Abstract
The Alphaproteobacterium Rhizobium radiobacter F4 (RrF4) was originally characterized as an endofungal bacterium in the beneficial endophytic Sebacinalean fungus Piriformospora indica. Although attempts to cure P. indica from RrF4 repeatedly failed, the bacterium can easily be grown in pure culture. Here, we report on RrF4's genome and the beneficial impact the free-living bacterium has on plants. In contrast to other endofungal bacteria, the genome size of RrF4 is not reduced. Instead, it shows a high degree of similarity to the plant pathogenic R. radiobacter (formerly: Agrobacterium tumefaciens) C58, except vibrant differences in both the tumor-inducing (pTi) and the accessor (pAt) plasmids, which can explain the loss of RrF4's pathogenicity. Similar to its fungal host, RrF4 colonizes plant roots without host preference and forms aggregates of attached cells and dense biofilms at the root surface of maturation zones. RrF4-colonized plants show increased biomass and enhanced resistance against bacterial leaf pathogens. Mutational analysis showed that, similar to P. indica, resistance mediated by RrF4 was dependent on the plant's jasmonate-based induced systemic resistance (ISR) pathway. Consistent with this, RrF4- and P. indica-induced pattern of defense gene expression were similar. In clear contrast to P. indica, but similar to plant growth-promoting rhizobacteria, RrF4 colonized not only the root outer cortex but also spread beyond the endodermis into the stele. On the basis of our findings, RrF4 is an efficient plant growth-promoting bacterium.
Introduction
Although soil-borne beneficial microbes are prevalent, their phylogenetic, spatial and functional diversity is widely unresolved (Berg et al., 2014). Complex symbiotic interactions including bacteria, fungi and their host plants are especially little understood, and their global prevalence is widely unknown (Lackner et al., 2009; Hoffman and Arnold 2010; Naumann et al., 2010). To address this issue, we have been studying the tripartite Sebacinalean symbiosis, comprising fungi of the order Sebacinales (Basidiomycota), phylogenetically diverse endofungal bacteria and a broad range of monocotyledonous and dicotyledonous host plants (for review, see Qiang et al., 2012a). A first analysis of the distribution on a world-wide scale claimed that Sebacinalean symbioses are prevalent in all continents (Weiss et al., 2011; Riess et al., 2014) potentially making them a vital part of global soil ecosystems. Although the molecular mechanisms by which the Sebacinalean symbiosis is established and by which the plant benefits from its fungal partners is largely understood, the role of the endofungal bacteria is still unclear (Peškan-Berghöfer et al., 2004; Waller et al., 2005; Deshmukh et al., 2006; Camehl et al., 2010; Zuccaro et al., 2011; Qiang et al., 2012b).
The root endophyte Piriformospora indica (syn. Serendipita indica) is a model fungus of the Serendipitaceae of the Sebacinales (Oberwinkler et al., 2014). Since its discovery in the Indian Thar dessert in 1996 (Varma et al., 1999), P. indica and related Sebacina vermifera strains were shown to promote biomass, yield and health of a broad spectrum of plants (Varma et al., 2012; Ye et al., 2014). Genetic and biochemical assessments of the resistance mechanism that is induced by P. indica in Arabidopsis thaliana (Arabidopsis) against a wide spectrum of leaf and root pathogens showed a requirement of jasmonate synthesis and signaling and thus an operable induced systemic resistance (ISR) pathway (Stein et al., 2008; Jacobs et al., 2011).
In 2008, Sharma and coworkers (Sharma et al., 2008) reported for the first time that members of the Serendipitaceae regularly undergo complex symbioses involving plants and endofungal bacteria of different genera. Endofungal bacteria were previously detected in Glomeromycotan arbuscular mycorrhiza symbioses (Bonfante and Anca, 2009; Naumann et al., 2010), in the ectomycorrhizal fungus Laccaria bicolor (Bertaux et al., 2003, 2005), in the rice pathogenic fungus Rhizopus microsporus (Partida-Martinez and Hertweck, 2005), in hyphae of phylogenetically diverse foliar fungal endophytes (Hoffman and Arnold, 2010), in the soil isolate Mortierella alpina (Kai et al., 2012) and the plant symbiotic Endogone Mucoromycotina fungi (Desirò et al., 2015). Endofungal bacteria, associated with fungi of the genera Piriformospora and Sebacina, belong to two genera of Gram-negative (Rhizobium and Acinetobacter) and two genera of Gram-positive (Paenibacillus and Rhodococcus) bacteria. The most comprehensively studied example of a tripartite Sebacinalean symbiosis is the association of P. indica with the Alphaproteobacterium Rhizobium radiobacter (syn. Agrobacterium radiobacter; syn. Agrobacterium tumefaciens) strain F4 (RrF4). Fluorescence in situ hybridization using a Rhizobium-specific probe confirmed the stable endocellular association of small numbers of RrF4 cells within P. indica chlamydospores and hyphae, and a ratio of 0.035 ng of bacterial DNA per 100 ng of P. indica DNA was determined by quantitative PCR (qPCR) analysis (Sharma et al., 2008). This result coincides well with the low number of different bacteria (1–20 per fungal cell) that were detected in the ectomycorrhiza fungus Laccaria bicolor (Bertaux et al., 2003; Bertaux et al., 2005). Intriguingly, RrF4 could be isolated from powdered fungal mycelia and propagated in axenic cultures, showing that the bacterium is not entirely dependent on its fungal host. However, various attempts to stably cure P. indica of its resident bacterial cells repeatedly failed, raising the possibility that RrF4 is essential for P. indica survival. Consistent with an intricate association, no other bacteria have been isolated from various laboratory cultures of P. indica.
In an attempt to further elucidate the plant growth-promoting function of the endofungal RrF4, its genome was sequenced and compared with closely related non-endofungal R. radiobacter and other related Agrobacterium strains. We also show that RrF4's root colonization pattern and beneficial activity are widely reminiscent of those induced by its fungal host P. indica. Our data suggest that the ubiquitous Sebacinalean symbiosis is a novel source for beneficial bacteria potentially useful for agronomic applications.
Materials and methods
RrF4 strain
Experiments were performed with Rhizobium radiobacter (syn. Agrobacterium radiobacter; syn. Agrobacterium tumefaciens) F4(RrF4), a subculture of strain PABac-DSM isolated from P. indica DSM 11827 (Sharma et al., 2008). Generation of RrF4 strains expressing ß-glucuronidase (GUS) and green fluorescent protein (GFP) is described in the Supplementary Materials and Methods.
RrF4 genome sequence analysis
Genomic DNA of strain RrF4 was isolated and shotgun libraries were prepared for sequencing. Emulsion PCR, emulsion breaking of DNA-enriched beads and sequencing of the shotgun libraries were performed on a second-generation pyrosequencer (454 GS FLX Titanium, Roche, Mannheim, Germany) using Titanium reagents and Titanium procedures as recommended by the developer following protocols for shotgun sequencing.
Quality filtering of the pyrosequencing reads was performed using the automatic standard signal processing pipeline of the GS Run Processor (Roche) to remove failed and low-quality reads from raw data and to remove adaptor sequences.
The initial assembly of the data from 454 pyrosequencing was performed using the GS FLX Newbler software 2.0.01 (Roche) with a minimum overlap length of 40 bp and a minimum overlap identity of 90% followed by a comparative alignment to the genome of R. radiobacter (syn. Agrobacterium tumefaciens, A. radiobacter) C58, which finally resulted in four distinct contigs. The data were uploaded into GenDB (Meyer et al., 2003) and subjected to an automatic annotation. Blast ring images were generated using BRIG (Alikhan et al., 2011). Genome comparisons were performed in EDGAR (Blom et al., 2009). This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession JZLL00000000. The version described in this paper is version JZLL01000000.
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0, N1092) and the following mutants were obtained from the Nottingham Arabidopsis Stock Centre (NASC): npr1-1 (Cao et al., 1994), npr1-3, jar1-1 (Staswick et al., 1992), ein2-1 (Guzman and Ecker, 1990), jin1 (Berger et al., 1996) and NahG (Gaffney et al., 1993). Seeds were germinated on half strength MS medium supplemented with 0.5% (w/v) sucrose and solidified with 0.4% (w/v) gelling agent (Gelrite, Duchefa, BH Haarlem, The Netherlands) under short day conditions (8 h light at 22 °C/18 °C (day/night) and a photon flux density of 183 μmol m−2 s−1) in 100 mm square Petri dishes. Alternatively, plants were grown in pots containing soil (Archut Fruhstorfer Erde, Type P, HAWITA Gruppe, Lauterbach, Germany) or a vermiculite (Ø 0–3 mm, Deutsche Vermiculite Dämmstoffe GmbH, Sprockhövel, Germany)—sand mixture (3:1).
Seeds of wheat cv. Bobwhite and barley cv. Golden Promise (GP) were germinated on sterilized wet filter paper in sterile glass jars for 3 days. Plants were grown in a growth chamber at 22 °C/18 °C (day/night cycle), with 60% relative humidity, and a photoperiod of 16 h (240 μmol m−2 s−1 photon flux density). Before germination, all seeds were surface-sterilized with ethanol 70% (v/v) for 3 min (wheat/barley) or 1 min (Arabidopsis) and 25% sodium hypochlorite for 90 min (wheat/barley) or 10 min (Arabidopsis) under continuous shaking, washed once with sterilized water (pH 3) and rinsed four times with sterile distilled water.
Inoculation of root with RrF4
RrF4 was grown overnight in modified LB broth (1% casamino hydrolysate, 0.5% yeast extract and 0.5% NaCl, pH 7.0 supplemented with 100 μg ml−1 gentamycin) at 28 °C and 150 r.p.m. GUS- and GFP-expressing RrF4 were cultured in the presence of 100 μg ml−1 spectinomycin. Bacterial cells were collected by centrifugation (3202 g, 10 min), washed and resuspended in 10 mM MgSO4 7H2O buffer. Roots of 3-day-old barley/wheat seedlings or 7-day-old Arabidopsis seedlings, respectively, were dip-inoculated for 30 min in RrF4 suspensions (OD600=1.0–1.4). Control seedlings were dipped into 10 mM MgSO4 7H2O.
Quantification of RrF4 in roots
The relative abundance of RrF4 in root tissues was quantified by qPCR targeting the internal transcribed spacer (ITS) between the 16S rRNA and 23S rRNA genes of the ribosomal RNA operon of RrF4 (Sharma et al., 2008; see Supplementary Table S1 and Supplementary Materials and Methods).
Visualization of root colonization by RrF4
The colonization of plant roots was visualized using GUS- and GFP-expressing RrF4 strains combined with light-, epifluorescence- and confocal laser scanning microscopy. Root cross-sections also were analyzed by transmission electron microcopy (TEM) (see Supplementary Materials and Methods).
Gene expression analysis in roots
Roots of 3-day-old barley seedlings were dip-inoculated with a solution of RrF4 (OD600=1.4) for 30 min and with a fungal spore solution (500 000 spore per ml) for 1.5 h. Seedlings were transferred to pots containing a 2:1 mixture of expanded clay (Seramis, Masterfoods, Verden, Germany) and Oil-Dri (Damolin, Mettmann, Germany) under long day condition. Fertilization was carried out weekly with 20 ml of a 0.1% WUXAL top N solution (N/P/K: 12/4/6; Aglukon, Düsseldorf, Germany) per pot containing one plant. Total RNA extraction, cDNA synthesis and qPCR analysis were performed according to Jacobs et al. (2011).
Plant infection with Pseudomonas
Pseudomonas syringae pv. tomato DC3000 (Pst; Preston, 2000) was obtained from Dr N. Schlaich, RWTH Aachen, Germany. Pst was grown on liquid King's B medium (King et al., 1954) supplemented with 50 μg ml−1 rifampicin and 50 μg ml−1 kanamycin overnight at 28 °C. Bacterial cells were collected by centrifugation (3202 g; 10 min), washed and suspended in 10 mM MgSO4 7H2O containing 0.02% Silwet L-77 to a concentration of 108 colony-forming units (CFU) per ml (OD600=0.2).
For the analysis of induced resistance, roots of 2-week-old Arabidopsis plants were dip-inoculated with RrF4 (OD600=1) for 30 min and grown in soil. After 2 weeks, leaves were sprayed with Pst (0.5 ml bacterial solution on each plant). One day before challenge inoculation, plants were placed in transparent boxes with closed lids to increase the relative humidity for optimal infection. Gene expression values were normalized to the housekeeping gene AtUBQ5-4 values using the 2−ΔCt method (Schmittgen and Livak, 2008). Primers used in this study are listed in Supplementary Table S1.
Statistical analysis
Statistical analyses were performed in SigmaPlot 12 (Systat Software) using one-way analysis of variance or Student's t-tests after data were tested for normality distribution (Shapiro-Wilk test) and equal variance.
Results and Discussion
Comparative genome analysis and phylogenetic placement of RrF4
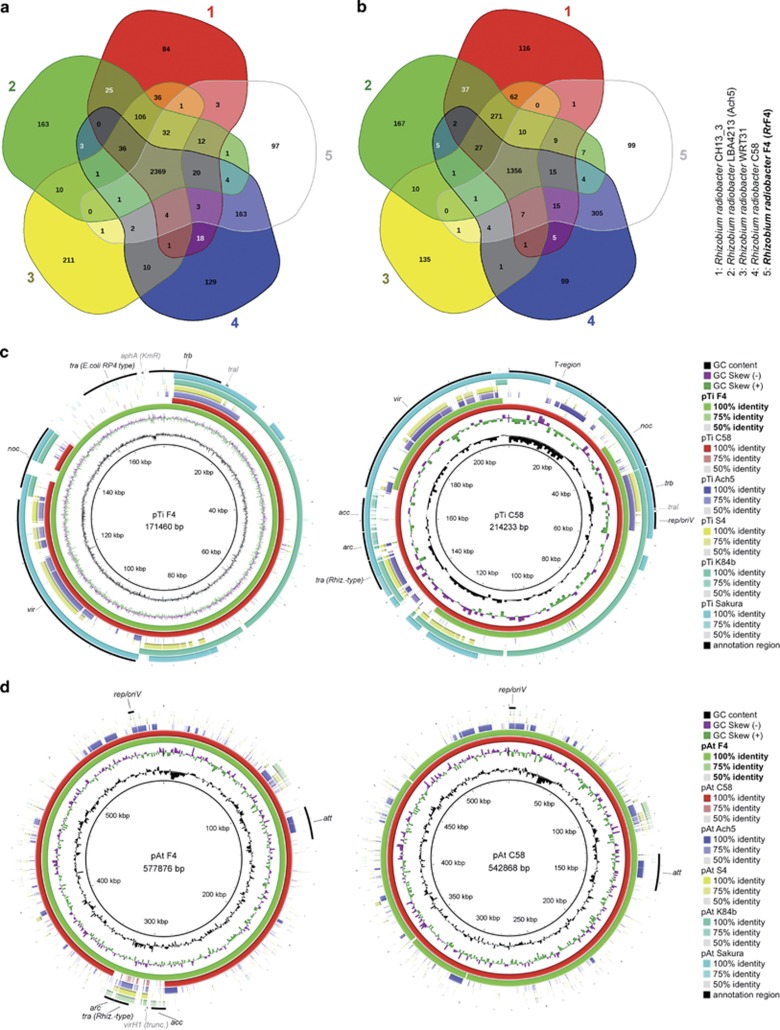
The genome of RrF4 is organized in a circular (2.8 Mbp) and a linear chromosome (2.06 Mbp), a tumor-inducing plasmid pTiF4 (0.21 Mbp), and an accessory plasmid pAtF4 (0.54 Mbp) in the same manner as R. radiobacter C58 (formerly: A. tumefaciens C58; Agrobacterium biovar I, genomovar G8) (Table 1; Goodner et al., 2001). This unusual genome structure, containing a linear chromosome, has been observed only for Agrobacterium biovar I strains (Slater et al., 2013). A phylogenetic tree calculated based on the core genome of RrF4 and Agrobacterium biovar I, II and III reference strains (Supplementary Figure S1a) and average amino acid identity analyses (Supplementary Figure S1b) showed the assignment of RrF4 to biovar I strains with very close relationship to C58 (amino acid identity=99.8%). The presence of all genomovar G8-specific gene cluster defined by Lassalle et al. (2011) indicated the assignment of RrF4 genomovar G8 in the same manner as C58 though some difference in C58-specific gene cluster indicated genetic distinction of both strains (Supplementary Table S2).
Table 1. Comparison of chromosome and plasmid sizes and GC contents of the genomes of RrF4 and C58 and comparison of the location and presence of gene regions on the two plasmids of RrF4 and C58.
| Circ. chr. | Lin. chr. | pTi | pAt | |
|---|---|---|---|---|
| Size and G+C content of chromosomes and plasmids | ||||
| RrF4 size (Mbp) | 2.80 | 2.06 | 0.17 | 0.58 |
| C58 size (Mbp) | 2.84 | 2.08 | 0.21 | 0.54 |
| RrF4 GC content (%) | 59.41 | 59.32 | 58.63 | 57.42 |
| C58 GC content (%) | 59.38 | 59.28 | 56.67 | 57.33 |
| Feature | RrF4 | C58 |
|---|---|---|
| Localization of gene regions on the pAt and pTi plasmids | ||
| T-region (incl. ipt) | missing | pTi |
| Acc | pAt | pTi |
| arc (incl. traR) | pAt | pTi |
| tra (Rhiz.-type) | pAt | pTi |
| tra (E. coli pRP4) type) | pTi | missing |
| virH1 | pAt (truncated by 200 bp) | pTi |
Abbreviations: Circ. chr., circular chromosome; Lin. chr., linear chromosome. Genome sequence of C58 was obtained from GenBank of NCBI (Genome Acc. number: AE007869.2-AE007872.2).
Comparison of the gene content of the circular chromosome of RrF4 and the four biovar I reference strains showed that RrF4 shared 87.3% of its genes (2369 of 2713 genes) with all biovar I strains, but 94.5% with strain C58 (2563 of 2713 genes), among those 163 genes (6.4% of the total genes) were only shared with C58 (Figure 1a). Moreover, 73.9% (1356 of 1834 genes) of the genes from the linear chromosome were shared with all biovar I strains but 98.2% (1702 of 1834 genes) with C58, among those 305 genes (17.9% of the total genes) were only shared with C58 (Figure 1b). Several of the genes encoded by the circular and linear chromosome of C58 and RrF4 are relevant not only for pathogenicity but also for the plant-microbe interaction in general (Goodner et al., 2001; Heindl et al., 2014). Among those, we detected homologs that were reported to be involved in the interaction of plant growth-promoting bacteria with host plants (Supplementary Table S3). In contrast to other, obligate endofungal bacteria that live in symbiosis with their fungal hosts, RrF4 does not have a reduced genome (Lackner et al., 2011; Ghignone et al., 2012; Fujimura et al., 2014; Torres-Cortés et al., 2015; Naito et al., 2015), which may indicate a facultative symbiosis of RrF4 with P. indica. The high similarity (in size and gene content) of the chromosomal genomes of RrF4 and C58 indicated that RrF4 did not lose any essential genes or partial genetic pathways. The circular and linear chromosomes of RrF4 had 100 and 80 singleton open reading frames, respectively, not present in C58. Most of these open reading frames were of unknown function and may be candidates for future studies to elucidate a potential role for the endofungal growth of RrF4 (Supplementary Table S1) and/or fitness of its fungal partner P. indica.
Figure 1.
Venn diagrams showing shared and unique genes of RrF4 and biovar I reference strains. (a) Circular chromosome, (b) linear chromosome. (c, d) BRIG-comparison of different Rhizobium pTi and pAt plasmids. The name of the reference plasmid sequence is given in the center of the rings, the concentric circles show the similarities of the compared plasmids as explained in the legend on the right of every picture. acc=agrocinopine A+B uptake and catabolism operon; arc=agrocinopine regulation of conjugation operon; noc=nopaline catabolism region; T-region=transfer T-DNA region at plant infection; tra (Rhizobium-type)=genes for conjugation/DNA metabolism homolog to Rhizobium spp.; tra (E. coli RP4 type)=genes for conjugation/DNA metabolism homolog to E. coli RP4 plasmid; trb=operon for plasmid transfer (mating pair formation), incl. traI; vir=virulence gene cluster; aphA=Kanamycin resistance gene; rep/oriV=replication genes repABC; att=gene cluster required for attachment to plant cells. Acc. numbers of reference genomes are listed in the legend of Supplementary Figure S1.
Whereas RrF4 and C58 showed a high degree of similarity based on the circular and linear chromosomes, the plasmids were more diverse (Figures 1c and d). These differences partly stem from gene translocations in RrF4 from the pTi to pAt plasmid, partly from unique sequences in both strains (Table 1). The acc operon, responsible for agrocinopine A+B uptake and catabolism, the arc operon for agrocinopine regulation of conjugation, a truncated version of the virH1 gene and the tra operon described by Piper et al. (1999) were translocated in RrF4 from plasmid pTi to pAt. The latter one also includes the quorum sensing regulator gene traR but not the AHL synthase traI, which remains upstream of the trb operon on pTi in both strains. Furthermore, RrF4 harbors an additional set of tra genes on the pTiF4 plasmid which shows highest similarity to the conjugation genes on the RP4 plasmid of Escherichia coli. Most interestingly, in contrast to C58, RrF4 lacks the complete transfer DNA region and some adjacent genes belonging to the nopalin catabolic (noc) region. It is hard to speculate which role the additional E. coli-like tra genes might have for the non-pathogenic RrF4. While the ‘Rhizobium-like' tra genes are usually involved in the transfer of the T-region to the infected plant cells and are regulated by a luxI/luxR type quorum sensing system (Fuqua and Winans, 1994), the E. coli tra genes are required for bacterial conjugation (Frost et al., 1994).
Although it shares nearly identical circular and linear chromosomes with the pathogenic A. tumefaciens C58, RrF4 is non-pathogenic. As shown previously, and consistent with our finding, curing C58 from its pTi plasmid results in a non-pathogenic strain with albeit weak plant growth-stimulating activity (Walker et al., 2013). Nevertheless, at present, we do not know all the factors needed by RrF4 to associate with its fungal host P. indica. Lackner et al. (2011) denoted the intrahyphal Burkholderia rhizoxinica as a bacterium with a ‘genome in transition' because it showed to some extent a reduced genome size compared with free-living Burkholderia species which was not as strongly reduced as found in several other endobacteria. Although RrF4 did not show a reduced genome size, changes in the structure and gene content of both, the pTi and pAt plasmids may hint to an adaptation to a specific ecological niche. The gene content alone, however, will not be sufficient to explain the endofungal lifestyle of RrF4; further studies must elucidate the molecular communication in the tripartite association between bacterium, fungus and plant.
RrF4 promotes plant growth
Treatment of barley roots with free-living RrF4 increased plant biomass and powdery mildew resistance (Sharma et al., 2008). To further expand this finding, we assessed the potential of RrF4 to mediate growth promotion in Arabidopsis. To this end, roots of 7-day-old seedlings were dip-inoculated with RrF4 (OD600=1.0) and subsequently transferred to the three different growth substrates soil-sand mixtures (3:1), vermiculite-sand mixtures (3:1) and half strength MS medium. Two to three weeks after inoculation, plants showed a significant increase in shoot and root fresh weight (FW), and strongly accelerated lateral root formation, regardless of the substrate in which they were grown (Supplementary Figures S2a–f). This finding adds another level of complexity to the yet controversial discussion as to whether P. indica induces biomass formation in Brassicaceae (Lahrmann et al., 2013). Our data doubtless show that the bacterial partner of P. indica is able to strongly induce growth promotion in Arabidopsis.
RrF4 colonizes roots and proliferates independent of its fungal host
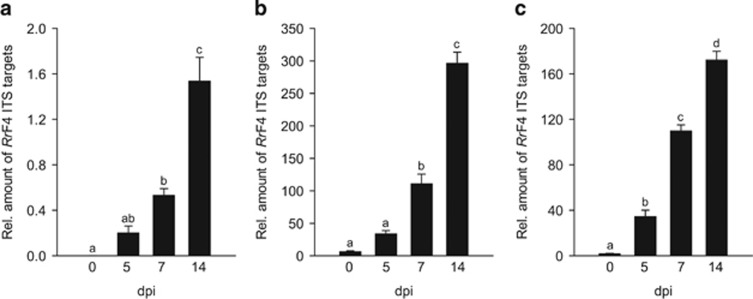
We asked whether free-living RrF4 can colonize plant roots and multiply independently of its fungal host. Because axenically grown plants are most suitable for bacterial quantification and microscopy, Arabidopsis seedlings grown on half strength MS medium were used for this analysis. At various time points after dip-inoculation, root material was collected and RrF4 was quantified by qPCR targeting the ITS of the ribosomal RNA operon. Before DNA extraction, roots were sequentially washed in 70% ethanol and distilled water and sonicated, to remove excess of bacteria from the surface. Upon this treatment (30 min after dip-inoculation =0 days post inoculation (dpi)), the amount of RrF4 cells that remained attached to the roots was below the detection limit. During 2 weeks, RrF4 cells proliferated in the apical 4 cm of the roots as indicated by a significant increase in RrF4 ITS targets relative to plant ubiquitin (Figure 2a). Consistent with an increase in the relative abundance, the absolute amounts of RrF4 cells per gram root FW increased (Supplementary Table S4). At 14 dpi, bacterial cell numbers reached 2.9 (±0.5) × 109 cells per gram Arabidopsis root FW.
Figure 2.
Proliferation of RrF4 in roots of Arabidopsis (a), barley (b) and wheat (c). Roots of 7-day-old seedlings (Arabidopsis) or 3-day-old seedlings (barley, wheat) were dip-inoculated with RrF4. Genomic DNA was extracted at the indicated time point and RrF4 ITS targets quantified by qPCR relative to Arabidopsis or barley ubiquitin or wheat alpha tubulin, respectively. Error bars indicate standard errors based on three independent biological replicates. Different letters on top of the bars indicate statistically different differences tested by one-way analysis of variance (all pairwise multiple comparison) performed with the Tukey test (P<0.05).
In parallel, the propagation of RrF4 was assessed in roots of barley and wheat seedlings (Figures 2b and c). Higher ratio values for ITS targets vs root gene targets suggests a more intense bacterial colonization of these graminaceous plants. In contrast to Arabidopsis, relatively high amounts of RrF4 cells remained attached to the root surface after dip-inoculation (0 dpi; barley: 2.1 (±0.4) × 108; wheat: 3.3 (±2.0) × 108 cells per gram FW of roots (Supplementary Table S4)), which may indicate that RrF4 bacteria interact more efficaciously with the surface of graminaceous plants. At 14 dpi, bacterial cell numbers reached 8.1 (±1.4) × 109 (barley) and 6.0 (±2.5) × 109 (wheat) per gram root FW, suggesting that RrF4 interacts and multiplies in graminaceous plants with high efficiency.
RrF4 multiplies at the root surface
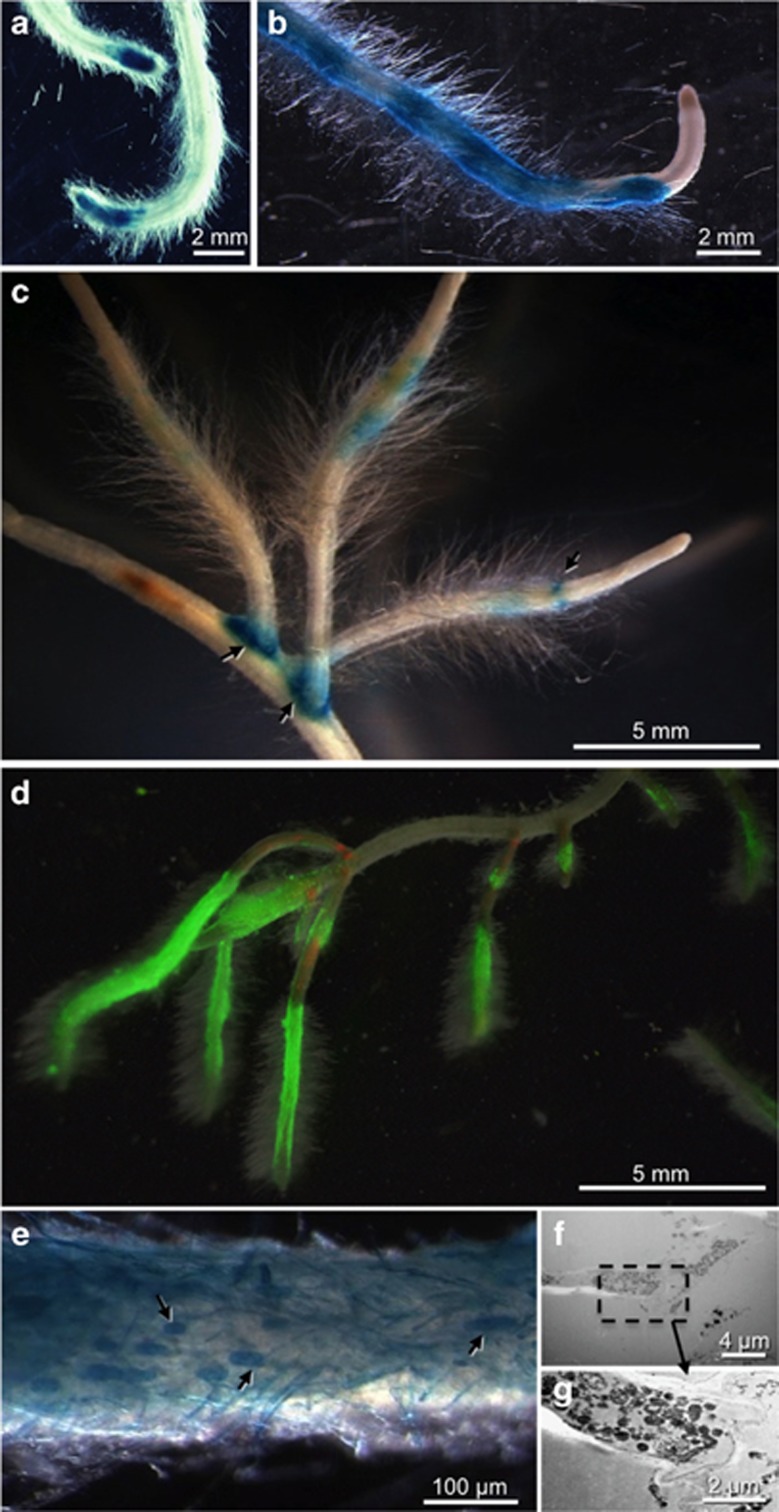
Next, colonization of barley roots was assessed microscopically with GUS- and GFP-tagged bacteria (Figure 3). At 5 dpi, GUS-expressing RrF4 cells were seen in the maturation zone I of primary roots in an area covering approximately 1 cm in length (Figure 3a). At later time points (14 dpi), bacteria had spread into the maturation zone II while the elongation and meristematic zones as well as the root cap remained virtually free of bacteria (Figure 3b). This colonization pattern conspicuously resembles the root colonization pattern of P. indica (Deshmukh et al., 2006). Microscopy also revealed a distinct pattern of dark and bright blue staining in the root hair zone suggesting specific sites of higher RrF4 proliferation at the root surface (Figure 3b). At later time points, root hair zones of lateral roots also were colonized with the same pattern as in primary roots. Bacterial conglomerates were particularly present at lateral root protrusions (Figure 3c) that probably serve as entry sites. Inoculation of roots with GFP-tagged RrF4 cells confirmed this pattern. Microscopic assessment at 30 dpi revealed strong bacterial colonization of primary and secondary roots (Figure 3d). Inoculation of wheat and Arabidopsis roots with RrF4 virtually showed the same colonization pattern (Supplementary Figure S3) and accordingly resembled the pattern observed with P. indica (Supplementary Figure S4; also see Jacobs et al., 2011, Figure 1).
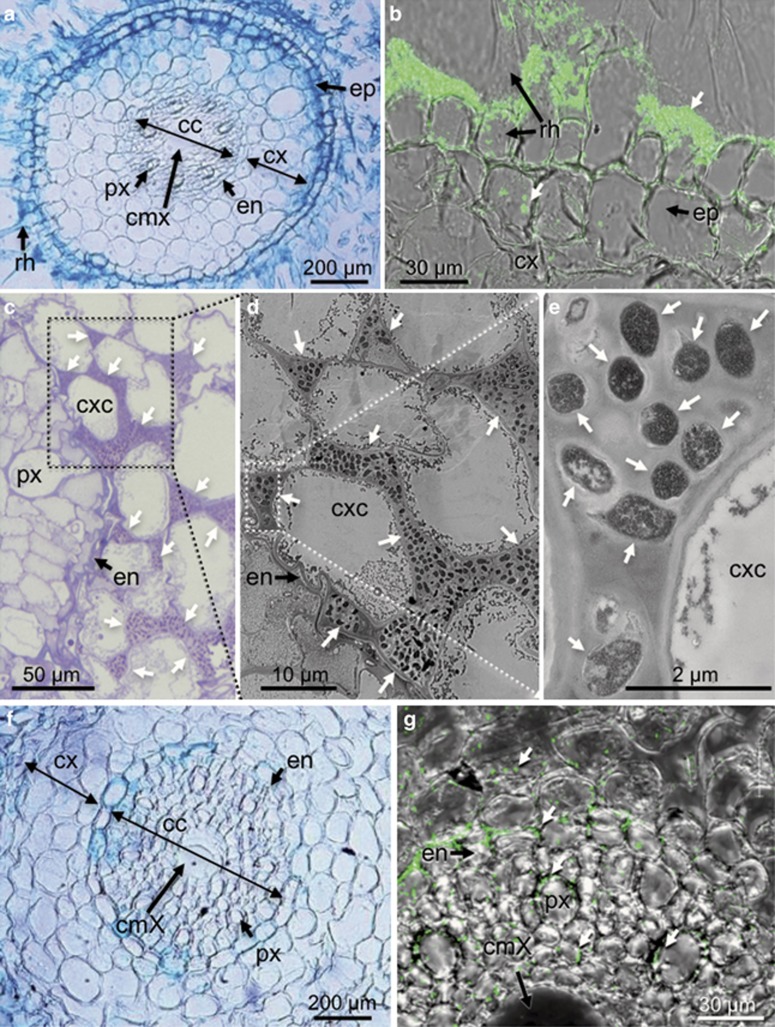
Figure 3.
Localization of GUS- and GFP-expressing RrF4 in barley roots. GUS-expressing RrF4 on a primary root at 5 (a) and 14 dpi (b), and on lateral protrusions and hair zones of secondary roots at 21 dpi (c). GFP-expressing RrF4 on hair zones (30 dpi) visualized by epifluorescence microscopy (d). (e–g) Single (dead) rhizodermal cells colonized by GUS-tagged RrF4 at 21 dpi. Light microscopy (e) and TEM (f, g) of GUS-tagged RrF4. Dark blue stained plant cells (arrows) are densely packed with bacteria. Dashed box: area heightened in (f); arrows: bacterial cells.
GUS staining showed single dark blue GUS-stained rhizodermal cells suggesting that these cells were heavily colonized by RrF4 (Figure 3e), which was further confirmed by TEM (Figures 3f and g). The high number of bacterial cells as well as the fact that intact cytoplasm and plant cell organelles were neither detected by TEM nor by light microscopy after 4',6-diamidino-2-phenylindole staining (data not shown) indicate that the colonized plant cells were dead. Whether these plant cells were dead before bacterial colonization remains to be resolved.
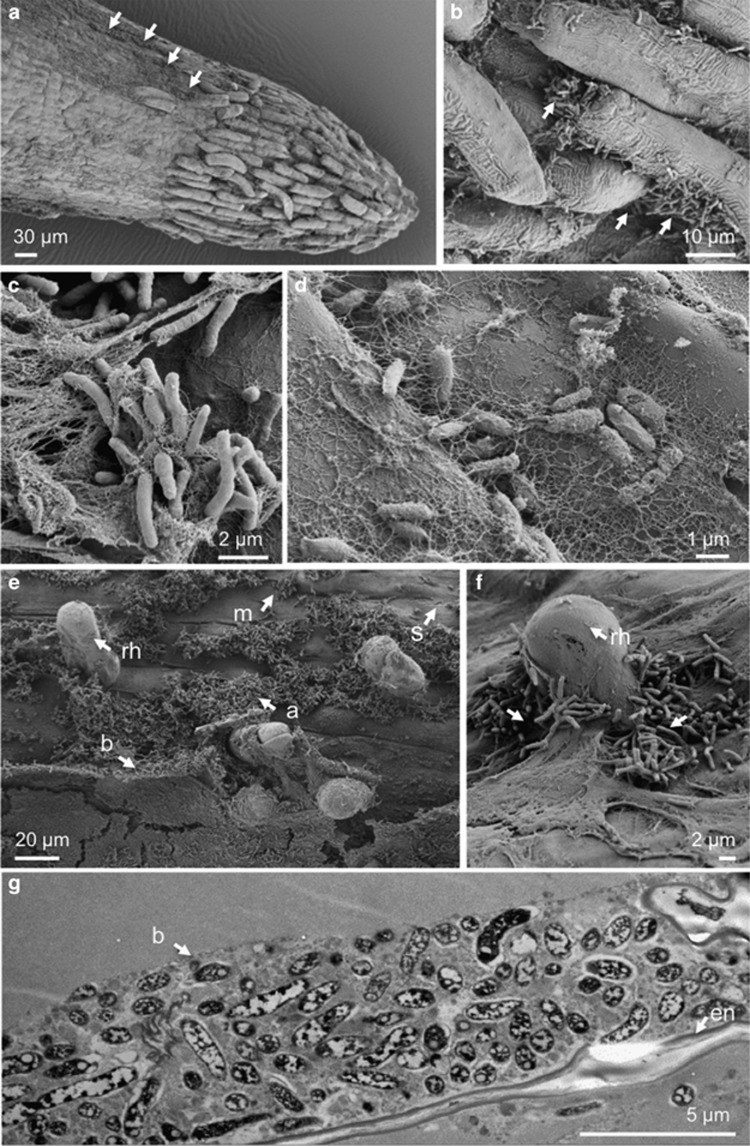
The colonization of barley primary roots was further investigated by TEM and scanning electron microscopy. Consistent with the above findings, root-cap and elongation zones were much less colonized by RrF4 (Figures 4a–d) compared with root hair zones, where RrF4 formed dense surface-attached biofilms (Figures 4e–g). Root cap-colonizing RrF4 cells formed cell aggregates located in cracks between root cap cells but did not fully cover the cell surface (Figures 4a and b). These cell aggregates and single surface-attached cells were cross-linked by fiber-like structures (Figures 4c and d). It was previously described that cellulose production leads to a loose aggregation of R. radiobacter cells (Matthysse, 1983) and is required for efficient root surface attachment of ‘Agrobacterium tumefaciens' (reviewed by Heindl et al., 2014; Matthysse, 2014). Because the two gene clusters required for cellulose production in R. radiobacter C58, celABCG and celDE, are present at the linear chromosome of RrF4 (Supplementary Table 3), we presumed that the fiber-like structures can be cellulose.
Figure 4.
Colonization of barley primary roots by RrF4 analyzed by scanning (a–f) and transmission (g) electron microscopy. (a, b) RrF4 at the root tip (b is a zoom out of a showing RrF4 cells at cracks of the root cap cells, arrows). (c) RrF4 cell aggregates at the root tip. (d) Single RrF4 cells attached to the root surface distal to the tip area. Bacterial cells are cross-linked by fiber-like structures. (e) Different stages in biofilm formation at the rhizoplane of the root hair zone: single RrF4 cells attached to the rhizoplane (s); micro-colonies formed through multiplication of single attached cells (m); larger cell aggregates (a), thick surface attached biofilm (b); root hairs (rh). (f) RrF4 cell aggregates around the root hair protrusion site; assumed area of penetration into the root tissue (arrow). (g) Surface-attached dense biofilm of RrF4 cells. Bacterial cells are embedded in an extracellular polysaccharide (EPS)-like structure forming a four to five cell layers thick biofilm at the root surface (b).
Larger cell aggregates (micro-colonies) often developed at the sites of root hair protrusion rather than being attached to fully developed root hairs (Figures 4e and f). Root hair protrusion sides serve as bacterial entrance into the inner root tissue (Compant et al., 2005). TEM showed a detailed picture of the approximately 5-μm-thick multilayer biofilm with RrF4 cells embedded by a dense matrix of extracellular polymeric substances (Figure 4g). This matrix may be formed by extracellular polysaccharides, extracellular DNA or (glyco)proteins (Flemming and Wingender, 2010). Like C58, the genome of RrF4 contains several genes for polysaccharide synthesis such as cellulose, succinoglycan, glucans and outer membrane lipopolysaccharides (Goodner et al., 2001; Supplementary Table S4), which are known to be involved in the formation of extracellular biofilm matrix of ‘Agrobacterium tumefaciens' strains (Heindl et al., 2014; Matthysse, 2014).
RrF4 colonizes the intercellular space of the root cortex and, unlike P. indica, proceeds into the central cylinder
Cross-sections of barley roots were analyzed for bacteria colonizing the inner root (Figure 5). At 7 dpi, GUS-tagged or GFP-tagged RrF4 were visible in the root cortex (Figures 5a and b). Bacteria were often detected at cell junctions of rhizodermal and cortical cells. Light microscopy and TEM of ultrathin root cross-sections showed a dense colonization of the intercellular spaces in the cortex tissue up to the endodermis (Figures 5c–e). Root hair cells and cortex cells were not colonized, which implies an extracellular colonization pattern.
Figure 5.
Localization of RrF4 in the root hair zone of barley primary roots. (a, b) Root cross-sections showing the colonization of the root surface and the cortical tissue 7 dpi with (a) GUS- and (b) GFP-tagged RrF4. (c–e) Light microscopic (c) and TEM (d, e) images of ultra-thin cross-sections of the primary root shown in (a) illustrating the intercellular localization of RrF4 in the cortex; (c) overview; (d) RrF4 cells in the intercellular space; (e) zoom to single intercellular rod-shaped RrF4 cells. (f, g) Cross-sections showing colonization of the central cylinder at 21 dpi by GUS- (f) or GFP-tagged RrF4 cells (g). (a, f) Light microscopic and (b, g) confocal laser scanning images. Epidermis (ep), root hair cells (rh), cortex (cx), cortex-cells (cxc), endodermis (en), central cylinder (cc), peripheral xylem vessels (pX) central metaxylem vessels (cmX). Right arrows highlight bacterial cells.
At later time points (21 dpi), RrF4 cells were detected at cell junctions of endodermal cells and in the central cylinder. While we cannot exclude that RrF4 is able to pass the endodermis, it is more likely that bacterial cells invaded the vascular tissue via the non-maturated tissue of the elongation zone where differentiation has not been completed yet. Inside the central cylinder, bacteria also were seen in the intercellular spaces, whereas intracellular colonization was not unambiguously detectable (Figures 5f and g). This finding is crucial insofar as colonization of the inner part of the root beyond the endodermis has not been observed with RrF4's fungal host P. indica (Deshmukh et al., 2006; Jacobs et al., 2011). At 21 dpi, very few single GFP-tagged RrF4 cells also were detected inside the stem but not in the leaves. Consistent with this, RrF4-specific ITS targets could not be amplified by qPCR from shoot and leaf extracts (at 14 and 21 dpi), and RrF4 cells also could not be cultured from surface-sterilized leaf tissue. The later data suggest that, in contrast to other endophytic bacteria (Rothballer et al., 2008; Hardoim et al., 2008; Van der Ent et al., 2009), RrF4 cells do not spread systemically into the upper plant tissue.
RrF4 colonizes the interior of Arabidopsis roots
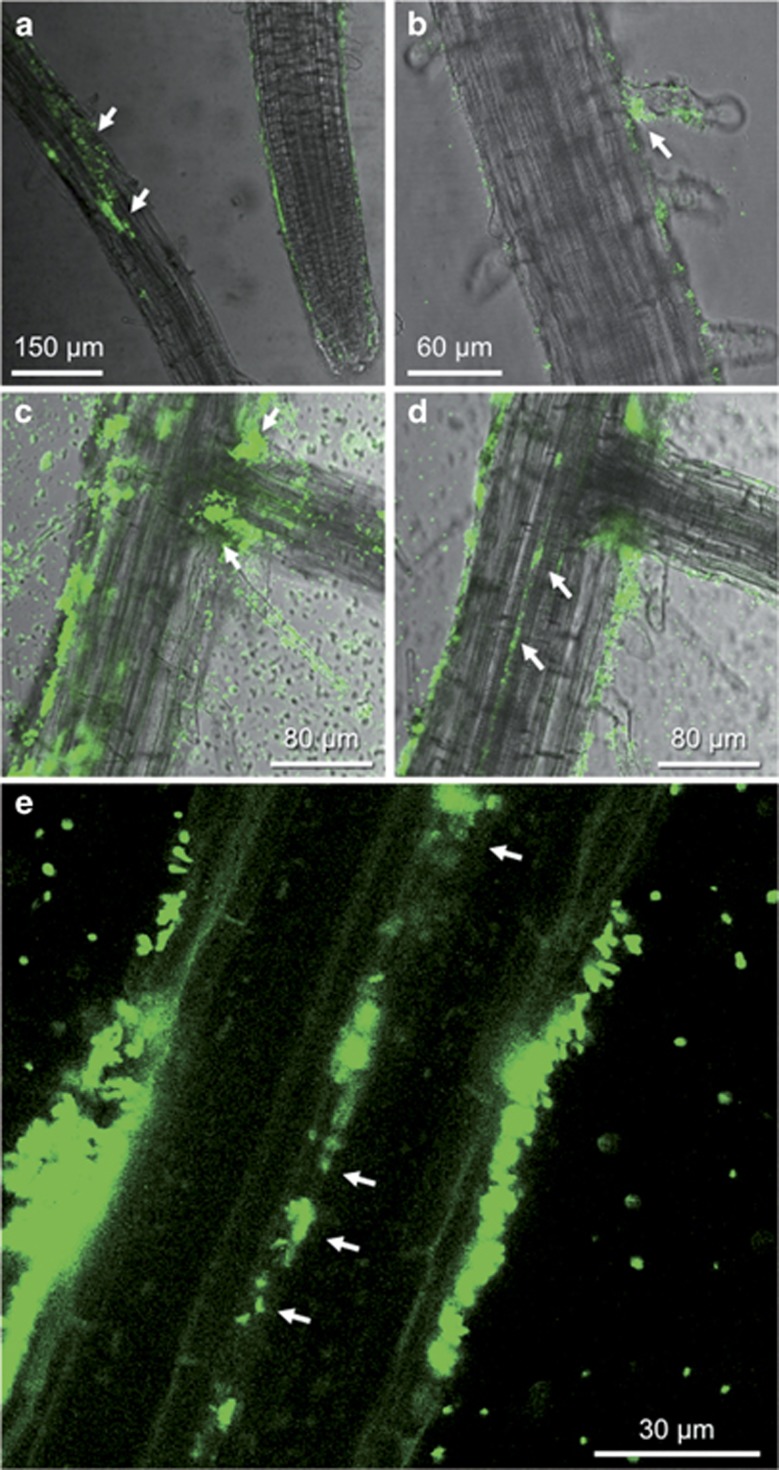
The colonization pattern in Arabidopsis roots was assessed using GFP-tagged bacteria. At 7 and 14 dpi, single RrF4 cells as well as dense, locally restricted aggregates were seen at the surface of root hair zones, the surface of root hairs and, in higher abundance, at the base of root hair cells (Figures 6a–d). Compared with barley, biofilms were less dense (data not shown), and bacterial cells were again not detected within root hair cells. At a later time point (21 dpi), RrF4 also colonized the sites of secondary root emergence. Cracks at secondary root emergence sites may be used by RrF4 for passive entrance into the inner root tissue as was suggested for several endophytic bacteria (Reinhold-Hurek and Hurek, 1998; Compant et al., 2005). In contrast to barley, the central vascular system (xylem) of primary (Figures 6d and e) and secondary roots (data not shown) were heavily colonized. However, like in barley, RrF4 cells were hardly detectable in the green parts of the plant.
Figure 6.
Colonization of Arabidopsis roots by GFP-tagged RrF4. Images were taken at 7 (a, b) and 21 (c-e) days after root dip-inoculation. (a) RrF4 colonization of the root surface in the root hair zone with single attached bacteria, thin biofilms and dense locally restricted aggregates (arrows). (b) Colonization of the root hairs mainly at the cell bottom (arrows). (c) Localization of bacteria forming biofilms and aggregates at the root surface of the primary root and at the sites of lateral root protrusions (arrows). (d, e) Bacterial cells inside the plant's vascular system (arrow: xylem). (c–e) Different layers of a confocal laser scanning microscopy screen through a root, (c) surface, (d, e) central layer showing the central vascular system.
Plant defense gene expression in response to RrF4
We addressed the question whether RrF4 activates genes that are indicative of the plant hormones salicylic acid (SA), jasmonate (JA) and gibberellin (GA), which are involved in plant defense (Spoel et al., 2003). The barley genes Pathogenesis-related 1b (PR1b), PR10, Jasmonate-induced protein 23 (JIP23), 1-Deoxy-D-xylulose-5-phospate synthase (DXS), Ent-kaurene synthase 1 (KS1) and Ent-kaurene synthase like 4 (KSL4) are responsive to P. indica (Deshmukh and Kogel 2007; Schäfer et al., 2009; Pedrotti et al., 2013) and were therefore used for our analysis. A direct qPCR-based comparison of gene expression in barley roots in response to RrF4 and P. indica, respectively, showed a similar activation pattern for all tested genes (Supplementary Figure 5). SA-induced (PR1b, PR10) and GA-induced (DXS, KS1, KSL4) genes were upregulated both by RrF4 and P. indica, whereas the JA-induced gene JIP23 was downregulated at 3 dpi as compared with controls. The only detectable difference in gene expression between RrF4 and P. indica concerns JIP23 at 7 dpi as the gene is induced by RrF4 but not by P. indica.
RrF4 mediates ISR to Pseudomonas syringae
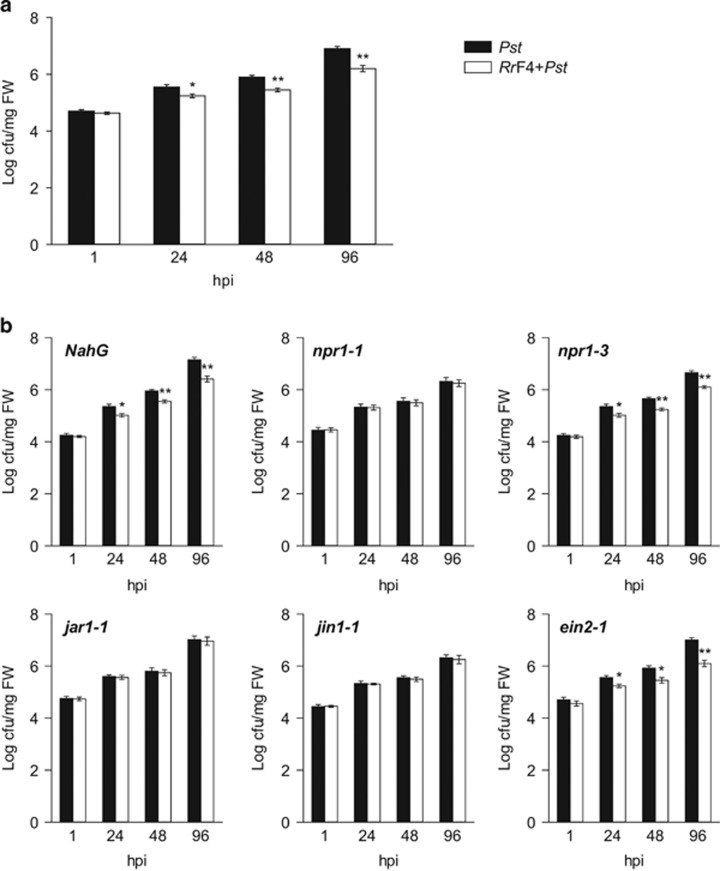
P. indica-induced resistance in Arabidopsis against the powdery mildew fungus Golovinomyces orontii involved the activation of the ISR pathway (Stein et al., 2008). To assess whether RrF4 induces ISR, Arabidopsis mutants indicative of SA signaling (SA-degrading SA hydroxylase (NahG), non-expressor of PR 1-3 (npr1-3)), JA signaling (jasmonate-responsive 1-1 (jar1-1), jasmonate-insensitive 1 (jin1), npr1-1), and ethylene signaling (ethylene insensitive 2-1 (ein2-1)) were assessed for RrF4-mediated resistance to Pseudomonas syringae pv. tomato DC3000 (Pst). Dip-inoculation of wild-type roots with RrF4 reduced the number of Pst bacteria in leaves showing that RrF4 induces systemic resistance to bacterial pathogens (Figure 7a). Similarly, and consistent with the report on P. indica, pretreated NahG, npr1-3 and ein2-1 mutants, like wild-type plants, displayed systemic resistance against Pst compared with non-pretreated plants (Figure 7b). In contrast, jar1-1, jin1 and npr1-1 plants were fully compromised for RrF4-mediated resistance (Figure 7b), showing that the jasmonate-based ISR pathway is also required for RrF4-mediated systemic resistance.
Figure 7.
RrF4-mediated ISR to Pseudomonas syringae pv. tomato DC3000 (Pst) in (a) Arabidopsis wild type and (b) mutants indicative of SA-, JA-, and ethylene based defense (b). Two-week-old roots were dip-inoculated with RrF4, and seedlings were grown in soil under short-day condition. Two weeks later, leaves were spray-inoculated with Pst, harvested at the indicated time points and assessed for Pst infection by determining colony-forming units (cfu) per milligram fresh leaf. Mutants jar1-1, jin1-1 and npr1-1, which are compromised in JA-based ISR, are also compromised for RrF4-induced Pst resistance, while SA-defence-associated NahG and npr1-3 as well as ethylene-associated ein2-1 are not. Error bars indicate standard errors based on three independent biological replicates. Asterisks indicate statistical significant difference (Student's t-test *P<0.05; **P<0.01).
To further substantiate this finding, expression of JA-, SA- and ET-responsive genes was assessed in Arabidopsis leaves in response to RrF4 (inducer) and Pst (challenger) inoculations. PR1, Vegetative storage protein2 (VSP2), Ethylene receptor factor1 (ERF1) and Plant defensin1.2 (PDF1.2) were determined at 24 and 48 h after inoculation by qPCR. JA marker genes VSP2 and PDF1.2 were induced while the SA marker gene PR1 and ethylene marker gene ERF1 were not induced as compared with plants that were challenged with Pst but not pretreated with RrF4 (Supplementary Figure S6). Together, these data show that RrF4, like its host P. indica, induces disease resistance via the ISR pathway.
RrF4 induces systemic resistance to Xanthomonas translucens pv. translucens in wheat
RrF4's activity also was assessed in wheat against the pathogenic bacterium Xanthomonas translucens pv. translucens (Xtt), the causal agent of bacterial leaf streak of cereals. Roots of 3-day-old wheat seedlings were dip-inoculated in a suspension of RrF4, and subsequently transferred to soil. Three weeks later, leaves were spray-inoculated with a suspension of Xtt, and assessed for infection symptoms at 5 and 7 dpi. Leaves from plants pretreated with RrF4 showed reduced bacterial leaf streak symptoms (43% at 5 dpi; 34% at 7 dpi) compared with plants not pretreated with RrF4 (Supplementary Figures S7a and b). Consistent with this, at 7 dpi, total chlorophyll content of leaves infected with Xtt was 20% lower when plants were not pretreated with RrF4 (Supplementary Figure S7c).
Conclusions
Our study shows that the non-pathogenic Alphaproteobacterium R. radiobacter RrF4, which is intricately associated with its fungal host Piriformospora indica, is genetically very similar to the well-studied plant pathogenic R. radiobacter biovar I strain C58 (genomovar G8). Highly similar chromosomal genetic content of RrF4 and C58 indicate the high potential for RrF4's interaction with host plants, whereas differences in the plasmid may explain its non-pathogenic nature. Further studies are needed to understand the basis of specific genomic features supporting RrF4's endofungal lifestyle. The failure to cure P. indica from RrF4 still hampers a conclusively prediction of the bacterium's role in the Sebacinalean symbiosis. Interestingly, however, induced resistance responses and defense gene expression in barley, wheat and Arabidopsis were hardly distinguishable when either induced by RrF4 or P. indica. Thus, our data support the possibility that the beneficial biological activity previously assigned to P. indica may be at least partly allotted to the bacterium RrF4. Finally, RrF4's biological activity was in several aspects comparable with other plant growth-promoting rhizobacteria indicating that endofungal bacteria of the Sebacinalean symbiosis are a valuable source of beneficial bacteria with agronomical potential.
Acknowledgments
We thank S Agel, B Hönig and A Moebus for technical assistance and Dr Jörn Pons-Kühnemann for his support on statistical analysis. This work was supported by the German Science Foundation (DFG) to KHK (KO 1208/24-1) and PK (DFG KA 875/8-1).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Grube M, Schloter M, Smalla K. (2014). Unraveling the plant microbiome: looking back and future perspectives. Front Microbiol 5: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Bell E, Mullet JE. (1996). Two methyl jasmonate-insensitive mutants show altered expression of Atvsp in response to methyl jasmonate and wounding. Plant Physiol 111: 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaux J, Schmid M, Prévost-Bourre NC, Churin JL, Hartmann A, Garbaye J et al. (2003). In situ identification of intracellular bacteria related to Paenibacillus spp. in the mycelium of the ectomycorrhizal fungus Laccaria bicolor S238N. App Environ Microbiol 69: 4243–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaux J, Schmid M, Hutzler P, Hartmann A, Garbaye J, Frey-Klett P. (2005). Occurrence and distribution of endobacteria in the plant-associated mycelium of the ectomycorrhizal fungus Laccaria bicolor S238N. Environ Microbiol 7: 1786–1795. [DOI] [PubMed] [Google Scholar]
- Blom J, Albaum SP, Doppmeier D, Pühler A, Vorhölter FJ, Jewski MZ et al. (2009). EDGAR: A software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics 10: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P, Anca IA. (2009). Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu Rev Microbiol 63: 363–383. [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camehl I, Sherameti I, Venus Y, Bethke G, Varma A, Lee J et al. (2010). Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytologist 185: 1062–1073. [DOI] [PubMed] [Google Scholar]
- Compant S, Reiter B, Sessitsch A, Nowak J, Clement C, Essaid AB. (2005). Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl Environ Microbiol 71: 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh SD, Kogel KH. (2007). Piriformospora indica protects barley from root rot caused by Fusarium graminearum. J Plant Dis Prot 114: 263–268. [Google Scholar]
- Deshmukh SD, Hückelhoven R, Schäfer P, Imani J, Sharma M, Weiss M et al. (2006). The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc Natl Acad Sci USA 103: 18450–18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desirò A, Faccio A, Kaech A, Bidartondo MI, Bonfante P. (2015). Endogone, one of the oldest plant-associated fungi, host unique Mollicutes-related endobacteria. New Phytol 205: 1464–1472. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. (2010). The biofilm matrix. Nat Rev Microbiol 8: 623–633. [DOI] [PubMed] [Google Scholar]
- Frost LS, Ippen-Ihler K, Skurray RA. (1994). Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev 58: 162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura R, Nishimura A, Ohshima S, Sato Y, Nishizawa T, Oshima K, Hattori M, Narisawa K, Ohta H. (2014). Draft genome sequence of the betaproteobacterial endosymbiont associated with the fungus Mortierella elongata FMR23-6. Genome Announc 2: pii: e01272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC. (1994). A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol 176: 2796–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S et al. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756. [DOI] [PubMed] [Google Scholar]
- Ghignone S, Salvioli A, Anca I, Lumini E, Ortu G, Petiti L et al. (2012). The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J 6: 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B et al. (2001). Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294: 2323–2328. [DOI] [PubMed] [Google Scholar]
- Guzman P, Ecker JR. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim PR, van Overbeek LS, Elsas JD. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16: 463–471. [DOI] [PubMed] [Google Scholar]
- Heindl JE, Wang Y, Heckel BC, Mohari B, Feirer N, Fuqua C. (2014). Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium. Front Plant Sci 5: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MT, Arnold AE. (2010). Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl Environ Microbiol 76: 4063–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S, Zechmann B, Molitor A, Trujillo M, Petutschnig E, Lipka V et al. (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol 156: 726–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai K, Furuyabu K, Tani A, Hayashi H. (2012). Production of the quorum-sensing molecules N-acylhomoserine lactones by endobacteria associated with Mortierella alpina A-178. Chembiochem 13: 1776–1784. [DOI] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE. (1954). Two simple media for the demonstration of phycocyanin and fluorescin. J Lab Clin Med 44: 301–307. [PubMed] [Google Scholar]
- Lackner G, Möbius N, Scherlach K, Partida-Martinez LP, Winkler R, Schmitt I et al. (2009). Global distribution and evolution of a toxinogenic Burkholderia-Rhizopus symbiosis. Appl Environ Microbiol 75: 2982–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner G, Moebius N, Partida-Martinez LP, Boland S, Hertweck C. (2011). Evolution of an endofungal lifestyle: Deductions from the Burkholderia rhizoxinica genome. BMC Genomics 12: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahrmann U, Ding Y, Banhara A, Rath M, Hajirezaei MR, Döhlemann S et al. (2013). Host-related metabolic cues affect colonization strategies of a root endophyte. Proc Natl Acad Sci USA 110: 13965–13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle F, Campillo T, Vial L, Baude J, Costechareyre D, Chapulliot D et al. (2011). Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens. Genome Biol Evol 3: 762–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG. (1983). Role of bacterial cellulose fibrils in A. tumefaciens infection. J Bacteriol 154: 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG. (2014). Attachment of Agrobacterium to plant surfaces. Front Plant Sci 5: 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J et al. (2003). GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res 31: 2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M, Morton JB, Pawlowska TE. (2015). Minimal genomes of mycoplasma-related endobacteria are plastic and contain host-derived genes for sustained life within Glomeromycota. Proc Natl Acad Sci USA 112: 7791–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M, Schüssler A, Bonfante P. (2010). The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes. ISME J 4: 862–871. [DOI] [PubMed] [Google Scholar]
- Oberwinkler F, Riess K, Bauer R, Garnica S. (2014). Morphology and molecules: the Sebacinales, a case study. Mycological Progress 13: 445–470. [Google Scholar]
- Partida-Martinez LP, Hertweck C. (2005). Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437: 884–888. [DOI] [PubMed] [Google Scholar]
- Pedrotti L, Mueller MJ, Waller F. (2013). Piriformospora indica root colonization triggers local and systemic root responses and inhibits secondary colonization of distal roots. PLoS One 8: e69352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peškan-Berghöfer T, Shahollari B, Giong PH, Hehl S, Markert C, Blanke V et al. (2004). Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant-microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol Plant 122: 465–477. [Google Scholar]
- Piper WE, Ogrodniczuk JS, Joyce AS, McCallum M, Rosie JS, O'Kelly JG et al. (1999). Prediction of dropping out in time-limited, interpretive individual psychotherapy. PsycNET 36: 114–122. [Google Scholar]
- Preston G. (2000). Pseudomonas syringae pv. tomato: the right pathogen, of the right plant, at the right time. Mol Plant Pathol 1: 263–275. [DOI] [PubMed] [Google Scholar]
- Qiang X, Weiss M, Kogel KH, Schäfer P. (2012. a). Piriformospora indica - a mutualistic basidiomycete with an exceptionally large plant host range. Mol Plant Pathol 13: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang X, Zechmann B, Reitz MU, Kogel KH, Schäfer P. (2012. b). The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an ER stress-triggered caspase-dependent cell death. Plant Cell 24: 794–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Hurek T. (1998). Life in grasses: diazotrophic endophytes. Trends Microbiol 6: 139–144. [DOI] [PubMed] [Google Scholar]
- Riess K, Oberwinkler F, Bauer R, Garnicaet S. (2014). Communities of endophytic sebacinales associated with roots of herbaceous plants in agricultural and grassland ecosystems are dominated by Serendipita herbamans sp. nov. PLoS One 9: e94676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer M, Eckert B, Schmid M, Fekete A, Schloter M, Lehner A et al. (2008). Endophytic root colonization of gramineous plants by Herbaspirillum frisingense. FEMS Microbiol Ecol 66: 85–95. [DOI] [PubMed] [Google Scholar]
- Schäfer P, Pfiffi S, Voll LM, Zajic D, Chandler P, Waller F et al. (2009). Phytohormones in plant root-Piriformospora indica mutualism. Plant Signal Behav 4: 669–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008). Analyzing real-time PCR data by the comparative C (T) method. Nat Protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Sharma M, Schmid M, Rothballer M, Hause G, Zuccaro A, Imani J et al. (2008). Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cell Microbiol 11: 2235–2246. [DOI] [PubMed] [Google Scholar]
- Slater S, Setubal JC, Goodner B, Houmiel K, Sun J, Kaul R et al. (2013). Reconciliation of sequence data and updated annotation of the genome of Agrobacterium tumefaciens C58, and distribution of a linear chromosome in the genus Agrobacterium. Appl Environ Microbiol 79: 1414–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ et al. (2003). NPR1 modulates crosstalk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. (1992). Methyl jasmonate inhibition of root growth and induction of leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89: 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein E, Molitor A, Kogel KH, Waller F. (2008). Systemic resistance conferred by the root endophyte Piriformospora indica to Arabidopsis requires jasmonic acid and the cytoplasmic function of NPR1. Plant Cell Physiol 49: 1747–1751. [DOI] [PubMed] [Google Scholar]
- Torres-Cortés G, Ghignone S, Bonfante P, Schüßler A. (2015). Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: Transkingdom gene transfer in an ancient mycoplasma-fungus association. Proc Natl Acad Sci USA 112: 7785–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent S, Van Wees SC, Pieterse CM. (2009). Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochem 70: 1581–1588. [DOI] [PubMed] [Google Scholar]
- Varma A, Bakshi M, Lou B, Hartmann A, Oelmueller R. (2012). Piriformospora indica: A novel plant growth-promoting mycorrhizal fungus. Agric Res 1: 117–131. [Google Scholar]
- Varma A, Verma S, Sudah SN, Franken P. (1999). Piriformospora indica, a cultivable plant growth-promoting root endophyte. Appl Environ Microbiol 65: 2741–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci USA 102: 13386–13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker V, Bruto M, Bellvert F, Bally R, Muller D, Prigent-Combaret C, Moënne-Loccoz Y, Comte G. (2013). Unexpected phytostimulatory behavior for Escherichia coli and Agrobacterium tumefaciens model strains. Mol Plant Microbe Interact 26: 495–502. [DOI] [PubMed] [Google Scholar]
- Weiss M, Selosse MA, Rexer KH, Urban A, Oberwinkler F. (2011). Sebacinales everywhere: previously overlooked ubiquitous fungal endophytes. PLoS One 108: 1003–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Shen CH, Lin Y, Chen PJ, Xu X, Oelmüller R et al. (2014). Growth promotion-related miRNAs in Oncidium orchid roots colonized by the endophytic fungus Piriformospora indica. PLoS One 9: e84920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccaro A, Lahrmann U, Güldener U, Langen G, Pfiffi S, Biedenkopf D et al. (2011). Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathogens 7: e1002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.