Abstract
Microorganisms can influence inorganic phosphate (Pi) in pore waters, and thus the saturation state of phosphatic minerals, by accumulating and hydrolyzing intracellular polyphosphate (poly-P). Here we used comparative metatranscriptomics to explore microbial poly-P utilization in marine sediments. Sulfidic marine sediments from methane seeps near Barbados and from the Santa Barbara Basin (SBB) oxygen minimum zone were incubated under oxic and anoxic sulfidic conditions. Pi was sequestered under oxic conditions and liberated under anoxic conditions. Transcripts homologous to poly-P kinase type 2 (ppk2) were 6–22 × more abundant in metatranscriptomes from the anoxic incubations, suggesting that reversible poly-P degradation by Ppk2 may be an important metabolic response to anoxia by marine microorganisms. Overall, diverse taxa differentially expressed homologues of genes for poly-P degradation (ppk2 and exopolyphosphatase) under different incubation conditions. Sulfur-oxidizing microorganisms appeared to preferentially express genes for poly-P degradation under anoxic conditions, which may impact phosphorus cycling in a wide range of oxygen-depleted marine settings.
Polyphosphate (poly-P) is a linear phosphate polymer that is produced by organisms from all domains of life. Microorganisms use intracellular poly-P for energy and nutrient storage, metal chelation, stress response and for certain regulatory functions (Rao et al., 2009). The ability to use poly-P as an energy reserve appears to be especially important in episodically anoxic settings. For example, poly-P is thought to act as an energy source during wastewater treatment by the enhanced biological phosphorus removal (EBPR) process. EBPR reactors cycle P-rich sludge through oxygenated and anoxic phases. Certain organoheterotrophic organisms accumulate poly-P in the oxygenated phase, and then hydrolyze that poly-P in the anoxic phase for energy to uptake and store organic carbon (Oehmen et al., 2007). In a similar manner, large marine sulfur-oxidizing bacteria in the family Beggiatoaceae also store poly-P, and have been shown to hydrolyze their poly-P stores upon exposure to anoxic or sulfidic conditions (Schulz and Schulz, 2005; Brock and Schulz-Vogt, 2011). Poly-P hydrolysis by large sulfur-oxidizing bacteria has been implicated in phosphate mineral formation in seasonally anoxic upwelling zones through the supersaturation of sediment pore waters, and might constitute an important sink in the global P cycle, both in modern sediments (Schulz and Schulz, 2005; Goldhammer et al., 2010; Brock and Schulz-Vogt, 2011) and in the past (Bailey et al., 2013; Crosby et al., 2014).
However, marine microbial communities are diverse, and beyond studies of large sulfur-oxidizing bacteria, little is known about the diversity and activity of poly-P utilizing microbes in marine sediments that experience fluctuating redox conditions. Therefore, we used a metatranscriptomic approach to compare the expression of poly-P-related genes in sediments that were experimentally exposed to anoxic and oxygenated conditions. We collected sediments from two marine environments: (1) sediments from a deep methane seep near Barbados (13°46.65′ N, 57°32.25′ W, 4743 m water depth) that included a white sulfur-oxidizing bacterial biofilm, and (2) sediments from the Santa Barbara Basin (SBB) oxygen minimum zone (34°14.493′ N, 120°06.932′ W, 573 m water depth) with no visible sulfur-oxidizing mat. Concentrations of authigenic phosphatic minerals do not occur in sediments at either site, and weight percent phosphorus is below 0.2% (Supplementary Table S1; Reimers et al., 1996). (For contrast, phosphorites are commonly defined as sediments with >18% P2O5 (Föllmi, 1996).) After collection, Barbados sediments were stored in plastic centrifuge tubes at 4 °C with 0.5 mM Pi added to core top water, whereas SBB sediments were stored at 4 °C in unamended core top water. Bulk sediments likely remained anoxic prior to incubation (see Supplementary Information and Supplementary Figure S1). Upon return to the laboratory, sediments were incubated in duplicate under oxic and anoxic conditions for 6–9 h, using the agar plug method of Schulz and Schulz (2005) to supply sulfide to anoxic incubations. Incubations were performed in low Pi artificial seawater to detect small changes associated with Pi release from cells and sediment, and sediments were gently washed in anoxic artificial seawater to remove excess Pi prior to incubation. Pi changes in the incubations were compared against sediment-free controls. Pi concentrations in the supernatants were measured using the method of Hansen and Koroleff (1999), and total cations in the initial and terminal supernatants were analyzed by inductively coupled plasma optical emission spectroscopy (ICP-OES). Following incubation experiments, sediments were immediately preserved in RNAlater (Thermo Fisher Scientific, Waltham, MA, USA) for metatranscriptomic analysis. cDNA libraries were generated from one of the oxic and anoxic incubation replicates from each location (Supplementary Table S2) using the Ovation RNA-Seq system V2 (NuGen Technologies, San Carlos, CA, USA) as in Orsi et al. (2013). A library was also generated from Barbados sediments that were preserved before the incubation experiments, after the Pi addition and collected immediately prior to the washing step (library ‘T0'). Paired-end sequencing was performed via HiSeq and MiSeq platforms (Illumina, San Diego, CA, USA), and reads were assembled de novo using IDBA-UD (Peng et al., 2012) following quality screening and rRNA removal by the methods of Jones et al. (2015). Complete details are provided in the Supplementary Methods. Raw metatranscriptomic and rRNA amplicon data sets have been deposited in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession SRP056300 and assembled contigs were uploaded to the MG-RAST server (http://metagenomics.anl.gov/) under Project ID 5305.
During all anoxic sulfidic incubations, Pi concentrations increased in the supernatant (Figure 1). Conversely, Pi concentrations decreased in oxic incubations and did not change in sediment-free controls (Figure 1). Pi release from SBB sediments was accompanied by a more than twofold increase in iron in the supernatant, but iron concentration did not change during the Barbados incubations (Supplementary Table S2).
Figure 1.
Pi release during (a) Barbados and (b) Santa Barbara Basin sediment incubations. The (*) symbols indicate samples for metatranscriptomic analysis. Barbados sample T0 (Figure 2) was collected from the pre-incubation sediment, before the incubations were initiated and prior to washing. Sediment-free controls were incubated under anoxic sulfidic conditions.
Based on rRNA gene and transcript sequencing, Barbados sediments were dominated by Gamma-, Epsilon-, Deltaproteobacteria, Bacteriodetes and anaerobic oxidation of methane group II (ANME-II) archaea (Methanosarcinales) (Supplementary Figure S2, Supplementary Table S4). The most abundant Gammaproteobacteria were close relatives of Photobacterium in the Vibrionales, and the majority of Epsilonproteobacteria were affiliated with clades of environmental sequences related to the genus Sulfurovum. The SBB sediment community was dominated by Gamma-, Epsilon-, Deltaproteobacteria, Bacteriodetes and Planktomyces (Supplementary Figure S3). SBB sediments also included abundant phytodetrital material as indicated by chloroplast and diatom rRNA sequences.
Differences in mRNA transcripts between anoxic and oxic incubations indicate short-term changes in activity (Supplementary Figure S4) that may have impacted Pi dynamics. Homologues of genes that encode enzymes for poly-P metabolism were differentially represented in the data sets (Figure 2). In prokaryotes, the synthesis and hydrolysis of intracellular poly-P is catalyzed by polyphosphate kinase (Ppk) and exopolyphosphatase (Ppx). Ppk catalyzes the reversible synthesis of poly-P from nucleoside triphosphates (NTP) (Reaction 1), and Ppx catalyzes the sequential hydrolysis of the terminal phosphate residue (Reaction 2).
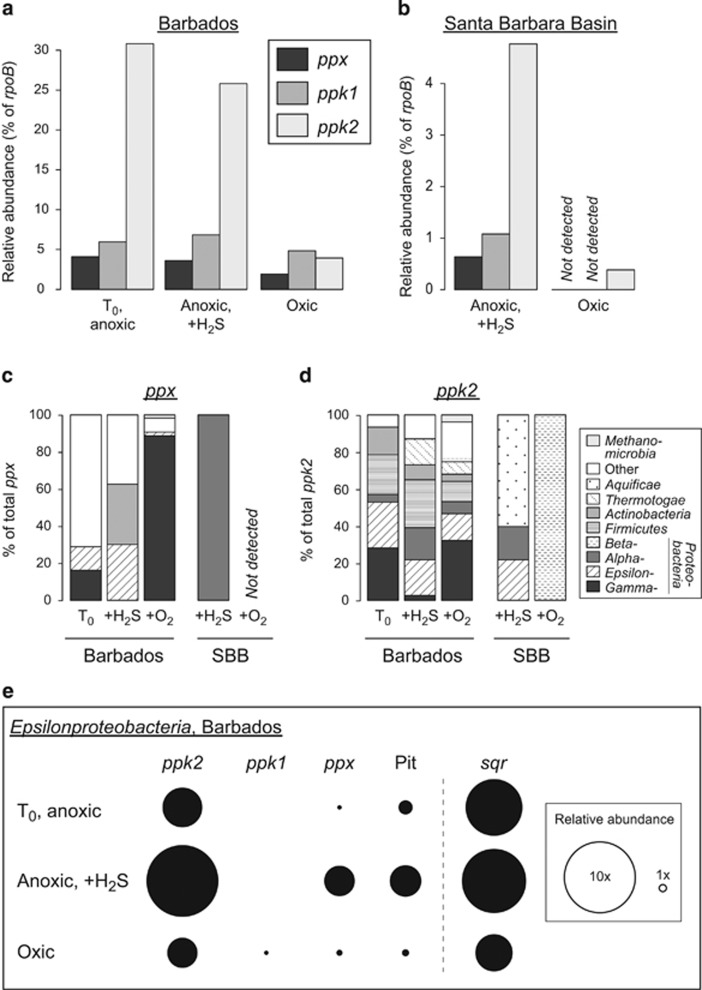
Figure 2.
(a, b) Relative abundance of ppk1, ppk2 and ppx homologues in the metatranscriptomes. The relative abundance of each transcript is expressed as a percentage of rpoB in each data set. Data set T0 was generated from the pre-incubation sediment from Barbados. (c, d) Taxonomic classification of ppx (c) and ppk2 (d) transcripts in each data set. ‘Other' includes unclassified sequences and clades that only occur in that sample (complete information in Supplementary Table S6). SBB, Santa Barbara Basin. (e) Relative abundance of Epsilonproteobacteria transcripts from Barbados, normalized to epsilonproteobacterial rpoB. sqr sequences are more abundant than ppx and ppk, and so are scaled relative to each other in (e) (see Supplementary Information for details).
Two types of Ppk are associated with Reaction 1. Ppk1 has a higher affinity for the forward reaction and is typically associated with poly-P synthesis, whereas Ppk2 generally has a higher affinity for the reverse reaction (Achbergerová and Nahálka, 2011).
Because Ppx catalyzes the liberation of Pi residues from the poly-P chain, we hypothesized that increased ppx expression would be associated with Pi release from cells in sediment pore waters. A role for Ppx in Pi release has been proposed in EBPR (Martín et al., 2006; Zafiriadis et al., 2013), and export of Pi liberated directly from poly-P has been proposed as a mechanism for proton motive force generation in some metabolic models of EBPR organisms (Saunders et al., 2007; Burow et al., 2008; Kawakoshi et al., 2012). Transcripts homologous to ppx were detected in the anoxic, but not the oxic, SBB data set, and were 1.4 × and 1.25 × more abundant in the anoxic sulfidic and T0 Barbados data sets, respectively, compared with the oxic data set. Furthermore, among the Barbados data sets, certain populations expressed ppx differently (Figure 2c). In particular, ppx from Epsilonproteobacteria were >5.8 × more abundant in the anoxic sulfidic Barbados data set than in the other two data sets (Figure 2e).
The low-affinity Pi transport system (Pit) is thought to have a role in energy generation from poly-P hydrolysis under anoxic conditions, perhaps facilitating the generation of a proton motive force (Saunders et al., 2007; Kawakoshi et al., 2012). Overall, transcripts homologous to Pit transporters seem to correlate with ppx expression (r2=0.78, P=0.045) (Supplementary Figure S5,Supplementary Table S5).
Transcripts of genes homologous to ppk1 were detected in the anoxic, but not the oxic, SBB data sets. In the Barbados data sets, ppk1 homologues were 1.9 × and 2.1 × higher in the anoxic sulfidic and T0 data sets compared with the oxic treatment (Figure 2). However, at both sites, ppk2 homologues were >6 × more abundant in the anoxic as compared with the oxygenated-treatment metatranscriptomes (Figure 2). This suggests that poly-P degradation by the reverse of Reaction 1 may be an important metabolic response to anoxia by certain sediment microorganisms. Accordingly, some models have proposed that poly-P is directly used for NTP generation by Ppk activity under anoxic EBPR conditions, and EBPR organisms often encode multiple ppk2 (Martín et al., 2006; Kawakoshi et al., 2012; Motomura et al., 2014). Phylogenetic analysis of transcripts homologous to ppk2 (Supplementary Figure S6) shows that the majority of ppk2 transcripts expressed in the anoxic treatments are ppk2 Type I (Motomura et al., 2014), which synthesize ATP and other NTPs from poly-P and nucleoside diphosphates (Nocek et al., 2008). Ppk2 may represent an important target for future characterization of poly-P metabolism in marine sediments.
Differences in treatment conditions between Barbados and SBB (Pi addition to Barbados, slightly different incubation times) may have impacted gene expression, and thus the incubation experiments from the two sites are not directly comparable. Despite these differences, we observe important consistencies between SBB and Barbados. Transcripts associated with poly-P metabolism were more highly expressed under anoxic versus oxic incubations from both sites, and Pi release was observed in all anoxic incubations. Pi flux observed in experiments from both sites could have been generated, at least in part, by poly-P hydrolysis. Indeed, higher abundances of poly-P-related transcripts in Barbados data sets, as well as a lack of increasing iron in Barbados incubation supernatants (Supplementary Table S2) suggests that poly-P hydrolysis may have been an important mechanism for Pi release from the Barbados sediments. Accordingly, Pi release from the reductive dissolution of iron oxides has been proposed as a source of Pi flux under anoxic conditions in the SBB (Reimers et al., 1996). Iron release from sediments in anoxic incubations may have also been masked by the precipitation of iron sulfides in our experiments.
Poly-P hydrolysis by large sulfur-oxidizing bacteria (Thiomargarita and other members of the family Beggiatoaceae) has been linked to the precipitation of apatite in marine sediments (Schulz and Schulz, 2005), and may be specifically triggered by exposure to sulfide (Brock and Schulz-Vogt, 2011). Interestingly, the Barbados sediments contained Thiomargarita-like bacteria ('Ca. Thiopilula' spp.), but no poly-P-related transcripts were associated with that population, perhaps because despite their large size, the Thiomargarita-like microbes are numerically less abundant than other bacteria in these sediments (Jones et al., 2015). However, expressed homologues of sulfide-quinone oxidoreductase (sqr) were affiliated with the Epsilonproteobacteria. Sqr catalyzes the first step in sulfide oxidation, which indicates that these smaller sulfur-oxidizing bacteria were likely oxidizing sulfide concurrently with poly-P degradation (Figure 2e). Cold seep sediments have steep redox gradients near the sediment surface, and perhaps as in EBPR reactors and marine upwelling zones, poly-P metabolism may also serve as an energy reserve in these environments. Our finding that multiple different taxa including sulfide-oxidizing epsilonproteobacteria (Figures 2c–e; Supplementary Tables S6 and S7) could represent a source of poly-P-liberated Pi under anoxic sulfidic conditions expands the diversity of organisms that are known to metabolize poly-P under sulfidic marine conditions and opens up the possibility that other taxa contribute to the formation of phosphatic mineral deposits. Although phosphorite is not forming at either location sampled here, authigenic calcium phosphate phases could be forming at lower concentrations, as is observed in other ocean environments (Ruttenberg and Berner, 1993), and poly-P hydrolysis could be involved in the formation of these less-concentrated, but more widespread, phosphates. Increased poly-P utilization under anoxic conditions may also indicate that poly-P has a role in phosphorus cycling in other hypoxic marine settings, including those that are expanding in response to eutrophication and climate change.
Acknowledgments
We thank three anonymous reviewers for their contributions that improved this manuscript. This work was supported by a Grant-in-Aid from the University of Minnesota and by National Science Foundation (NSF) grant EAR-1057119 to JB. The Barbados cruise was supported by NSF grant OCE-1031050 to C. Van-Dover and C. Cunningham at Duke University, and the SBB cruise was supported by NSF grant OCE-1230900 to C. Reimers. We acknowledge the Agouron Institute for a generous postdoctoral fellowship to DJ. C. Nguyen and the Minnesota Supercomputing Institute provided computing support and the use of facilities. Special thanks to V. Edgecomb for insightful discussion regarding metatranscriptomics, and to A. Becker, K. Beckman, A. Hague and D. Gohl at the UMGC for assistance and advice with metatranscriptome and amplicon sequencing. We also thank C. Van-Dover and the crew of the R/V Atlantis and ROV Jason (AT 21-02) for assisting with sample collection in Barbados and to the crew of the R/V New Horizon, C. Reimers, and the participants of the UNOLS Early Career Chief Scientist Training Cruise for sample collection in SBB.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Achbergerová L, Nahálka J. (2011). Polyphosphate – an ancient energy source and active metabolic regulator. Microb Cell Fact 10: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J, Corsetti F, Greene S, Crosby C, Liu P, Orphan V. (2013). Filamentous sulfur bacteria preserved in modern and ancient phosphatic sediments: implications for the role of oxygen and bacteria in phosphogenesis. Geobiology 11: 397–405. [DOI] [PubMed] [Google Scholar]
- Brock J, Schulz-Vogt HN. (2011). Sulfide induces phosphate release from polyphosphate in cultures of a marine Beggiatoa strain. ISME J 5: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burow LC, Mabbett AN, McEwan AG, Bond PL, Blackall LL. (2008). Bioenergetic models for acetate and phosphate transport in bacteria important in enhanced biological phosphorus removal. Environ Microbiol 10: 87–98. [DOI] [PubMed] [Google Scholar]
- Crosby CH, Bailey JV, Sharma M. (2014). Fossil evidence of iron-oxidizing chemolithotrophy linked to phosphogenesis in the wake of the Great Oxidation Event. Geology 42: 1015–1018. [Google Scholar]
- Föllmi K. (1996). The phosphorus cycle, phosphogenesis and marine phosphate-rich deposits. Earth Sci Rev 40: 55–124. [Google Scholar]
- Goldhammer T, Brüchert V, Ferdelman TG, Zabel M. (2010). Microbial sequestration of phosphorus in anoxic upwelling sediments. Nat Geosci 3: 557–561. [Google Scholar]
- Hansen H, Koroleff F. (1999) Determination of nutrients. In: Grasshoff K, Kremling K, Ehrhardt M (eds). Methods of Seawater Analysis. Wiley-VCH: Weinheim, pp 159–226. [Google Scholar]
- Jones DS, Flood BE, Bailey JV. (2015). Metatranscriptomic analysis of diminutive Thiomargarita-like bacteria (“Candidatus Thiopilula spp.”) from abyssal cold seeps of the Barbados Accretionary Prism. Appl Environ Microbiol 81: 3142–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakoshi A, Nakazawa H, Fukada J, Sasagawa M, Katano Y, Nakamura S et al. (2012). Deciphering the genome of polyphosphate accumulating actinobacterium Microlunatus phosphovorus. DNA Res 19: 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín HG, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC et al. (2006). Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol 24: 1263–1269. [DOI] [PubMed] [Google Scholar]
- Motomura K, Hirota R, Okada M, Ikeda T, Ishida T, Kuroda A. (2014). A new subfamily of polyphosphate kinase 2 (class III PPK2) catalyzes both nucleoside monophosphate phosphorylation and nucleoside diphosphate phosphorylation. Appl Environ Microbiol 80: 2602–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocek B, Kochinyan S, Proudfoot M, Brown G, Evdokimova E, Osipiuk J et al. (2008). Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria. Proc Natl Acad Sci USA 105: 17730–17735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehmen A, Lemos PC, Carvalho G, Yuan Z, Keller J, Blackall LL et al. (2007). Advances in enhanced biological phosphorus removal: from micro to macro scale. Water Res 41: 2271–2300. [DOI] [PubMed] [Google Scholar]
- Orsi WD, Edgcomb VP, Christman GD, Biddle JF. (2013). Gene expression in the deep biosphere. Nature 499: 205–208. [DOI] [PubMed] [Google Scholar]
- Peng Y, Leung HC, Yiu S, Chin FY. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Rao NN, Gómez-García MR, Kornberg A. (2009). Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem 78: 605–647. [DOI] [PubMed] [Google Scholar]
- Reimers CE, Ruttenberg KC, Canfield DE, Christiansen MB, Martin JB. (1996). Porewater pH and authigenic phases formed in the uppermost sediments of the Santa Barbara Basin. Geochim Cosmochim Acta 60: 4037–4057. [Google Scholar]
- Ruttenberg KC, Berner RA. (1993). Authigenic apatite formation and burial in sediments from non-upwelling, continental margin environments. Geochim Cosmochim Acta 57: 991–1007. [Google Scholar]
- Saunders AM, Mabbett AN, McEwan AG, Blackall LL. (2007). Proton motive force generation from stored polymers for the uptake of acetate under anaerobic conditions. FEMS Microbiol Lett 274: 245–251. [DOI] [PubMed] [Google Scholar]
- Schulz HN, Schulz HD. (2005). Large sulfur bacteria and the formation of phosphorite. Science 307: 416–418. [DOI] [PubMed] [Google Scholar]
- Zafiriadis I, Ntougias S, Kapagiannidis AG, Aivasidis A. (2013). Metabolic behavior and enzymatic aspects of denitrifying EBPR sludge in a continuous-flow anaerobic–anoxic system. Appl Biochem Biotechnol 171: 939–953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.