Abstract
The symbiotic gut microbial community is generally known to have a strong impact on the fitness of its host. Nevertheless, it is less clear how the impact of symbiotic interactions on the hosts' fitness varies according to environmental circumstances such as changes in the diet. This study aims to get a better understanding of host–microbiota interactions under different levels of food availability. We conducted experiments with the invertebrate, experimental model organism Daphnia magna and compared growth, survival and reproduction of conventionalized symbiotic Daphnia with germ-free individuals given varying quantities of food. Our experiments revealed that the relative importance of the microbiota for the hosts' fitness varied according to dietary conditions. The presence of the microbiota had strong positive effects on Daphnia when food was sufficient or abundant, but had weaker effects under food limitation. Our results indicate that the microbiota can be a potentially important factor in determining host responses to changes in dietary conditions. Characterization of the host-associated microbiota further showed that Aeromonas sp. was the most prevalent taxon in the digestive tract of Daphnia.
Introduction
The digestive tract of animals is host to a diverse community of symbiotic microorganisms, collectively called the gut microbiota. It is well known that the gut microbiota can have a positive impact on the fitness of its host through various mechanisms like producing essential nutrients (Tokuda et al., 2013), enhancing resistance to disease (Belden & Harris, 2007) or breaking down indigestible substances (Ohkuma, 2008) and toxic compounds (Kohl & Dearing, 2012). In some cases, however, host fitness can also be reduced by the microbiota through competition for resources (Douglas, 2010) or via a dysfunctional symbiotic microbial community causing disease (Clemente et al., 2012).
The fitness impact of services provided by symbiotic interactions on the host is often strongly dependent on the ecological context (Daskin & Alford, 2012). Several studies have shown that the hosts' diet, especially food quality, can be an important influencing factor on interactions between the host and its gut microbiota. For example, de Vries et al. (2004) found that bacteria belonging to the genus Erwinia, present in the gut of western flower thrips, were parasitic under a high-quality diet but mutualistic when the thrips were given a nutrient-poor diet, indicating that these symbionts benefit their host only in nutritionally poor environments. In contrast, Ben-Yosef et al. (2008) found that the gut microbiota of Mediterranean fruit flies (Ceratitis capitata) can negatively affect longevity and egg deposition, but only when the hosts feed on a low-quality diet. These contrasting results imply that the strength and direction in which host–microbiota interactions change under different dietary conditions cannot be easily predicted.
Although most research has focused on food quality, several studies found that food quantity can affect the composition and functionality of the gut microbiota (Dillon et al., 2010; Freese & Schink, 2011). In mice, for example, starvation is known to alter the gut microbiota in a way that it confers health benefits to its host (Zhang et al., 2013). Furthermore, the gut microbiota is known to positively influence physiological responses of the host to starvation (Crawford et al., 2009). As strong fluctuations in food availability commonly occur in natural populations (Müller-Navarra & Lampert, 1996), this factor might be an important driver of host–microbiota interactions and can thus be highly relevant from an ecological perspective. Here we address this issue using the water flea Daphnia magna and its symbiotic microbial community as a model system. D. magna is a keystone species in many freshwater habitats and has been widely used as an experimental system to study ecological and evolutionary interactions (for example, Decaestecker et al., 2007; Miner et al., 2012). Daphnia is known to harbor high numbers of bacteria on the surface of its body and in its gut (Grossart et al., 2010; Eckert & Pernthaler, 2014), and Qi et al. (2009) showed that several bacterial taxa are consistently found in affiliation with Daphnia, even in geographically separated populations, indicating a highly specific association between Daphnia and its microbiota. Notably, the β-proteobacterial genus Limnohabitans (Comamonadaceae) seems to be a major constituent of the Daphnia microbiota (Freese & Schink, 2011; Eckert & Pernthaler, 2014; Peerakietkhajorn et al., 2015). Sison-Mangus et al. (2014) showed, by comparing bacteria-free Daphnia with conventional ones, that the relationship between D. magna and its microbiota is mutualistic, as removing the microbiota results in decreased growth, survival and reproduction of the host. It is, however, not known how this interaction is affected by variation in environmental conditions, such as food availability.
Food availability is known to profoundly affect life-history characteristics and population dynamics in D. magna (Vanoverbeke, 2008). Food limitation has been shown to decrease growth and reproduction while increasing longevity (Pietrzak et al., 2010). Here, we investigate the effects of variation in food availability on host–microbiota interactions in Daphnia by comparing symbiotic with germ-free hosts raised in environments with different levels of food availability. Dependent on the specific mode-of-action of the microbiota and how this will interact with variation in food availability, different outcomes are possible. If, for example, the microbiota helps its host to acquire nutrients, increasing the amount of food will not correspond to an equal increase in nutrient uptake when the microbiota is removed. Under this hypothesis, we expect that germ-free Daphnia will show a lower response to an increase in food availability than symbiotic Daphnia. It is, however, also possible that the microbiota competes with its host for nutrients when food availability is limited. If this is the case, we expect symbiotic Daphnia to have a lower fitness than germ-free Daphnia under food limitation. To identify bacterial taxa possibly responsible for the observed effects on their host, we characterized the Daphnia microbiota by sequencing 16S rDNA.
Materials and methods
Cultivation of algae
An axenic batch culture of Scenedesmus obliquus was grown in Wright's cryptophyte medium (Guillard & Lorenzen, 1972) for 1 week, after which the algae were harvested. The presence of bacteria in the cultures was checked by 4',6-diamidino-2-phenylindole-stained preparations under a fluorescence microscope at × 1000 magnification and by plating 100 μl on nutrient agar in triplicate and checking for bacterial growth after 5 days of incubation at 37 °C. Ash-free dry weight (expressed as mg C) of the culture was determined following Moheimani et al. (2013).
Daphnia cultivation and obtaining of germ-free animals
We used D. magna clone KNO15.04 in all experiments. This clone was isolated from a pond in Knokke, Belgium (51°20'05.62” N, 03°20'53.63” E) and has been maintained in the lab for multiple generations. Stock cultures were kept in 500 ml jars filled with Aachener Daphnien medium (ADaM; CaCl2·2H2O: 273 mg l−1; NaHCO3: 56 mg l−1; sea-salt: 333 mg l−1; Klüttgen et al., 1994) in a temperature-controlled room at 19±2 °C, under a light regime of 16:8 h light:dark, and on a diet of saturating amounts of S. obliquus. Before the start of each experiment, reproduction of the animals was synchronized by maintaining individuals of the same age for at least two generations under identical conditions.
To obtain germ-free Daphnia individuals, a protocol that we recently developed and which is published for the first time was applied. Females carrying parthenogenetic eggs were dissected under a stereo-microscope and the eggs were collected in a petri dish containing ADaM. Only recently deposited eggs, which are characterized by the presence of an external membrane, were isolated. Preliminary experiments showed that the timing of egg isolation is very important for hatching success: eggs isolated <12 h after deposition had a significant lower hatching rate than those who were isolated later (but still surrounded by an external membrane; personal observation). From here onwards, all experimental manipulations were performed under sterile conditions in a laminar flow hood. To remove all microbial organisms from the eggs, they were placed in a petri dish containing 10 ml of a 0.01% peracetic acid (PAA) solution. These were gently agitated for 10 min, after which the eggs were transferred to another petri dish containing sterile ADaM to remove any PAA residues. Afterwards, the disinfected eggs were transferred to six-well (cell culture) plates. Each well contained ~50 eggs in 8 ml of sterile ADaM. Well plates were incubated at 20±0.5 °C. Eggs were allowed to hatch during 48 h, and the resulting germ-free individuals were used in the experiment.
Verification of symbiont removal
Because we applied a newly developed method for obtaining germ-free Daphnia, removal of bacteria from the experimental animals needed to be verified after applying the disinfection protocol. To do this, the presence of bacterial 16S rDNA on disinfected Daphnia was checked using PCR. This was tested on pooled samples of 20 neonate juveniles hatched from PAA-disinfected eggs. As a positive control, 20 neonate juveniles from a stock laboratory culture were used. Both the disinfection treatment and the positive control were tested in triplicate. A blank sample was also included in the analysis as negative control. Preliminary tests on samples containing a known number of E. coli cells showed that when applying the following protocol, the detection threshold is 104 bacterial cells.
To lyse the cells, Daphnia were first homogenized in 0.5 ml of double-distilled H2O. Then, 30 μl of lysozyme (50 mg ml−1) was added to each sample and these were incubated for 1 h at 37 °C. To each tube, a small amount of glass beads (0.1 mm diameter, acid washed) was added, and they were placed on a bead-beater (2 min, 30 m s−1). Afterwards, 50 μl of proteinase K (1 mg ml−1) was added and the samples were incubated for 30 min at 55 °C. Finally, 25 μl lysis buffer (10% sodium dodecyl sulfate; 0.5 m ethylenediaminetetraacetic acid) was added and samples were incubated for 1 h at 55 °C.
DNA was extracted from the lysate by adding 500 μl of phenol–chloroform–isoamylalcohol (25–24–1) to the samples, followed by centrifugation (13 200 rpm, 10 min) and transfer of the aqueous supernatant to a new micro-centrifuge tube. Phenol–chloroform extraction was repeated once more on the supernatant. DNA was precipitated by adding 0.1 vol of sodium acetate (3m) and an equal volume of isopropanol to the tube, followed by a centrifugation step (25 min; 14 000 rpm; 4 °C). The supernatant was discarded and the pellet was washed with 70% ice-cold ethanol, centrifuged (10 min; 14 000 rpm; 4 °C), dried and dissolved in 15 μl tris-EDTA buffer. Extracted DNA was stored at −20 °C until further processing.
PCRs were performed on 5 μl of extracted DNA using Platinum Taq DNA polymerase (Life Technologies, Waltham, MA, USA) according to the manufacturers' instructions. Universal bacterial primers 8 F and 1492R (Weisburg et al., 1991) were used to amplify a 1484 kp fragment of the 16S rDNA. PCR reactions were run for 30 cycles (94 °C 30 s, 50 °C 30 s and 72 °C 60 s). The presence of amplified 16S rDNA was checked under UV light on 10 μl of PCR product loaded on a 1% agarose gel stained with GelRed (Biotium, Hayward, CA, USA).
Experimental setup
Germ-free Daphnia were placed individually in a 50-ml falcon tube containing 5 ml of sterile ADaM. Each tube was randomly assigned to either the symbiotic (n=240) or germ-free (n=240) treatment. A microbiota inoculum was prepared by homogenizing one adult Daphnia per ml in sterile ADaM. In the symbiotic treatment, axenic juveniles were re-colonized with Daphnia microbiota by adding 100 μl of this inoculum. Individuals in the germ-free treatment also received 100 μl of the inoculum, but which had been autoclaved prior to administration. All Daphnia were incubated for another 24 h, after which sterile ADaM was added to obtain a final volume of 40 ml medium in all tubes.
To test for the effects of different food quantities on host–microbiota interactions, both symbiotic and germ-free animals were administered daily one of four different food amounts: 2 mg C l−1 (high), 0.5 mg C l−1 (intermediate), 0.1 mg C l−1 (low) or no food (starvation). This resulted in a total of eight treatments (presence or absence of symbionts × four different food amounts, with 30 individuals per treatment). All experimental units were kept in an incubator at 20±0.5 °C and a 16:8 h light:dark regime. Every 24 h during 3 weeks, survival and reproduction (time of egg deposition and number of juveniles per brood) were registered and released juveniles were removed.
To measure the effect of the different treatments on Daphnia body size, a similar experiment was performed with circa 30 Daphnia per treatment. After 10 days, all surviving animals were fixated with 4% formaldehyde and mounted in glycerol on microscope slides. Carapace size (from the underside of the head to the base of the caudal spine) was measured under a stereo-microscope using the Infinity Analyze 6.3.0 software (Lumenera, Ottawa, ON, Canada).
Identification of symbiotic bacteria
To identify the bacteria present on Daphnia in our laboratory cultures, we characterized the composition of symbiotic bacterial communities of both entire Daphnia and their guts by sequencing 16S rDNA. For each sample, 20 neonate KNO15.04 Daphnia were kept in 2 l containers filled with ADaM medium and given a daily amount of 2 mg C l−1 S. obliquus. After 10 days, adult Daphnia were collected from each culture and kept for 24 h in 50 ml of 0.22 μl filtered water originating from their respective container to remove any non-symbiotic bacteria. To characterize the entire community of host-associated microbiota, 15 Daphnia were pooled in a micro-centrifuge tube. To specifically characterize the gut microbiota, the digestive tracts of 15 Daphnia were pooled after dissecting them under a stereo-microscope using sterilized dissecting needles. Both communities were characterized in triplicate.
DNA was extracted using the protocol described above and 16 rDNA fragments were amplified using a nested primer design. PCR reactions were performed using high-fidelity Platinum Pfx DNA polymerase (Life technologies) according to the manufacturer's instructions. For the external amplification, a PCR reaction was run for 25 cycles (95 °C 30 s, 50 °C 30 s, 68 °C 60 s) using primers Eub8F and 984yR (Bakke et al., 2011). The PCR products were subsequently purified and amplified in a second PCR reaction for 25 cycles (94 °C 30 s, 45 °C 30 s, 68 °C 60 s) using primers 343 F (Wilson et al., 1990) and 798 R (Rochelle et al., 1995) modified by adding ligation adaptors and sample identification sequences to the 5'-end. PCR products were purified and forward 16S rDNA amplicons were sequenced using 454 pyrosequencing.
Data analysis
Normal distribution and equal variance of data were tested using a Shapiro–Wilk and a Bartlett's test, respectively. Differences in size and production of juveniles between treatments were analyzed using a two-way analysis of variance with microbiota, food quantity and their interaction as fixed factors. A Tukey honest significant difference test was used to make post hoc pairwise comparisons. The proportions of reproducing individuals were tested using a χ2-test. All these analyses were performed with R 3.1.0.
The intrinsic rate of natural increase (rm) was calculated using the equation ∑e-rxlxmx=1, with x=time in days, lx=age-specific survival (number of living females on day x/number of females alive at the start) and mx=age-specific reproduction (number of offspring released at day x/number of females alive at day x) (Birch, 1948).
Longevity and age at first reproduction were analyzed with a Cox proportional-hazards model regression using the SAS 9.4 software (PHREG procedure). Microbiota, food quantity and their interaction were declared as fixed factors. The survival times of individuals that were still alive at the end of the experiment and the time at first reproduction of individuals that died before reproducing were coded as censored. As ties in survival and reproduction times were numerous, we used the Efron approximate likelihood. Pairwise comparisons were performed using the CONTRAST statement, which provided both the hazard ratios (HR) between groups for the variable of interest, and the associated P-values.
Sequences of 16S rDNA were demultiplexed, quality-filtered and clustered into operational taxonomic units (OTUs) at 97% pairwise sequence similarity using the QIIME pipeline (Caporaso et al., 2010). As some samples contained a low number of sequences, only operational taxonomic units with a mean relative abundance of >1% in either the entire or gut microbiome were used to assess community composition. Operational taxonomic units were taxonomically classified by searching for the sequence with the highest similarity using the SINA aligner (Pruesse et al., 2012). Nucleotide sequences obtained in this study have been submitted to the European Nucleotide Archive (accession numbers LN869908-LN869916).
Results
Removal of symbionts from Daphnia
In all samples containing PAA-disinfected Daphnia, no fragments of amplified 16S rDNA were observed on an agarose gel after PCR. In all positive controls a band of the amplified 16S rDNA fragment was clearly visible (Supplementary Figure 1). This indicates that the PAA treatment was capable of removing all bacteria from Daphnia, or at least reducing them to an amount undetectable by PCR.
Effects of symbiotic bacteria and food quantity on daphnia survival
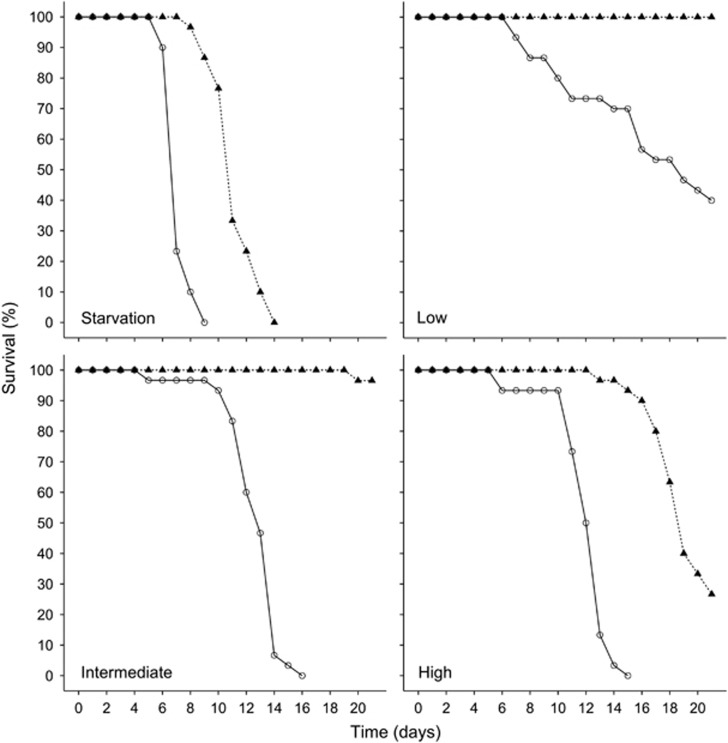
Both food quantity (X23=111.3, P<0.001) and the microbiota (X21=50.9, P<0.001) had a significant main effect on survival, and there was a significant interaction between these two factors (X23=9.9, P<0.05). Within each food treatment, symbiotic Daphnia always survived significantly longer than germ-free ones and the effect of the microbiota was the strongest under intermediate food quantities (starvation: HR=18.8, P<0.001; low: HR=35.5, P<0.001; intermediate: HR=214.3, P<0.001; high: HR=19.1, P<0.001) (Figure 1, Table 1). For symbiotic Daphnia, survival was significantly higher when they received low or intermediate food quantities compared with starved individuals or those receiving high food quantities (all pairwise comparisons, P<0.001) and symbiotic Daphnia in the high food quantity treatment showed a higher survival than starved ones (HR=0.03, P<0.001). For germ-free Daphnia, survival in the low food quantity treatment was significantly higher compared with all other treatments (all pairwise comparisons, P<0.001). Germ-free individuals receiving an intermediate or high food quantity showed no difference in survival (P=0.11). Daphnia from the germ-free/starvation treatment showed the lowest survival of all (all pairwise comparisons, P<0.001).
Figure 1.
Percentage of surviving Daphnia exposed to different microbiota- (open circles=germ-free, black triangles=symbiotic) and food quantity (starvation=no food, low=0.1 mg C l−1 per day, intermediate=0.5 mg C l−1 per day, high=2 mg C l−1 per day) treatments.
Table 1. Summary of life-history parameters.
| Parameter | Food quantity | Symbiotic | Germ-free |
|---|---|---|---|
| Mean survival time (days) | High | 18.9 d (s.d.=2.0 d; n=30) | 12.1 d (s.d.=1.9 d; n=30)* |
| Intermediate | 20.9 d (s.d.=0.4 d; n=29) | 12.8 d (s.d.=2.0 d; n=30)* | |
| Low | 21.0 d (s.d.=0.0 d; n=30) | 16.6 d (s.d.=5.1 d; n=30)* | |
| Starvation | 11.3 d (s.d.=1.5 d; n=30) | 7.2 d (s.d.=0.8 d; n=29)* | |
| Mean body size (mm) | High | 1.78 mm (s.d=0.11; n=29) | 1.13 mm (s.d.=0.15; n=29)* |
| Intermediate | 1.68 mm (s.d.=0.10; n=29) | 1.18 mm (s.d.=0.14; n=29)* | |
| Low | 1.13 mm (s.d.=0.07; n=29) | 1.04 mm (s.d.=0.09; n=26)* | |
| Starvation | NA | NA | |
| Observed egg deposition (%) | High | 100% (n=30) | 6.70% (n=30)* |
| Intermediate | 100% (n=29) | 3.30% (n=30)* | |
| Low | 0% (n=30) | 0% (n=30) | |
| Starvation | 0% (n=30) | 0% (n=29) | |
| Mean time to first clutch (days) | High | 6.5 d (s.d.=1.0 d; n=30) | 7.0 d (s.d.=0.0 d; n=2)* |
| Intermediate | 7.5 d (s.d.=1.0 d; n=29) | 9.0 d (s.d.=0.0 d; n=1)* | |
| Low | NA | NA | |
| Starvation | NA | NA | |
| Mean number of juveniles | High | 4.6 (s.d.=3.0; n=30) | 0.0 (s.d.=0.0; n=30)* |
| Intermediate | 7.3 (s.d.=3.2; n=28) | 0.0 (s.d.=0.0; n=30)* | |
| Low | 0.0 (s.d.=0.0; n=30) | 0.0 (s.d.=0.0; n=30) | |
| Starvation | 0.0 (s.d.=0.0; n=30) | 0.0 (s.d.=0.0; n=30) |
An asterisk indicates a significant difference between symbiotic and germ-free Daphnia within a food treatment.
Effects of symbiotic bacteria and food quantity on body size
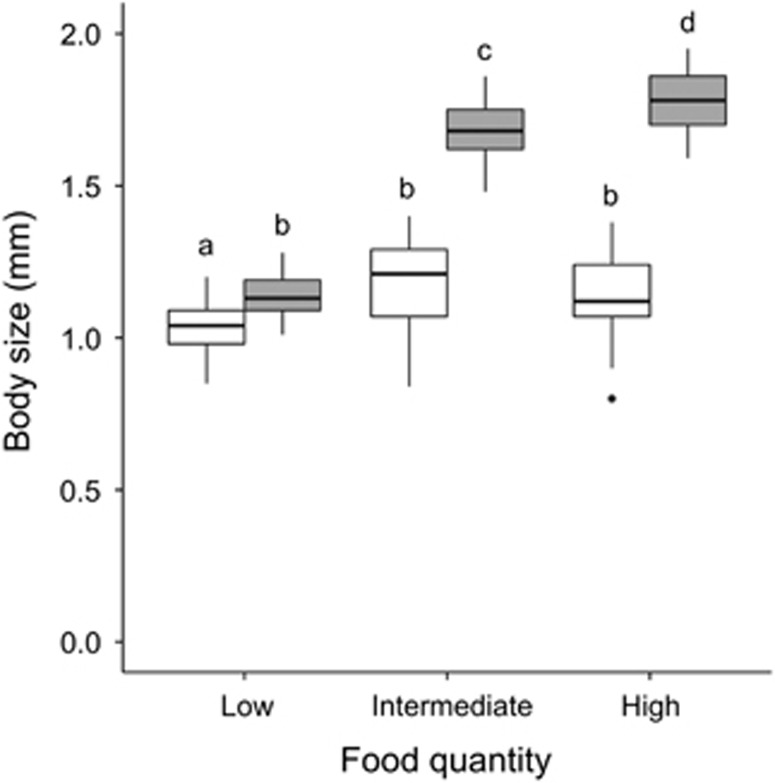
Both food quantity (F2,169=180; P<0.001) and microbiota (F1,170=587.13; P<0.001) had a significant main effect on growth, and there was a significant interaction between these two factors (F2,169=88.28; P<0.001) (Figure 2). For symbiotic animals, body size increased significantly with increasing food quantity (Tukey post hoc, P-adj.<0.05). For germ-free animals, growth was slightly (but significantly) higher when food quantity increased from low to intermediate (Tukey post hoc, P-adj.<0.05). However, no difference in size was observed between animals fed with intermediate and high food quantities. Within a particular food treatment, symbiotic animals were always significantly larger than germ-free ones (Tukey post hoc, P-adj.<0.05), but the difference between symbiotic and germ-free animals was more pronounced under intermediate and high food levels than under low food levels.
Figure 2.
Boxplot of Daphnia body size exposed to different microbiota- (white=germ-free, gray=symbiotic) and food quantity (low=0.1 mg C l−1 per day, intermediate=0.5 mg C l−1 per day, high=2 mg C l−1 per day) treatments. Letters above samples represent differences as determined by a Tukey HSD post hoc test. Different letters are significant at 5%.
Effects of symbiotic bacteria and food quantity on reproduction
The combined food- and microbiota treatments had a significant effect on egg production (X2=223.29, P<0.001). Both symbiotic and germ-free Daphnia receiving a low food quantity or starvation treatment never deposited eggs during the experiment (Table 1). In all treatments receiving a high- or intermediate food quantity, egg production was observed at least once. Within the high- and intermediate food quantity treatments, egg deposition was observed in 100% of symbiotic Daphnia during the experiment. However, only a small fraction of the germ-free individuals produced eggs (Table 1; high food quantity: 6.70% intermediate food quantity: 3.30%).
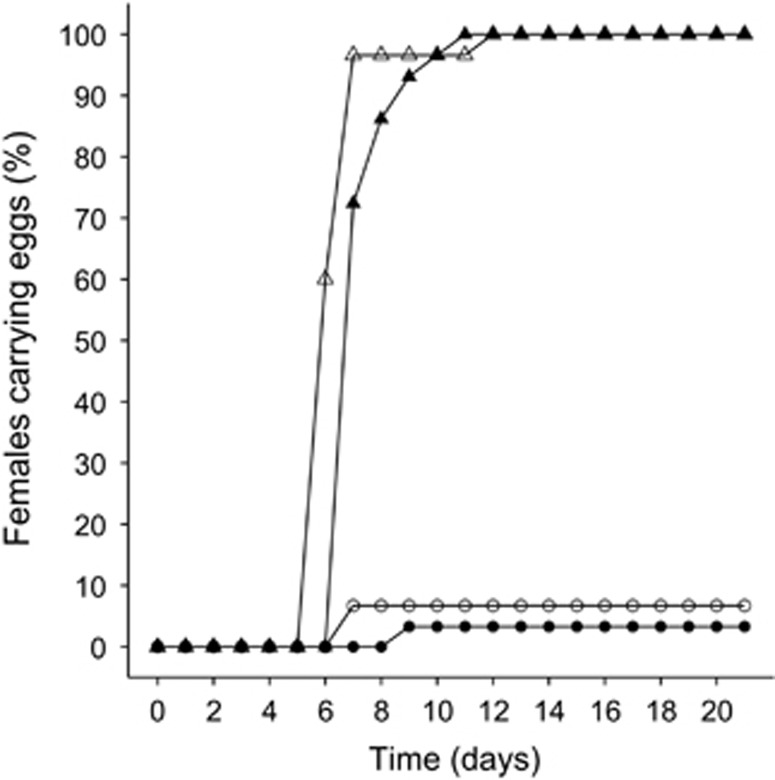
There was a significant effect of both food availability (X21=11.7, P<0.001) and the microbiota (X21=18.3, P<0.001) on the age at first reproduction, the interaction between these two factors was not significant (X21=0.01, P=0.91). Symbiotic Daphnia receiving a high food quantity reproduced significantly earlier than those receiving an intermediate food quantity (HR=2.5, P<0.001). Furthermore, Daphnia receiving the same amount of food always reproduced significantly earlier in the presence of symbionts than when germ-free (high: HR=0.01, P<0.001; intermediate: HR=0.01, P<0.001) (Figure 3).
Figure 3.
Percentage of Daphnia where production of first clutch eggs was observed in different microbiota (triangles=symbiotic; circles=germ-free) and food quantity (open=high (2 mg C l−1 per day); filled=intermediate (0.5 mg C l−1 per day)) treatments. Data from the starvation- and low food quantity treatments are omitted from the graphs because here no egg deposition was observed.
Only in symbiotic Daphnia did the eggs fully develop and were the juveniles released from the brood chamber (Table 1). Over the course of the experiment, individuals receiving an intermediate food quantity produced a significantly higher number of juveniles than those receiving high amounts of food (t28,30=3.27, P<0.05). However, the intrinsic rate of natural increase was very similar in the high food (rm=0.16) and intermediate food treatment (rm=0.15). This is attributable to an early peak in reproduction in the high food treatment compared with a later and more spread-out reproduction in the intermediate food treatment.
Identification of symbiotic bacteria
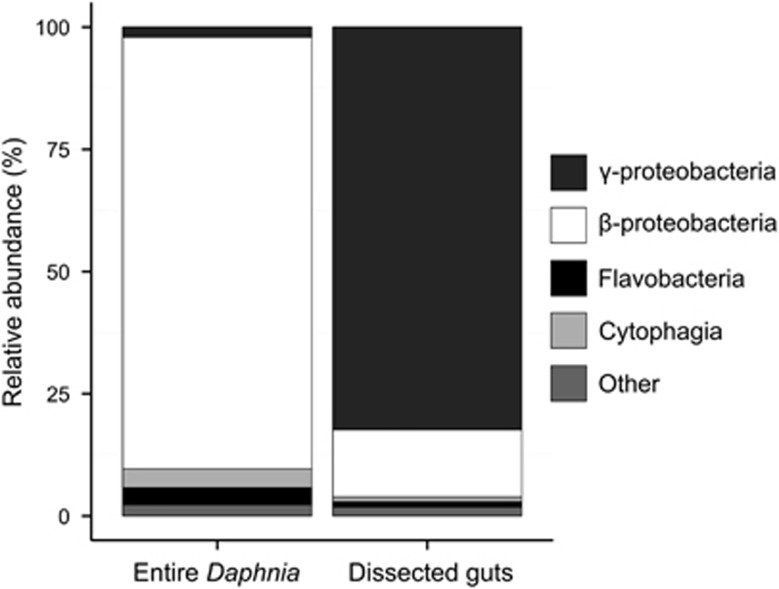
The entire bacterial community on Daphnia was found to be dominated by β-proteobacteria (mainly Limnohabitans sp. and Hydrogenophaga sp.), whereas γ-proteobacteria, flavobacteria and cytophagia occurred at lower relative abundances (Figure 4,Table 2). The gut microbiota, on the other hand, was found to be consistently dominated by γ-proteobacteria (mainly Aeromonas sp.), whereas the occurrence of other taxa was lower and more variable (Figure 4, Table 3).
Figure 4.
Class-level taxonomic composition of the bacterial communities found on entire Daphnia (left) and in dissected guts of Daphnia (right). The relative abundance of each class was calculated from mean values over three samples.
Table 2. List of bacteria found in the microbiome of entire Daphnia.
| Phylum | Class | Order | Family | Bacterium | Accession number | Identity | Rel. ab. (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Proteobacteria | β-proteobacteria | Burkholderiales | Comamonadaceae | uncultured Chlorochromatium sp. | HM128814 | 95.27% | 2.4 (0.7) % | Zhang et al., 2013 |
| uncultured Hydrogenophaga sp. | EU801511 | 95.80% | 14.7 (2.7) % | Shaw et al., 2008 | ||||
| uncultured Limnohabitans sp. | HM561445 | 96.56% | 65.8 (0.6) % | Freese & Schink, 2011 | ||||
| Limnohabitans sp. 2KL-7 | HE600663 | 95.79% | 1.1 (0.5) % | Kasalicky et al., 2013 | ||||
| γ-proteobacteria | Aeromonadales | Aeromonadaceae | uncultured Aeromonas sp. | HM778882 | 94.15% | 2.0 (1.9) % | Roeselers et al., 2011 | |
| Bacteroidetes | Cytophagia | Cytophagales | Arcicella | Bacteroidetes bacterium 3–6 | AB578880 | 96.99% | 2.6 (2.3) % | – |
| Flavobacteriia | Flavobacteriales | Flavobacteriaceae | Uncultured Flavobacterium sp. | DQ264455 | 97.54% | 1.3 (0.4) % | DeSantis et al., 2007 | |
| Flavobacterium sp. HME6126 | HM776984 | 100% | 1.2 (1.0) % | – |
Only OTUs with a mean relative abundance >1% are included.
Table 3. List of bacteria found in the gut microbiome of Daphnia.
| Phylum | Class | Order | Family | Bacterium | Accession number | Identity | Rel. abundance (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Proteobacteria | β-proteobacteria | Burkholderiales | Comamonadaceae | uncultured Acidovorax sp. | JX120471 | 89.73% | 1.4 (0.8) % | Handley et al., 2013 |
| uncultured Limnohabitans sp. | HM561445 | 96.56% | 13.6 (12.3) % | Freese & Schink, 2011 | ||||
| γ-proteobacteria | Aeromonadales | Aeromonadaceae | uncultured Aeromonas sp. | HM778882 | 94.15% | 80.3 (15.2) % | Roeselers et al., 2011 | |
| Bacteroidetes | Flavobacteriia | Flavobacteriales | Flavobacteriaceae | uncultured Flavobacterium sp. | DQ264455 | 97.54% | 1.1 (0.9) % | DeSantis et al., 2007 |
Only OTUs with a mean relative abundance >1% are included.
Discussion
In this study we investigated the interaction between Daphnia and its microbiota under different levels of food availability. We observed a consistent mutualistic effect of the microbiota on its host. However, the relative impact of this mutualistic interaction on the hosts' fitness was dependent on food availability. This is reflected by the smaller difference in host growth and reproduction between symbiotic and germ-free Daphnia when food was limited compared with treatments where food availability was higher. The impact of the microbiota on host survival was also dependent on food availability and was the strongest under intermediate food quantities.
Our experiments clearly showed that removing the microbiota of D. magna had an overall negative effect on host growth, survival and reproduction. Although in this study we used a newly developed method to obtain germ-free Daphnia without the use of antibiotics, results from non-nutritionally stressed treatments are similar to what Sison-Mangus et al. (2014) found with respect to growth, survival and reproduction. This confirms the generality of the mutualistic relationship between Daphnia and its microbiota.
Characterization of the symbiotic bacterial community showed Limnohabitans sp. to be the dominant taxon, this is in accordance with earlier studies who also found this taxon to be a major constituent of the Daphnia microbiota (Freese & Schink, 2011; Eckert & Pernthaler, 2014; Peerakietkhajorn et al., 2015; Gorokhova et al., 2015). However, by specifically characterizing the gut microbiota, we found that in our cultures Aeromonas sp. is the dominant taxon in the digestive tract. Previous mono-association experiments with either Limnohabitans sp. or Aeromonas sp. showed that inoculating germ-free Daphnia with these bacteria had a positive effect on host fitness, although for Limnohabitans sp. this was found to be restricted to specific strains (Sison-Mangus et al., 2014; Peerakietkhajorn et al., 2015b). Interestingly, the dominant Aeromonas sp. sequence found in our Daphnia guts most closely matched a zebrafish gut symbiont (Roeselers et al., 2011). In zebrafish, Aeromonas sp. is known to be a major constituent of the gut microbiota and mono-association experiments with this bacterium showed that it increases the expression of digestive enzymes and abundance of secretory cells in the gut epithelium. Its presence furthermore enabled the uptake of protein macromolecules and influenced gut motility (Bates et al., 2006). Interestingly, Gorokhova et al. (2015) found a strong decrease in feeding activity, digestion efficiency and carbon uptake in Daphnia of which the gut microbiota was affected by antibiotic exposure. Possibly, Aeromonas sp. or Limnohabitans sp. present in the Daphnia microbiota had similar effects on gut maturation as those found in zebrafish. These findings suggest that in our experiment the presence of the microbiota positively affected Daphnia through an enhanced nutrient acquisition, a hypothesis that is supported by the fact that the effect of the microbiota on host fitness was greater when food availability increased. Alternatively, the microbiota could also be important for neutralizing effects from harmful substances produced by the hosts' metabolism. Higher food intake can result in the release of higher concentrations of such harmful substances and reduce survival if these are not removed. For example, nitrogenous metabolic waste products are known to be efficiently degraded by symbiotic microbial communities in different organisms (Hoffmann et al., 2009; Sabree et al., 2009). This could also explain the stronger impact of the microbiota on the host when food availability increases.
In the starvation treatment, however, we observed a pronounced effect of the microbiota on host survival, suggesting that the microbiota have an additional role. Symbiotic Daphnia might make more efficient use of nutrients obtained via the egg than germ-free Daphnia, as they survived for a longer period in the absence of food. In mice, for example, the microbiota is known to shape metabolic and physiological adaptations of the host to periods of nutrient deprivation by enhancing utilization of stored fat reserves (Crawford et al., 2009). It is also known that bacteria can act as a supplementary food source for Daphnia. These are, however, nutritionally inferior to algae (Martin-Creuzburg et al., 2011; Taipale et al., 2012). In our experiment, we controlled for the effect of feeding on bacteria by inoculating autoclaved microbiota to germ-free treatments. The starvation treatment is of particular interest here, as no food was added, which can support further bacterial growth. Nevertheless, we still found a pronounced effect on host survival, indicating that the presence of a live, metabolically active microbiota is essential.
Both symbiotic and germ-free Daphnia showed a clear response to food availability. However, an increase in food availability from intermediate to high was found to have less-pronounced effects on Daphnia growth and reproduction then when food availability increased from low to intermediate. In our experiment the saturating food concentration, at which an increase in food availability does not correspond to an increase in food intake, was probably situated somewhere between the intermediate and high concentration of algae, resulting in a lower response. Nevertheless, an increase in caloric intake was associated with an overall increase in growth and reproduction. Survival, on the other hand, decreased with increasing food quantities. This discrepancy can be associated with a trade-off between growth and reproduction on one side, and survival on the other. Such a trade-off is commonly observed in Daphnia (Glazier & Calow, 1992; Dudycha, 2003). For example, Pietrzak et al. (2010) found that investments in early reproduction were costly in Daphnia and result in a shortened lifespan. In our experiment a similar pattern was observed. This was especially clear in the symbiotic treatments where under high food conditions Daphnia showed an early peak in reproduction and a reduced longevity compared with those receiving intermediate amounts of food. The intrinsic rate of natural increase remained, however, almost identical in both treatments.
In conclusion, we showed a consistent mutualistic effect of the Daphnia microbiota on its host, but food availability was found to affect the strength of these host–microbiota interactions. This shows that the effect of the microbiota on Daphnia can vary according to dietary conditions. The microbiota can thus be an important factor in determining responses of Daphnia to changing environmental circumstances. Further research is needed on both the response of host–microbiota interactions to other environmental factors and the precise nature of this interaction to be able to accurately quantify the functional importance of the microbiota for Daphnia.
Acknowledgments
We thank Luc De Meester and Dieter Ebert for stimulating discussions. Funding was provided by the research projects: FWO G.0643.13, Belspo IAP project SPEEDY, P7/4 and the Centre of Excellence SEEDS PF/2010/007 of the KULeuven Research Fund.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bakke I, De Schryver P, Boon N, Vadstein O. (2011). PCR-based community structure studies of Bacteria associated with eukaryotic organisms: a simple PCR strategy to avoid co-amplification of eukaryotic DNA. J Microbiol Methods 84: 349–351. [DOI] [PubMed] [Google Scholar]
- Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. (2006). Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol 297: 374–386. [DOI] [PubMed] [Google Scholar]
- Belden LK, Harris RN. (2007). Infectious disease in wildlife: the community ecology context. Front Ecol Environ 5: 533–539. [Google Scholar]
- Ben-Yosef M, Behar A, Jurkevitch E, Yuval B. (2008). Bacteria-diet interactions affect longevity in the medfly – Ceratitis capitata. J Appl Entomol 132: 690–694. [Google Scholar]
- Birch LC. (1948). The intrinsic rate of natural increase of an insect population. J Anim Ecol 17: 15–26. [Google Scholar]
- Caporaso GJ, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell 148: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M et al. (2009). Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. PNAS 106: 11276–11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskin JH, Alford RA. (2012). Context-dependent symbioses and their potential roles in wildlife diseases. Proc R Soc Lond B Biol Sci 279: 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaestecker E, Gaba S, Raeymaekers JAM, Stoks R, Van Kerckhoven L, Ebert D, De Meester L. (2007). Host-parasite “Red Queen” dynamics archived in pond sediment. Nature 450: 870–873. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. (2007). High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol 53: 371–383. [DOI] [PubMed] [Google Scholar]
- de Vries EJ, Jacobs G, Sabelis MW, Menken SBJ, Breeuwer JAJ. (2004). Diet-dependent effects of gut bacteria on their insect host: the symbiosis of Erwinia sp. and western flower thrips. Proc R Soc Lond Biol Sci 271: 2171–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RJ, Webster G, Weightman J, Charnley AK. (2010). Diversity of gut microbiota increases with aging and starvation in the desert locust. Antonie van Leeuwenhoek 97: 69–77. [DOI] [PubMed] [Google Scholar]
- Douglas AE. (2010) The symbiotic habit. Princeton University Press: Princeton, NJ, USA.. [Google Scholar]
- Dudycha JL. (2003). A multi-environment comparison of senescence between sister species of Daphnia. Oecologia 135: 555–563. [DOI] [PubMed] [Google Scholar]
- Eckert EM, Pernthaler J. (2014). Bacterial epibionts of Daphnia: a potential route for the transfer of dissolved organic carbon in freshwater food webs. ISME J 8: 1808–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese HM, Schink B. (2011). Composition and stability of the microbial community inside the digestive tract of the aquatic crustacean Daphnia magna. Microb Ecol 62: 882–894. [DOI] [PubMed] [Google Scholar]
- Glazier DS, Calow P. (1992). Energy allocation rules in Daphnia magna: clonal and age differences in the effects of food limitation. Oecologia 90: 540–549. [DOI] [PubMed] [Google Scholar]
- Gorokhova E, Rivetti C, Furuhagen S, Edlund A, Ek K, Breitholtz M. (2015). Bacteria-mediated effects of antibiotics on Daphnia nutrition. Environ Sci Technol 49: 5779–5787. [DOI] [PubMed] [Google Scholar]
- Grossart HP, Dziallas C, Leunert F, Tang KW. (2010). Bacteria dispersal by hitchhiking on zooplankton. PNAS 107: 11958–11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard RRL, Lorenzen CJ. (1972). Yellow-green algae with chlorophyllide C. J Phycol 8: 10–14. [Google Scholar]
- Handley KM, VerBerkmoes NC, Steefel CI, Williams KH, Sharon I, Miller CS et al. (2013). Biostimulation induces syntrophic interactions that impact C, S and N cycling in a sediment microbial community. ISME J 7: 800–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, Rapp HT et al. (2009). Complex nitrogen cycling in the sponge Geodia barretti. Environ Microbiol 11: 2228–2243. [DOI] [PubMed] [Google Scholar]
- Kasalický V, Jezbera J, Hahn MW, Šimek K. (2013). The diversity of the Limnohabitans genus, an important group of freshwater bacterioplankton, by characterization of 35 isolated strains. PLoS one 8: e58209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klüttgen BU, Dulmer U, Engels M, Ratte HT. (1994). ADaM, an artificial freshwater for the culture of zooplankton. Water Res 28: 743–746. [Google Scholar]
- Kohl KD, Dearing MD. (2012). Experience matters: prior exposure to plant toxins enhances diversity of gut microbes in herbivores. Ecol Lett 15: 1008–1015. [DOI] [PubMed] [Google Scholar]
- Martin-Creuzburg D, Beck B, Freese HM. (2011). Food quality of heterotrophic bacteria for Daphnia magna: evidence for a limitation by sterols. FEMS Microbiol Ecol 76: 592–601. [DOI] [PubMed] [Google Scholar]
- Miner BE, De Meester L, Pfrender ME, Lampert W, Hairston NG. (2012). Linking genes to communities and ecosystems: Daphnia as an ecogenomic model. Proc R Soc Lond Biol Sci 279: 1873–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moheimani NR, Borowitzka MA, Isdepsky A, Sing FS. (2013) Standard methods for measuring growth of algae and their composition. In: Borowitzka MA, Moheimani MR (ed). Algae for biofuels and Energy, Developments in applied Phycology 5. Springer Science+Business media: Dordrecht, the Netherlands, pp 265–284.
- Müller-Navarra D, Lampert W. (1996). Seasonal patterns of food limitation in Daphnia galeata: separating food quantity and food quality effects. J Plankton Res 18: 1137–1157. [Google Scholar]
- Ohkuma M. (2008). Symbioses of flagellates and prokaryotes in the gut of lower termites. Trends Microbiol 16: 345–352. [DOI] [PubMed] [Google Scholar]
- Peerakietkhajorn S, Tsukada K, Kato Y, Matsuura T, Watanabe H. (2015). Symbiotic bacteria contribute to increasing the population size of a freshwater crustacean, Daphnia magna. Environ Microbiol Rep 7: 364–372. [DOI] [PubMed] [Google Scholar]
- Peerakietkhajorn S, Kato Y, Kasalicky V, Matsuura T, Watanabe H. (2015. b). Betaproteobacteria Limnohabitans strains increase fecundity in the crustacean Daphnia magna: Symbiotic relationship between major bacterioplankton and zooplankton in freshwater ecosystem. Environ Microbiol 27: 2015; doi:10.1111/1462-2920.12919. [DOI] [PubMed] [Google Scholar]
- Pietrzak B, Grzesiuk M, Bednarska A. (2010). Food quantity shapes life history and survival strategies in Daphnia magna (Cladocera). Hydrobiologia 643: 51–54. [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. (2012) . SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Nong G, Preston JF, Ben-Ami F, Ebert D. (2009). Comparative metagenomics of Daphnia symbionts. BMC Genomics 10: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochelle PA, Will JAK, Fry JC, Jenkins GJS, Parkes RJ, Turley CM, Weightman AJ. (1995) and amplification of 16S rRNA genes from deep marine sediments and seawater to assess bacterial community diversity. In: van Elsas JD, Trevors JT (ed). Nucleic Acids in the Environment. Springer: Berlin, Germany, pp, 219–239. [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. (2011). Evidence for a core gut microbiota in the zebrafish. ISME J 5: 1595–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AK, Halpern AL, Beeson K, Tran B, Venter JC, Martiny JB et al. (2008). It's all relative: ranking the diversity of aquatic bacterial communities. Environ Microbiol 10: 2200–2210. [DOI] [PubMed] [Google Scholar]
- Sabree ZL, Kambhampati S, Moran NA. (2009). Nitrogen recycling and nutritional provisionig by Blattabacterium, the cockroach endosymbiont. PNAS 106: 19521–19526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sison-Mangus MP, Mushegian AA, Ebert D. (2014). Water fleas require microbiota for survival, growth and reproduction. ISME J 9: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale SJ, Brett MT, Pulkkinen K, Kainz MJ. (2012). The influence of bacteria-dominated diets in Daphnia magna somatic growth, reproduction, and lipid composition. FEMS Microbiol Ecol 82: 50–62. [DOI] [PubMed] [Google Scholar]
- Tokuda G, Elbourne LDH, Kinjo Y, Saitoh S, Sabree Z, Hojo M et al. (2013). Maintenance of essential amino acid synthesis pathways in the Blattabacterium cuenoti symbiont of a wood-feeding cockroach. Biol Lett 9: 20121153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoverbeke J. (2008). Modeling individual and population dynamics in a consumer-resource system: Behavior under food limitation and crowding and the effect on population cycling in Daphnia. Ecol Model 216: 385–401. [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. (1991). 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KH, Blitchington R, Greene R. (1990). Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol 28: 1942–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li S, Yang L, Huang P, Li W, Wang S et al. (2013). Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat Comm 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.