Abstract
Previous studies on enhanced biological phosphorus removal (EBPR) have focused on reconstructing genomic blueprints for the model polyphosphate-accumulating organism Candidatus Accumulibacter phosphatis. Here, a time series metatranscriptome generated from enrichment cultures of Accumulibacter was used to gain insight into anerobic/aerobic metabolism and regulatory mechanisms within an EBPR cycle. Co-expressed gene clusters were identified displaying ecologically relevant trends consistent with batch cycle phases. Transcripts displaying increased abundance during anerobic acetate contact were functionally enriched in energy production and conversion, including upregulation of both cytoplasmic and membrane-bound hydrogenases demonstrating the importance of transcriptional regulation to manage energy and electron flux during anerobic acetate contact. We hypothesized and demonstrated hydrogen production after anerobic acetate contact, a previously unknown strategy for Accumulibacter to maintain redox balance. Genes involved in anerobic glycine utilization were identified and phosphorus release after anerobic glycine contact demonstrated, suggesting that Accumulibacter routes diverse carbon sources to acetyl-CoA formation via previously unrecognized pathways. A comparative genomics analysis of sequences upstream of co-expressed genes identified two statistically significant putative regulatory motifs. One palindromic motif was identified upstream of genes involved in PHA synthesis and acetate activation and is hypothesized to be a phaR binding site, hence representing a hypothetical PHA modulon. A second motif was identified ~35 base pairs (bp) upstream of a large and diverse array of genes and hence may represent a sigma factor binding site. This analysis provides a basis and framework for further investigations into Accumulibacter metabolism and the reconstruction of regulatory networks in uncultured organisms.
Introduction
Enhanced biological phosphorus removal (EBPR) is a widespread environmental biotechnology that exploits microorganisms capable of polyphosphate (polyP) accumulation to remove phosphorus (P) from wastewater (Bond et al., 1995; Hesselmann et al., 1999). The most widely studied organism responsible for EBPR in many wastewater treatment plants is named Candidatus Accumulibacter phosphatis (henceforth Accumulibacter; Nielsen et al., 2012). Although not yet isolated, a great deal has been learned about Accumulibacter physiology by studying enrichment cultures in laboratory scale bioreactors. Engineers have used this information to build quantitative metabolic models to predict how carbon, P, energy and reducing equivalents move through the wastewater ecosystem (Comeau et al., 1986; Oehmen et al., 2010). The accuracy and utility of these models depends heavily on an accurate understanding of Accumulibacter physiology.
It is well established that alternating cycles of carbon rich (feast) anerobic and carbon poor (famine) aerobic environments are essential for successful EBPR operation (Oehmen et al., 2007). Under anerobic conditions, Accumulibacter transports short-chain fatty acids (for example, acetate and propionate) into the cell and stores the carbon as polyhydroxyalkanoates (PHA). Current metabolic models generally assume that the energy for this process is obtained from ATP generated through polyP and glycogen degradation as well as from reducing equivalents generated through the degradation of glycogen and the anerobic operation of the TCA cycle (Filipe and Daigger, 1998; Wexler et al., 2009; Zhou et al., 2009; Oehmen et al., 2010). Under subsequent aerobic conditions, PHA degradation supplies carbon and energy for growth and replenishment of glycogen and polyP storage molecules (Comeau et al., 1986; Mino et al., 1998).
The ability to store large quantities of polyP has led researchers to refer to organisms that display the aforementioned phenotype as polyphosphate-accumulating organisms. Although the ability to produce polyP and carbon storage polymers such as PHA and glycogen are phylogenetically dispersed traits (Wood and Clark, 1988; Reddy et al., 2003), by linking these metabolic processes and synchronizing them with key environmental conditions, Accumulibacter and other polyphosphate-accumulating organisms have a highly specialized and biotechnologically important phenotype that is the foundation of EBPR. To validate the underlying assumptions embedded in the metabolic models used by engineers, it is necessary to dissect the molecular mechanisms responsible for this synchronization.
The sequencing and completion of the first Accumulibacter genome (García Martín et al., 2006) has facilitated numerous transcriptional investigations, with the hypothesis that Accumulibacter's highly coordinated physiology is the result of dynamic gene expression. Changes in transcript abundances have been previously investigated with reverse transcriptase quantitative PCR, microarrays, and RNA-seq under both stable and perturbed conditions (He et al., 2010; He and McMahon, 2011; Mao et al., 2014). However, these previous studies either targeted a handful of specific genes or examined limited time points during the anerobic–aerobic cycle. Here we used high-resolution time series metatranscriptomics with next-generation RNA-seq to identify highly expressed/dynamic genes and to identify putative co-regulated gene clusters. We then used comparative genomics to identify putative regulatory sequences and explore the underlying control mechanisms of the EBPR phenotype. Our results further validated some previously hypothesized aspects of Accumulibacter metabolism, uncovered important metabolic pathways that have been previously overlooked, and identified two putative sequence motifs providing the first step in determining gene expression regulatory mechanisms in Accumulibacter.
Materials and methods
Reactor maintenance
A single bioreactor was used in this study. Detailed reactor description and operating conditions are provided in García Martín et al. (2006). Briefly, the sequencing batch reactor was operated with a 2-l working volume and was fed with a mineral medium with acetate as a primary carbon source. The hydraulic retention time was 12 h and the sludge retention time was 4 days. The anerobic/aerobic cycle time was 6 h with 140 min anerobic contact (sparging with N2 gas), 190 min aerobic contact (sparging with air) and 30 min settling time. Nitrification was inhibited using allylthiourea. For the experiment described herein, the cycle differed from normal operation in that acetate was fed over a 60-min period to elongate acetate contact. Representative phosphate, PHB and acetate profiles across the cycle are shown in Supplementary Figure 1A. Steady state operation is demonstrated by characteristic high anerobic and low aerobic P concentrations for the month before the experiment (Supplementary Figure 1B).
Chemical analysis
All chemical analyses were conducted during the same reactor cycle used for transcriptomic analysis (on 28 May 2013), except for hydrogen production assays, which were conducted after RNA-seq results were analyzed. To monitor the EBPR cycle, soluble phosphate, total suspended solids, volatile suspended solids and acetate were measured using previously described methods (Flowers et al., 2009). Polyhydroxyalkanoate analysis was performed using a gas chromatography–mass spectrometry as outlined previously (Comeau et al., 1988). Hydrogen production was measured under anerobic conditions using six batch tests conducted in 150-ml septum bottles with 25 ml of sludge. Three were negative controls in which no acetate was fed to get background hydrogen production rates and three were fed with 0.18 mmol of acetate. Hydrogen production was measured using a reduction gas analyzer (ta3000 Gas Analyzer, Trace Analytical, Ametek, Newark, DE, USA). To test the viability of glycine as a carbon source, anerobic batch tests were conducted in triplicate with negative control (no carbon addition), positive control (acetate addition) and glycine for a total of nine batch tests conducted in 60-ml septum bottles with 50 ml of sludge. Approximately 0.06 mmol of acetate and glycine were added respectively and phosphorus release was measured as previously described (Flowers et al., 2009).
Biomass sample collection and RNA extraction
Six biomass samples were collected across a single reactor cycle to capture key transition points in the EBPR cycle (Supplementary Figure 1A and Supplementary Table 1). Bulk biomass (2 ml) was collected in microcentrifuge tubes. Samples were centrifuged, supernatant removed and cell pellets flash frozen in dry ice and ethanol bath within 3 min of collection. RNA was extracted from the samples using an RNeasy kit (Qiagen, Valencia, CA, USA) with a DNase digestion step. RNA integrity and DNA contamination was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Community characterization
Fluorescence in situ hybridization was conducted using PAOMIX probes to target all Accumulibacter clades (Crocetti et al., 2000), Acc-I-444 to target clade IA, and Acc-II-444 to target clade IIA, as previously described (Flowers et al., 2009). Cells were counter-stained with 4′,6-diamidino-2-phenylindole (DAPI). A Zeiss Imager.Z2 equipped with an AxioCam MRm camera was used to image fluorescing cells, which were then enumerated using ImageJ software (Abràmoff et al., 2004).
Library
Ribosomal RNA (rRNA) was removed from 1 μg of total RNA using Ribo-Zero rRNA Removal Kit (Bacteria) (Epicentre, Madison, WI, USA). Libraries were generated using the Truseq Stranded mRNA sample preparation kit (Illumina, San Diego, CA, USA). Briefly, the rRNA-depleted RNA was fragmented and reversed transcribed using Superscript II (Invitrogen, Carlsbad, CA, USA), followed by second-strand synthesis. The fragmented cDNA was treated with end-repair, A-tailing, adapter ligation and 10 cycles of PCR amplification.
Sequencing
The libraries were quantified using KAPA Biosystem's next-generation sequencing library quantitative PCR kit and run on a Roche LightCycler 480 real-time PCR instrument. The quantified libraries were then prepared for sequencing on the Illumina HiSeq 2000 sequencing platform utilizing a TruSeq paired-end cluster kit, v3, and Illumina's cBot instrument to generate a clustered flowcell for sequencing. Sequencing of the flowcell was performed on the Illumina HiSeq 2000 sequencer using a TruSeq SBS sequencing kit 200 cycles, v3, following a 2 × 150 indexed run recipe. Sequence data were deposited at IMG/M under Taxon Object IDs 3300002341-3300002346.
Bioinformatics
Reads were quality trimmed and quality statistics were calculated using FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/; Supplementary Figure 2). Ribosomal RNA sequences were removed with SortMeRNA using six built in databases for bacterial, archaeal and eukaryotic small and large subunits (Kopylova et al., 2012). Reads that passed filtering were then mapped to the Accumulibacter clade IIA strain UW-1 (CAP2UW1; García Martín et al., 2006) chromosome and plasmids using the BWA mem algorithm with default parameters (Li and Durbin, 2009). Read counts were then calculated using HTseq with the ‘intersection strict' parameter (Anders et al., 2014). Read counts were normalized by total reads in the sequencing run, the number of reads that remained after rRNA filtering, and the fraction of total reads that aligned to the Accumulibacter genome (Supplementary Table 1). Non-rRNA reads represented between 35 and 65 percent of all reads. Reads were then converted to log base two reads per kilo base per million (RPKM; Mortazavi et al., 2008; Supplementary Tables 4 and 5). Before clustering, all genes that did not have at least one observation with a log2(RPKM) read count of one or greater from the minimum observation were removed from the data set. Of 4735 genes in strain UW-1, 3893 passed this filter. These genes were further binned into clusters of co-expressed genes (Supplementary Table 6, Supplementary Figures S3–S7) based on an uncentered Pearson similarity metric of expression profiles followed by a centroid linkage clustering method using Java Treeview (Saldanha, 2004; Supplementary Figure 8). Clusters of co-expressed genes were then manually curated in Java TreeView. The resulting clusters are henceforth referred to as trend categories and are named with characters (for example, Trend Category A). Trend categories were then classified into patterns (Table 1) through visual inspection and comparison with known solute and biopolymer transformations that occur during an EBPR cycle.
Table 1. A summary of the trend categories identified in this study and the patterns they display.
| Pattern | No. of genes | Trend categories | No. of trend categories |
|---|---|---|---|
| Anerobic acetate contact | 126 | Q,R | 2 |
| Redox transition | 697 | AA,BB,CC,DD,EE,FF,GG,HH,NN | 9 |
| Aerobic | 1844 | II,JJ,KK,PP,QQ,RR,SS,TT,VV,WW,XX,YY,AAA,BBB,CCC,HHH,III,JJJ,KKK,LLL,MMM | 21 |
| High phosphorus | 40 | O,P | 2 |
| Low phosphorus | 438 | F,EEE,GGG,NNN,OOO,PPP,QQQ,RRR,SSS,TTT,UUU,VVV | 12 |
| Sum | 3145 | — | 46 |
Identification of highly expressed and highly dynamic genes
Genes that displayed the highest relative transcript abundance and those that displayed the largest changes in relative transcript abundance were identified as follows: each gene was represented as a vector of six RNA relative transcript abundance values. The maximum value of this vector represents the maximum relative expression of that gene. The maximum minus the minimum value of this vector represents the relative change in abundance over the entire cycle. Using these statistics, the genes may be ranked by those that show the highest relative expression and largest relative change. On the basis of the distribution of maximum expression values, a cut-off of 350 was determined to identify the highly expressed and highly dynamic genes (Supplementary Figure 9).
Functional enrichment analysis
Numerous subsets of genes were identified in this investigation including highly expressed/dynamic genes and trend categories. To determine whether these subsets were enriched in specific functions, a bootstrap method was used to determine how the distribution of COG functions compared with a null model produced from 1000 randomly generated gene subsets of equal size to the gene subset in question. The randomly generated null models for each gene subset was then compared with the observed abundance using a one sided t-test.
Operons and upstream motif identification
Putative operons were determined using the following set of criteria: (1) genes must have the same orientation; (2) adjacent genes were co-expressed with a cut-off correlation of 0.7; and (3) there was an intergenic region of 1000 base pairs or less (Supplementary Table 7). For each identified trend category, upstream sequences of called operons were analyzed for putative upstream motifs using MEME (Bailey et al., 2009; meme input.fasta -bfile background.txt –mod zoops –evt 0.05 –dna –nmotifs 10 –minsites 3 –o output), with one motif identified in a specific trend category (Supplementary Table 7). Additional motif sites were identified on the basis of sequence homology using MAST (Bailey et al., 1998; mast meme.txt input.fasta -bfile background.txt -oc. -nostatus -remcorr -ev 10 -norc -m 1). In addition, a motif search was conducted on the highly dynamic genes that displayed the anerobic acetate contact (AAC) pattern.
Results
Community composition, chemical analysis and total raw read statistics
On the date samples were collected for transcriptomics, P removal exceeded 99% and carbon and P dynamics characteristic of EBPR systems were observed (Supplementary Figure 1A). Accumulibacter relative abundance measured by fluorescence in situ hybridization was 80% of total DAPI-stained cells and Clade IIA accounted for 99% of the total Accumulibacter cells. P measurements at the end of aerobic and anerobic phases during the month of the investigation indicated stable state operation (Supplementary Figure 1B).
Illumina sequencing of ribosomal-depleted total RNA resulted in 1 461 769 869 reads across six samples (Supplementary Table 1). Quality filtering of reads removed 695 865 184, resulting in 765 904 685 for downstream analysis. Resulting reads were then mapped to the finished Accumulibacter clade IIA strain UW-1 reference genome (García Martín et al., 2006; Flowers et al., 2013), where 104 844 897 reads were aligned.
Co-expression patterns during a single EBPR cycle
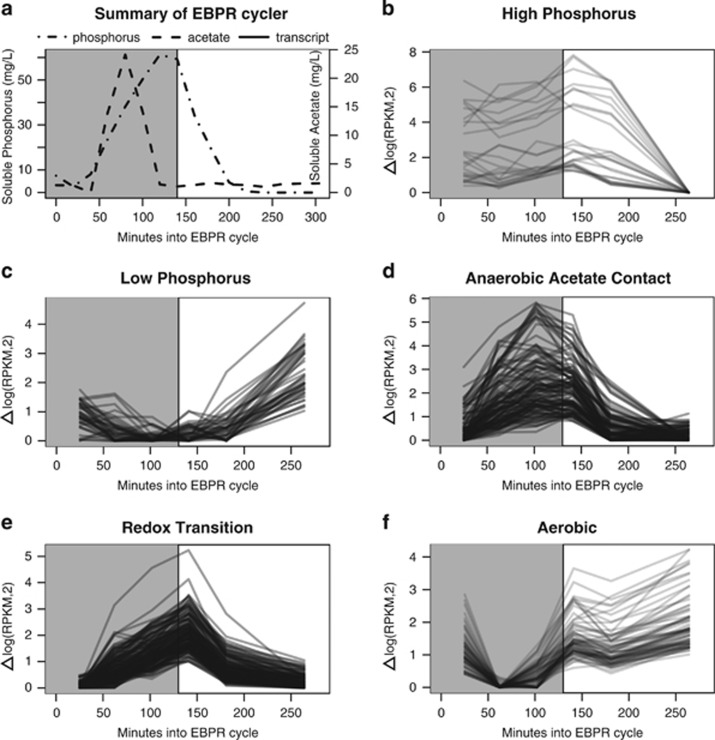
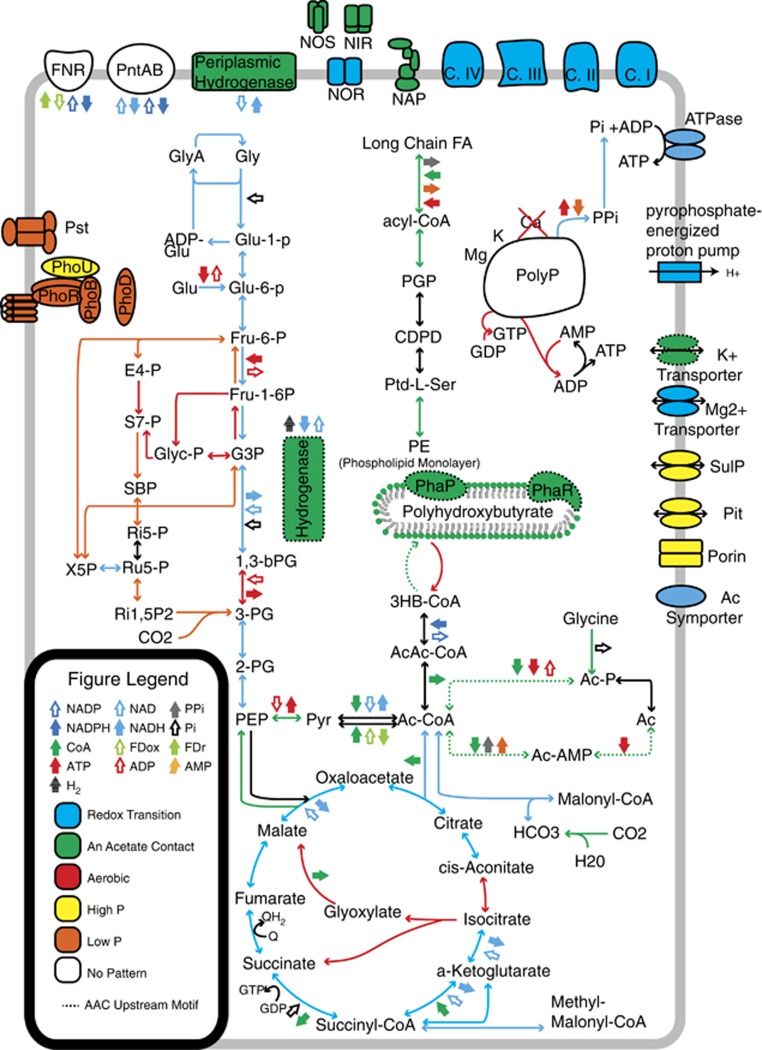
Sampling across a single EBPR cycle and subsequent hierarchical clustering analysis allowed the identification of clusters of co-expressed transcripts henceforth referred to as trend categories. Trend categories had an average Pearson correlation of 0.96 and an average size of ~50 genes (Supplementary Table 8,Supplementary Figure 3). Ecologically relevant patterns of transcript abundance were identified that corresponded with the following important EBPR stages: high phosphorus concentration (Figure 1b), low phosphorus concentration (Figure 1c), AAC (Figure 1d), redox transition (Figure 1e), aerobic (Figure 1f). The largest number of transcripts displayed the aerobic pattern, followed by redox transition, low phosphorus, AAC and finally high-phosphorus patterns. A summary of the number of genes, trend categories and which trend categories were assigned to each pattern are given in Table 1. Ecologically relevant expression profile patterns were overlaid on a map of central Accumulibacter metabolic processes to enable interpretation of how they might relate to regulation of key pathways (Figure 2, see Supplementary Figure 11 for model with locus tags). Biochemical transformations were color-coded on the basis of the expression pattern to which the corresponding gene was assigned.
Figure 1.
Time-series representation of a single EBPR cycle including soluble phosphorus and acetate (a) and gene expression profile patterns (b–f). Gray and white backgrounds represent anerobic and aerobic phases respectively. (b–f) Each panel depicts a single trend category that is representative of an ecologically relevant pattern. Genes were assigned to trend categories on the basis of co-expression analysis using hierarchical clustering, as explained in the Materials and methods. Trend categories were then binned into pattern groups with putative ecological relevance by manually inspecting the gene expression profiles relative to soluble phosphorus, acetate, PHB profiles as well as redox state (aerobic/anerobic). Each solid line represents the change in relative transcript abundance (measured as log(RPKM,2)) compared with its minimum value. (b) Transcripts displaying the high-phosphorus pattern had transcript abundance that were relatively high until the end of the aerobic phase when phosphorus was low. In this panel, they are represented by Trend Category P. (c) Transcripts displaying the low phosphorus patterns had transcript abundance that were relatively low until the end of the anerobic phase when phosphorus levels are low. In this panel, the transcripts within Trend Category PPP are representative of this pattern. (d) Transcripts displaying the anerobic acetate contact pattern increased drastically after acetate contact and peaked before oxygen contact. In this panel, the transcripts within Trend Category Q are representative of this pattern. (e) Transcripts displaying the redox transition pattern displayed a pattern of increasing abundance throughout the anerobic period, peaking after oxygen contact. In this panel, the transcripts within Trend Category DD are representative of this pattern. (f) Transcript displaying the aerobic trend category increased in relative abundance during the aerobic phase. In this panel, the transcripts within Trend Category RR are representative of this pattern.
Figure 2.
Updated metabolic model with biochemical reactions color-coded based on the expression profile pattern to which the corresponding gene was assigned. Genes involved in PHB formation demonstrate the anerobic acetate contact pattern and are colored green. Genes involved in the TCA cycle/glycolysis generally demonstrated high expression levels across the redox tansition (RT) and are colored blue. Genes involved in the Calvin Cycle demonstrated either the aerobic or low P patterns and are colored red and orange, respectively. Genes grouped into the high-phosphorus pattern are colored in yellow. These include low-affinity phosphate transporters. Ac, acetate; AcAc-CoA, acetoacetyl-CoA; Ac-CoA, acyl-CoA; Ac-AMP, acetyl AMP; Ac-P, acetyl-P; ADP-Glu, adenosine 5-diphosphoglucose; CDPD, cytidine diphosphate diacylglycerol; C.I, complex I oxidative phosphorylation; C.II, complex II oxidative phosphorylation; C.III, complex III oxidative phosphorylation; C.IV, complex IV oxidative phosphorylation; E4-P, erythrose 4-phosphate; FNR, NADPH-ferredoxin reductase; Fru-1-6P, fructose 1,6-bisphosphate; Fru-6-P, fructose 6-phosphate; G3P, glyceraldehyde 3-phosphate; Glu, glucose; Glu-1-p, glucose 1-phosphate; Glu-6-P, glucose 6-phosphate; Gly, glycogen; GlyA, glycogen amylose; Glyc-P, glycerone-P; Long Chain FA, long chain fatty acid; PE, phosphatidylethanolamine; PEP, phosphoenolpyruvate; PGP, 1,2-diacyl-sn-glycerol-3p; pntAB, proton-translocating transhydrogenase; PolyP, polyphosphate; PPP, pyrophosphate-energized proton pump; Ptd-L-Ser, phosphatidylserine; Pyr, pyruvate; 1,3-bPG, 1,3-bisphosphoglyceric acid; Ri15P2, ribulose 1,5P2; Ri5-P, ribose 5-phosphate; Ru5P, ribulose 5-phosphate; S7-P, sedoheptulose-7-phosphate; SBP, sedoheptulose 1,7-bisphosphate; X5P, xylulose 5-phosphate; 3HB-CoA, (R)-3-hydroxy-butanoyl-CoA; 2-PG, 2-phosphoglycerate; 3-PG, 3-phosphoglyceric acid.
Key genes identified as upregulated after AAC include those involved in acetate activation, PHB synthesis (CAP2UW1_3191) and regulation (phasins, CAP2UW1_0642-CAP2UW1_0643), glycine cleavage (CAP2UW1_1955-CAP2UW1_1960), phospholipid monolayer formation (CAP2UW1_3192, CAP2UW1_3266, CAP2UW1_3702, CAP2UW1_0341, CAP2UW1_2586), carbonic anhydrase (CAP2UW1_1967) and hydrogenases (CAP2UW1_0998-CAP2UW1_0999, CAP2UW1_2286). The presence of high P was accompanied by relatively high expression rates of various transporters such as low-affinity P transporters (Pit, CAP2UW1_2085), sulfur transporters (SulP, CAP2UW1_2094) as well as porins (CAP2UW1_1151, CAP2UW1_1152) and the regulatory phoU (CAP2UW1_2086, CAP2UW1_2093, CAP2UW1_3728). Low P conditions corresponded to increased relative transcript abundance of Calvin cycle genes as well as those involved in high-affinity P transporters (Pst, CAP2UW1_2002-CAP2UW1_2008) numerous regulatory genes including phoR (CAP2UW1_1995), phoB (CAP2UW1_1996) and phoD (CAP2UW1_1732).
Differential transcript abundances across COG categories in a single EBPR cycle
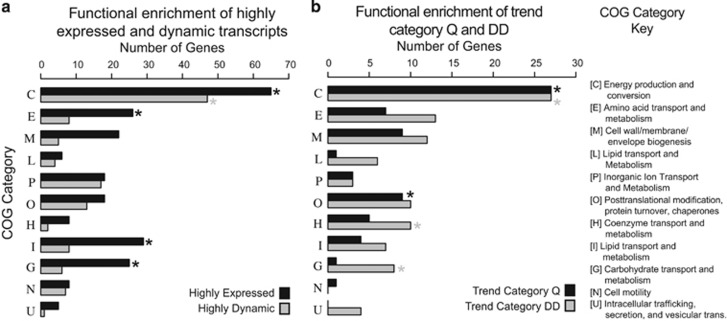
Numerous subsets of genes (transcripts) were identified in this investigation including highly expressed/dynamic genes (Supplementary Table S10) as well as Trend Category Q and DD (Supplementary Table S6), in which genes related to Energy Production and Conversion were enriched (P-values 5.7e−26, 1.6e−10, 1.9e−13 and 2.3e−08, respectively) (Figures 3a and b). Furthermore, in each of these gene subsets, energy production and conversion represented the largest fraction of genes with predicted functions (Figures 3a and b). Additional details are located in the Supplementary Material.
Figure 3.
Bar plots of the number of genes from each COG category in various gene subsets. Stars indicate statistically significant enrichment of a COG category over the expected number given the background abundance of each COG category in the CAP2UW1 genome. (a) The top 350 most highly expressed and dynamic genes. (b) Trend Categories Q and DD.
Hydrogen gas production and glycine utilization in Accumulibacter
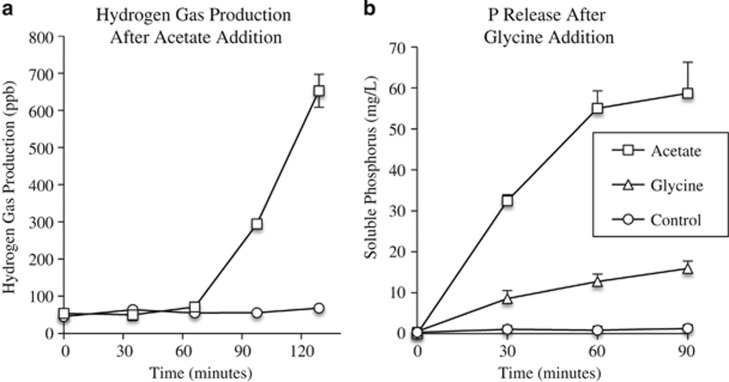
On the basis of the transcriptional profile of hydrogenases and a glycine cleavage operon detected in this study, we hypothesized that hydrogen gas production would occur during anerobic conditions after acetate addition and that glycine is a viable carbon source and would anerobic P release. To test these hypotheses, two sets of batch tests were conducted. Hydrogen gas production after acetate addition was measured and confirmed above background anerobic hydrogen gas production levels (Figure 4a). In addition, anerobic glycine addition resulted in P release, albeit at a lower rate than achieved by acetate contact (Figure 4b).
Figure 4.
(a) Hydrogen production assay demonstrating low background levels of anerobic hydrogen production without any carbon addition. Acetate addition produces elevated hydrogen production. Hydrogen production after acetate addition may be owing to the activity of a cytoplasmic hydrogen dehydrogenase restoring the NADH/NAD imbalance caused by glycogen degradation anerobically. (b) Batch tests were conducted to test the viability of glycine as a carbon source for Accumulibacter. Phosphorus release after carbon contact was measured for acetate, glycine and a no carbon addition control. These results demonstrate that glycine addition stimulates phosphorus release and is therefore a viable carbon source for Accumulibacter.
Upstream sequence motif identification
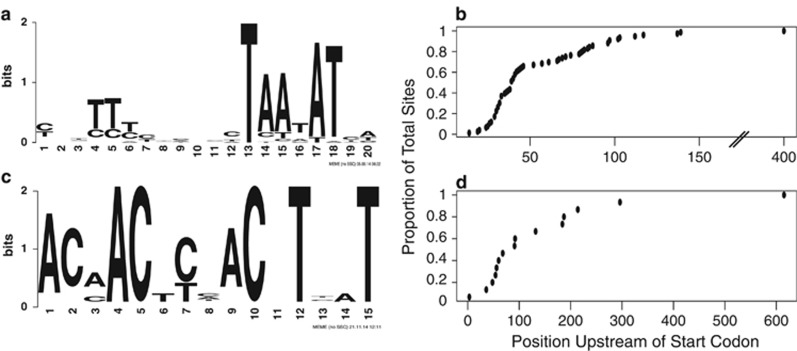
To identify genes putatively co-regulated by cis-regulatory elements, an upstream motif analysis was conducted. A sequence motif was identified upstream of 51 sequences within Trend Category DD using MEME (each with a P-value <9.07e−04) and upstream of 25 additional genes using MAST (each with a P-value <1.44e−04; Figure 5a,Supplementary Table S9; Bailey et al., 2009, 1998). A majority of the sequence motifs (~60%) were found between 25 and 45 base pairs upstream of the start codon (Figure 5b). In addition, a motif analysis using MEME conducted on the subset of genes within the AAC that were designated as highly dynamic revealed a palindromic motif upstream of 10 genes (each with a P-value <1.58 e-06) and an additional 5 genes were identified with MAST (each with a P-value <1.20e-04; Figure 5c, Supplementary Table S9; Bailey et al., 2009, 1998). The genes found to share this upstream motif were generally (~50%) between 45 and 95 base pairs downstream; however, the spacing ranged considerably (Figure 5d).
Figure 5.
(a) Motif diagram showing a putative sigma-binding site identified from a subset of genes within Trend Category DD. (b) Positions of putative sigma-binding site motif from a. (c) Motif diagram showing a palindromic motif identified from a subset of the highly dynamic genes from Trend Category Q. This motif may represent a binding site for PhoR, a known regulatory protein involved in PHA synthesis. (d) Positions of palindromic motif identified from highly dynamic genes displaying from Trend Category Q.
Discussion
Carbon metabolism in accumulibacter
One of the defining features of Accumulibacter metabolism is anerobic intracellular carbon flux. Bulk analysis of Accumulibacter-enriched cultures consistently demonstrates that as VFAs are transported into the cell, PHAs are synthesized and glycogen is degraded (Oehman et al., 2007). Therefore, we expected to find genes involved in the flux of carbon through acetyl-CoA and then to PHB to be upregulated during AAC. Interestingly, no evidence for an immediate transcriptome-level response upregulating genes involved in acetate acquisition via active (acetate permease, actP, CAP2UW1_1608) or passive (porins, CAP2UW1_1151 and CAP2UW1_1152) transport was identified upon initial acetate contact (that is, early in the cycle). However, acetate uptake triggered the upregulation of numerous other genes directly related to intracellular acetate and PHA processing. Once inside the cell, acetate is activated via the low-affinity acetate phosphotransferase (CAP2UW1_1002) and high-affinity acyl-coenzyme A (CoA) synthetase (CAP2UW1_3266) to acetyl-CoA, both of these transcripts exhibit the AAC pattern (Figures 1d and 2). Other pathways to acetyl-CoA also exhibited the AAC pattern including pyruvate kinase (CAP2UW1_0821), phosphoenolpyruvate carboxykinase (CAP2UW1_1298) and an anerobic glycine cleavage system operon (CAP2UW1_1955-CAP2UW1_1960; Figures 1d and 2). These findings suggest that Accumulibacter transcriptionally regulates gene expression to route as much carbon as possible toward acetyl-CoA formation via several different pathways. This is discordant with conventional metabolic models for Accumulibacter that aim to identify a single (or primary) route.
For example, the anerobic expression of the glycine cleavage system suggests an important role for glycine, and potentially other non-VFAs carbon sources, in Accumulibacter metabolism. Some previously reported experimental evidence from full scale systems provides support for this hypothesis: glycine addition resulted in the highest P release of any tested amino acid in batch tests with activated sludge (Wilinński, 2009). Furthermore, we conducted batch tests using Accumulibacter-enriched sludge and confirmed that glycine addition results in P release (Figure 4b). Thus, although free glycine contributes to the carbon budget of Accumulibacter, it remains unclear whether Accumulibacter may be able to liberate glycine from more complex sources. For example, one possible complex source of glycine would be collagen, which has an important role in bacterial biofilm formation (Oliver-Kozup et al., 2013, 2011). Every third amino acid in bacterial collagen is composed of glycine (Yu et al., 2014). Interestingly, a predicted collagenase/peptidase operon (CAP2UW1_0989-CAP2UW1_0991) was highly expressed and clustered within the AAC. Together the glycine cleavage and collagenase/peptidase operon represent a possible mechanism for the release and acquisition of glycine via peptidase/collagenase activity and subsequent anerobic glycine oxidization, thus providing additional acetate and reducing equivalents through Strickland reactions (Sagers and Gunsalus, 1960; Okamura-Ikeda et al., 1993; Andreesen, 1994). The specificity of this predicted collagenase/peptidase should be further investigated to understand its role during anerobic carbon metabolism in Accumulibacter. No mechanism for collagen synthesis was identified in Accumulibacter; however, collagen and other peptides produced by either Accumulibacter or the other bacterial community members may provide an important and previously unrecognized carbon source anerobically.
Regardless of its initial form, once the carbon source has been transformed into acetyl-CoA, three additional enzymes are required for PHA synthesis: β-ketothiolase (PhaA), acetoacetyl-CoA reductase (PhaB) and PHA synthase (PhaC), with PhaC acting as the key enzyme catalyzing the polymerization of hydroxyacyl-CoA (Peoples and Sinskey, 1989). In addition, PHA granule formation requires the synthesis of a phospholipid monolayer as well as the presence of PHA-associated proteins (phasins) to stabilize and guide PHA granule formation (Jendrossek, 2009). Indeed, PhaC (CAP2UW1_3191), phasins (PhaP, CAP2UW1_0642-CAP2UW1_0643) and numerous genes involved in the formation of phospholipids including the initial activation step (Mashek et al., 2007; long chain fatty acid CoA ligase; CAP2UW1_3192, CAP2UW1_3266), an intermediate step (1-acyl-sn-glycerol-3-phosphate acyltransferase; CAP2UW1_3702, CAP2UW1_0341) and the final step in phosphatidylethanolamine synthesis (phosphatidylserine decarboxylase; CAP2UW1_2586), all followed the AAC pattern (Figures 1d and 2). Phosphatidyl-ethanolamine is the most common phospholipid in E. coli (Raetz, 1978), and its synthesis in Accumulibacter under anerobic conditions may explain the net increase of fatty acids previously reported during the anerobic phase (Wexler et al., 2009). Intriguingly, a known PHA synthesis regulatory protein PhaR (Jendrossek, 2009) displayed the AAC pattern and its potential role regulating genes within the AAC is discussed below in the section describing regulatory sequence motif detection.
Another intriguing finding within the AAC pattern related to carbon metabolism is a carbonic anhydrase (CAP2UW1_1967) that shows homology with recently characterized carboxysome shell carbonic anhydrases (Heinhorst et al., 2006). Other carbonic anhydrases are also expressed (CAP2UW1_1300, CAP2UW1_4260, CAP2UW1_2752, CAP2UW1_3656, CAP2UW1_4334, CAP2UW1_1398, CAP2UW1_1977, CAP2UW1_2924) and show varying expression profiles. However, only CAP2UW1_1967 is highly expressed, dynamic and within the AAC. The biological relevance of carbonic anhydrase in Accumulibacter is unclear; however, carbonate produced via carbonic anhydrase activity may be used for the carboxylation of acetyl-CoA producing malonyl-CoA via an acetyl-CoA carboxylase (CAP2UW1_1136-CAP2UW1_1137) that clusters within the redox transition expression pattern (Supplementary Table S6). Interestingly, this operon also contains a methylmalonyl-CoA mutase (CAP2UW1_1139), which may link fatty acids synthesis and degradation to the TCA cycle. Further investigations using labeled substrates under diverse conditions could help elucidate whether Accumulibacter fixes carbon using acetyl-CoA carboxylase and under what conditions.
Anerobic reducing equivalents and energy metabolism
During anerobic carbon metabolism, energy generation and conversion reactions must provide the reducing equivalents and ATP needed to drive the specialized metabolism of Accumulibacter, and much research effort has been directed at quantifying these (Comeau et al., 1986; Mino et al., 1998; Oehmen et al., 2007). Indeed, a salient characteristic of the highly expressed and highly dynamic gene subsets as well as Trend Category Q and DD was the enrichment of genes found in the Energy Production and Conversion COG Category (Figures 3a and b). We were particularly intrigued by the fact that multiple oxidoreductases were among the most highly expressed and most dynamic genes assigned to these patterns, since oxidoreductases are key to managing the movement of reducing equivalents through the cell. This prompted us to look more closely at their potential role in Accumulibacter metabolism as revealed through expression patterns.
Current EBPR models explicitly require that anerobic reducing equivalents for PHA synthesis be produced through glycogen degradation (during oxidation of glyceraldehyde-3-phosphate) and/or anerobic operation of the TCA cycle (oxidation of isocitrate, alpha-ketoglutarate and malate), in the form of NADH (Hesselmann et al., 2000; Zhou et al., 2009). When intracellular stores of glycogen are limiting, acetate flux through the TCA cycle increases via the oxidation of isocitrate, alpha-ketoglutarate and malate (Zhou et al., 2009). However, reducing equivalents provided by glycolysis and the TCA cycle are in the form of NADH while PHA synthesis generally requires NADPH (Peoples and Sinskey, 1989; Steinbüchel et al., 1993; Madison and Huisman, 1999; Kim et al., 2014). Thus, existing models include an implicit assumption that an NAD(P)H transhydrogenase converts NADH to NADPH as needed to maintain redox homeostasis.
An increase in transcript abundance of cytoplasmic Ni–Fe hydrogen dehydrogenase (CAP2UW1_0998-CAP2UW1_0999) and membrane-bound hydrogenase (CAP2UW1_2286) (Figure 2) within the AAC suggested a potential role of hydrogen production for balancing the redox state of the cells during anerobic acetate uptake and storage. We investigated this possibility by measuring hydrogen gas production after anerobic carbon contact. Acetate addition resulted in quantifiable hydrogen gas production in comparison to background experiments that did not receive anerobic acetate addition (Figure 4a).
Therefore, we hypothesize that upon glycogen degradation (1) the NADH/NAD+ ratio increases and recycling of NAD+ is accomplished by hydrogenase activity that produces free H2, (2) NADPH is the cofactor required for PhaB activity, and transhydrogenase activity is involved in the conversion of NADH to NADPH, (3) the supply of NADH is greater than what is required or cannot be converted to NADPH quickly enough and (4) in addition to requiring reducing equivalents, other factors such as anerobic demand for glycolysis-derived ATP (Saunders et al., 2007) drive glycogen degradation. In support of (2), we confirmed that NAD(P)H transhydrogenase (CAP2UW1_4179, CAP2UW1_4180) was highly expressed. However, curiously the two subunits show slight relative upregulation in the aerobic phase. Enzymatic assays testing the NADH/NADPH specificity of Accumulibacter PhaB and measurements of NAD(P)H/NAD(P) ratios throughout an EBPR cycle should also help test these hypotheses.
Low/high soluble phosphorus and carbon correlated gene expression
During aerobic metabolism within an EBPR cycle both nutrient rich (feast) and poor (famine) states exist. Immediately after first oxygen contact, the environment is nutrient rich; P and C are abundant extracellularly and intracellularly (as stored PHB) respectively. As the aerobic phase continues, P is transported and stored intracellularly while PHB is degraded to drive P uptake, polyP/glycogen synthesis and growth. Thus, at the end of the aerobic phase, both P and C may be considered limiting as extracellular P and PHB are depleted.
The high-phosphorus pattern (Figure 1b) included the most highly transcribed gene in Accumulibacter, a porin operon (CAP2UW1_1151, CAP2UW1_1152), as well as inorganic phosphate transporters (Pit, CAP2UW1_2085) and sulfur transporters within two operons (CAP2UW1_2085-CAP2UW1_2087, CAP2UW1_2092-2094) (Figure 2). The high relative abundance of Pit, SulP and porin transcripts during periods of high soluble P concentrations is consistent with the high flux of metabolites, such as P and counter cations, into and out of the cell across the anerobic/aerobic phases of an EBPR cycle. The Pit/SulP operons also contained multiple phoU-like genes (Liu et al., 2005; Oganesyan et al., 2005). Co-expression of PhoU and Pit during periods with high soluble P concentrations are consistent with PhoU's hypothesized role as a negative regulator for the PhoR-PhoB two-component system (Baek et al., 2007). During periods with low P, phoU transcripts decreased and the transcription of the high-affinity P transporter system occurred (Pst, CAP2UW1_2002-CAP2UW1_2008) as well as phoR (CAP2UW1_1995), phoB (CAP2UW1_1996) and phoD (CAP2UW1_1732) (Figure 2).
When P levels are low at the end of the aerobic phase, measured PHB levels are also below detection (Supplementary Figure 1). During this low phosphorus (Figure 1C) or ‘carbon starved' state, we identified an increase in the relative abundance of Ribulose 1,5-bisphosphate carboxylase (Rubisco; CAP2UW1_0825). Multiple genes involved in the Calvin cycle exhibited a similar expression pattern (CAP2UW1_0959, CAP2UW1_0958, CAP2UW1_0957, CAP2UW1_0823) (Figure 2). The potential for carbon fixation by the Accumulibacter lineage has been hypothesized since the original metagenome was sequenced (García Martín et al., 2006) and further supported by the sequencing of additional draft genomes (Skennerton et al., 2014). In addition, experiments have shown that Accumulibacter enrichments may be sustained in the absence of organic substrates in the medium (Kang and Noguera, 2014). Investigations into the ability of Accumulibacter to fix carbon during aerobic conditions when PHAs have been exhausted must be conducted to confirm this potential. Furthermore, perturbation experiments decoupling low P and low C conditions will be important in further distinguishing co-expression patterns.
New insights into regulatory mechanisms in accumulibacter
Upstream sequence motif identification
Dynamic gene expression is an adaptive mechanism that allows organisms to respond to changing environmental conditions (Seshasayee et al., 2009). By regulating genes at the level of transcription, organisms may prevent the energy waste associated with the synthesis of unnecessary proteins (Stoebel et al., 2008), or the negative effects of certain proteins when synthesized under the inappropriate conditions (Eames and Kortemme, 2012). Co-expression clusters determined through transcriptomic analysis may be used in conjunction with comparative genomics methods to identify sequence motifs that represent putative regulatory features (Kellis et al., 2003; Imam et al., 2014). We hypothesized that genes following a particular expression profile would be under the same regulatory mechanisms and may therefore share upstream sequence motifs.
To explore such regulatory features, we analyzed the upstream sequences of operons within each trend category. A sequence motif was identified upstream of 51 sequences within Trend Category DD and a sequence homology search identified an additional 25 locations of the motif. (Figure 5a,Supplementary Table S9). We hypothesize that this motif is a putative sigma factor binding site because a large number of the motif occurrences are found ~35 bp upstream (Figure 5b) of the transcriptional start site (Harley and Reynolds, 1987). No strong -10 site was identified in this study, suggesting that the promoter associated with this putative DNA binding domain may have a reduced requirement for a -10 binding region (Thouvenot et al., 2004; Hook-Barnard and Hinton, 2007). However, the biological relevance of this motif must be tested using additional methods such as DNase-seq (He et al., 2014).
We also identified a palindromic sequence motif upstream of 10 genes binned in the AAC pattern and subsequent homology searches identified an additional six locations of the motif (Figure 5c). Genes found to have this upstream motif included those involved directly in PHA synthesis, such as phaC (CAP2UW1_3191), phaP (CAP2UW1_0642-CAP2UW1_0643), phaR (CAP2UW1_3918), both low and high-affinity acetate activation enzymes (CAP2UW1_1002, CAP2UW1_3266), a potassium transporter (CAP2UW1_2100), and an operon containing the cytoplasmic hydrogenase (CAP2UW1_1001- CAP2UW1_0998). These observations provide yet additional evidence for the co-regulation of hydrogen gas production with acetate uptake and PHA synthesis. Therefore, we hypothesize that this group of genes may represent a co-regulated PHA synthesis modulon. The inclusion of phaR within this modulon is especially interesting as PhaR is found to bind upstream of both itself and phaP and thus negatively regulates transcription in the model PHA accumulating organism Ralstonia eutropha (Potter et al., 2002). When PHA synthesis is occurring, PhaR is recruited away from DNA to the growing PHA granules, releasing transcriptional inhibition (Jendrossek, 2009). DNA footprinting (Hampshire et al., 2007) using a purified PhaR protein is necessary to confirm if the motif identified here is indeed a PhaR binding site.
The identification of the first putative regulatory motifs in Accumulibacter represents a milestone that will invite further investigation to (1) determine whether the sequence motifs are true transcription factor binding sites, (2) identify the transcriptional regulatory protein associated with these sites, (3) determine whether regulation is via activation or repression. Transcriptomic analysis of Accumulibacter under diverse conditions will result in further differentiating clusters of co-expressed genes, giving additional power to comparative methods to identify putative regulatory mechanisms.
Previous proteomics-based research has demonstrated that relative protein abundances do not change markedly across an EBPR cycle (Wilmes, et al., 2008, 2008; Wexler et al., 2009). In contrast, we demonstrated that the stable protein abundances observed through the cycle are maintained via bursts of high mRNA productivity at specific, environmentally triggered times during the cycle rather than sustained levels of expression. Further, we showed that particular functions, such as those related to energy production and conversion (Figures 3a and b) display highly dynamic transcripts over an EBPR cycle.
Conclusion
Metatranscriptomic sequencing was conducted on six samples collected across an aerobic/anerobic EBPR cycle in a bioreactor enriched in Accumulibacter clade IIA. Despite relatively stable protein abundance identified in previous studies (Wexler et al., 2009), the identification of numerous co-expressed gene sets provides strong evidence that transcriptional regulation is critical for the anerobic/aerobic metabolism of Accumulibacter. AAC triggered the expression of genes related to acetyl-CoA and PHB formation, including genes involved with anerobic glycine metabolism. These findings suggest that Accumulibacter routes more diverse carbon sources through acetyl-CoA to PHB than previously recognized.
The discovery of hydrogenase expression and the demonstration of hydrogen gas production highlight previously unknown components of the EBPR cycle, and indicate that a redox imbalance may exist during AAC. We suggest that reducing equivalents in the form of NADH from glycogen degradation cannot be used directly for PHB synthesis, but rather NADPH is required by PhaB for the reduction of acetoacetyl-CoA to (R)-3-hydroxybutyryl-CoA, as commonly observed in other organisms. Thus, efficient conversion of NADH to NADPH may be a rate-limiting step that has not been adequately recognized in metabolic models of Accumulibacter (Niederholtmeyer et al., 2010; Angermayr et al., 2012). This is an excellent example of how discovery-based sequencing can reveal new metabolic features.
Comparative genomics of upstream sequences in co-regulated gene sets allowed, for the first time, the identification of two sequence motifs in Accumulibacter. The first was a palindromic motif upstream of genes showing upregulation during AAC and involved in acetate activation, PHA granule formation, fatty acid synthesis, counter cation transport and reducing equivalent balance through hydrogen gas production. The second motif was identified as a putative sigma factor binding site upstream of many genes/operons within the redox transition pattern. The discovery of this putative regulatory motif suggests that portions of acetate uptake, PHA synthesis, and P metabolism (counter cation transport) are co-regulated. Additional metatranscriptomic analyses may further identify regulatory mechanisms within Accumulibacter and the regulons associated with the unique Accumulibacter metabolism.
Acknowledgments
We thank the following individuals for insightful discussion and friendly reviews of draft manuscripts: Christopher Lawson, Francisco Moya and Travis Korosh. We thank Alisha Truman, Mitch Heffernan, Antonio Garcia and Lianne Estrella for assistance with bioreactor operation. KDM acknowledges funding from the US National Science Foundation (CBET-0967646) and the UW-Madison Graduate School. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abràmoff MD, Magalhães PJ, Ram RJ. (2004). Image Processing with ImageJ. Biophotonics 11: 36–43. [Google Scholar]
- Anders S, Pyl PT, Huber W. (2014). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreesen JR. (1994). Glycine metabolism in anaerobes. Antonie Van Leeuwenhoek 66: 223–237. [DOI] [PubMed] [Google Scholar]
- Angermayr SA, Paszota M, Hellingwerf KJ. (2012). Engineering a cyanobacterial cell factory for production of lactic acid. Appl Environ Microbiol 78: 7098–7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek JH, Kang YJ, Lee SY. (2007). Transcript and protein level analyses of the interactions among PhoB, PhoR, PhoU and CreC in response to phosphate starvation in Escherichia coli. FEMS Microbiol Lett 277: 254–259. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske Fa, Frith M, Grant CE, Clementi L et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M, Diego S, Box PO. (1998). Combining evidence using P-values: application to sequence homology searches. Bioinformatics 14: 48–54. [DOI] [PubMed] [Google Scholar]
- Bond PL, Hugenholtz P, Keller J, Blackall LL, Keller RG, Blackall LL. (1995). Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol 61: 1910–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau Y, Hall KJ, Hancock REW, Oldham WK. (1986). Biochemical model for enhanced biological phosphorus removal. Water Res 20: 1511–1521. [Google Scholar]
- Comeau Y, Hall KJ, Oldham WK. (1988). Determination of poly-3-hydroxybutyrate and poly-3-hydroxyvalerate in activated sludge by gas-liquid chromatography. Appl Environ Microbiol 54: 2325–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocetti GR, Hugenholtz P, Bond PL, Schuler A, Keller J, Jenkins D et al. (2000). Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl Environ Microbiol 66: 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames M, Kortemme T. (2012). Cost-benefit tradeoffs in engineered lac operons. Science 336: 911–915. [DOI] [PubMed] [Google Scholar]
- Filipe DMC, Daigger GTD. (1998). Filipe 1998 WR.pdf. Water Environ Res 70: 67–79. [Google Scholar]
- Flowers JJ, He S, Malfatti S, del Rio TG, Tringe SG, Hugenholtz P et al. (2013). Comparative genomics of two ‘Candidatus Accumulibacter' clades performing biological phosphorus removal. ISME J 7: 2301–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JJ, He S, Yilmaz S, Noguera DR, McMahon KD. (2009). Denitrification capabilities of two biological phosphorus removal sludges dominated by different ‘Candidatus Accumulibacter' clades. Environ Microbiol Rep 1: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Martín H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC et al. (2006). Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol 24: 1263–1269. [DOI] [PubMed] [Google Scholar]
- Hampshire AJ, Rusling DA, Broughton-Head VJ, Fox KR. (2007). Footprinting: a method for determining the sequence selectivity, affinity and kinetics of DNA-binding ligands. Methods 42: 128–140. [DOI] [PubMed] [Google Scholar]
- Harley CB, Reynolds RP. (1987). Analysis of E. coli promoter sequences. Nucleic Acids Res 15: 2343–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Hu SS, Chen M-W, Zang C, Liu Y et al. (2014). Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods 11: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Kunin V, Haynes M, Martin HG, Ivanova N, Rohwer F et al. (2010). Metatranscriptomic array analysis of ‘Candidatus Accumulibacter phosphatis'-enriched enhanced biological phosphorus removal sludge. Environ Microbiol 12: 1205–1217. [DOI] [PubMed] [Google Scholar]
- He S, McMahon KD. (2011). ‘Candidatus Accumulibacter' gene expression in response to dynamic EBPR conditions. ISME J 5: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinhorst S, Williams EB, Cai F, Murin CD, Shively JM, Cannon GC. (2006). Characterization of the carboxysomal carbonic anhydrase CsoSCA from Halothiobacillus neapolitanus. J Bacteriol 188: 8087–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselmann RPX, Rummell R, Von, Resnick SM, Hany R, Zehnder JB. (2000). Anaerobic metabolism of bacteria performing enhanced biological phosphate removal. Water Res 34: 3487–3494. [Google Scholar]
- Hesselmann RPX, Werlen C, Hahn D, Roelof, Van Der Meer J, Zehnder AJB. (1999). Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst Appl Microbiol 22: 454–465. [DOI] [PubMed] [Google Scholar]
- Hook-Barnard IG, Hinton DM. (2007). Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters the multi-step process of transcription initiation. Gene Regul Syst Biol 1: 275–293. [PMC free article] [PubMed] [Google Scholar]
- Imam S, Noguera DR, Donohue TJ. (2014). Global analysis of photosynthesis transcriptional regulatory networks viollier, PH (ed). PLoS Genet 10: e1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D. (2009). Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191: 3195–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Noguera DR. (2014). Candidatus Accumulibacter phosphatis: elusive bacterium responsible for enhanced biological phosphorus removal. J Environ Eng 140: 2–10. [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. (2003). Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254. [DOI] [PubMed] [Google Scholar]
- Kim J, Chang JH, Kim E-J, Kim K-J. (2014). Crystal structure of (R)-3-hydroxybutyryl-CoA dehydrogenase PhaB from Ralstonia eutropha. Biochem Biophys Res Commun 443: 783–788. [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H. (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28: 3211–3217. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lou Y, Yokota H, Adams PD, Kim R, Kim S-H. (2005). Crystal structure of a PhoU protein homologue: a new class of metalloprotein containing multinuclear iron clusters. J Biol Chem 280: 15960–15966. [DOI] [PubMed] [Google Scholar]
- Madison LL, Huisman GW. (1999). Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev 63: 21–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Yu K, Xia Y, Chao Y, Zhang T. (2014). Genome Reconstruction and Gene Expression of ‘Candidatus Accumulibacter phosphatis' Clade IB Performing Biological Phosphorus Removal. Environ Sci Technol 48: 10363–10371. [DOI] [PubMed] [Google Scholar]
- Mashek DG, Li LO, Coleman RA. (2007). Long-chain acyl-CoA synthetases and fatty acid channeling. Future Lipidol 2: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino T, van Loosdrecht MCM, Heijnen JJ. (1998). Microbiology and biochemistry of the enhanced biological phosphate removal. Water Res 32: 3193–3207. [Google Scholar]
- Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Niederholtmeyer H, Wolfstädter BT, Savage DF, Silver PA, Way JC. (2010). Engineering cyanobacteria to synthesize and export hydrophilic products. Appl Environ Microbiol 76: 3462–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PH, Saunders AM, Hansen AA, Larsen P, Nielsen JL. (2012). Microbial communities involved in enhanced biological phosphorus removal from wastewater—a model system in environmental biotechnology. Curr Opin Biotechnol 23: 452–459. [DOI] [PubMed] [Google Scholar]
- Oehmen A, Carvalho G, Lopez-Vazquez CM, van Loosdrecht MCM, Reis MAM. (2010). Incorporating microbial ecology into the metabolic modelling of polyphosphate accumulating organisms and glycogen accumulating organisms. Water Res 44: 4992–5004. [DOI] [PubMed] [Google Scholar]
- Oehmen A, Lemos PC, Carvalho G, Yuan Z, Blackall LL, Reis MAM. (2007). Advances in enhanced biological phosphorus removal: from micro to macro scale. Microbiology 41: 2271–2300. [DOI] [PubMed] [Google Scholar]
- Oganesyan V, Oganesyan N, Adams PD, Jancarik J, Yokota Ha, Kim R et al. (2005). Crystal structure of the ‘PhoU-like' phosphate uptake regulator from Aquifex aeolicus. J Bacteriol 187: 4238–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura-Ikeda K, Ohmura Y, Fujiwara K, Motokawa Y. (1993). Cloning and nucleotide sequence of the gcv operon encoding the Escherichia coli glycine-cleavage system. Eur J Biochem 216: 539–548. [DOI] [PubMed] [Google Scholar]
- Oliver-Kozup HA, Elliott M, Bachert BA, Martin KH, Reid SD, Schwegler-Berry DE et al. (2011). The streptococcal collagen-like protein-1 (Scl1) is a significant determinant for biofilm formation by group A Streptococcus. BMC Microbiol 11: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Kozup H, Martin KH, Schwegler-Berry D, Green BJ, Betts C, Shinde AV et al. (2013). The group A streptococcal collagen-like protein-1, Scl1, mediates biofilm formation by targeting the extra domain A-containing variant of cellular fibronectin expressed in wounded tissue. Mol Microbiol 87: 672–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples OP, Sinskey AJ. (1989). Poly-P-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16 identification and characterization of the PHB polymerase gene (phbC). J Biol Chem 264: 15298–15303. [PubMed] [Google Scholar]
- Potter M, Madkour MH, Mayer F, Steinbuchel A. (2002). Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 148: 2413–2426. [DOI] [PubMed] [Google Scholar]
- Raetz CRH. (1978). Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol Rev 42: 614–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy CSK, Ghai R, Rashmi, Kalia VC. (2003). Polyhydroxyalkanoates: an overview. Bioresour Technol 87: 137–146. [DOI] [PubMed] [Google Scholar]
- Sagers RD, Gunsalus IC. (1960). Intermediary metabolism of diplococcus glycinophilus glycine cleavage interconversions and one-carbon. J Bacteriol 81: 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. (2004). Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Mabbett AN, McEwan AG, Blackall LL. (2007). Proton motive force generation from stored polymers for the uptake of acetate under anaerobic conditions. FEMS Microbiol Lett 274: 245–251. [DOI] [PubMed] [Google Scholar]
- Seshasayee ASN, Fraser GM, Babu MM, Luscombe NM. (2009). Principles of transcriptional regulation and evolution of the metabolic system in E. coli. Genome Res 19: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skennerton CT, Barr JJ, Slater FR, Bond PL, Tyson GW. (2014). Expanding our view of genomic diversity in Candidatus Accumulibacter clades. Environ Microbiol 17: 1574–1585. [DOI] [PubMed] [Google Scholar]
- Steinbüchel A, Hustede E, Liebergesell M, Pieper U, Timm A, Valentin H. (1993). Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids in bacteria. FEMS Microbiol Rev 10: 347–350. [DOI] [PubMed] [Google Scholar]
- Stoebel DM, Dean AM, Dykhuizen DE. (2008). The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics 178: 1653–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouvenot B, Charpentier B, Branlant C. (2004). The strong efficiency of the Escherichia coli gapA P1 promoter depends on a complex combination of functional determinants. Biochem J 383: 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler M, Richardson DJ, Bond PL. (2009). Radiolabelled proteomics to determine differential functioning of Accumulibacter during the anaerobic and aerobic phases of a bioreactor operating for enhanced biological phosphorus removal. Environ Microbiol 11: 3029–3044. [DOI] [PubMed] [Google Scholar]
- Wilinński PR. (2009). Kinetics and stoichiometry of P-release with different carbon sources in the anaerobic phase of the biological phosphorus removal process in activated sludge wastewater treatment plants. Masters Dissertation, Aalborg University, Denmark.
- Wilmes P, Andersson AF, Lefsrud MG, Wexler M, Shah M, Zhang B et al. (2008). Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME J 2: 853–864. [DOI] [PubMed] [Google Scholar]
- Wilmes P, Wexler M, Bond PL. (2008). Metaproteomics provides functional insight into activated sludge wastewater treatment. PLoS One 3: e1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood HG, Clark JE. (1988). Biological aspects of inorganic polyphosphates. Annu Rev Biochem 57: 235–260. [DOI] [PubMed] [Google Scholar]
- Yu Z, An B, Ramshaw JAM, Brodsky B. (2014). Bacterial collagen-like proteins that form triple-helical structures. J Struct Biol 186: 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Pijuan M, Zeng RJ, Yuan Z. (2009). Involvement of the TCA cycle in the anaerobic metabolism of polyphosphate accumulating organisms (PAOs). Water Res 43: 1330–1340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.