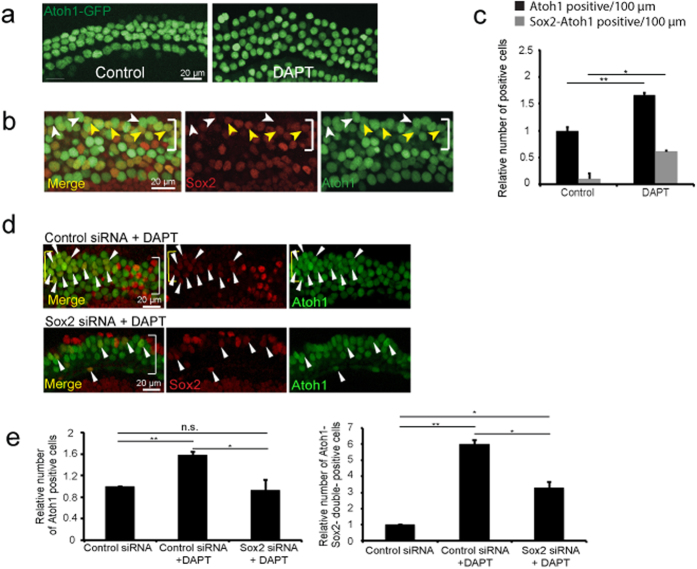
Sox2 is expressed in inner ear progenitor cells33 and, even though the cultures are heterogeneous, we have previously demonstrated an overall decrease in progenitor markers and an increase in hair cell markers when inner ear progenitors are transferred from a proliferative self-renewing culture (as floating neurospheres in growth factors) to a differentiating culture (as adherent cells in the absence of growth factors). Atoh1 activation was followed by GFP expression in spheres from Atoh1-nGFP mice. At 3 days of differentiation, groups of Sox2-positive cells were observed in the cultures (Fig. 4a,c). After 7 days in culture, immature hair cells (myosin VIIa and Atoh1 staining cells) differentiated and showed strong staining for Sox2 (Fig. 4b,c). The total number of hair cells (Atoh1-GFP positive cells) increased during hair cell differentiation between day 7 and 10, whereas the percentage of immature hair cells (Sox2 and Atoh1 double-positive) decreased upon maturation (Fig. 4c).
Figure 4. Requirement of Sox2 for the Formation of New Hair Cells from Inner Ear Spheres.
(a) Distinct patches of Sox2-positive cells were seen in differentiating spheres at 3 days (dotted line). (b) In progenitors differentiated for 7 days, all myosin VIIa-positive cells (green) co-expressed Sox2 (asterisks). Many myosin VIIa-positive cells had lost Sox2 expression at 10 days. (c) Quantification of Sox2-positive cells (top graph) at 3,7 and 10 days of differentiation (**P < 0.01). Bottom graph shows quantification of Atoh1-positive cells (black columns) and the fraction of Atoh1-Sox2 double positive cells (patterned columns). Atoh1-positive hair cells and Sox2- Atoh1 double-positive hair cells were not seen during early differentiation and increased significantly at later time points (**P < 0.01). (d) The number of newly formed hair cells after treatment with DAPT increased and hair cells were Atoh1 (green) and myosin VIIa (red)-positive (insert). (e) DAPT treatment increased the proportion of myosin VIIa-positive cells compared to control (**P < 0.01). (f) Neurospheres under differentiating conditions in the presence of DAPT and treated with siRNA for Sox2 (100 nM total) had a reduced number of myosin VIIa (top panel) or Atoh1-nGFP (bottom panel) positive cells (arrows) compared to control siRNA after differentiation for 7 days. The transfection efficiency was 40–50%. (g) The reduction in the number of hair cells was significant (*P < 0.05).
Effect of Sox2 on Differentiation of Hair Cells from Inner Ear Progenitors
Inhibition of Notch signaling increases differentiation of hair cells from inner ear progenitor cells isolated as neurospheres27 by altering the expression of Atoh1. It may exert this influence through an effect on Sox2, in addition to the effects mediated by Notch effectors, Hes and Hey12. The level of Sox2 was decreased by 30% in cells treated with γ-secretase inhibitor, DAPT (Fig. S4a), but the treatment did not alter the number of Sox2-positive cells (Fig. S4b), indicating a reduction of Sox2 levels in preexisting progenitors upon differentiation into hair cells. Treatment with DAPT increased the percentage of Atoh1-single-positive and Atoh1-Sox2-double-positive cells obtained from inner ear progenitor cells almost 4-fold (Fig. S4c).
A significant increase in myosin VIIa-positive cells was apparent after treatment of inner ear progenitor cells with DAPT for 7 days (Fig. 4d,e). To evaluate whether hair cell differentiation induced by a γ-secretase inhibitor depended on expression of Sox2, we silenced Sox2 with siRNA. Effective reductions in Sox2 RNA levels (70%; Fig. S4d) were observed in inner ear progenitor cells. Sox2 silencing with siRNA decreased the number of DAPT-induced Atoh1- and myosin VIIa-positive cells (Fig. 4f), and the decrease was significant (Fig. 4g). Thus, Sox2 expression at the correct level was important for both the upregulation of Atoh1 and hair cell differentiation that resulted from γ-secretase inhibition.
Sox2 in Hair Cell Differentiation
To determine whether Sox2 levels played a role in accumulation of new hair cells after Notch inhibition in the newborn organ of Corti, Sox2 was followed in explants treated with DAPT, which reduced Sox2 levels in supporting cells34,35. At 48 hours, additional Atoh1-nGFP-positive outer hair cells were seen in the apex (Fig. 5a,c). New Atoh1-nGFP-positive hair cells derived from supporting cells expressed Sox2 (Fig. 5b,c). The original hair cells expressed high levels of Atoh1 but no detectable Sox2, while new hair cells expressed a lower level of Sox2 than adjacent supporting cells (Fig. 5d). Expression of Sox2 was lost later during maturation of hair cells. Sox2 silencing with siRNA decreased the number of both Atoh1-single-positive and Atoh1/Sox2-double-positive cells induced by DAPT (Fig. 5d,e).
Figure 5. Requirement of Sox2 for Hair Cell Differentiation in γ-Secretase-Treated Organ of Corti.
(a) Organ of Corti explants from neonatal Atoh1-nGFP mice (P1-P2) were kept in culture for 48 hours and treated with DAPT (2.5 μM) or carrier (DMSO, 0.1%) as a control. An increased number of hair cells was seen (mid-apical region is shown). (b) At 48 hours of DAPT treatment, Sox2-positive supporting cells that upregulated Atoh1-nGFP were observed in the hair cell layer at a position below the original hair cells (bracket). New supporting cell-derived hair cells showed a reduced level of Sox2 compared to supporting cells and were weakly Atoh1-nGFP-positive (yellow arrowheads), whereas original hair cells showed stronger Atoh1-nGFP expression and no Sox2 expression (white arrowheads). (c) Treatment with DAPT reduced Sox2 in supporting cells and increased the percentage of Atoh1-nGFP positive cells/100 μm and new Atoh1-nGFP positive cells (black columns) that expressed Sox2 (grey columns) after 72 hours (*P < 0.05). (d) Control siRNA (100 nM) with DAPT (2.5 μM) treatment resulted in an increased number of Atoh1-positive cells. Treatment with DAPT (2.5 μM) and Sox2 siRNA (100 nM total) partly suppressed the increase in new hair cells. White arrowheads mark Sox2-Atoh1 double-positive hair cells. (e) Atoh1-nGFP positive and Sox2-Atoh1 double-positive outer hair cells (OHC) per 100 μm in the apical region of organ of Corti explants treated with control siRNA, control siRNA and DAPT, or Sox2 siRNA and DAPT revealed that DAPT treatment significantly increased the number of hair cells in control siRNA-treated explants. Sox2 siRNA reduced the number of newly formed hair cells (**P < 0.01).
Discussion
We show here that Sox2 plays an important role in the upregulation of transcription factor, Atoh1, in both embryonic and postnatal cochlear progenitor cells, thus triggering the differentiation of hair cells. This is an important auxiliary role of Sox2, which also functions to establish prosensory cells in the cochlea, and is indicative of a proposed role for Sox2 throughout the body, not only in the maintenance of stem/progenitor cells, but in the specification of mature cell fate. However, this is a role for Sox2 that is seemingly at odds with its inhibition of differentiation factor expression. Since Atoh1 is a key gene for specifying hair cell fate, upregulation of Atoh1 expression by Sox2 binding to DNA in the regulatory region is a key upstream event in differentiation.
Sox2 inhibits the expression of differentiation factors1,7,8,9, and functions as a core factor for stem cell pluripotency along with Oct4 and Nanog1. Sox2 is, in fact, one of four transcription factors that reprogram various cell types to induced pluripotent stem cells36. Sox2 plays a role in the development of neural progenitors in the CNS37 and acts to inhibit the expression of neural transcription factors. Sox2 downregulation is thought to be an essential step for progenitors to undergo commitment to neuronal differentiation via expression of proneural genes7,29,38, and a similar mechanism, in which Sox2 had to be downregulated4,5 to permit hair cell genesis was proposed for the inner ear4.
Our data suggest a role for Sox2 in the activation of proneural transcription factors and, thus, in cell fate determination. Deletion of Sox2, after the prosensory cells had generated the prosensory domain, but before they had become hair cells, prevented hair cell differentiation. Such a role for Sox2 would suggest that it both maintained progenitor cell potency and initiated differentiation. This apparent paradox may be resolved by the timing of expression, i.e. the function of Sox2 would be, first, to establish the progenitor cells and, subsequently, to specify cell fate by a positive effect on a differentiation factor. The switch in roles could be due to an altered level of Sox2 or to the changing status of the progenitor cells during the course of inner ear development. Sox2 expression comes up at the earliest stages of cochlear development, when the otic placode first appears, and during the formation of the otocyst and the prosensory domain, for which its expression is necessary5. These progenitor cells are the source of both sensory cells and neurons, both of which require Sox2 expression for normal development5,14. We traced the expression of Sox2 throughout the formation of hair cells and found that it was expressed not only in hair cell progenitors, but also in newly born hair cells. Its expression diminished in a gradient from base to apex, and it was permanently downregulated in hair cells of the cochlea soon after birth. The importance of Sox2 for bHLH transcription factor expression has also been observed in neurons, in which Sox2 expression upregulates the expression of Neurog127, and in Xenopus, where Sox2 was implicated in the conversion from proliferation to differentiation of neural progenitors28. Subsequent to differentiation, Sox2 appeared to be inhibited by downstream transcription factors in a feedback loop that prevented excess neurogenesis7,9. Interaction of Sox2 with other transcription factors is another potential mechanism allowing Sox2 to play different roles in development10,39.
The role of Sox2 as a differentiation factor is at odds with a global antagonism of proneural transcription factor activity, but is not inconsistent with an eventual antagonistic role. We found that a reduction in Sox2 level was necessary for proneural transcription factors to be expressed in vitro and correlated with hair cell differentiation from supporting cells brought about by treatment of cochlear explants in vitro with DAPT. We think that this reduction is necessary for an active role of Sox2 in differentiation, not simply to release an inhibitory influence. Decreased Atoh1 expression at higher levels of Sox2 could be due to insufficient levels of the DNA-binding protein complex or to competition with other DNA-binding factors1. This criticality of concentration for Sox2 is consistent with the CNS40, where gene dosage leads to alternate activities of Sox24,41, including cell differentiation. The level of Sox2 in hair cells subsides further and is eventually extinguished after hair cell differentiation.
Sox2 is downstream of Notch and its expression is decreased by treatment with a γ-secretase inhibitor27,42. Notch signaling is necessary for the early stages of cochlear sensory development and may act in part by upregulating Sox2 in progenitor cells, whereas loss of Notch signaling due to lateral inhibition could drop Sox2 levels in a progenitor cell that becomes a hair cell. We show here that Sox2 is necessary for the effects of Notch inhibition on hair cell differentiation in newborn explants and cochlear spheres. The antagonism between Atoh1 and Sox2 may be attributable to an effect of Sox2 on Notch effectors such as Hes1 and Hes5 that antagonize bHLH transcription factor activity12. However, effects of Notch on proliferation were independent of Sox242.
Heterozygous mutations in Sox2 in humans result in eye defects that can be accompanied by deafness43. Sox2 knockout animals have severe deficiencies in the development of the inner ear, but, because Sox2 is expressed early in development of the mouse inner ear - well before Atoh1 - and because its loss resulted in incomplete initial formation of the sensory epithelium25,26,44,45, it was difficult to assess any role in hair cell differentiation prior to this work. The current results with conditional knockout show that Sox2 plays a critical role in the differentiation of hair cells from prosensory epithelial progenitor cells.
Methods
Chromatin Immunoprecipitation (ChIP)
Consensus sequences for Sox2 binding in the Atoh1 3′-enhancer were initially identified using the MatInspector software tool for transcription factor binding sites (Genomatix). OC-1 and 293T (HEK, Thermo Fisher) cells were transfected at 30% confluency using Lipofectamine (3 μl/μg DNA/ml) and Sox2-HA cDNA (0.01–200 ng/40,000 cells) in Opti-MEM. After 48 hours, cells were harvested and processed for ChIP (Active Motif, ChIP-IT Express Enzymatic). Chromatin was precipitated with mouse anti-HA antibody (3 μg), or normal rabbit IgG (3 μg).
Quantitative RT-PCR
Total RNA was extracted with the RNeasy Maxi Kit (Qiagen) according to the manufacturer’s instructions from embryonic tissue harvested in RNAlater (Ambion) or from OC-1 cells or spheres transfected using Lipofectamine with different concentrations of Sox2 cDNA (in ng/40,000 cells) for 48 hours or treated with DAPT or DAPT and siRNA. RNA was denatured at 65 °C for 5 min. For reverse transcription, ImProm II (Promega) was used with random hexamers. The reverse transcription conditions were 25 °C for 5 min followed by 42 °C for 60 min. The reaction was terminated at 70 °C for 15 min. The cDNAs were mixed with Platinum quantitative PCR Supermix ROX with UDG (Invitrogen) and primers for 18S RNA, Atoh1 or Sox2 (ABI) according to the manufacturer’s instructions (primers are listed in Table S1). Gene expression was measured relative to 18S RNA. Samples were analyzed in 96 well plates in triplicate by quantitative PCR (Applied Biosystems 7900HT) using the following conditions: initial denaturation at 95 °C for 2 min, denaturation at 95 °C for 40 s and annealing/extension at 60 °C for 35 s for 45 cycles. All statistical analysis was performed with the Student’s t-test.
Plasmids
A vector containing a 1.4 kb enhancer for Atoh1, sufficient for endogenous Atoh1 expression46, cloned upstream of luciferase27, was used in luciferase assays. Sox2-HA (pCAG-HA-Sox2-IP) was from Addgene. A 4x repeat of a 41 base pair enhancer sequence (acccaaacaaacaaagagtcagcacttcttaaagtaatgaa) spanning the binding site at position 301–315 was synthesized (GenScript) and cloned into pGL4 (Promega).
Luciferase Assay
IEC6 cells (ATCC) were seeded onto 96-well plates 1 day before transfection. IEC6 cells are an intestinal cell line that shows high levels of Atoh1 and increased responsiveness to induction. At 40–50% confluency, 100 ng of luciferase reporter construct (intact Atoh1 enhancer or 4x-binding site reporter), 10 ng of Renilla luciferase construct and different amounts of Sox2 effector (from 0.1 ng–100 ng) were transfected with Lipofectamine 3000 (3 μl/μg DNA/ml, Life Technologies) into the cells for 6 h. Cells were lysed after 48 h, and luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions in a Wallac Victor2 1420 Multilabel Counter (Perkin-Elmer).
Isolation of Organ of Corti Cells for Neurosphere Culture
For each experiment, cochleae of 4–6 neonatal C57BL/6 or Atoh1-nGFP pups47 that express GFP under the control of the Atoh1 enhancer (a generous gift from Jane E. Johnson, University of Texas) were dissected in HBSS and the organ of Corti was separated from the stria vascularis and the spiral ganglion neurons. The tissues were dissociated in trypsin (0.05%) for 13 min in PBS at 37 °C. 10% FBS in DMEM-high glucose medium was used to stop the reaction. After washing, the tissue was manually dissociated. The triturated cells were then passed through a 70 μm cell strainer (BD Labware) to remove tissue debris. Single cells were cultured in DMEM/F12 (1:1) supplemented with N2 and B27 (Invitrogen), and EGF (20 ng/ml; Chemicon), bFGF (10 ng/ml; Chemicon), IGF-1 (50 ng/ml;Chemicon), and heparan sulfate (50 ng/ml; Sigma). Single cells were maintained in ultra-low cluster plates (Costar) for several days in culture to obtain neurospheres. For passage, neurospheres of the first generation were dissociated with a 27G needle and syringe (BD Labware) 6–8 times. Single cell suspensions were cultured in fresh medium F12/DMEM (1:1) with the same growth factors to form neurospheres until use at the 4th to 5th generation.
Immunohistochemistry of Neurospheres
Differentiated neurospheres were fixed at room temperature in 4% paraformaldehyde/PBS for 10 min and then washed in PBS. Permeabilization and blocking was performed for spheres or sections with blocking solution (0.3% Triton X-100, 15% heat-inactivated goat or donkey serum in PBS) for 1 h. Diluted primary antibody (0.1% Triton X-100, and 10% heat inactivated goat or donkey serum in PBS) was applied overnight at 4 °C. Secondary antibodies were applied for 2 hours at room temperature. Nuclei were visualized with 4,6-diamidino-2-phenylindole (Vector Laboratories). Staining was analyzed with epifluorescence microscopy (Axioskop2 Mot Axiocam, Zeiss) and confocal microscopy (TCD, Leica).
Antibodies
The antibodies used were monoclonal mouse antibody against myosin VIIa (used at 1:100; Developmental Studies Hybridoma Bank), polyclonal rabbit antibody against myosin VIIa (used at 1:250; Proteus), polyclonal goat antibody against Sox2 (used at 1:300; Santa Cruz). Secondary antibodies for detection of primary antibodies: Alexa Fluor 488, 568, and 647-conjugated (all used at 1:500; Invitrogen). Mouse monoclonal antibody against HA (Sigma, used for ChIP).
Mice for in Vivo Experiments
Sox2-Cre-ER48 and Sox2flox/flox mice49 (Jackson Labs, stock numbers 013093 and 017593, respectively) were used for in vivo knockout experiments. All animal studies were approved by the Institutional Animal Care and Use Committee of Massachusetts Eye and Ear Infirmary according to National Institutes of Health guidelines. Homozygous Sox2flox/flox mice were mated with hemizygous Sox2-Cre-ER mice. Tamoxifen (100 μl, 50 mg/ml, Sigma) was injected intraperitoneally for 2 consecutive days (E12 and E13, or E14 and E15) to obtain hemizygous Sox2-Cre-ER animals with deleted Sox2flox. Sox2-Cre-ER; Sox2flox/+ mice without exposure to tamoxifen were examined at E13 and P0 to exclude potential effects of the undeleted transgenes. Embryos were genotyped and Cre-negative embryos were used as controls. Immunohistochemistry was performed at E16 and E18 in tamoxifen-treated animals.
Tissue Preparation of Embryos
Embryos of Sox2-Cre-ER; Sox2flox/+ mice were collected at E13, E16 or E18 and Atoh1-nGFP mice were collected at E13, E15 and E18 (with identification of a positive plug in the morning counted as E0.5), or postnatally at P0 and P2, and fixed in 4% paraformaldehyde for 4 hours at 4 °C. After dehydration with sucrose (5% and 30%), embryos were embedded in OCT and kept at −80 °C. For immunohistochemistry, tissue was cut (12 μm) and then stained as described for neurospheres. Midbasal regions were imaged in the in vivo knockout and explant experiments unless otherwise specified.
Dissection of Organ of Corti Explants
For each experiment, tissue of neonatal pups with C57BL/6 (Jackson Labs) or Atoh1-nGFP background were dissected on ice in HBSS and the organ of Corti was harvested after removal of spiral ganglion neurons and stria vascularis as previously described50.
Differentiation and Treatment of Neurospheres or Organ of Corti Explants with DAPT
4th–5th generation neurospheres or organ of Corti explants (P1-P2) were plated without growth factors in 4-well plates (Greiner) on round 10 mm glass coverslips coated with poly-L-lysine (Cultrex) and attachment took place overnight in 10% FBS/DMEM-high glucose (GIBCO). Attachment was ensured with microscopic inspection and the medium was changed to serum-free DMEM-high glucose/F12 (mixed 1:1, GIBCO) and N2 and B27 (Invitrogen). Neuropheres were differentiated for 3, 7 or 10 days. For treatment, DAPT (CalBiochem) (2.5 μM) or control medium containing only DMSO (0.1%) were applied for 7 days on spheres and 48 hours on organ of Corti explants. Cells were harvested and further analyzed by immunohistochemistry as described for cultured neurospheres. Axiovision 4.3 was used for data acquisition and the number of cells was quantified with Metamorph software. Cell counts were expressed as mean ± standard deviation. An average of 1,000 cells were counted for neurospheres or 100 μm for OC explants. Origin software was used for statistical evaluation.
SiRNA Silencing
Cochlear neurospheres or organ of Corti explants were transfected with siRNA (100 nM ON-TARGET plus, Smart Pool from Dharmacon for Sox2, or non-targeting siRNA), using Gene Silencer (Genlantis) for 24 hours according to the manufacturer’s instructions. Transfection efficiency was monitored using siGlo Red Transfection Indicator (Dharmacon). In addition, 2.5 μM DAPT was added during differentiation culture. After transfection, the cells were washed and cultured in DMEM/F12 with DAPT (1:1) and N2/B27 (spheres and explants) or DMEM/10% FBS and DAPT (IEC6 cells and OC-1 cells). For quantitative PCR experiments after siRNA treatment, cells were differentiated for additional 24 hours after transfection. Organ of Corti explants were kept in culture for an additional 24 hours (48 hours total) and differentiating neurospheres were cultured for up to 6 days after siRNA treatment.
Statistics
Statistical analysis was performed using an unpaired, two-tailed t-test using Origin software. Data generated with cell lines was repeated at least 3 times, data collected from sphere and explant tissue was repeated at least 4 times. Bonferroni correction was used for comparison of multiple variables. Experimental values obtained for each antibody by ChIP were compared to a NIgG control, and luciferase expression values were compared to sample without effector. Brackets are used to indicate the compared values in all other figures with significance indicated by asterisks.
Additional Information
How to cite this article: Kempfle, J. S. et al. Sox2 in the differentiation of cochlear progenitor cells. Sci. Rep. 6, 23293; doi: 10.1038/srep23293 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute on Deafness and other Communicative Disorders, RO1 DC007174 and P30 DC05209, by a grant from the Deafness Research Foundation, a cooperative grant from the European Commission (FP7 Health 2013 Innovation), by the Shulsky Foundation, David H. Koch, and Robert Boucai.
Footnotes
Author Contributions J.S.K., J.L.T. and A.S.B.E. designed experiments; J.S.K. and J.L.T. performed experiments; J.S.K., J.L.T. and A.S.B.E. analyzed data; and J.S.K. and A.S.B.E. wrote the manuscript.
References
- Boyer L. A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosetti D. C., Basilico C. & Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol 17, 6321–6329 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer B. et al. Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2:Oct-3/4 target genes. Nucleic Acids Res 35, 1773–1786 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabdoub A. et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci USA 105, 18396–18401 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan A. E. et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature 434, 1031–1035 (2005). [DOI] [PubMed] [Google Scholar]
- Hume C. R., Bratt D. L. & Oesterle E. C. Expression of LHX3 and SOX2 during mouse inner ear development. Gene Expr Patterns 7, 798–807 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund M., Andersson E., Novitch B. G. & Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci 6, 1162–1168 (2003). [DOI] [PubMed] [Google Scholar]
- Episkopou V. SOX2 functions in adult neural stem cells. Trends Neurosci 28, 219–221 (2005). [DOI] [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P. & Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron 39, 749–765 (2003). [DOI] [PubMed] [Google Scholar]
- Ahmed M. et al. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell 22, 377–390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardot E. S. et al. Polycomb subunits Ezh1 and Ezh2 regulate the Merkel cell differentiation program in skin stem cells. EMBO J 32, 1990–2000 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves J., Uchikawa M., Bigas A. & Giraldez F. The prosensory function of Sox2 in the chicken inner ear relies on the direct regulation of Atoh1. PLoS One 7, e30871 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigoto C. N., Bardot E. S., Valdes V. J., Santoriello F. J. & Ezhkova E. Embryonic maturation of epidermal Merkel cells is controlled by a redundant transcription factor network. Development 141, 4690–4696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puligilla C., Dabdoub A., Brenowitz S. D. & Kelley M. W. Sox2 induces neuronal formation in the developing mammalian cochlea. J Neurosci 30, 714–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhall N. F., Shi F., Arnold K., Hochedlinger K. & Edge A. S. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Reports 2, 311–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B. C. et al. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 141, 816–829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley M. W., Talreja D. R. & Corwin J. T. Replacement of hair cells after laser microbeam irradiation in cultured organs of corti from embryonic and neonatal mice. J Neurosci 15, 3013–3026 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge A. S. & Chen Z. Y. Hair cell regeneration. Curr Opin Neurobiol 18, 377–382 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F., Kempfle J. S. & Edge A. S. Wnt-responsive lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci 32, 9639–9648 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham N. A. et al. Math1: an essential gene for the generation of inner ear hair cells. Science 284, 1837–1841 (1999). [DOI] [PubMed] [Google Scholar]
- Zheng J. L. & Gao W. Q. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci 3, 580–586 (2000). [DOI] [PubMed] [Google Scholar]
- Izumikawa M. et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med 11, 271–276 (2005). [DOI] [PubMed] [Google Scholar]
- Gubbels S. P., Woessner D. W., Mitchell J. C., Ricci A. J. & Brigande J. V. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature 455, 537–541 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F., Cheng Y. F., Wang X. L. & Edge A. S. Beta-catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3′ enhancer. J Biol Chem 285, 392–400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Q., Cole L. K., Mulheisen M., Chang W. & Wu D. K. The role of Pax2 in mouse inner ear development. Dev Biol 272, 161–175 (2004). [DOI] [PubMed] [Google Scholar]
- Lawoko-Kerali G., Rivolta M. N. & Holley M. Expression of the transcription factors GATA3 and Pax2 during development of the mammalian inner ear. J Comp Neurol 442, 378–391 (2002). [DOI] [PubMed] [Google Scholar]
- Jeon S. J., Fujioka M., Kim S. C. & Edge A. S. B. Notch signaling alters sensory or neuronal cell fate specification of inner ear stem cells. J. Neurosci. 31, 8351–8358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathocleous M. et al. A directional Wnt/beta-catenin-Sox2-proneural pathway regulates the transition from proliferation to differentiation in the Xenopus retina. Development 136, 3289–3299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp J. L., Ormsbee B. D., Desler M. & Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 26, 903–911 (2008). [DOI] [PubMed] [Google Scholar]
- Maruyama M., Ichisaka T., Nakagawa M. & Yamanaka S. Differential roles for Sox15 and Sox2 in transcriptional control in mouse embryonic stem cells. J Biol Chem 280, 24371–24379 (2005). [DOI] [PubMed] [Google Scholar]
- Kalinec F., Kalinec G., Boukhvalova M. & Kachar B. Establishment and characterization of conditionally immortalized organ of corti cell lines. Cell Biol Int 23, 175–184 (1999). [DOI] [PubMed] [Google Scholar]
- Kelley M. W. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci 7, 837–849 (2006). [DOI] [PubMed] [Google Scholar]
- Martinez-Monedero R., Yi E., Oshima K., Glowatzki E. & Edge A. S. Differentiation of inner ear stem cells to functional sensory neurons. Dev Neurobiol 68, 669–684 (2008). [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A. et al. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell 16, 58–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N. et al. Inhibition of Notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J Mol Med (Berl) 84, 37–45 (2006). [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell 1, 39–49 (2007). [DOI] [PubMed] [Google Scholar]
- Bani-Yaghoub M. et al. Role of Sox2 in the development of the mouse neocortex. Dev Biol 295, 52–66 (2006). [DOI] [PubMed] [Google Scholar]
- Taranova O. V. et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev 20, 1187–1202 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y., Uchikawa M. & Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet 16, 182–187 (2000). [DOI] [PubMed] [Google Scholar]
- Pevny L. H. & Nicolis S. K. Sox2 roles in neural stem cells. Int J Biochem Cell Biol 42, 421–424 (2010). [DOI] [PubMed] [Google Scholar]
- Mallanna S. K. & Rizzino A. Systems biology provides new insights into the molecular mechanisms that control the fate of embryonic stem cells. J Cell Physiol 227, 27–34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. et al. Ectopic expression of activated notch or SOX2 reveals similar and unique roles in the development of the sensory cell progenitors in the mammalian inner ear. J Neurosci 33, 16146–16157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelberman D. et al. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J Clin Invest 116, 2442–2455 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T., Mohamed O. A., Taketo M. M., Dufort D. & Groves A. K. Wnt signals mediate a fate decision between otic placode and epidermis. Development 133, 865–875 (2006). [DOI] [PubMed] [Google Scholar]
- Torres M., Gomez-Pardo E. & Gruss P. Pax2 contributes to inner ear patterning and optic nerve trajectory. Development 122, 3381–3391 (1996). [DOI] [PubMed] [Google Scholar]
- Helms A. W., Abney A. L., Ben-Arie N., Zoghbi H. Y. & Johnson J. E. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127, 1185–1196 (2000). [DOI] [PubMed] [Google Scholar]
- Lumpkin E. A. et al. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns 3, 389–395 (2003). [DOI] [PubMed] [Google Scholar]
- Arnold K. et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317–329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O. et al. Pax6 is essential for lens fiber cell differentiation. Development 136, 2567–2578 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M., Brugeaud A. & Edge A. S. Primary culture and plasmid electroporation of the murine organ of Corti. J Vis Exp (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.