Abstract
Interpretation of 16S ribosomal RNA (rRNA) to 16S rRNA gene ratios (rRNA:rDNA) is based on a limited number of studies with rapidly growing copiotrophic bacteria. The most abundant bacteria in the ocean are oligotrophs, which probably grow more slowly than those bacteria whose rRNA:rDNA versus growth rate relationships are known. To examine whether rRNA:rDNA varies differently in oligotrophic marine bacteria than in copiotrophic bacteria, we used quantitative PCR and reverse transcriptase quantitative PCR to measure rRNA:rDNA in two marine copiotrophs and in two marine oligotrophs, including Candidatus Pelagibacter ubique HTCC1062, a coastal isolate of SAR11, the most abundant bacterial clade in the ocean. The rRNA:rDNA ratios for the two copiotrophs were similar to those expected on the basis of an analysis of previously studied copiotrophic bacteria, while the ratios for the two oligotrophs were substantially lower than predicted even given their slow growth rates. The rRNA:rDNA ratios determined along a transect in the Delaware estuary suggested that SAR11 bacteria grow at rates close to the growth rate in culture, while rates of the two copiotrophs were far below those observed in laboratory cultures. Our results have implications for interpreting rRNA:rDNA from natural communities, understanding growth strategies and comparing regulatory mechanisms in copiotrophs and oligotrophs.
Introduction
Growth rates of bacteria and other microbes are important for understanding marine food web processes and biogeochemical cycles. The abundance and activity of bacterial taxonomic groups depend on the growth rate as well as top-down control by grazing and viral lysis. Survival strategies using rapid growth might co-exist with strategies of slow growth owing to tradeoffs between growth rate and defensive specialization (Våge et al., 2013, 2014). Bacterial taxonomic groups that grow rapidly likely transfer more carbon up the food chain than other slow-growing taxa (Azam et al., 1983). These rapidly growing bacteria may also respire more rapidly, contributing more to CO2 fluxes than slow growers (del Giorgio and Cole, 1998).
There are few estimates of growth rates of specific taxonomic groups in natural communities, although average rates for the entire community have been explored extensively. Growth rates of entire natural communities are estimated to range from 0.05 per day in oligotrophic systems to 0.3 per day in coastal waters, as calculated from leucine incorporation and cellular abundance (Ducklow, 2000; Kirchman, in press). These rates are unlikely to represent rates for individual taxa due to the broad diversity of survival strategies among bacteria, leading to various combinations of activity and abundance in natural communities (Jones and Lennon, 2010; Campbell et al., 2011; Lennon and Jones, 2011). Estimates of growth rates of individual taxonomic groups range from 0.13 to 0.73 per day in the North Atlantic and as high as 6.1 per day in communities from estuaries (Malmstrom et al., 2004; Yokokawa et al., 2004). These estimates are based on leucine incorporation detected by microautoradiography coupled with FISH (fluorescence in situ hybridization) or on net changes in abundance detected by FISH in incubations that exclude grazers. Both methods rely on incubations, which may alter the growth environment, leading to changes in the community. In addition, the phylogenetic resolution of these estimates is limited because the oligonucleotide probes used for FISH in previous studies recognize broad phylogenetic groups at the class or even phylum level (Amann et al., 1995).
Ratios of rRNA to rRNA genes (rRNA:rDNA) for bacterial taxa could potentially yield growth rate estimates at higher phylogenetic resolution (Kemp et al., 1993; Lee and Kemp, 1994). As ribosome content is positively correlated with growth rate for many bacteria, and because 16S rDNA sequences are used to define taxonomic groups (Delong et al., 1989), rRNA:rDNA could be used to estimate growth of specific bacterial taxa (Kerkhof and Ward, 1993). Early studies on ribosome content and growth focused on model enteric bacteria such as Escherichia coli and Salmonella typhimurium (Kjeldgaard et al., 1958; Schaechter et al., 1958; Harvey, 1970; Bremer and Dennis, 1987), whereas later studies examined more diverse bacteria such as Desulfovibrio vulagris and Rickettsia prowazekii (Poulsen et al., 1993; Pang and Winkler, 1994). All of these bacteria are copiotrophs that grow at rates more than 100-fold faster than estimates of the mean growth rates of natural bacterial communities in the ocean (Ducklow, 2000). Some studies have linked rRNA content of marine bacteria to growth rate in culture, but these bacteria are not closely related to the most abundant types of marine bacteria (Kemp et al., 1993; Kerkhof and Ward, 1993; Fegatella et al., 1998). The growth rates of these marine copiotrophic bacteria are also high compared with the estimated growth rates of most natural bacterial communities (Ducklow, 2000; Kirchman, in press). The relationship between rRNA:rDNA and growth rates has not been examined for oligotrophic bacteria thought to be abundant in the ocean.
The most abundant group of bacteria in the ocean is the SAR11 clade of Alphaproteobacteria (Morris et al., 2002; Brown et al., 2012). The growth rate of this clade of oligotrophic bacteria and the role of growth in setting its abundance are unclear. Previous studies found that the number of SAR11 sequences in the rRNA pool is low relative to its representation in the rDNA pool (Campbell et al., 2009, 2011; Campbell and Kirchman, 2013; Hunt et al., 2013; Zhang et al., 2014) and that its rRNA:rDNA ratio is lower than that for other bacterial groups in coastal waters, implying lower than average growth rates for this oligotrophic clade of bacteria (Kirchman, in press). Low growth rates for SAR11 would be consistent with the hypothesis that the clade is abundant because it is a defense specialist (Våge et al., 2014). However, interpretation of the rRNA:rDNA ratios for SAR11 and other oligotrophic bacteria depends on extrapolation from studies of copiotrophic bacteria. In fact, it may not be even possible to extrapolate from one copiotrophic organism to another (Blazewicz et al., 2013); Kemp et al. (1993) found that the relationship between rRNA and growth rate varied among four unidentified isolates of marine bacteria. In contrast to the rRNA:rDNA studies, other approaches have indicated that SAR11 grows as fast or faster than other bacteria in the ocean (Malmstrom et al., 2004; Teira et al., 2009; Campbell et al., 2011; Salter et al., 2014). The growth rate of SAR11 in marine waters remains unresolved.
The goal of this study was to examine rRNA:rDNA of oligotrophic and copiotrophic bacteria in culture and then to use the results to explore growth rates of bacteria in the Delaware estuary. We chose four taxa that are abundant in coastal Delaware and elsewhere, are genetically diverse and are thought to use contrasting growth strategies. Two of these isolates are oligotrophs that grow more slowly than bacterial isolates with known rRNA:rDNA versus growth rate relationships. Candidatus Pelagibacter ubique is a representative of the SAR11 clade (Morris et al., 2002; Rappé et al., 2002; Brown et al., 2012). Ca. P. ubique grows more slowly than other bacterial isolates in culture and has a streamlined genome (1.3 Mbp), characteristic of an oligotroph (Rappé et al., 2002; Giovannoni et al., 2005, 2014). Another oligotroph, the gammaproteobacterium SAR92 HTCC2207, has the second smallest genome of the four bacteria (2.0 Mbp), and grows slowly relative to most isolates but faster than Ca. P. ubique (Cho and Giovannoni, 2004). Ruegeria pomeroyi DSS-3 is a copiotroph in the Alphaproteobacteria class with a relatively large genome (4.1 Mbp) and one megaplasmid (0.5 Mbp) (Moran et al., 2004). R. pomeroyi has genes for rapid growth, attachment to particles and several other metabolic strategies that suggest it is a copiotroph (Gonzalez et al., 2003; Moran et al., 2004). The other copiotroph, Polaribacter sp. MED152, is a coastal member of the Flavobacteria, a class that has been implicated in the degradation of high molecular weight organic matter (Cottrell and Kirchman, 2000). The genome (3.0 Mbp) of MED152 predicts copiotrophic attributes as it encodes for genes involved in attachment to particles and polymer degradation (Gonzalez et al., 2008). Our results indicate that the oligotrophic bacteria had lower rRNA:rDNA ratios and grew more slowly in cultures than the copiotrophic bacteria, but not in the Delaware estuary.
Materials and methods
Batch culture growth conditions and sampling
Batch cultures were grown in acid-washed polycarbonate bottles, in triplicate, in the dark and at 18 °C. This temperature was chosen because it is in the middle of the range of isolation temperatures for the four isolates examined here; Ca. P. ubique was isolated at the lowest temperature of the four strains (15 °C) while R. pomeroyi was isolated at the highest (room temperature) (Rappé et al., 2002; Gonzalez et al., 2003). To monitor cellular abundance, cells stained for 1 h with 5x SYBR Green I (Life Technologies, Carlsbad, CA, USA) were counted using a FACSCaliber flow cytometer (BD Biosciences, San Jose, CA, USA). Samples for DNA and RNA were preserved in RNAlater (Life Technologies), filtered onto 0.22 μM GVWP membranes (Millipore, Billerica, MA, USA) and stored in Buffer RLT (Qiagen, Valencia, CA, USA) at −80 °C. Nucleic acids were extracted using the All Prep DNA and RNA extraction kit (Qiagen). Contaminating DNA was removed from RNA samples by digestion with Turbo DNase (Life Technologies), and RNA samples were checked for DNA using PCR with general primers for the bacterial 16S rRNA gene.
Ca. P. ubique and SAR92 were grown on variations of the medium based on AMS1 salts described by Carini et al. (2013). The medium for SAR92 consisted of AMS1 salts with the organic carbon and vitamin additions described by Steindler et al. (2011). This medium was modified for Ca. P. ubique by lowering the bicarbonate concentration to 100 μM and by adding 10 mM HEPES (pH 7.5) buffer. Ca. P. ubique and SAR92 were started from glycerol stocks, and after repeated growth to high density, each experimental culture was started with 300 ml of exponentially growing cells. For the experiment, 20 l Ca. P. ubique and SAR92 cultures were mixed by bubbling with 0.22 μM filtered air. Sample volumes for nucleic acids (1 liter) were identical for Ca. P. ubique and SAR92.
Liquid cultures of MED152 were started from colonies on an agar plate, and R. pomeroyi (obtained from ATCC) was rehydrated from a freeze-dried stock in YTSS medium. After transferring R. pomeroyi several times, 3 l cultures on Difco YTSS (BD Biosciences) were started with 100 ml of exponentially growing cells. Volumes for nucleic acid samples for R. pomeroyi ranged from 3 to 50 ml, depending on cell abundance. R. pomeroyi and MED152 cultures were shaken continuously at 100 r.p.m. for aeration and mixing. The liquid media for MED152 included the organic constituents of Difco Marine Broth 2216 (BD Biosciences) combined with AMS1 salts (Gonzalez et al., 2008; Carini et al., 2013). The volume of MED152 cultures for starting the experiments was 50 ml which was added to 1.5 l. The nucleic acid sample volume for MED152 ranged from 10 to 100 ml. The differences among nucleic acid sample volumes were owing to the variable maximum density of each strain and the predicted DNA yields.
Quantifying rDNA and rRNA by quantitative PCR and reverse transcriptase quantitative PCR
Quantitative PCR (qPCR) and reverse transcriptase quantitative PCR (RT-qPCR) were used to quantify copies of genes and transcripts of 16S rRNA. We measured template DNA and RNA concentrations using PicoGreen and RiboGreen assays, respectively (Life Technologies). Template nucleic acids were then diluted to below 500 pg μl−1 and re-quantified. All amplification reactions were done on a RotorGene 6000 (Corbett Robotics, San Francisco, CA, USA), using SYBR Green qPCR and RT-qPCR reaction kits (Qiagen). Reactions were done in duplicate but were repeated if duplicates varied by >10%. Standards were prepared linearized plasmids from clones of known sequence. Reaction conditions for thermal cycles were 55 °C for 10 min (RT-qPCR only), 95 °C for 10 min, 95 °C for 15 s, X °C for 15 s, 72 °C for 15 s, followed by 72 °C for 10 min, and a melt analysis ramping from 72 to 100 °C. The 15 s steps were repeated for 45 cycles, and X varied by primer set. For Ca. P. ubique X=62 °C, SAR92 X=60 °C, R. pomeroyi X=61 °C, and MED152 X=60 °C. Reaction efficiencies ranged from 93 to 102%, and reaction efficiencies and y-intercepts for standard curves did not vary between corresponding qPCR and RT-qPCR reactions. Data from qPCR and RT-qPCR are reported as copies per milliliter in Supplementary Table S1.
Primer design and specificity
Taxon-specific primers were designed for the 16S rRNA gene sequences of Ca. P. ubique (GenBank accession no. NR_074224.1), SAR92 (GenBank accession no. AY386335.1), R. pomeroyi (GenBank accession no. NR_028727.1) and MED152 (GenBank accession no. DQ481463.1) (Table 1). We designed primers in Oligo 7 (Molecular Biology Insights, Colorado Springs, CO, USA) using the GreenGenes database to minimize amplifying 16S rRNA gene sequences of other bacteria (DeSantis and Hugenholtz, 2006). Specificity was confirmed in silico using ARB-SILVA TestPrime (Klindworth et al., 2013) and tested using DNA extracted from a mixed community in the Delaware estuary as template. The single product resulting from each PCR reaction was sequenced. BLAST analysis of the cloned sequences indicated that each primer set retrieved its intended 16S rRNA target. The mean percent identity of the amplicons to their target sequence was 98.9% for Ca. P. ubique (number of clones (n)=12), 98.5% for SAR92 (n=12), 93.7% for R. pomeroyi (n=36) and 97.4% for MED152 (n=36).
Table 1. Sequences of primers for qPCR and RT-qPCR of the 16S rRNA gene. The sequences are listed 5′ to 3′.
| Forward | Reverse | |
|---|---|---|
| Ca. P ubique | GGCCTGGAATAACACGAGGAA | GGGCTCATCCAATGGTGCATA |
| SAR92 | GCGGCCACCTGGACTAAT | TGCGCCACTAAGAGATCAAGT |
| MED152 | GCGGATTAGAAAGTTAGGGGTG | TCGCCACTGGTGTTCTTCC |
| R. pomeroyi | TGGGCAATGGAGGTAACTCT | AGCCGGTCCTTATTCTTACAG |
| All bacteriaa | CGGTCCAGACTCCTACGGG | TTACCGCGGCTGCTGGCAC |
Sequences from Lee et al. (1996) and Delbes et al. (2000).
To compare the data from our culture experiments to previous studies, we converted RNA and DNA concentrations given in the historical data sets to rRNA and rDNA. Previous studies reported RNA and DNA concentrations determined by various methods, including mass, calorimetric assays and fluorometric assays. We assumed that 90% of the total cellular RNA was rRNA for all bacteria tested (Chen and Duan, 2011). In addition, we assumed that the fraction of rDNA in the total DNA pool was equal to the number of base pairs in a bacterial 16S rRNA gene, multiplied by the copy number of the 16S rRNA gene and divided by the total base pairs in the genome. For bacteria with unknown 16S rRNA copy number or genome size, we assumed a copy number or genome size based on the closest phylogenetic relative (Herbert, 1961; Kemp et al., 1993). The data for the following bacterial taxa were analyzed: E. coli (Rosset et al., 1966; Harvey, 1970; Bremer and Dennis, 1987), S. typhimurium (Kjeldgaard and Kurland, 1963; Rosset et al., 1966; Bremer and Dennis, 1987), Aerobacter aerogenes (Caldwell et al., 1950; Neidhardt and Magasanik, 1960; Rosset et al., 1966; Sykes and Young, 1968), Delsulfovibrio vulgaris (Poulsen et al., 1993), Rickettsia prowazekii (Pang and Winkler, 1994), Sphingomonas sp. strain RB2256 (Fegatella et al., 1998), Pseudomonas stutzeri Zobell (Kerkhof and Ward, 1993) and several unidentified marine isolates (Kemp et al., 1993).
Transect of the Delaware estuary
The Delaware estuary was sampled during 18–22 November 2013. We sampled 24 stations with salinities ranging from 0.3 to 31.5 PSU and temperatures varying from 9.2 °C to 11.2 °C. Samples for nucleic acids were collected at each station by filtering whole seawater onto 0.22 μM GVWP membrane filters (Millipore), immersing these in 1 ml of CTAB buffer, and immediately freezing them at −80 °C. Extractions of DNA and RNA were performed as described by Dempster et al. (1999). Total prokaryotic abundance was estimated using epifluorescence microscopy with 4′, 6-diamidino-2-phenylindole (Porter and Feig, 1980). Bacterial production was estimated using 3H-leucine incorporation (Kirchman et al., 1985; Kirchman, 2001) and the centrifugation protocol described by Smith and Azam (1992). A conversion factor of 3.1 kg C mol−1 was used to convert 3H-leucine incorporation into bacterial production (Simon and Azam, 1989). We estimated the growth rates of natural bacterial communities by dividing bacterial production by total bacterial biomass (Kirchman, 2001). Bacterial biomass was estimated from abundance assuming 15 fg per cell (Fukuda et al., 1998). The word ‘bacteria' is used throughout this paper because archaeal abundance was relatively low (<1% of metagenomic sequences) in these communities (B. Campbell, unpublished data).
Results
rRNA:rDNA ratios and growth rates in culture
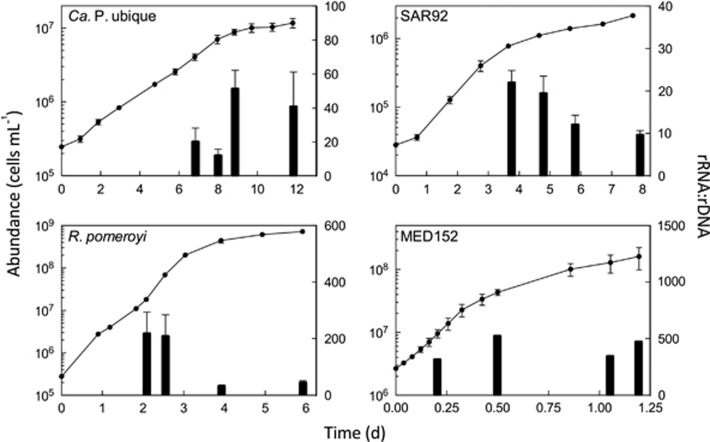
Four cultured representatives of abundant bacterial taxa were grown in batch cultures in which cell abundance and rRNA content were followed over time (Figure 1). All growth curves were divided into exponential phase and near-stationary phase. Oligotrophs Ca. P. ubique and SAR92 grew more slowly than the copiotrophs R. pomeroyi and MED152. The maximum growth rate in exponential phase was 0.4 per day for Ca. P. ubique and 1.0 per day for SAR92, much lower than the rates for R. pomeroyi (2.1 per day) and MED152 (6.2 per day). Growth rates in near-stationary phase also varied among the isolates. The growth rate in this phase was 0.3 per day for R. pomeroyi and 1.5 per day for MED152, while the near-stationary phase rates of oligotrophs Ca. P. ubique (0.06 per day) and SAR92 (0.2 per day) were even lower.
Figure 1.
Cellular abundance and rRNA:rDNA ratios for Ca. P. ubique HTCC1062, SAR92, R. pomeroyi and MED152 over time in triplicate batch cultures. The points are the abundance data and the bars are the rRNA:rDNA ratio data. The error bars are s.d. of biological replicates (n=3), except in the case of MED152 where one culture did not grow (n=2).
Ratios of 16S rRNA to rDNA (rRNA:rDNA) varied between isolates and also varied over time within experiments (Figure 1). The rRNA:rDNA ratios in fast-growing R. pomeroyi and MED152, regardless of growth rate, were 12-fold higher than the rRNA:rDNA ratios in Ca. P. ubique and SAR92. The sharpest contrast between rRNA:rDNA ratios over time was seen for R. pomeroyi. Ratios in exponentially growing R. pomeroyi were 5.5-fold greater than in near-stationary phase R. pomeroyi. Ratios in SAR92 decreased from 20 to 16 when growth slowed from 1.0 per day to 0.2 per day. The rRNA:rDNA ratios in Ca. P. ubique approximately doubled (from 22 to 45) when transitioning from exponential growth to the near-stationary phase. There was no significant change in rRNA:rDNA in MED152 during batch growth. One MED152 culture did not grow at all; rRNA:rDNA in this culture was significantly lower than in growing MED152 cultures (13.9 versus 417.3; Student's t-test, P<0.05).
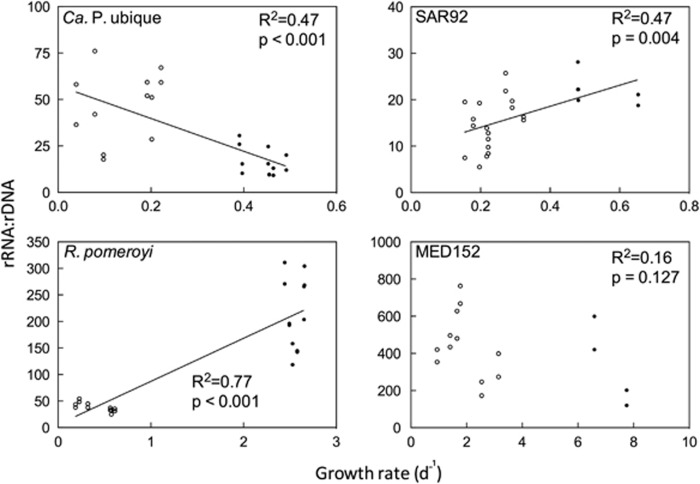
To determine the relationship between rRNA:rDNA and growth, we plotted rRNA:rDNA ratios against growth rate for each isolate (Figure 2). Growth rates for this analysis were calculated through the three time points immediately before a sampling point for nucleic acids. The rRNA:rDNA versus growth rate relationships of Ca. P. ubique, SAR92 and R. pomeroyi were statistically significant according to linear regression analyses (Figure 2). These relationships were positive for R. pomeroyi (R2=0.77, P<0.001) and SAR92 (R2=0.32, P<0.01) but negative for Ca. P. ubique (R2=0.46, P<0.001). There was no significant relationship for MED152 (P>0.05). However, even when statistically significant, the relationships were not simple linear ones. Data from the near-stationary phase were separate from those from the exponential phase, especially for R. pomeroyi. For SAR92, rRNA:rDNA varied between 5 and 20 when growth rate was 0.2 per day or lower (near-stationary phase), but then remained relatively constant at 20±3.9 when growth rate varied from 0.3 to 0.65 per day (exponential phase).
Figure 2.
Ratios of rRNA:rDNA versus growth rate for four bacterial taxa. Lines were calculated from Model II linear regression analyses (P<0.05). The slopes of all three lines are different from one another (Student's t-test, P<0.05). For each sample, two sets of matching technical replicates are plotted. Empty points represent near-stationary phase, while solid points represent exponential growth.
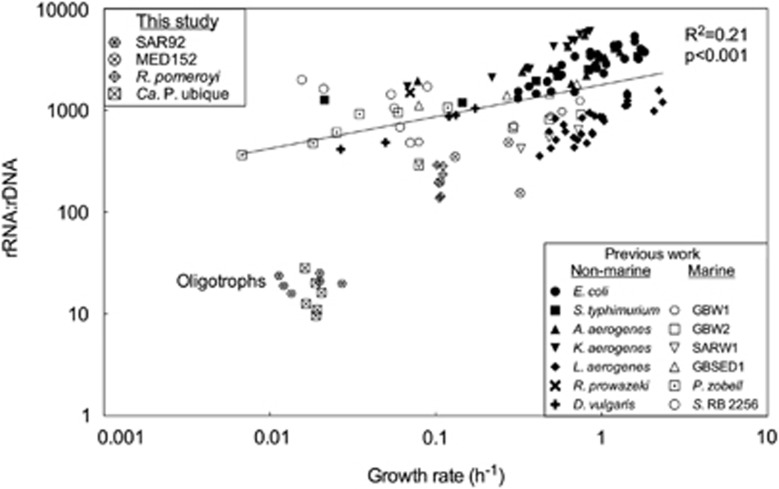
We compared our rRNA:rDNA data with a historical data set of rRNA:rDNA ratios and growth rates in other cultured bacteria (Figure 3). Only the exponential phase data were used because that growth phase was the one examined by previous studies. Ratios from MED152 and R. pomeroyi in exponential phase fell within the range of values in the historical data set (rRNA:rDNA >100). In contrast, ratios and growth rates for Ca. P. ubique and SAR92 were substantially lower than those in the historical data set regardless of growth phase (rRNA:rDNA <100).
Figure 3.
Ratios of rRNA:rDNA versus growth rate for the four isolates in this study and for the historical data set. The line was calculated from linear regression analysis of all copiotrophic organisms in this study and in the historical data set (R2=0.21, P<0.001).
To explore differences between oligotrophic and copiotrophic bacteria in more detail, all of the data from copiotrophic bacteria were used in a linear regression analysis which was then used to examine the two oligotrophic bacteria. Linear regression analysis indicated that the rRNA:rDNA versus growth rate relationship for all copiotrophic bacteria was statistically significant (slope=1170±220, intercept =1078±184, R2=0.21, P<0.001; Figure 3). The points for the oligotrophic bacteria are substantially below the regression line. The growth rate for the oligotrophic bacteria predicted from the regression line is ~0.0002 h−1, 40–60-fold slower than their actual growth rate of ~0.01 h−1.
rRNA:rDNA ratios in the Delaware estuary
We examined rRNA:rDNA ratios of the four chosen taxa as well as environmental parameters during sampling of the Delaware estuary in November 2013. Ratios of rRNA:rDNA in the estuary were similar to ratios from cultures for some taxa but not for others (Table 2). Ratios for R. pomeroyi and MED152 were significantly lower in the estuary than in culture, regardless of the culture growth phase (Student's t-test, P<0.05). Ratios for SAR92 in the estuary were not significantly different from ratios for this bacterium in culture (Student's t-test, P>0.05). SAR92 was the least abundant of the groups examined in the estuary and was present in only 4 of 24 transect samples (data not shown). Ratios for Ca. P. ubique in the estuary were not significantly different from exponentially growing Ca. P. ubique in culture (Student's t-test, P>0.05), but were statistically different from ratios in near-stationary Ca. P. ubique cultures (Student's t-test, P<0.05). The mean community growth rate in the estuary, calculated from bacterial production and abundance, was 0.2±0.3 per day (s.d., n=23).
Table 2. Ratios of rRNA:rDNA for taxonomic groups in this study.
| Exponential | Near-stationary | Transect | |
|---|---|---|---|
| Ca. P. ubique | 22±16 | 45±17a | 27±17 |
| SAR92 | 20±2 | 16±6 | 9±6 |
| R. pomeroyi | 215±67a | 39±8a | 4±3 |
| MED152 | 330±167a | 470±191a | 15±14 |
| All bacteria | — | — | 40±20 |
Culture samples were divided into two periods of growth. Transect samples were from the Delaware estuary. Errors are s.d. For Pelagibacter, SAR92 and Ruegeria, n=6 in each phase of culture growth. For MED152 growth phases, n=4. For transect samples: SAR92, n=4; MED152, n=18; Ruegeria, n=21; Pelagibacter, n=24; and all bacteria, n=24.
Differed significantly from transect ratios (Student's t-test, P<0.05).
Discussion
The ratio of 16S rRNA to 16S rRNA genes (rRNA:rDNA) is one basis for linking taxonomic identity and growth activity in bacterial communities (Kemp et al., 1993; Lee and Kemp, 1994; Blazewicz et al., 2013). There is a positive relationship between rRNA:rDNA and growth rate in many bacteria, but the bacteria with known relationships are not representative of dominant members of marine bacterial communities (Kemp et al., 1993; Kerkhof and Ward, 1993; Fegatella et al., 1998). In particular, it is not clear whether the relationship between rRNA:rDNA and growth rate for oligotrophic bacteria, most notably the SAR11 clade, is similar to the relation known for copiotrophic bacteria. We determined the rRNA:rDNA ratio versus growth rate relationship in culture for four diverse marine bacteria that are representative of abundant clades in marine waters. We then determined rRNA:rDNA ratios in the Delaware estuary for taxonomic groups closely related to the cultured isolates.
We found the anticipated positive relationship between rRNA:rDNA and growth rate for R. pomeroyi and SAR92, but not for Ca. P. ubique and MED152. The rRNA:rDNA ratio is expected to depend on growth rate because increased ribosome content enables more protein synthesis and faster growth (Kjeldgaard et al., 1958; Schaechter et al., 1958; Neidhardt and Magasanik, 1960; Rosset et al., 1966; Sykes and Young, 1968). In spite of the lack of a positive relationship for Ca. P. ubique and MED152, the positive relationship still held when all strains were compared together. Oligotrophs SAR92 and Ca. P. ubique grew more slowly than copiotrophs MED152 and R. pomeroyi, and the rRNA:rDNA ratios for the oligotrophic taxa were correspondingly lower than ratios for copiotrophic taxa.
The rRNA:rDNA to growth rate relationship varied among the isolates, even for those with the expected positive relationship. Kemp et al. (1993) also found varying rRNA:rDNA versus growth rate relationships for the four isolates they examined. A possible physiological explanation for this variation is that protein translation is more efficient in some taxa than in others owing to codon usage bias (Andersson and Kurland, 1990; Novoa and Ribas de Pouplana, 2012). Differences in translational efficiencies lead to difference in ribosome numbers per cell (Klumpp et al., 2012) and could explain variance among rRNA:rDNA versus growth rate relationships. But these possible explanations do not address why some rRNA:rDNA versus growth rate relationships are not positive, which was the case for Ca. P. ubique and MED152. In contrast, Salter et al. (2014) recently found a positive correlation between leucine incorporation by SAR11 and rRNA:rDNA for SAR11 in the Mediterranean Sea.
We suspect that the complicated rRNA:rDNA-growth relationships seen for these isolates are caused in part from using data from the near-stationary phase. The physiological limitations that cause slow growth in the near-stationary phase are different from the limitations that are responsible for slow growth in the exponential phase of batch cultures. The most common response of bacteria entering stationary phase, is to degrade rRNA (Deutscher, 2003). However, there is evidence that some marine Vibrio may not immediately degrade their rRNA during the onset of starvation depending on the conditions inducing the stationary phase (Kramer and Singleton, 1992). That some bacteria may not decrease their rRNA under starvation conditions could help to explain the lack of a positive relationship between rRNA:rDNA and growth rate for Ca. P. ubique and MED152.
Ratios from oligotrophic Ca. P. ubique and SAR92 were much lower than expected based on the regression analysis of rRNA:rDNA versus growth rate data for copiotrophic bacteria, which has implications for understanding the physiology underlying their survival strategy and for estimating growth rates of these organisms in the environment. The ratio data indicate that oligotrophs need fewer ribosomes (30 versus >100) than a copiotroph would require if it could grow at a rate of ~0.1 per day, which implies that translation in oligotrophs is more efficient than in copiotrophs. The number of ribosomes does not change greatly with growth state for Ca. P. ubique, suggesting that transcriptional regulation of rRNA is not an important regulatory mechanism in this oligotroph. Ca. P. ubique may regulate gene expression at a post-transcriptional level with riboswitches (Tripp et al., 2009; Smith et al., 2010, 2013). Perhaps similar regulatory mechanisms operate in the other oligotroph, SAR92, as its rRNA:rDNA ratios also did not vary much. Ratios for both oligotrophs varied much less than for the two copiotrophs examined here (5.51–76.0 versus 24.9–763).
Assuming that Ca. P. ubique is an adequate representative of the SAR11 clade, our data help to resolve the discrepancy between the rRNA studies, which suggested that SAR11 grows slower than the average (Campbell et al., 2009; Campbell and Kirchman, 2013; Hunt et al., 2013; Zhang et al., 2014) and the studies using other approaches that suggest relatively fast growth for SAR11 (Malmstrom et al., 2004; Teira et al., 2009; Campbell et al., 2011; Salter et al., 2014). The discrepancy is resolved by using the ratio to growth relationship for Ca. P. ubique to convert the observed rRNA:rDNA ratio for SAR11 in the ocean to a growth rate. Using the Ca. P. ubique relationship yields higher estimates of SAR11 growth than when the relationship between the ratio and growth for copiotrophs is used. In short, studies using 16S rRNA levels and other approaches both now indicate that growth rates of SAR11 are as high as or even higher than the average bacterium in marine waters.
To estimate the growth rates of SAR11 and the other three taxa in a natural community, we compared the rRNA:rDNA ratios measured in the culture experiments with ratios measured in bacterial communities along a transect in the Delaware estuary. Ratios for Ca. P. ubique in the Delaware were similar to ratios in cells growing in pure cultures at 0.4 per day, but not similar to ratios in cultured Ca. P. ubique growing at 0.06 per day (Student's t-test, P<0.05). This growth rate of Ca. P. ubique (0.4 per day) is similar to growth rates of SAR11 determined by Malmstrom et al. (2004) in the North Atlantic (0.13–0.72 per day), Teira et al. (2009) in coastal mesocosm experiments (0.46–0.59 per day) and Ferrera et al. (2011) in the NW Mediterranean Sea (0.1–1.8 per day). Our results suggest that the type of Ca. P. ubique we examined grows faster than the mean growth rate of the total community (0.2 per day) in the Delaware estuary, calculated from bacterial production and abundance. This result contributes to the discussion of SAR11 survival strategy (Zhao et al., 2013; Våge et al., 2013, 2014). Assuming that our results for this Ca. P. ubique strain are applicable to other SAR11 ecotypes, these data suggest that the high abundance of SAR11 is the result of superior competitive strategy rather than a defensive strategy.
The other bacteria grew more slowly than Ca. P. ubique in the Delaware and also more slowly than previous measurements of growth rates for bacteria in related phylogenetic groups. The average rRNA:rDNA for SAR92 in the estuary was only 9, nearly half the ratio measured for these bacteria in near-stationary phase when their growth rate was 0.2 per day. Slow growth may explain the low abundance of SAR92 in the estuary; it was found in only four of 24 samples. SAR92 does not appear to be adapted to the estuarine environment, probably reflecting that it was isolated from Pacific coastal waters (Cho and Giovannoni, 2004). Growth rates of even the two copiotrophic bacteria appeared to be slower than growth of Ca. P. ubique. The rRNA:rDNA ratios for R. pomeroyi in the Delaware were 50-fold lower than ratios from fast-growing R. pomeroyi in culture and 10-fold lower than slow-growing R. pomeroyi in culture. Bacteria in the broader phylogenetic group Roseobacter, which contains R. pomeroyi (Gonzalez et al., 2003), grow more rapidly; growth rates of Roseobacter range from 0.3 to 1.5 per day in the Delaware estuary and Mediterranean Sea (Yokokawa et al. 2004; Teira et al. 2009). Ratios for MED152 in natural communities of the Delaware were more than 10-fold lower than ratios of MED152 growing in culture, suggesting slow growth in the estuary. In fact, ratios in the estuary (15.3) were similar to the ratios from the experiment in which MED152 did not grow (13.9). Previous studies indicate that some members of Flavobacteria, which includes MED152, can grow at rates from 0.5 to 5.1 per day in the coastal and estuarine environments (Yokokawa et al. 2004; Ferrera et al. 2011), much faster than the rates we estimate for MED152.
Although the copiotrophs we examined appeared to grow slowly in the Delaware, they are likely adapted to the estuary; both copiotrophs were present in over 20 of 24 samples, only slightly fewer than the apparently rapidly growing Ca. P. ubique (24 of 24 samples). The persistence of the two copiotrophs in the estuary may be owing to short bursts of growth that occurred at other times of the year when we did not sample or were not captured by our sampling approach. Copiotrophs likely grow rapidly when associated with particles and other patches where nutrients are temporarily replete (Koch, 2001; Lauro et al., 2009; Kirchman, in press). The patchy distribution of resources means that the periods of growth only happen over short time periods and/or over small spatial scales. Incidents of fast growth by copiotrophs may be captured by simply taking more standard large samples over time or by focusing sampling efforts on particles and other small scale patches (Long and Azam, 2001).
Of course growth rates and thus rRNA:rDNA ratios will vary greatly in the Delaware and elsewhere because of variation in potentially many biogeochemical properties, such as temperature and organic material. A key question is how variation in these properties affects the relationship between the ratios and growth and the extrapolation from laboratory experiments to natural environments. Temperature in the Delaware estuary ranged from 9.2 °C to 11.2 °C, which was cooler than in our batch cultures (18 °C). In Salmonella, however, there is no effect of temperature on cellular rRNA content beyond the effect temperature has on the growth rate (Schaechter et al., 1958). Another difference between our culture experiments and the growth of these organisms in the environment is the type and varying amount of carbon available for growth. Organic carbon sources used to grow bacteria in culture are less complex and more labile than those in the environment. However, Schaechter et al. (1958) found no effect of differing carbon sources on rRNA content and thus on rRNA:rDNA.
The rRNA:rDNA of the marine oligotrophic bacteria examined in this study differed from the ratios in marine and non-marine copiotrophs examined by previous studies and this study. The rRNA:rDNA of the two oligotrophic bacteria was lower than predicted from work with copiotrophic bacteria, which has implications both for understanding the physiology of these organisms and for measuring their growth rate in the environment. We suggest that ribosomes of oligotrophic bacteria are more efficient, which enables them to achieve faster growth with fewer ribosomes than seen for copiotrophic bacteria. It may be evolutionarily advantageous for bacteria to have as few ribosomes as possible to succeed in the ocean and other oligotrophic environments, as ribosomes are costly to synthesize in terms of both rRNA and ribosomal proteins. The regulation of ribosomes has implications for using rRNA to explore growth rates in the environment. Further data on rRNA:rDNA under different growth conditions will strengthen the utility of the rRNA:rDNA tool for exploring growth rates in natural environments. Ratios of rRNA:rDNA currently still seem the best option for determining growth rates of marine bacteria at a high phylogenetic resolution and for exploring the contribution of specific taxa to the carbon cycle.
Acknowledgments
We thank Steve Giovannoni who supplied us with Ca. P. ubique HTCC1062 and Gammaproteobacteria SAR92 HTCC2207, and Jarone Panhassi who supplied us with Polaribacter MED152. Thanks to Liying Yu for her assistance in the lab, and to Tamara Huete who helped with the fieldwork in the Delaware estuary. This work was supported by grants from the National Science Foundation (OCE 1030306, 1261359, 1343773).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amann R, Ludwig W, Schleifer K. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S, Kurland C. (1990). Codon preferences in free-living microorganisms. Microbiol Rev 54: 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam F, Fenchel T, Field J, Gray J, Meyer-Reil L, Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar Ecol 10: 257–263. [Google Scholar]
- Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. (2013). Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J 7: 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H, Dennis P. (1987) Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F, Ingrahm J, Low K, Magasanik B, Schaechter M, Umbarger H (eds) Escherichia coli and Salmonella Typhimunum: Cellular and Molecular Biology. ASM Press: Washington, DC, USA, pp 1527–1542. [Google Scholar]
- Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T et al. (2012). Global biogeography of SAR11 marine bacteria. Mol Syst Biol 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell P, Mackor E, Hinshelwood C. (1950). The ribose nucleic acid content and cell growth of Bact. lactis ærogenes. J Chem Soc 1: 3151–3155. [Google Scholar]
- Campbell B, Yu L, Straza T, Kirchman D. (2009). Temporal changes in bacterial rRNA and rRNA genes in Delaware (USA) coastal waters. Aquat Microb Ecol 57: 123–135. [Google Scholar]
- Campbell BJ, Kirchman DL. (2013). Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J 7: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carini P, Steindler L, Beszteri S, Giovannoni SJ. (2013). Nutrient requirements for growth of the extreme oligotroph ‘Candidatus Pelagibacter ubique' HTCC1062 on a defined medium. ISME J 7: 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Duan X. (2011). Ribosomal RNA depletion for massively parallel bacterial RNA-sequencing applications. Methods Mol Biol 733: 93–103. [DOI] [PubMed] [Google Scholar]
- Cho J, Giovannoni SJ. (2004). Cultivation and growth characteristics of a diverse group of oligotrophic marine gammaproteobacteria. Appl Environ Microbiol 70: 432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell MT, Kirchman DL. (2000). Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol 66: 1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbes C, Moletta R, Godon J. (2000). Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction-single-strand conformation polymorphism. Environ Microbiol 2: 506–515. [DOI] [PubMed] [Google Scholar]
- del Giorgio P, Cole JJ. (1998). Bacterial growth efficiency in natural aquatic systems. Annu Rev Ecol Syst 29: 503–541. [Google Scholar]
- Delong EF, Wickham G, Pace NR. (1989). Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243: 1360–1363. [DOI] [PubMed] [Google Scholar]
- Dempster E, Pryor K, Francis D, Yound J, Rogers H. (1999). Rapid DNA extraction from ferns for PCR-based analysis. Biotechniques 68: 66–68. [DOI] [PubMed] [Google Scholar]
- DeSantis T, Hugenholtz P. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. (2003). Degradation of stable RNA in bacteria. J Biol Chem 278: 45041–45044. [DOI] [PubMed] [Google Scholar]
- Ducklow HW. (2000) Bacterial production and biomass in the oceans. In: Kirchman DL (ed) Microbial Ecology of the Oceans. Wiley-Liss, Inc, pp 85–120. [Google Scholar]
- Fegatella F, Lim J, Kjelleberg S, Cavicchioli R. (1998). Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol 64: 4433–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrera I, Gasol JM, Sebastián M, Hojerová E, Koblízek M. (2011). Comparison of growth rates of aerobic anoxygenic phototrophic bacteria and other bacterioplankton groups in coastal Mediterranean waters. Appl Environ Microbiol 77: 7451–7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, Ogawa H, Toshi N, Isao K. (1998). Direct determination of carbon and nitrogen contents of natural bacterial assemblages in marine environments. Appl Environ Microbiol 64: 3352–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Cameron Thrash J, Temperton B. (2014). Implications of streamlining theory for microbial ecology. ISME J 8: 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245. [DOI] [PubMed] [Google Scholar]
- Gonzalez JM, Covert JS, Whitman WB, Henriksen JR, Mayer F, Scharf B et al. (2003). Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int J Syst Evol Microbiol 53: 1261–1269. [DOI] [PubMed] [Google Scholar]
- Gonzalez JM, Fernadez-Gomez B, Fernandez-Geurra A, Gomez-Consarnau L, SAnchex O, Coll-Llado M et al. (2008). Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc Natl Acad Sci USA 105: 8724–8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RJ. (1970). Metabolic regulation in glucose-limited chemostat cultures of escherichia coli. J Bacteriol 104: 698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert D. (1961). The chemical composition of micro-organisms as a function of their environment. Microb React to Environ 11: 391–416. [Google Scholar]
- Hunt DE, Lin Y, Church MJ, Karl DM, Tringe SG, Izzo LK et al. (2013). Relationship between abundance and specific activity of bacterioplankton in open ocean surface waters. Appl Environ Microbiol 79: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. (2010). Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA 107: 5881–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp PF, Lee S, LaRoche J. (1993). Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol 59: 2594–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhof L, Ward BB. (1993). Comparison of nucleic acid hybridization and fluorometry for measurement of the relationship between RNA/DNA ratio and growth rate in a marine bacterium. Appl Environ Microbiol 59: 1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman D. (2001). Measuring bacterial biomass production and growth rates from leucine incorporation in natural aquatic environments. Methods Microbiol 30: 227–237. [Google Scholar]
- Kirchman D, K'nees E, Hodson R. (1985). Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol 49: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman DL. (in press). Growth rates of microbes in the oceans. Annu Rev Mar Sci doi:10.1146/annurev-marine-122414-033938. [DOI] [PubMed]
- Kjeldgaard NO, Kurland CG. (1963). The distribution of soluble and ribosomal RNA as a function of growth rate. J Mol Biol 6: 341–348. [Google Scholar]
- Kjeldgaard NO, Maaloe O, Schaechter M. (1958). The transition between different physiological states during balanced growth of Salmonella typhimurium. J Gen Microbiol 19: 607–616. [DOI] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, Dong J, Hwa T. (2012). On ribosome load, codon bias and protein abundance. PLoS One 7: e48542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL. (2001). Oligotrophs versus copiotrophs. Bioessays 23: 657–661. [DOI] [PubMed] [Google Scholar]
- Kramer J, Singleton F. (1992). Variations in rRNA content of marine Vibrio spp. during starvation-survival and recovery. Appl Environ Microbiol 58: 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S et al. (2009). The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA 106: 15527–15533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kemp PF. (1994). Single-cell RNA content of natural marine planktonic bacteria measured by hybridization with multiple 16S rRNA-targeted fluorescent probes. Limnol Oceanogr 39: 869–879. [Google Scholar]
- Lee D, Zo Y, Kim S. (1996). Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand-conformation polymorphism. Appl Environ Microbiol 62: 3112–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon JT, Jones SE. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9: 119–130. [DOI] [PubMed] [Google Scholar]
- Long R a., Azam F. (2001). Microscale patchiness of bacterioplankton assemblage richness in seawater. Aquat Microb Ecol 26: 103–113. [Google Scholar]
- Malmstrom RR, Cottrell MT, Elifantz H, Kirchman DL. (2004). Biomass production andassimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean. Appl Environ Microbiol 71: 2979–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Buchan A, González JM, Heidelberg JF, Whitman WB, Kiene RP et al. (2004). Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432: 910–913. [DOI] [PubMed] [Google Scholar]
- Morris RM, Rappe MS, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810. [DOI] [PubMed] [Google Scholar]
- Neidhardt F, Magasanik B. (1960). Studies on the role of ribonucleic acid in the growth of bacteria. Biochem Biophys Acta 42: 99–116. [DOI] [PubMed] [Google Scholar]
- Novoa EM, Ribas de Pouplana L. (2012). Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet 28: 574–581. [DOI] [PubMed] [Google Scholar]
- Pang H, Winkler HH. (1994). The concentrations of stable RNA and ribosomes in Rickettsia prowazekii. Mol Microbiol 12: 115–120. [DOI] [PubMed] [Google Scholar]
- Porter KG, Feig YS. (1980). The use of DAPI for identifying and counting aquatic microfloral. Limnol Oceanogr 25: 943–948. [Google Scholar]
- Poulsen LK, Ballard G, Stahl DA. (1993). Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol 59: 1354–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418: 630–633. [DOI] [PubMed] [Google Scholar]
- Rosset R, Julien J, Monier R. (1966). Ribonucleic acid composition of bacteria as a function of growth rate. J Mol Biol 18: 308–320. [DOI] [PubMed] [Google Scholar]
- Salter I, Galand PE, Fagervold SK, Lebaron P, Obernosterer I, Oliver MJ et al. (2014). Seasonal dynamics of active SAR11 ecotypes in the oligotrophic Northwest Mediterranean Sea. ISME J, 1–14. [DOI] [PMC free article] [PubMed]
- Schaechter M, Maaloe O, Kjeldgaard NO. (1958). Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J Gen Bacteriol 19: 592–606. [DOI] [PubMed] [Google Scholar]
- Simon M, Azam F. (1989). Protein content and protein synethesis rates of planktonic marine bacteria. Mar Ecol Prog Ser 51: 201–213. [Google Scholar]
- Smith D, Thrash J, Nicora C. (2013). Proteomic and transcriptomic analyses of ‘Candidatus Pelagibacter ubique' describe the first PII-independent response to nitrogen limitation in a free-living Alphaproteobacterium. MBio 4: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DC, Azam F. (1992). A simple, economical method for measuring bacterial protein synthesis rates in seawater using tritiated-leucine. Mar Microb Food Webs 6: 107–114. [Google Scholar]
- Smith DP, Kitner JB, Norbeck AD, Clauss TR, Lipton MS, Schwalbach MS et al. (2010). Transcriptional and translational regulatory responses to iron limitation in the globally distributed marine bacterium Candidatus pelagibacter ubique. PLoS One 5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. (2011). Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes J, Young T. (1968). Studies on the ribosomes and ribonucleic acids of Aerobacter aerogenes grown at different rates in carbon-limited continuous culture. Biochem Biophys Acta 169: 103–116. [DOI] [PubMed] [Google Scholar]
- Teira E, Martínez-García S, Lønborg C, Alvarez-Salgado XA. (2009). Growth rates of different phylogenetic bacterioplankton groups in a coastal upwelling system. Environ Microbiol Rep 1: 545–554. [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Schwalbach MS, Meyer MM, Kitner JB, Breaker RR, Giovannoni SJ. (2009). Unique glycine-activated riboswitch linked to glycine-serine auxotrophy in SAR11. Environ Microbiol 11: 230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Våge S, Storesund JE, Giske J, Thingstad TF. (2014). Optimal defense strategies in an idealized microbial food web under trade-off between competition and defense. PLoS One 9: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Våge S, Storesund JE, Thingstad TF. (2013). SAR11 viruses and defensive host strains. Nature 499: E3–E5. [DOI] [PubMed] [Google Scholar]
- Yokokawa T, Nagata T, Cottrell MT, Kirchman DL. (2004). Growth rate of the major phylogenetic bacterial groups in the Delaware estuary. Limnol Oceanogr 49: 1620–1629. [Google Scholar]
- Zhang Y, Zhao Z, Dai M, Jiao N, Herndl GJ. (2014). Drivers shaping the diversity and biogeography of total and active bacterial communities in the South China Sea. Mol Ecol 23: 2260–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC et al. (2013). Abundant SAR11 viruses in the ocean. Nature 494: 357–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.