Abstract
Bacterial CRISPR-Cas systems provide insight into recent population history because they rapidly incorporate, in a unidirectional manner, short fragments (spacers) from coexisting infective virus populations into host chromosomes. Immunity is achieved by sequence identity between transcripts of spacers and their targets. Here, we used metagenomics to study the stability and dynamics of the type I-E CRISPR-Cas locus of Leptospirillum group II bacteria in biofilms sampled over 5 years from an acid mine drainage (AMD) system. Despite recovery of 452 686 spacers from CRISPR amplicons and metagenomic data, rarefaction curves of spacers show no saturation. The vast repertoire of spacers is attributed to phage/plasmid population diversity and retention of old spacers, despite rapid evolution of the targeted phage/plasmid genome regions (proto-spacers). The oldest spacers (spacers found at the trailer end) are conserved for at least 5 years, and 12% of these retain perfect or near-perfect matches to proto-spacer targets. The majority of proto-spacer regions contain an AAG proto-spacer adjacent motif (PAM). Spacers throughout the locus target the same phage population (AMDV1), but there are blocks of consecutive spacers without AMDV1 target sequences. Results suggest long-term coexistence of Leptospirillum with AMDV1 and periods when AMDV1 was less dominant. Metagenomics can be applied to millions of cells in a single sample to provide an extremely large spacer inventory, allow identification of phage/plasmids and enable analysis of previous phage/plasmid exposure. Thus, this approach can provide insights into prior bacterial environment and genetic interplay between hosts and their viruses.
Introduction
The biology of natural ecosystems is shaped by interactions between microorganisms and their phage (Chibani-Chennoufi et al., 2004). However, cultivation has usually been required to determine phage host range and to study the interaction dynamics (Hyman and Abedon, 2010). Cultivation-independent genomic methods provide new approaches to these problems and can provide insight into the impacts of phage on population and community structures (Allen and Banfield, 2005). Genomic analysis can also elucidate the roles of phage and mobile elements in genome evolution (Allen et al., 2007). When applied to time series samples, these methods may also be able to constrain the rates of evolutionary processes (Denef and Banfield, 2012).
Many bacterial and archaeal genomes encode one or more CRISPR loci, named for the clustered regularly interspaced short palindromic repeats that separate spacer sequences that are transcribed and processed into small interfering RNAs (crRNAs) to confer immunity to phage, plasmids and transposons (reviewed extensively in Horvath and Barrangou, 2010; Sorek et al., 2013; Barrangou and Marraffini, 2014; van der Oost et al., 2014; Westra et al., 2014). New spacers are incorporated at the leader end of CRISPR loci (where transcription begins), whereas older spacers remain at the trailer end (Barrangou et al., 2007; Tyson and Banfield, 2008; Sun et al., 2012; Paez-Espino et al., 2013). The crRNA silencing requires identity with targeted sequences, and immunity may be lost by mutation in either the target region or an associated proto-spacer adjacent motif (PAM) that is required for CRISPR function ((Deveau et al., 2008; Horvath et al., 2008), reviewed in van der Oost et al., 2014)). Proto-spacers are the regions in phage and plasmid sequences flanked by PAMs that give rise to spacer sequences during adaptation; they also refer to the regions targeted by the CRISPR spacers during interference. Whereas some mechanistic aspects remain unclear, cells that contain at least one CRISPR spacer that perfectly matches a region of the invading phage or plasmid with a flanking PAM will be immune (Deveau et al., 2008; Horvath and Barrangou, 2010). However, studies in a certain type of CRISPR-Cas system have shown that mutations in the proto-spacer, nearest the PAM, allows the phage to escape, whereas mutations in other regions of the proto-spacer have no impact on immunity (Deveau et al., 2008; Semenova et al., 2011; Wiedenheft et al., 2011; Sun et al., 2012).
An important challenge in ecological studies is to detect and recover genome sequences from uncultivated phage and to link phage to their hosts. The CRISPR locus provides a means to address these two issues. First, spacer sequences extracted from CRISPR loci can be used to identify phage genome fragments in sequence datasets and initiate phage genome reconstruction (Andersson and Banfield, 2008). Second, assuming that hosts only incorporate spacer sequences from phage that infect them, CRISPR spacer sequences can be used to define the host range (Andersson and Banfield, 2008). Metagenomic datasets are a powerful way to approach these tasks because they simultaneously sample host CRISPR loci and the mobile elements they target.
Metagenomic datasets provide inventories of spacer sequences for a population. Identifying spacer targets can provide insight into the variety of phage and plasmids that target the host (Snyder et al., 2010). Metagenomic datasets also provide inventories of sequencing reads from the CRISPR locus. These can be used to compare the spacer complements of different host cells (for example, to determine whether different cells have the same immune potential), and provide insights into genetic diversity within a bacterial population (Held et al., 2013; Levin et al., 2013; Robles-Sikisaka et al., 2013). Importantly, the spacer complements of loci from coexisting individuals can be compared in a position-specific way to provide insight into population history. For example, conserved spacer sequences and spacer order in the older part of the locus may be interpreted to indicate origin and derivation from a common ancestral population or locus transfer (Tyson and Banfield, 2008), and transition from clonal to non-clonal loci has been suggested to indicate a recent selective sweep (Andersson and Banfield, 2008).
A limitation with metagenomic studies that target whole community DNA is that the sequencing is spread over entire genomes, so the number of reads recovered from a specific CRISPR locus may be insufficient to fully sample the spacer diversity of each host population. This problem is particularly significant if the loci are large and rapidly expanding. One approach to building a deeper inventory of CRISPR spacer sequences is to amplify the loci using PCR primers that target the repeat sequences (Pride et al., 2011). This relies upon knowledge of the repeat sequence, and can provide only very limited information about locus structure. Here, we combined metagenomic reconstruction with the PCR-based approach so as to take advantage of both methods. Using a metagenomic sequence, we reconstructed CRISPR locus architecture from natural populations of bacteria from the genus Leptospirillum and recovered sequences for their dominant phage. In addition, we used high-throughput sequencing to sample the spacer inventory of Leptospirillum deeply enough to assess population diversity and evaluate the phage/mobile elements they target. The analysis targeted biofilm samples collected over a 5-year period. The results show that population-level analyses of CRISPR loci can provide insight into phage-host interaction dynamics and the recent history of bacteria in natural systems.
Materials and methods
Identification of Cas proteins and construction of phylogenetic trees for Cas1
Cas proteins were identified using the CRISPR-Cas classification system developed by Makarova et al. (2011). Genes flanking the CRISPR loci in Leptospirillum group II and group II genomes were evaluated for conserved domains attributed to Cas proteins (Makarova et al., 2011). The order and identity of cas genes are shown in Supplementary Figure S2. The Cas1 proteins (translated in silico) from Leptospirillum group II and group II were then aligned with the 228 Cas1 proteins used in phylogenetic tree found in Figure 3 of Makarova et al. (2011). The re-alignment was completed using MUSCLE (Edgar, 2004), with some manual curation. A tree was generated with the final alignment by using FastTree (Price et al., 2009) (Supplementary Figure S3).
Preparation and sequencing of metagenomic and CRISPR amplicon samples
Extraction of community DNA and sequencing of biofilms sampled from the 5way (March 2002), UBA (June 2005 and November 2005) and C75 (June 2006, August 2006, November 2006, May 2007 and August 2007) locations in Richmond Mine, Iron Mountain, CA, USA (Table 1 and Supplementary Figure S1) have been previously described (Tyson et al., 2004; Lo et al., 2007; Andersson and Banfield, 2008; Denef and Banfield, 2012). Primers were designed to target the entire CRISPR locus in 5way- and UBA-type Leptospirillum group II genomes: 5′-GCTCTTTCAGCCAAGATGGT-3′ and 5′-TGGGGACCCTCCTTAGAAAT-3′. The primers target the regions immediately flanking the CRISPR locus (outside of the repeat-spacer arrays). Specifically, the primers target the putative transcriptional regulator and the region upstream of the cas2 (see Figure 1, Tyson and Banfield, 2008). CRISPR loci were amplified with these primers using the Hot Start Herculase (Stratagene, Agilent, La Jolla, CA, USA) with the an initial denaturation of 95 °C, then followed by 31 cycles of 92 °C for 30 s, 45 °C for 30 s and 72 °C for 8 min, and a final extension at 72 °C for 12 min. Agarose gel visualization of amplicons from both samples revealed a smear of fragments. Replicate PCR reactions were combined for 454 GS FLX sequencing, which was completed by the Joint Genome Institute (Walnut Creek, CA, USA). The PCR fragments were not size-selected as the amplicons were not expected to be an exact length. Thus, there may have been preferential sequencing of shorter fragments. The amplicon data from 5way and UBA have been deposited in SRA, with the accession numbers SRR2063344 (5way) and SRR2063507 (UBA).
Table 1. AMD sample information.
| Date | Location | pH | Temperature | Type | Platform | Seq. Data | LII Spacers | LII Groups | LIII Spacers | LIII Groups |
|---|---|---|---|---|---|---|---|---|---|---|
| Mar 2002 | 5way | 0.83 | 42.0 °C | MG PCR | Sanger 454 | 136 MB 52 MB | 389991 | 3516 | 157 | 91 |
| Jun 2005 | A Drift (UBA) | 1.1 | 41.0 °C | MG | Sanger | 114 MB | 1871 | 277 | 237 | 187 |
| Jul 2005 | A Drift (UBA) | 1.23 | 38.0 °C | PCR | 454 | 13 MB | 59764 | 716 | n/a | n/a |
| Nov 2005 | A Drift (UBA) | 1.50 | 38.0 °C | MG | Sanger | 106 MB | 41 | 41 | 48 | 44 |
| Jun 2006 | C Drift (C75) | 0.70 | 43.0 °C | MG | 454 | 80 MB | 143 | 75 | 0 | 0 |
| Aug 2006 | C Drift (C75) | 1.00 | 43.0 °C | MG | 454 | 88 MB | 79 | 61 | 0 | 0 |
| Nov 2006 | C Drift (C75) | 1.18 | 42.7 °C | MG | 454 | 90 MB | 261 | 115 | 8 | 8 |
| May 2007 | C Drift (C75) | 1.17 | 44.4 °C | MG | 454 | 96 MB | 304 | 100 | 7 | 7 |
| Aug 2007 | C Drift (C75) | 1.12 | 40.2 °C | MG | 454 | 95 MB | 232 | 99 | 0 | 0 |
Abbreviations: AMD, acid mine drainage; n/a, not applicable. Sampling conditions, sequencing information and the number of CRISPR spacers and CRISPR spacer groups recovered from Leptospirillum groups II (LII) and III (LIII) for each AMD biofilm collected for this study. Each line represents a separate sample. ‘Type' refers to the source of the sequencing, either metagenomic dataset (MG) or CRISPR amplicon dataset (PCR). In certain cases, spacers and spacer groups may be shared across multiple samples.
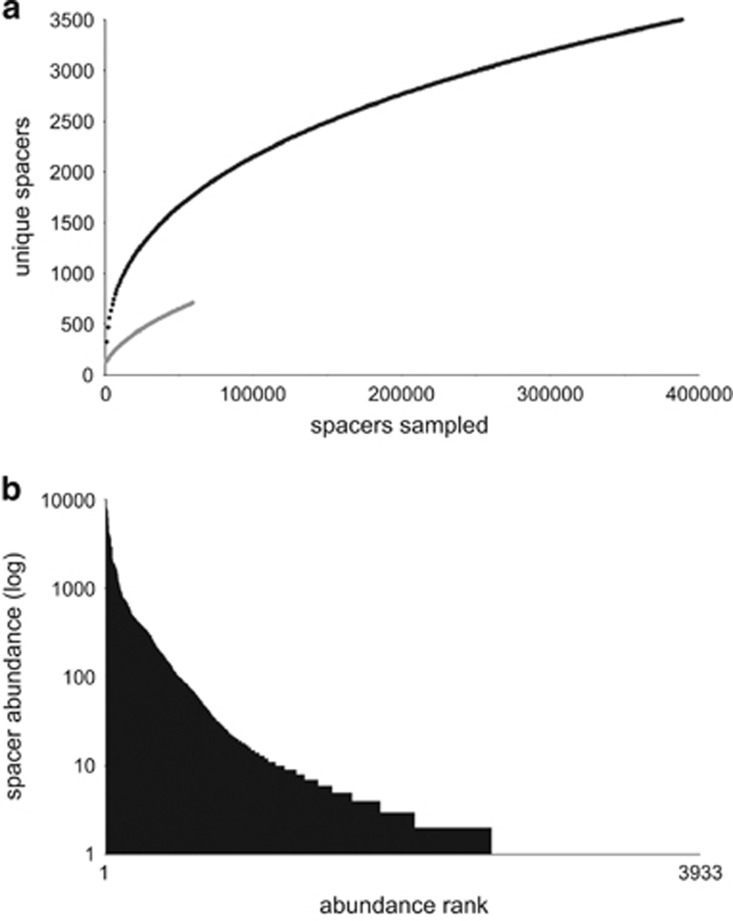
Figure 1.
CRISPR spacer diversity in Leptospirillum group II. (a) Rarefaction curve for spacer groups recovered from the 5way March 2002 sample (black line) and UBA July 2005 sample (grey line) datasets. Note that neither curve is approaching saturation, despite deep sampling. (b) Rank abundance graph for the 5way CRISPR showing that only a few spacer groups were highly sampled (>1000 counts).
Trimming and filtering of 454 sequencing reads from CRISPR amplicon datasets
In addition to the standard quality clipping of the 454 GS FLX read sequences performed by JGI, the SFF files were rescored using sffrescore (from the Genome Sequencer FLX System off-instrument software package) to generate the new phred-like quality scores. Because analysis of CRISPR spacers was conducted on the read level without assembly, extra filtering was performed to ensure good quality sequence. Reads containing at least one ambiguous base (‘N') were automatically removed (Huse et al., 2007). In addition, the ends of each reads were trimmed until a base passed 20/15 NQS (neighborhood quality standard) (Altshuler et al., 2000), with a variation described in Brockman et al. (2008). The program Cross_match (developed by P. Green, University of Washington) was used to remove any remaining B adaptor sequences (from the 454 library construction process) from the trimmed reads.
Screening of Sanger and 454 sequencing reads from metagenomic datasets
Prior to in silico spacer extraction from sequencing reads, we further screened the metagenomic data to remove reads that do not contain a CRISPR repeat. For individual Sanger reads, we required at least one instance of exact Leptospirillum group II (repeat) or group III (repeat) repeat sequence. For individual 454 reads, we required at least one instance of a Leptospirillum group II or group II repeat sequence, allowing for homopolymer errors in each position.
Extraction of CRISPR spacer sequences and code availability
We developed a custom Ruby script used to extract CRISPR spacer sequences from both CRISPR amplicons and metagenomic sequencing reads. The script has not been publicly released. Briefly, the spacers were extracted sequentially along each sequencing read. Generally, a sequence was only called a spacer only if the sequence was 29–36 bases long and flanked by two perfect Leptospirillum group II repeat sequences. Two exceptions occurred regarding the flanking repeat sequences: (i) If the spacer occurred near either end of a read, only a perfect match of at least six bases was required, and (ii) if the read was short (not long enough for repeat/spacer/repeat), at least a perfect match of six bases of the repeat flanking the potential spacer was required. Any read containing repeats in both directions was automatically removed from analysis. All spacers were deposited into a database, which stores read information, primer information and so on. Each unique spacer sequence is converted into a unique identification number (that is, spacer_4984) used in this text and in the Supplementary Materials.
We developed a suite of tools for analyzing CRISPR sequences from DNA sequence reads. The tools process raw reads, produce a summarized output and finally align CRISPR groups for analysis.
Initially, sequencing reads are processed using software that recursively scans the read for a repeat sequence. Starting at the 5′ end of the sequence and working towards the 3′ end, the software looks for the repeat, and if found, breaks the sequence on the repeat, creating a pre- and a post-fragment. The pre-fragment is analyzed to see whether it is of minimum length to be a spacer sequence as well to determine whether it contains a significant partial match to the repeat sequence. If it does, the pre-fragment is inventoried as a ‘matching spacer.' Alternatively, if the pre-fragment is too short or does not contain a partial repeat sequence, it is inventoried as an ‘unknown.' The post-fragment is then treated as a new sequencing read and analyzed again, but without the partial repeat scanning (because the original sequence was broken on a legitimate repeat sequence). This recursive analysis continues until the input read has been completely processed. The last fragment of the process is treated in a similar manner as the initial fragment and scanned for partial repeats. If a partial repeat is found, the spacer is inventoried. Otherwise, if the last fragment is too small too analyze, it is treated as an ‘unknown.'
Every step in the process is inventoried in a mysql database. This allows us to reconstruct the processing events of every read as it gets analyzed by the repeat matching software. Additionally, it simplifies report generation for the final step in the analysis. After processing all the sequencing reads for an amplification experiment, a report is generated that contains a summary of the unique spacers and every configuration that is found in the data set. For example, if a sequence contains the spacer configuration A-B-C, it is treated as distinct from A-C-B or A-B or A-C and so on. These are termed spacer groups, and the basis of the following alignment step.
Clustering of spacer sequences into groups and construction of rarefaction curves
To prevent an overestimation of CRISPR spacer diversity by accounting for 454 GS FLX sequencing read errors, the extracted spacers were grouped via BLASTclust, with parameters of 95% length overlap and 90% identity. Each group of spacers was considered one species and the abundance of each group was considered the abundance of each species, calculated from the total of the abundance of each spacer within the group. Rarefaction curves were created using Analytic Rarefaction 1.3 (developed by S. M. Holland, University of Georgia; program freely available at http://www.uga.edu/strata/software/).
Reconstruction of CRISPR loci variants
Spacer orders for each sequencing read (that contained at least two spacers) was obtained by listing the sequential order of each spacer (while ignoring the repeat sequence). The spacer order was then converted into a group order (all spacers were converted into groups), reducing the total overall amount of data used to assemble each CRISPR loci. The group orders were imported into Microsoft Excel and arrayed manually. Notably, the sequence of each spacer was listed as well to resolve any ambiguities.
Spacer matches to host genome and non-CRISPR reads
Spacer matches against the host genome and non-CRISPR-containing sequencing reads were determined by using blastn to detect perfect (100% match across 100% spacer length) and imperfect (90% identity over 85% length spacer length) nucleotide matches. For host genome matches, spacers were searched against the Leptospirillum group II and Leptospirillum group III genome sequences (Goltsman et al., 2009). For further analysis, only reads with proto-spacer sequences flanking an accurate PAM sequence (see below) were considered. For non-CRISPR-containing sequencing reads, the spacers were searched against all metagenomic read datasets listed in Table 1, with both Leptospirillum group II and Leptospirillum group III CRISPR reads removed.
Identification and frequency of PAM sequences
PAM sequences for both Leptospirillum group II and Leptospirillum group III were identified by obtaining all the 5′ and 3′ flanking sequences flanking a proto-spacer sequence that matched perfectly to a spacer. These flanking sequences from both ends were then used as input for the program Weblogo to examine the frequency of each nucleotide for every position. Notably for Leptospirillum group II, the 5′ flanking sequence contained three conserved nucleotides (‘AAG') and for Leptospirillum group III, the 5′ flanking sequence contained two conserved nucleotides (‘AA'). Both PAM sequences were immediately flanking the proto-spacer sequence. For all perfect and imperfect spacer/proto-spacer matches, the two and three bases immediately upstream of each proto-spacer sequence was obtained to determine the frequency of accurate and inaccurate PAM sequences for Leptospirillum group II and Leptospirillum group III, respectively.
Results
Sampling and genomic data
We studied nine microbial biofilm communities sampled from the air-water interface in the Richmond Mine (40° 40′ 38.42” N and 122” 31′ 19.90” W) at the 5way, A Drift (UBA) and C Drift (C75m) locations (Table 1 and Supplementary Figure S1). Analysis included Sanger-sequenced metagenomic data from the 5way (March 2002) and UBA location (June and November 2005) (Tyson et al., 2004; Lo et al., 2007; Andersson and Banfield, 2008). 454 FLX sequencing was applied to community DNA extracted from five samples from the C75 location (June 2006, August 2006, November 2006, May 2007 and August 2007) (Denef et al., 2010; Denef and Banfield, 2012).
Previous analysis of the Leptospirillum group II spacer complement indicated high levels of spacer diversity, especially in the 5way population (Tyson and Banfield, 2008), but sampling depth was insufficient to assess this inference in detail. Thus, for 5way (March 2002) and UBA (July 2005) samples, the entire Leptospirillum group II CRISPR locus was amplified with specific CRISPR primers and sequenced via 454 FLX (see Materials and methods).
CRISPR-Cas systems
From the metagenomic datasets (5way and UBA), we recovered spacers and associated Cas protein sequences from CRISPR regions in Leptospirillum group II and III genomes (Simmons et al., 2008; Tyson and Banfield, 2008; Goltsman et al., 2009). The two closely related species of Leptospirillum group II (5way and UBA type) both have a single type I-E CRISPR system (Supplementary Figure S2), whereas the Leptospirillum group III encodes two systems—one type I-E (Supplementary Figure S2) and one similar to type III (not discussed because of insufficient sequence coverage). The Cas1 proteins, present in all CRISPR-Cas systems, of the 5way and UBA types of Leptospirillum group II as well as Leptospirillum group III cluster closely with the Cas1 proteins of other type I-E systems (Supplementary Figure S3).
Spacer richness and diversity
Spacers were extracted in silico from single amplicon and metagenomic sequencing reads based on the detection of the Leptospirillum group II and III repeats (see Materials and methods). The average G+C content of the Leptospirillum group II and III spacers is 55% and 56% respectively, similar to G+C content of the genomes (55 and 58%). The average length of Leptospirillum group II spacers is 32.7±1.2 nucleotides while 33.1±0.4 is the average for Leptospirillum group III spacers. We detected 452 686 total and 18 187 unique Leptospirillum group II spacer sequences, and 457 total and 318 unique Leptospirillum group III spacer sequences (Table 1). High error rates in the individual 454 sequencing reads inflate the unique spacer count. Thus, the spacer sequences across all datasets were clustered into groups using blastclust, with the parameters of 85% length and 90% identity within each group. We identified a total of 3933 unique groups from Leptospirillum group II and 296 unique groups from Leptospirillum group III across all samples (Table 1).
For the two deeply sequenced Leptospirillum group II CRISPR amplicons datasets for the 5way and UBA samples, rarefaction curves were generated using spacer counts for each group found within each sample (Supplementary Table S1). Both curves demonstrate no approach to saturation, despite deep sampling, implying a large diversity of spacers in each CRISPR locus (Figure 1a). Most of the unique groups occur in the dataset only once or twice, for example, 35% of unique Leptospirillum group II groups are only found once across all datasets (Figure 1b). In contrast, a few spacers occur over 1000 times (Figure 1b).
Locus reconstruction
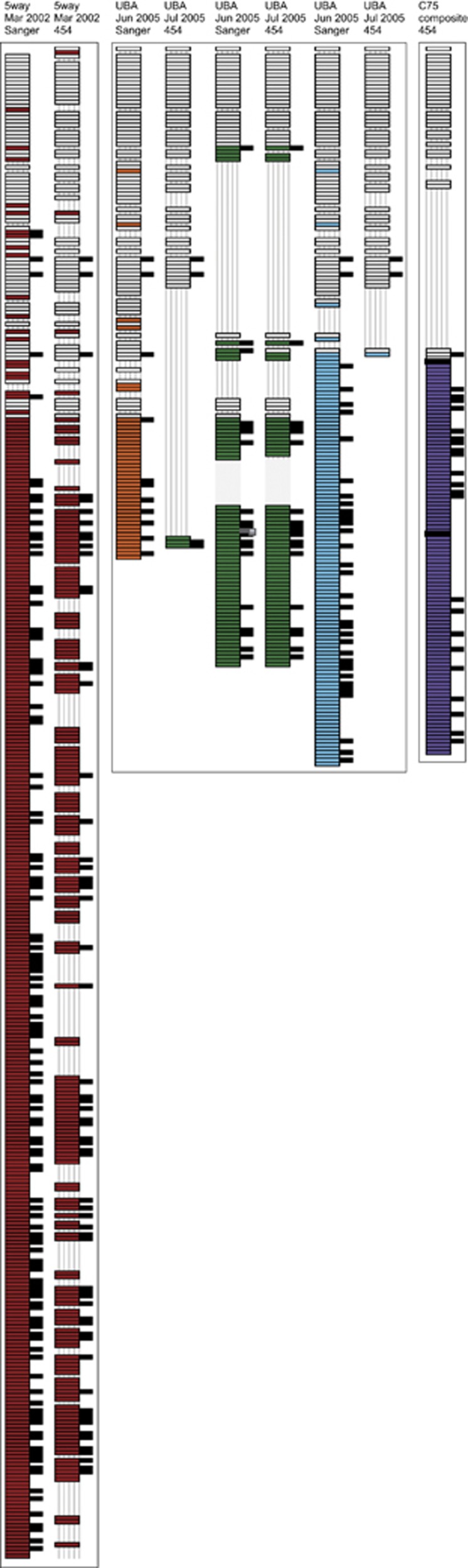
We determined the order of spacers and thus, the local locus spacer arrangement, by identifying two to five sequential spacers in individual reads from the metagenomic and amplicon datasets for Leptospirillum group II and group III CRISPR loci (Material and methods). By arraying overlapping patterns of spacers in different reads, we reconstructed the dominant variants of the CRISPR loci. The spacer sequence pattern in Leptospirillum group II was highly conserved at the trailer end (older end of the CRISPR locus) (Figure 2).
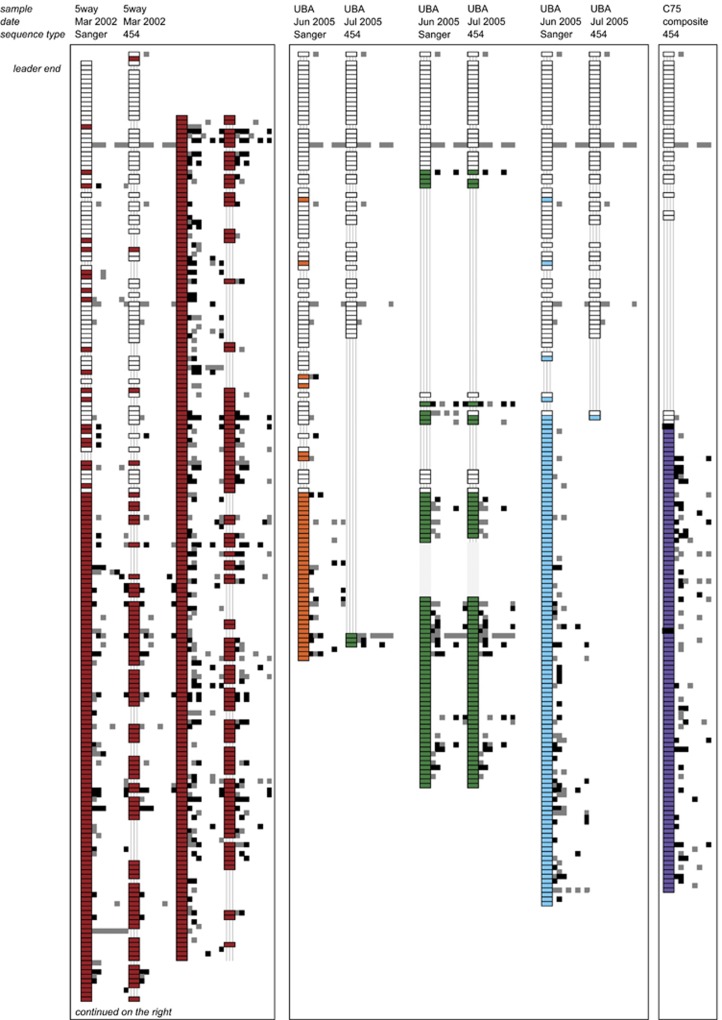
Figure 2.
Reconstruction of Leptospirillum group II CRISPR loci variants from 5way, UBA and C75 datasets. CRISPRs are shown vertically from trailer to leader end, with spacers represented as wide rectangles. White rectangles represent spacers shared between at least two CRISPR loci variants while colored rectangles represent spacers unique to a specific locus. Stripped lines show spacer loss. The two black rectangles in the C75 strain denote genes (transposase and hypothetical, from top to bottom) that interrupt the CRISPR locus. Note that the 5way variants are shown split in half owing to space constraints. In the eight columns right of each reconstructed CRISPR variant, the placement of squares indicates the sample that contained the matching mobile element sequence. The eight columns represent the following samples (from left to right): 5way-Mar 2002, UBA-Jun 2005, UBA-Nov 2005, C75-Jun 2006, C75-Aug 2006, C75-Nov 2006, C75-May 2007 and C75-Aug 2007. Perfect spacer matches with a PAM are shown as black squares while perfect spacer match without a PAM and imperfect spacer match with or without a PAM are shown as grey squares.
The CRISPR loci variants reconstructed from 454 sequenced amplicons were compared with variants reconstructed from Sanger metagenomic datasets from samples collected in 2002 and 2005 from the 5way and UBA locations, respectively (Tyson and Banfield, 2008). Figure 2 shows the trailer ends of reconstructed Leptospirillum group II CRISPR loci variants are conserved in all samples collected between 2002 and 2007. Consequently, spacer abundance correlates strongly with spacer position in the locus (Supplementary Figure S4). Within the shared block, excision of a single or multiple spacers are evident. There are locus-specific spacers found in the block of conserved shared spacers. These spacers may have been present in ancestral strains, but lost from all but one locus through excision.
We detect variation in Leptospirillum group II in the complement of spacers in coexisting individuals as well as variation in spacer complement in samples collected at different times. Lineage variants are defined by the spacer content at the leader end of the locus. When we compare the CRISPR locus structure inferred from the Sanger sequenced sample and that reconstructed from the 454 sequenced 5way sample, there is evidence of excision events throughout the locus. However, these samples were collected simultaneously. Although these may be real excisions occurring in the genomes, the possibility that missing spacers are PCR artifacts cannot be ruled out, so this is a potential limitation of the amplification approach.
In the two UBA samples used for Sanger and 454 sequencing that were collected only a month apart (Table 1), we could detect several distinct CRISPR loci variants (locus-specific series of spacers, colored in orange, green and blue). Interestingly, these variants display evidence for different patterns of spacer loss. There is evidence for differences in sub-strain (defined by spacer content) abundance between the samples. For example, the third UBA strain (blue in Figure 2) is essentially unrepresented in the sample collected 1 month later. Interestingly, there is evidence of locus recombination between UBA variants, as evidenced by the switch from trailer end spacers that are conserved and shared with the orange variant, followed by spacers unique to the green variant (Figure 2).
Spacers in the trailer end are also generally conserved in reconstructed Leptospirillum group III CRISPR loci (Supplementary Figure S5). Interesting, the number of spacers shared between CRISPR loci variants of Leptospirillum group III in the 5way (2002) and UBA (2005) samples is much lower than the number shared among the Leptospirillum group II loci across all time points (Figure 2).
PAM sequence
To search for the PAM for Leptospirillum group II and III CRISPR loci, we compared short stretches of sequence immediately flanking the targeted proto-spacer region of non-CRISPR read sequences (likely phage, plasmids). Flanking sequences of proto-spacers that perfectly matched spacers from all datasets were compared using WebLogo (Material and methods). For Leptospirillum group II, we detected a conserved tri-nucleotide ‘AAG' immediately flanking the 5′ end of the proto-spacers (Figure 3). For Leptospirillum group III, we identified the conserved di-nucleotide ‘AA', also flanking the 5′ end. We conclude that these are the PAM sequences required for CRISPR sampling and function. Notably, ‘AAG' is similar to the ‘AA' PAM of Escherichia coli, which is also a type I-E system (Mojica et al., 2009; Makarova et al., 2011).
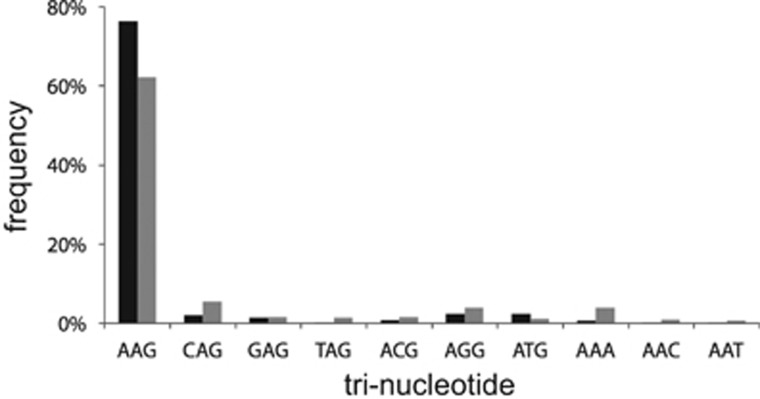
Figure 3.
Frequency of tri-nucleotide sequences in the PAM position of proto-spacers found across all samples. Only the accurate Leptospirillum group II PAM sequence (‘AAG') and imperfect PAMs (allowing for one polymorphism in any position) are shown. Relative abundances of perfect (black) and imperfect (grey) matches are shown.
We examined the frequency with which PAMs could be identified adjacent to protospacer regions targeted by spacers with perfect and imperfect matches. For Leptospirillum group II, we found that 76% of the 7643 spacers with perfect matches and 62% of the 27 559 spacers with imperfect matches have a PAM (Figure 3). The percentages increase to 86% and 83%, respectively, with the allowance of one polymorphism in any position in the tri-nucleotide for Leptospirillum group II (Figure 3). For Leptospirillum group III CRISPR loci, 94% of the 1046 spacers with a perfect match and 85% of the 1079 spacers with imperfect matches have a PAM.
Spacer matches to genome
We evaluated host genome self-targeting by all the Leptospirillum group II and group III spacers, considering both perfect (100% identity across entire length) and imperfect matches (90% identity over 85% length) (Supplementary Table S2,Supplementary Table S3). We limited the analysis to host genome regions with the PAM. This analysis used all the spacers within a group, not a representative sequence for the group. Leptospirillum group III spacers had only one match (perfect) to the host genome, and this targeted an intergenic region. For Leptospirillum group II, the majority of genes targeted by spacers with PAMs are transposases, hypothetical genes and other phage or plasmid genes (Supplementary Table S2).
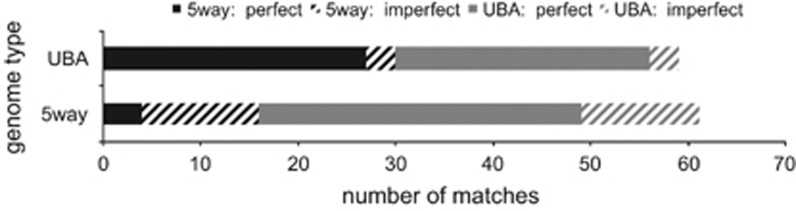
Overall, there are six genes in the Leptospirillum group II 5way-type genome and 26 genes in the UBA-type genome that have spacer matches (Supplementary Table S2). Spacers derived from the Leptospirillum group II 5way-type more often exactly match genes and intergenic regions of the UBA-type genome (27 matches) than its own genome (4 matches) (Figure 4). Similarly, spacers derived from the UBA-type more often exactly match the 5way-type genome (33 matches) than itself (26 matches) (Figure 4). However, this trend is not seen with the imperfect matches to genes or intergenic regions (Figure 4). Notably, one spacer group (group3548) is responsible for 90% of all matches of UBA-type CRISPR spacers to intergenic regions in both genomes types.
Figure 4.
Abundance of Leptospirillum group II spacers from 5way and UBA samples matching to non-CRISPR host genomic regions. Perfect (solid) and imperfect (striped) spacer matches to intergenic and intragenic regions in the 5way type Leptospirillum group II genome (black) and in the UBA type Leptospirillum group II genome (grey).
Spacer matches to non-genome DNA (phage and mobile elements)
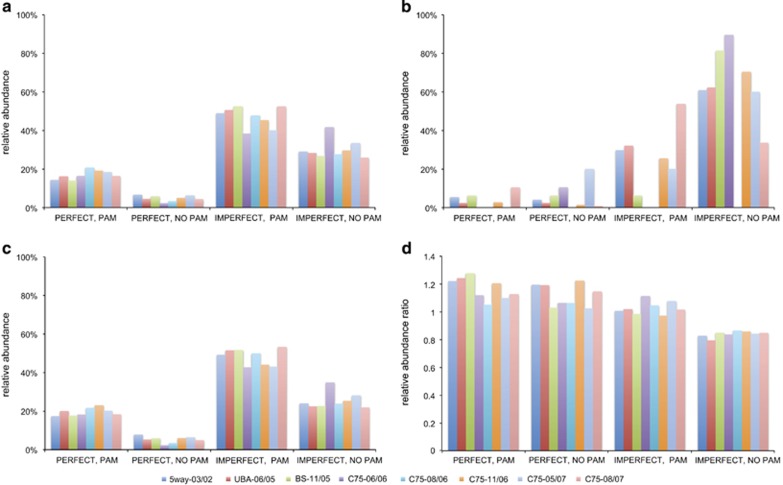
It is anticipated that reads with perfect or imperfect matches to spacers that are neither CRISPR nor host genome involve phage, plasmids or other mobile elements. We examined spacer matches to all reads in this study without clustering spacers into groups because, although clustering removes common sequencing errors, it also hides real sequence variants. The results are summarized in Figure 5; for full details, refer to Supplementary Table S4. For Leptospirillum group II, there were 35 564 matches (representing 7659 unique spacers from 1792 unique groups) to non-CRISPR, non-host genome read sequences. For Leptospirillum group III, there were 2125 (representing 199 unique spacers from 188 unique groups) (Supplementary Table S4). We categorized perfect and imperfect matches to non-CRISPR, non-host genome reads into four categories: perfect spacer matches with a PAM, imperfect spacer matches with a PAM, perfect spacer matches without a PAM and imperfect spacer matches without a PAM (Figure 5).
Figure 5.
Summary of Leptospirillum group II and group III spacer matches to non-CRISPR, non-host genome reads across all datasets. Matches are separated into four categories, as listed in the legend.
For each Leptospirillum group II CRISPR variant in each sample, the relative abundance of spacer matches is fairly consistent across the different match categories. The same is true for Leptospirillum group III CRISPR loci, though the patterns in Leptospirillum group II and group III differ. Imperfect matches with a PAM represented the most abundant category for Leptospirillum group II, whereas perfect matches with a PAM and imperfect matches with a PAM were the most abundant match types for Leptospirillum group III (Figure 5). Thus, the main difference between Leptospirillum group II and group III match types is the higher proportion of perfect matches with PAMs for Leptospirillum group III spacers. Notably, regardless of the spacer match type, there are consistently more matches with PAMs than without PAMs.
To determine the extent to which older spacers can silence phage and mobile elements, we tested for perfect and imperfect matches (black and grey boxes in Figure 2) as a function of spacer position within the CRISPR loci. In Figure 2, ‘imperfect matches' include every match except perfect spacer matches with a PAM. In Leptospirillum group II and III loci (Figure 2), we found that shared conserved spacers (found in more than one time point in same locus location in both genome types) have either no match or imperfect matches, with one exception (found in 5way Leptospirillum group II locus). This exception involves a spacer that occurs in multiple different locus contexts (attributed to sampling of the proto-spacer region in independent events). In contrast, there are numerous matches of locus-specific spacers (shown as colored boxes) to putative mobile elements.
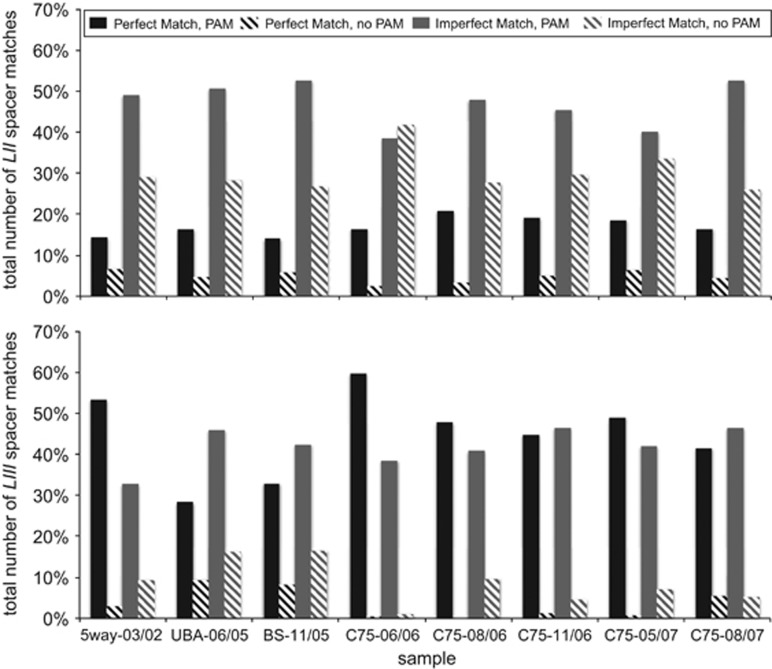
We also examined the relative abundance of the four different matches types as a function of spacer position within the Leptospirillum group II locus (Figure 6). The first panel shows match types regardless of locus position (Figure 6a), and is included for comparison with match types associated with the old (Figure 6b) and new (Figure 6c) end spacers. As noted above and in Figure 5, when including all spacers in the analysis regardless of their locus position, the most abundant category is an imperfect match with a PAM while the least common is a perfect spacer match without a PAM (Figure 6a). For the trailer end, which features spacers conserved across time, the most abundant type is imperfect matches without a PAM (Figure 6b). When only examining the spacers closer to the leader end (spacers not shown in Figure 2), the trend resembles that for all spacers (that is, Figure 6c resembles Figure 6a). To highlight subtle differences between Figures 6a and 6c, we plotted the ratio of the abundances (Figure 6d) and found a slightly elevated level of perfect matches with PAMs associated with protospacers matching spacers located at the leader end. The relatively small degree of elevation in perfect matches with PAMs is somewhat surprising, because trailer-end (newer) spacers are more likely to target co-existing phage and mobile elements.
Figure 6.
Different types of Leptospirillum group II spacer matches to all targets in the metagenomic datasets (excluding CRISPR and the host genome). The four types of matches include: perfect spacer matches with a PAM, imperfect spacer matches with a PAM, perfect spacer matches without a PAM and imperfect spacer matches without a PAM. (a) Plot shows relative abundance of matches from all spacers. (b) Plot shows relative abundance of matches from all spacers from the trailer end, limited to the region containing shared spacers (Figure 2). (c) Plot shows relative abundance of all matches from spacers from the leader end, limited to the spacers not shown in Figure 2. (d) The ratio of matches from spacers at the leader end relative to matches from all spacers.
The large number of spacer matches obtained for Leptospirillum group II (35 664) allowed us to test whether the relative frequency of mutations in the spacer sequences and PAMs is predicted by random mutation. If random, a simple expectation is that the ratio of mutation frequency in spacers vs PAMs should be predicted by the ratio of the lengths of the spacer and PAM (for Leptospirillum group II, this ratio is 10.4). Across the entire Leptospirillum group II CRISPR locus (Figure 5a), spacers with mutations (and a perfect PAM) are 9.2 times more common than PAMs with mutations (associated with a perfect spacer).
History of targeting of the known Leptospirillum phage, AMDV1
On the basis of the analysis of all CRISPR loci, spacers have been sampled from sites throughout the AMDV1 genome. For example, there are on average about nine different spacers that perfectly match the consensus sequence of the longest AMDV1 contig (Contig209; Supplementary Figure S6). The extent of sampling is likely much more extensive, as spacers that match AMDV1 strain variants are not included in this analysis.
To seek evidence for the persistence of the only well-defined Leptospirillum group II phage, AMDV1, in the acid mine drainage (AMD) ecosystem, we investigated the location of spacers within the CRISPR locus that target this phage population. The analysis included all spacers with perfect and imperfect matches to phage reads (Methods). Across all time points, spacers with matches to AMDV1 occur, and are associated with spacers found throughout most of the loci (black boxes in Figure 7). However, the oldest spacers shared among all loci do not contain detectable matches to any sequences (Figure 2).
Figure 7.
Spacers with matches to phage AMDV1 in Leptospirillum group II CRISPR loci from 5way, UBA and C75 datasets. Reconstructed loci are represented in the same manner as in Figure 2. In the column right of each reconstructed locus, the placement of black squares indicates the spacer has a perfect or imperfect match to phage AMDV1.
Notably, within the locus-specific spacers (colored boxes in Figure 7), there are several blocks of spacers within the first and second UBA June 2005 as well as C75 composite loci that that do not contain matches to phage AMDV1.
Discussion
We analyzed the targeting of the phage and mobile element populations by CRISPR spacers from Leptospirillum bacteria using data collected over a 5-year period. The sampling strategy, in combination with the high depth of sequencing achieved by targeting the CRISPR loci specifically, enabled us to identify an unprecedented number of spacer targets. This allowed us to detect changes in immune potential and in the effectiveness of spacers, and provided insight into the usefulness of locus reconstruction for recovery of information about population history.
The trailer end Leptospirillum group II spacers were largely conserved over the 5-year study period. Given low observed rates of trailer end change over the study period, the oldest spacers in the earliest sample were probably incorporated long before our first sampling, so the locus could potentially record information about phage community composition for well over 5 years. Trailer end conservation might indicate shared ancestry, although locus lateral transfer can complicate this interpretation (Tyson and Banfield, 2008). Similarly, the spacer complement can distinguish populations sampled only months apart, though changes may be due to environmental proliferation of strains with different CRISPR loci, spacer addition or both.
Most spacer diversity occurs at the leader end, as expected. Amplification and sequencing of the CRISPR region uncovered a vast variety of immune potential in one population. In fact, rarefaction curves derived from the recovered spacers show a lack of saturation, despite the unprecedented depth of sequencing. This finding provides support for prior speculation that, in some cases, most cells can contain different CRISPR loci (Tyson and Banfield, 2008; Paez-Espino et al., 2013). For loci with highly variable spacer complements, it may be inferred that the population has not experienced a strong bottleneck recently. In this circumstance, spacer sequences can be used to evaluate the diversity of coexisting phage and mobile elements. Two possible cases illustrate the potential utility of this approach for analysis of the recent growth environment of a host—Case one: a locus has spacers that target a single clonal phage; Case two: a locus has spacers that target a diverse phage population, as well as many different phage and plasmid types. In the first instance, we might infer recent growth in a simple ‘closed' environment, such as a laboratory culture; in the second, we might infer growth in a more complex, ‘open' diverse natural system. When attempting to recover information about the recent environment of an unknown bacterial strain population of medical or other significance, the effectiveness of CRISPR-based analyses will be higher if a large database of known phage types is available. Analysis of metagenomic sequences from coexisting phage can greatly augment this database.
Locus reconstruction provides a way of increasing the power of the CRISPR locus to provide information about recent population history. Specifically, if the spacer sequences can be classified into groups of new, older and old (based on where they occur on the locus), the targets for each group could be evaluated separately. In the current study, we find that older (but perhaps not the oldest) and new spacers target essentially the same phage population, a result that points to the persistence of Leptospirillum in an environment with the same phage population over the time period represented by the locus (>5 years). Notably, absence of targeting of AMDV1 by some mid-locus spacer blocks suggests short periods of fluctuation in phage exposure. Similarly, multiple strains (distinguished based on their CRISPR locus reconstruction) may record different exposure patterns. For example, the block of consecutive spacers in the UBA locus of sub-strain 2 without targets in our dataset, flanked by blocks with many targets, may record a period of time when that strain was exposed to a phage/mobile element pool not detected within the 5 years of our study. Lack of AMDV1 targeting by the oldest spacers may be due to virus evolution rather than the absence of the ancestral AMDV1 population. Regardless of the explanation in this case, phage evolutionary rates rather than spacer retention timescales, may determine the timespan for useful CRISPR-based tracking.
Because we generated a large dataset of CRISPR spacers, we could evaluate factors that determine the total spacer pool. The relevant parameters are the diversity of the phage/mobile element target populations and constrains on phage regions that can serve as spacer targets (proto-spacers). The most important consideration is apparently the requirement for a PAM. For phage AMDV1, there are 1445 detected PAMs for Leptospirillum group II (implying 1445 potential spacer sequences). Despite this, we found 3933 spacer groups. The great excess relative to the predicted spacer inventory, combined with the evidence of single mutations in PAM and spacer sequences, indicates that some spacer diversity is the result of resampling of rapidly evolving phage populations. Even after considering this effect, other targets (for example, as yet undetected other phage and plasmids) are likely required to explain the size of the inventory.
A few CRISPR spacers have matches to the host genome sequence. Notably, these almost only target mobile elements integrated into the host genome, not core functional genes. Self-targeting (chromosomal proto-spacer sequences with PAMs), regardless of the target type, should be a problem for the host if, as expected, the Cas machinery targets DNA (reviewed in Westra et al. (2012)). It is possible that this finding indicates that the target in this system is RNA, though most characterized type I systems target DNA. Alternatively, the spacer and genomic target may not coexist in the same genome (Vercoe et al., 2013; Gomaa et al., 2014).This is plausible, because many comparative genomic studies of closely related strains have shown that gain and loss of mobile element genes is a major contributor to divergence of coexisting individuals (for example, Allen et al. (2007)). This cannot be resolved in the current study because analyses involve short reads from innumerable coexisting individuals. However, if RNA targeting could be ruled out, apparent self-targeting or targeting of a gene recognized in other strains/species may indicate loss of the targeted gene (and the location of the spacer in the locus may distinguish recent from more gene ancient loss). Alternatively, these Leptospirillum group II with self-targeting spacers may have non-functional CRISPR-Cas systems.
The correspondence between spacer sequences and their targets (or lack thereof) can provide information about the factors that shape locus evolution. Generally, spacers in the ‘leader end' exactly match co-existing targets while those in the ‘trailer end' match imperfectly or even contain no detectable match at all. We infer that trailer end (inherited) spacers specific to only one population (for example, at one time point) were lost from all other populations sampled at other time points. In other words, there is pressure to maintain useful trailer end spacers if the element it targets is present, so detection of sample-specific trailer end spacer might imply the presence of the target only in that sample.
In addition to locus position-dependence of the degree to which spacers in a population match to co-existing targets (Figure 6), there are differences in the likelihood that a spacer will target a proto-spacer region with a PAM. Interestingly, the spacer region tends to mutate before the PAM, with frequencies approximately as expected for random mutation, and only spacers close to the old typically lack matches to sites with PAMs. This suggests that, on average, spacers transition through the locus until the probability is high that they are ineffective. This, and the balance between the spacer addition rate and phage mutation rate, may be important determinants of locus length.
Currently, sequencing of CRISPR repeats and analysis of spacer order in isolates is used for strain tracking (Liu et al., 2011; Shariat et al., 2013). The present study illustrates that metagenomic sampling of natural communities of bacteria, phage, plasmids and other mobile elements provides additional information. Specifically, comparison of loci in coexisting individuals provides insight into population diversity. In addition to uncovering evidence for recent bottlenecks, spacer inventory analysis can constrain the complexity of the current and past environments in which a bacterial population has grown. When applied to environmental populations, spacer matches to mobile elements provide insights into the recent history of exposure of host strains to the phage and plasmid pool.
Acknowledgments
We thank the late Mr T. W. Arman (President, Iron Mountain Mines Inc.) and Mr R. Sugarek (U.S. Environmental Protection Agency) for site access, and Mr R. Carver for on-site assistance. We thank Banfield laboratory members for their contributions to sample collection. We thank Dr A. Andersson for general and preliminary work and Dr G. Tyson for Leptospirillum group II CRISPR primer design. This work was supported by grant #DE-FG02-07ER64505 from the Department of Energy and #W911NF-10-0046 from the Army Research Office. 454 GS FLX sequencing of CRISPR amplicons was supported by a Community Sequence Project (CSP) from the Department of Energy Joint Genome Institute.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Rodolphe Barrangou is an inventor on several patents related to CRISPR-Cas systems and their various uses. The other authors declare no conflict of interest.
Supplementary Material
References
- Allen EE, Banfield JF. (2005). Community genomics in microbial ecology and evolution. Nat Rev Microbiol 3: 489–498. [DOI] [PubMed] [Google Scholar]
- Allen EE, Tyson GW, Whitaker RJ, Detter JC, Richardson PM, Banfield JF. (2007). Genome dynamics in a natural archaeal population. Proc Natl Acad Sci USA 104: 1883–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Pollara VJ, Van Etten WJ, Baldwin J, Linton L et al. (2000). An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407: 513–516. [DOI] [PubMed] [Google Scholar]
- Andersson AF, Banfield JF. (2008). Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA. (2014). CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell 54: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman W, Alvarez P, Young S, Garber M, Giannoukos G, Lee WL et al. (2008). Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res 18: 763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H. (2004). Phage-host interaction: an ecological perspective. J Bacteriol 186: 3677–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef VJ, Banfield JF. (2012). In situ evolutionary rate measurements show ecological success of recently emerged bacterial hybrids. Science 336: 462–466. [DOI] [PubMed] [Google Scholar]
- Denef VJ, Kalnejais LH, Mueller RS, Wilmes P, Baker BJ, Thomas BC et al. (2010). Inaugural Article: Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc Natl Acad Sci USA 107: 2383–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P et al. (2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190: 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltsman DS, Denef VJ, Singer SW, VerBerkmoes NC, Lefsrud M, Mueller RS et al. (2009). Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing ‘Leptospirillum rubarum' (Group II) and ‘Leptospirillum ferrodiazotrophum' (Group III) bacteria in acid mine drainage biofilms. Appl Environ Microbiol 75: 4599–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. (2014). Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio 5: e00928–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held NL, Herrera A, Whitaker RJ. (2013). Reassortment of CRISPR repeat-spacer loci in Sulfolobus islandicus - Held - 2013 - Environmental Microbiology - Wiley Online Library. Environ Microbiol 15: 3065–3076. [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170. [DOI] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S et al. (2008). Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190: 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. (2007). Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8: R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman P, Abedon ST. (2010). Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70: 217–248; Available at http://linkinghub.elsevier.com/retrieve/pii/S0065216410700071. [DOI] [PubMed] [Google Scholar]
- Levin BR, Moineau S, Bushman M, Barrangou R. (2013). The population and evolutionary dynamics of phage and bacteria with CRISPR–mediated immunity. PLoS Genet 9: e1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. (2011). Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl Environ Microbiol 77: 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo I, Denef VJ, Verberkmoes NC, Shah MB, Goltsman D, DiBartolo G et al. (2007). Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446: 537–541. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P et al. (2011). Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol 9: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Almendros C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733–740. [DOI] [PubMed] [Google Scholar]
- Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K-I, Stahl B et al. (2013). Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun 4: 1430. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride DT, Sun CL, Salzman J, Rao N, Loomer P, Armitage GC et al. (2011). Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time. Genome Res 21: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles-Sikisaka R, Ly M, Boehm T, Naidu M, Salzman J, Pride DT. (2013). Association between living environment and human oral viral ecology. ISME J 7: 1710–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B et al. (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA 108: 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariat N, Kirchner MK, Sandt CH, Trees E, Barrangou R, Dudley EG. (2013). Subtyping of Salmonella enterica serovar Newport outbreak isolates by CRISPR-MVLST and determination of the relationship between CRISPR-MVLST and PFGE results. J Clin Microbiol 51: 2328–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons SL, DiBartolo G, Denef VJ, Goltsman DS, Thelen MP, Banfield JF. (2008). Population genomic analysis of strain variation in Leptospirillum Group II bacteria involved in acid mine drainage formation. PLoS Biol 6: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JC, Bateson MM, Lavin M, Young MJ. (2010). Use of cellular CRISPR (clusters of regularly interspaced short palindromic repeats) spacer-based microarrays for detection of viruses in environmental samples. Appl Environ Microbiol 76: 7251–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. (2013). CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82: 237–266. [DOI] [PubMed] [Google Scholar]
- Sun CL, Barrangou R, Thomas BC, Horvath P, Fremaux C, Banfield JF. (2012). Phage mutations in response to CRISPR diversification in a bacterial population. Environ Microbiol 15: 463–470. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Banfield JF. (2008). Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol 10: 200–207. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43. [DOI] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. (2014). Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol 12: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS et al. (2013). Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet 9: e1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Buckling A, Fineran PC. (2014). CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol 12: 317–326. [DOI] [PubMed] [Google Scholar]
- Westra ER, Swarts DC, Staals RHJ, Jore MM, Brouns SJJ, van der Oost J. (2012). The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet 46: 311–339. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Bultema J, Waghmare SP, Waghmare S et al. (2011). RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA 108: 10092–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.