Abstract
Background
One strategy to increase the stability of proteins is to reduce the area of water-accessible hydrophobic surface.
Results
In order to test it, we replaced 14 solvent-exposed hydrophobic residues of acetylcholinesterase by arginine. The stabilities of the resulting proteins were tested using denaturation by high temperature, organic solvents, urea and by proteolytic digestion.
Conclusion
Altough the mutational effects were rather small, this strategy proved to be successful since half of the mutants showed an increased stability. This stability may originate from the suppression of unfavorable interactions of nonpolar residues with water or from addition of new hydrogen bonds with the solvent. Other mechanisms may also contribute to the increased stability observed with some mutants. For example, introduction of a charge at the surface of the protein may provide a new coulombic interaction on the protein surface.
Background
Acetylcholinesterase (AChE, EC 3.1.1.7) is a serine hydrolase, which catalyzes the hydrolysis of the neurotransmitter acetylcholine. This enzyme is irreversibly inhibited by organophosphate and carbamate pesticides leading to its use in biosensors to detect traces of these compounds in environment. Drosophila AChE was found to be the most sensitive enzyme when compared to enzymes of non-insect origin and in-vitro-mutagenesis was used to select enzymes up to 300-fold more sensitive [1,2]. But like most enzymes from mesophilic organisms, Drosophila AChE is not stable, and this instability precludes its utilization in biosensors. It can be stabilized by adding some molecules in the solution such as reversible inhibitors, polyethylene glycol or protein, provoking protein-protein interactions. Alternatively, stabilization may also be achieved by encapsulation in liposomes [3,7]. Another way to stabilize the enzyme is to use in vitro mutagenesis to modify the primary structure of the protein [8]. This method could have the additional advantage of stabilizing the enzyme during its synthesis leading to higher production and higher purification yields.
Irreversible denaturation of AChE at room temperature can be minimized by increasing the protein concentration in the sample, either by increasing the enzyme concentration or by addition of another protein such as BSA [6]. This suggested that denaturation occurred by interaction of the hydrophobic region at the surface of AChE with tube walls or air-solvent interface. Addition of protein in the solution would compete with hydrophobic surfaces and protect the enzyme. We thus hypothesized that decreasing the hydrophobicity at the surface of the protein may have some stabilizing effects.
Several examples showed that the change of hydrophobicity to hydrophilicity of amino-acid residues exposed to the solvent at the surface of proteins is an efficient stategy to stabilize proteins: i) Analysis of protein sequences showed a strong bias for hydrophilic residues and against large hydrophobic residues at most surface positions [9], ii) Mutagenesis showed that hydrophilic amino acids at surface positions is stabilizing Mutagenesis showed that hydrophilic amino acids at surface positions have stabilizing effect, while placing a hydrophobic residue in a solvent-exposed position causes destabilization [10-12], iii) The proportion of hydrophobic residues at the surface of proteins from hyperthermophilic species was found to be reduced compared to the proportion in their mesophilic counterparts [13-16], however, this observation is under debate [17].
Here we tested this strategy by mutating several hydrophobic residues scattered at the surface of Drosophila AChE to arginine. Hydrophobic residues were chosen by visual examination of the structure and arginine was chosen because the guanidinium group is the most polar of all the common amino-acid residues found in proteins.
Results and discussion
Effect of mutations on protein production
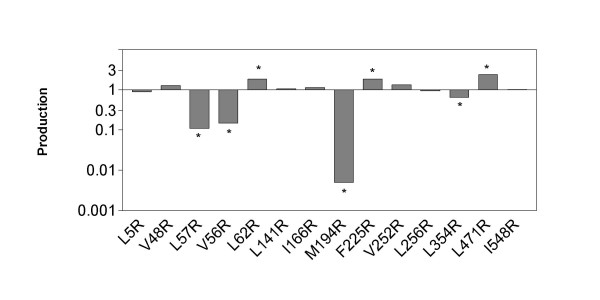
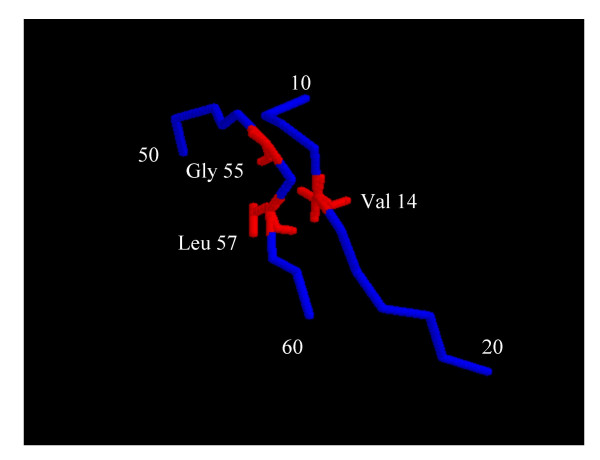
Production of the wild type was 60 nmoles per liter (5 mg/L). Several mutations affected the production of the protein (Fig. 1), most probably reflecting their influence on folding efficiency. One mutation drastically decreased production: the mutation M194R decreased production to 0.5% i.e. a level which was barely detectable. This amino-acid is partly buried behind proline 443 and we can assume that its hydrophobicity is important for the folding of the protein. The mutations V56R and L57R decreased production to 10%. It seems that these two residues are engaged in a hydrophobic bond with Val 14 helping to position the loop from Ala 50 to Thr 60 in connection with the N-terminus peptide of the protein (Fig. 2). Mutations to hydrophilic residue may disrupt the bond and thus decrease the probability of the protein reaching its active conformation. On the other hand, three mutations significantly increased the production: the mutations L62R, F225R and L471R. This increase of folding efficiency can be attributed to the reverse hydrophobic effect: if a hydrophobic residue is less exposed to the solvent in the denatured form than in the native form, it will oppose folding [18].
Figure 1.
Effect of replacement of hydrophobic residues at the surface by arginine on relative production. Ratio of production represents the relative production of each mutant.
Figure 2.
Position of Gly 55 and Leu 57, near Val 14.
Effect of mutations on protein stability
Stability was assayed with four denaturing agents. In all cases, denaturation was irreversible and followed apparent first order kinetics. Stability was then characterized by the half-life (t50), the time at which 50% of an initial enzymatic activity is preserved. The half life of the wild type protein is shown in Table 1. Stability was analyzed for all mutants except M194R for which the production was too low.
Table 1.
half-life (t50) of the wild type protein.
| Denaturing agent | t50 (min) |
| 50°C | 8 +/- 1.7 |
| 4 M Urea | 11 +/- 1.6 |
| 20% acetonitrile | 1.45 +/- 0.18 |
| 0.1 mg/ml pronase | 14.4 +/- 2.2 |
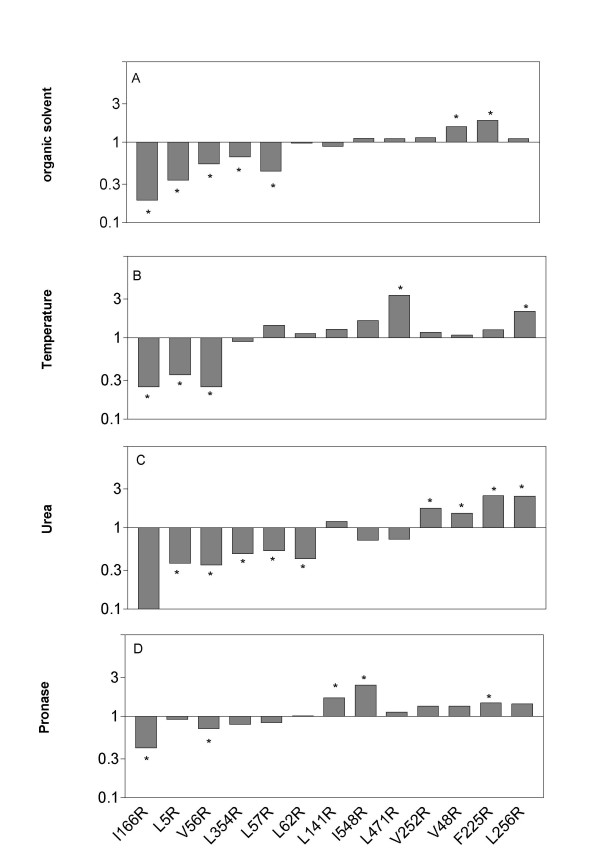
The effect of mutations on stability was homogeneous, a mutation either destabilizes or stabilizes the protein since we never found a mutation which significantly stabilizes the protein for one agent and significantly destabilizes it for another.
Most of the mutations significantly affect the stability of the protein (Fig. 3). However, differences were rather small. This is in accordance with literature, amino acid substitution usually does not significantly affect the stability [10,19], although important improvements of stability by mutagenesis of a single solvent-exposed residue have been reported [20,21].
Figure 3.
Effect of replacement of hydrophobic residues at the surface by arginine on relative stability. For each mutation, ratio of t50 (t50 mutant/t50 wild type) was calculated for each denaturation agent. * indicates a significant difference with the wild type protein with P < 0.05. Mutations are arranged from the less to the more stable, considering a mean stability for the four parameters analyzed.
Several mutations stabilize the protein. A possible explanation could be that the interactions of nonpolar residues with water present a thermodynamic disadvantage caused by the side chain being more exposed to solvent in the native than in the denatured state [18]. In addition, polar residues at the surface may provide additional hydrogen bonds with the solvent and then increase protein stability [16]. This explanation seems satisfactory for some mutations: when the hydrophobic residue is located inside a hydrophobic area made up of several residues. Thus, Valine 48 forms a small hydrophobic area at the surface of the protein with Valine 42, Phenylalanine 225 forms a hydrophobic region with Alanine 100, Alanine 224, Valine 6, Valine 7 and Isoleucine 174 (Fig. 4). This hydrophobic area is broken when the hydrophobic Phe 225 is replaced with a hydrophilic arginine. Similarly, Val 252 forms a hydrophobic area with Gly 338, Ala 339 and Ala 212.
Figure 4.
Position of Phe 225 inside an hydrophobic area at the surface of the protein. (hydrophobic residues have been colored in blue).
Some mutations destabilized the protein. The presence of hydrophobic residues at the surface may have stabilization properties by providing a shield from penetrating water molecules [22]. Or mutation to Arg may disrupt a hydrophobic bond. V56 and L57 are in hydrophobic contact with Val 14, mutation to Arg may disrupt this bond. Likewise, Ile 5 faces Gly 16 and this interaction may maintain the loop conformation.
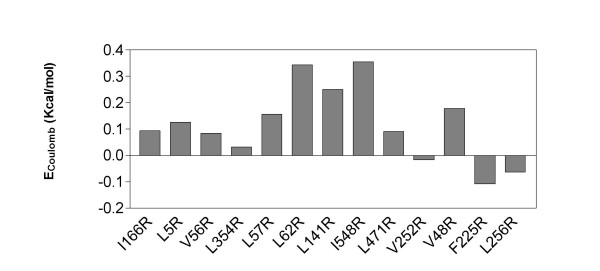
But, other mechanisms may contribute to the increased or decreased stability observed with some mutants. Introduction of a charge at the surface of the protein may provide a new coulombic interaction on the protein surface. This strategy seems efficient since it is used by proteins from thermophiles [23-25] and several groups showed significant stabilization of proteins with substitutions of the suface charges [20,26,27]. On the contrary, addition of a positive charge may result to the addition of a charge repulsion which decreases stability [28]. To test if introduction of a new positive charge may be responsible for the change of stability observed, we estimated the electrostatic stabilization expected for the addition of a charge using Coulomb's Law (Fig. 5). The stabilization provided by some mutations such as I548R may originate from the formation of new coulombic interaction, in that case between Arginine 548 and Glutamate 546.
Figure 5.
Electrostatic stabilization expected for the charge addition according to the Coulomb's Law (E = q1q2/Dr) with a dielectric constant of 80 by summing the interactions of the new charge with all the other charges of the protein.
Methods
Protein production and purification
cDNA encoding wild type drosophila AChE and mutant were expressed with the baculovirus system [29]. This enzyme is a dimer anchor to the membrane via a glycolipid [30]. We expressed a soluble dimeric form obtained by deletion of the hydrophobic peptide, precursor of the glycolipid anchor at the C-terminal end [31]. Furthermore, the enzyme was deleted of the loop from amino-acids 103 to 136 which has been replaced by 3 histidines. Secreted AChE was purified to homogeneity using the following steps: ammonium sulfate precipitation, ultrafiltration with a 10 kDa cutoff membrane, NTA-nickel chromatography, affinity chromatography with procainamide as ligand and gel filtration [6]. Activity was recorded at 25°C in 25 mM phosphate buffer pH 7, with 1 mM acetylthiocholine iodide as substrate using the method of Ellman et al. [32]. Mutations did not alter the specific activity of the proteins. Molecular modeling of mutated residues was performed from the structure solved by Harel et al. [[33], ref 1QO9] by homology using Swiss model server available at http://expasy.org/swissmod/. Residue numbering followed that of the mature protein.
Production
One liter containing 106 Sf9 cells was infected with more than 107 virus. After four days incubation at 28°C, the cells were lyzed by adding Triton X-100 (0.1%). Amount of AChE was estimated by active site titration using an irreversible inhibitor, chlorpyriphos oxon [34]. The mean of at least five independent productions was compared to the production of the wild type enzyme.
Denaturation
All denaturation experiments were performed with 10 picomoles of enzyme in one ml 25 mM phosphate buffer pH7 at 25°C. AChE was incubated in denaturing conditions. Aliquots were taken at regular intervals, diluted 20 x to stop the action of the denaturing agent and the remaining activity was measured.
To analyze thermosensitivity, enzymes were incubated at 50°C with 1 mg/ml Bovine Serum Albumin in the buffer. Before recording the remaining activity, aliquots were mixed with cold buffer chilled on ice and the solution was incubated at 25°C for ten minutes to eliminate the reversible component [35]. For urea denaturation, unfolding of AChE was induced by adding 4 M urea into the incubation buffer. The effect of organic solvent was followed by incubation of the enzyme in 20% acetonitrile. The effect of protease sensitivity was determined by incubation of AChE with 0.1 mg/ml pronase.
Three to nine batches of each mutant were produced and purified. Three repeats were performed for each batch and each denaturing agent. Significance of difference observed in stability was tested using the Mann Whitney test.
Coulombic interaction estimation
In order to estimate the effects of charge distribution on each of the mutants, the contributions from electrostatic interactions were extracted from the total energy of the minimized structures of the molecules. The minimization was performed with the GROMACS software [36], using the simulated annealing protocol. Position restraints were imposed on the whole molecule with the exception of the mutated residue, which was allowed to adopt the energetically favorable configuration. The Coulomb's interactions were normalized by subtracting the value obtained for the wild type protein and by rescaling the numbers by 80, to take into account the dielectric constant of water.
List of abbreviations
AChE, acetylcholinesterase
Authors' contributions
CS and CA purified the protein and performed biochemical analysis, AL performed in vitro mutagenesis, CA produced proteins, JC performed estimations of electrostatic interactions and DF conceived and coordinated the study. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This research was supported by grants from the European Community (ACHEB, QLK3-CT-2000-00650 and SAFEGUARD, QLK3-CT-2000-000481) and from DGA (PEA 99CO029).
Contributor Information
Caroline Strub, Email: caroline_strub@hotmail.com.
Carole Alies, Email: CaroleALIES@aol.com.
Andrée Lougarre, Email: Lougarre@ipbs.fr.
Caroline Ladurantie, Email: Ladurantie@ipbs.fr.
Jerzy Czaplicki, Email: cgeorges@ipbs.fr.
Didier Fournier, Email: fournier@ipbs.fr.
References
- Villatte F, Marcel V, Estrada-Mondaca S, Fournier D. Engineering sensitive acetylcholinesterase for detection of organophosphate and carbamate insecticides. Biosens Bioelectron. 1998;13:157–162. doi: 10.1016/S0956-5663(97)00108-5. [DOI] [PubMed] [Google Scholar]
- Boublik Y, Saint-Aguet P, Lougarre A, Arnaud M, Villatte F, Estrada-Mondaca S, Fournier D. Acetylcholinesterase engineering for detection of insecticide residues. Protein Engin. 2002;15:43–50. doi: 10.1093/protein/15.1.43. [DOI] [PubMed] [Google Scholar]
- Payne CS, Saeed M, Wolfe AD. Ligand stabilization of cholinesterases. Biochim Biophys Acta. 1989;999:46–51. doi: 10.1016/0167-4838(89)90028-9. [DOI] [PubMed] [Google Scholar]
- Burgess SK, Oxendine SL. Thermal inactivation of butyrylcholinesterase and acetylcholinesterase. J Prot Chem. 1983;12:651–658. doi: 10.1007/BF01024923. [DOI] [PubMed] [Google Scholar]
- Wilson EJ, Massoulié J, Bon S, Rosenberry TL. The rate of thermal inactivation of Torpedo acetylcholinesterase is not reduced in the C231S mutant. FEBS Lett. 1996;379:161–164. doi: 10.1016/0014-5793(95)01504-3. [DOI] [PubMed] [Google Scholar]
- Estrada-Mondaca S, Fournier D. Stabilization of recombinant Drosophila acetylcholinesterase. Prot Exp Purif. 1998;12:166–172. doi: 10.1006/prep.1997.0831. [DOI] [PubMed] [Google Scholar]
- Nasseau M, Boublik Y, Meier W, Winterhalter M, Fournier D. Substrate-permeable encapsulation of enzymes maintains effective activity, stabilizes against denaturation and protects against proteolytic degradation. Biotech Bioeng. 2001;75:615–618. doi: 10.1002/bit.10074. [DOI] [PubMed] [Google Scholar]
- Fremaux I, Mazères S, Brisson-Lougarre A, Arnaud M, Ladurantie C, Fournier D. Improvement of Drosophila acetylcholinesterase stability by elimination of a free cysteine. BMC Biochemistry. 2002;3:21. doi: 10.1186/1472-2091-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashford D, Chothia C, Lesk AM. Determinants of a protein fold. Unique features of the globin amino acid sequences. J Mol Biol. 1987;196:199–216. doi: 10.1016/0022-2836(87)90521-3. [DOI] [PubMed] [Google Scholar]
- Reidhaar-Olson JF, Sauer RT. Functionally acceptable substitutions in two alpha-helical regions of lambda repressor. Proteins. 1990;7:306–316. doi: 10.1002/prot.340070403. [DOI] [PubMed] [Google Scholar]
- Schwehm JM, Kristyanne ES, Biggers CC, Stites WE. Stability effects of increasing the hydrophobicity of solvent-exposed side chains in staphylococcal nuclease. Biochemistry. 1998;37:6939–6948. doi: 10.1021/bi9725069. [DOI] [PubMed] [Google Scholar]
- Wigley DB, Clarke AR, Dunn CR, Barstow DA, Atkinson T, Chia WN, Muirhead H, Holbrook JJ. The engineering of a more thermally stable lactate dehydrogenase by reduction of the area of a water-accessible hydrophobic surface. Biochim Biophys Acta. 1987;916:145–148. doi: 10.1016/0167-4838(87)90221-4. [DOI] [PubMed] [Google Scholar]
- Auerbach G, Huber R, Grättinger M, Zaiss K, Schurig H, Jaenicke R, Jacob U. Lactate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima: the crystal structure at 2.1 Å resolution reveals strategies for intrinsic protein stabilization. Structure. 1997;5:1475–1483. doi: 10.1016/S0969-2126(97)00297-9. [DOI] [PubMed] [Google Scholar]
- Argos P, Rossman RG, Grau , Zuber H, Franck G, Tratschin JD. Thermal stability and protein structure. Biochemistry. 1979;18:5698–5703. doi: 10.1021/bi00592a028. [DOI] [PubMed] [Google Scholar]
- Vogt G, Argos P. Protein thermal stability: hydrogen bonds or internal packing? Fold Des. 1997;2:S40–S46. doi: 10.1016/s1359-0278(97)00062-x. [DOI] [PubMed] [Google Scholar]
- Vogt G, Woell S, Argos P. Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol. 1997;269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tsai CJ, Nussinov R. Factors enhancing protein thermostability. Protein Eng. 2000;13:179–91. doi: 10.1093/protein/13.3.179. [DOI] [PubMed] [Google Scholar]
- Pakula AA, Sauer RT. Reverse hydrophobic effects relieved by amino-acid substitution at a protein surface. Nature. 1990;344:363–364. doi: 10.1038/344363a0. [DOI] [PubMed] [Google Scholar]
- Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol. 1991;222:67–88. doi: 10.1016/0022-2836(91)90738-r. [DOI] [PubMed] [Google Scholar]
- Perl D, Mueller U, Heinemann U, Schmid FX. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat Struct Biol. 2000;7:380–383. doi: 10.1038/75151. [DOI] [PubMed] [Google Scholar]
- Pedone E, Saviano M, Rossi M, Bartolucci S. A single point mutation (Glu85Arg) increases the stability of the thioredoxin from Escherichia coli. Protein Eng. 2001;14:255–260. doi: 10.1093/protein/14.4.255. [DOI] [PubMed] [Google Scholar]
- Van den Burg B, Dijkstra BW, Vriend G, Van Der Vinne B, Venema G, Eijsink VGH. Protein stabilization by hydrophobic interactions at the surface. Eur J Biochem. 1994;220:981–985. doi: 10.1111/j.1432-1033.1994.tb18702.x. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Raidt S. Stereochemical basis of heat stability in bacterial ferredoxins and in haemoglobin A2. Nature. 1975;255:256–259. doi: 10.1038/255256a0. [DOI] [PubMed] [Google Scholar]
- Jaenicke R, Bohm G. The stability of proteins in extreme environments. Curr Opin Struct Biol. 1998;8:738–748. doi: 10.1016/S0959-440X(98)80094-8. [DOI] [PubMed] [Google Scholar]
- Pace CN. Single surface stabilizer. Nat Struct Biol. 2000;7:345–346. doi: 10.1038/75100. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ruiz JM, Makhatadze GI. To charge or not to charge? Trends Biotechnol. 2001;19:132–135. doi: 10.1016/S0167-7799(00)01548-1. [DOI] [PubMed] [Google Scholar]
- Spector S, Wang M, Carp SA, Robblee J, Hendsch ZS, Fairman R, Tidor B, Raleigh DP. Rational modification of protein stability by the mutation of charged surface residues. Biochemistry. 2000;39:872–879. doi: 10.1021/bi992091m. [DOI] [PubMed] [Google Scholar]
- Grimsley GR, Shaw KL, Fee LR, Alston RW, Huyghues-Despointes BM, Thurlkill RL, Scholtz JM, Pace CN. Increasing protein stability by altering long-range coulombic interactions. Protein Sci. 1999;8:1843–1849. doi: 10.1110/ps.8.9.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabihi H, Fournier D, Fedon Y, Bossy JP, Devauchelle G, Cerutti M. Biochemical characterization of Drosophila melanogaster acetylcholinesterase expressed by recombinant baculovirus. Biochem Biophys Res Commun. 1994;203:734–742. doi: 10.1006/bbrc.1994.2243. [DOI] [PubMed] [Google Scholar]
- Fournier D, Berge JB, Cardoso de Almeida ML, Bordier C. Acetylcholinesterases from Musca domestica and Drosophila melanogaster brain are linked to membranes by a glycophospholipid anchor sensitive to an endogenous phospholipase. J Neurochem. 1988;50:1158–1163. doi: 10.1111/j.1471-4159.1988.tb10587.x. [DOI] [PubMed] [Google Scholar]
- Fournier D, Mutero A, Rungger D. Drosophila acetylcholinesterase. Expression of a functional precursor in Xenopus oocytes. Eur J Biochem. 1992;203:513–519. doi: 10.1111/j.1432-1033.1992.tb16577.x. [DOI] [PubMed] [Google Scholar]
- Ellman GL, Courtney KD, Andres KD, Featherstone RM. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmac. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- Harel M, Kryger G, Rosenberry TL, Mallender WD, Lewis T, Fletcher RJ, Guss JM, Silman I, Sussman JL. Three-dimensional structures of Drosophila melanogaster acetylcholinesterase and of its complexes with two potent inhibitors. Protein Sci. 2000;9:1063–1072. doi: 10.1110/ps.9.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier A, Menozzi P, Marcel V, Villatte F, Fournier D. A method to estimate acetylcholinesterase active sites and turnover in insects. Anal Biochem. 2000;285:76–81. doi: 10.1006/abio.2000.4738. [DOI] [PubMed] [Google Scholar]
- Pavlic MR, Alif M, Bozic M, Stojan J. On the stability of some enzymes under various experimental conditions. Period Biolog. 1991;93:211–215. [Google Scholar]
- Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–317. [Google Scholar]