Key Points
A new Gata2 reporter indicates that all HSCs express Gata2 and corroborates findings that Gata2 is not required for generation of all HPCs.
Isolatable non–Gata2-expressing HPCs show less potency and a distinct genetic program, thus having implications for reprogramming strategies.
Abstract
The Gata2 transcription factor is a pivotal regulator of hematopoietic cell development and maintenance, highlighted by the fact that Gata2 haploinsufficiency has been identified as the cause of some familial cases of acute myelogenous leukemia/myelodysplastic syndrome and in MonoMac syndrome. Genetic deletion in mice has shown that Gata2 is pivotal to the embryonic generation of hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs). It functions in the embryo during endothelial cell to hematopoietic cell transition to affect hematopoietic cluster, HPC, and HSC formation. Gata2 conditional deletion and overexpression studies show the importance of Gata2 levels in hematopoiesis, during all developmental stages. Although previous studies of cell populations phenotypically enriched in HPCs and HSCs show expression of Gata2, there has been no direct study of Gata2 expressing cells during normal hematopoiesis. In this study, we generate a Gata2Venus reporter mouse model with unperturbed Gata2 expression to examine the hematopoietic function and transcriptome of Gata2 expressing and nonexpressing cells. We show that all the HSCs are Gata2 expressing. However, not all HPCs in the aorta, vitelline and umbilical arteries, and fetal liver require or express Gata2. These Gata2-independent HPCs exhibit a different functional output and genetic program, including Ras and cyclic AMP response element-binding protein pathways and other Gata factors, compared with Gata2-dependent HPCs. Our results, indicating that Gata2 is of major importance in programming toward HSC fate but not in all cells with HPC fate, have implications for current reprogramming strategies.
Introduction
Gata2 is one of the “heptad” transcription factors that acts on regulatory regions of hematopoietic genes.1 It is upregulated in vivo in Ly6aGFP+ cells undergoing endothelial-to-hematopoietic cell transition (EHT), a process by which definitive hematopoietic progenitors (HPCs) and hematopoietic stem cells (HSCs) are generated in the embryo.2,3 As one of the major regulators of HPC and HSC generation, germline deficiency of Gata2 results in embryonic lethality between embryonic day (E)10 and E10.5 and an anemic phenotype, with a decreased number of primitive and definitive HPCs in the yolk sac (YS) and in Gata2−/− embryonic stem (ES) cell hematopoietic differentiation cultures.4-6 Chimeric embryo generation with Gata2−/− ES cells revealed defective production of all hematopoietic lineages.5 The E10.5 lethality of Gata2−/− embryos precludes the study of HSC generation in the aorta-gonad-mesonephros (AGM) region, the first site of de novo HSC production. Gata2+/− embryos contain greatly reduced number of HSCs in the AGM region.7,8 Gata2 haploinsufficiency perturbs adult HSC homeostasis in mice9 and, in humans, leads to MonoMac syndrome,10 which is associated with sporadic myelodysplasia and myeloid leukemia. Also, rearrangement of the remote Gata2 enhancer drives acute myeloid leukemogenesis by activating Evi1 expression.11,12 Overexpression studies also reveal that levels of Gata2 expression are important for its hematopoietic function.13-15 In situ hybridization studies localize Gata2 expression to aortic endothelial cells, intra-aortic hematopoietic cluster cells, placenta (PL), and fetal liver (FL) in the midgestation mouse.16-18 Conditional knockout of Gata2 or Gata2 regulatory elements in vascular endothelial cells indicates that Gata2 is essential for hematopoietic cluster formation and HSC generation.7,19,20 Gata2 plays a role in the emergence of cKit-expressing hematopoietic cells from the endothelium.7 Later, as shown in VavCre conditional knockout mice, Gata2 is essential for HSC maintenance,7 thus demonstrating a role for Gata2 as previously recognized in bone marrow LSK HSCs.21
To date, the correlation between Gata2 and hematopoietic cell generation in the embryo has been made in the absence of prospective isolation of viable Gata2-expressing cells.16 Although some hematopoietic cells remain in the embryo in the absence of Gata2,5-8 the identity of these cells is unknown. In this study, to further understand the requirement for Gata2 in normal hematopoietic development, we create and use a mouse model in which a fluorescent reporter for Gata2 (IRES-Venus knock-in gene) does not affect the normal level or function of Gata2. We demonstrate that all long-term repopulating HSCs and a large percentage of HPCs in the midgestation mouse embryo are Venus+. We isolate and characterize a Venus− HPC population that corresponds to the HPCs found in Gata2-null embryos. Gata2-independent hematopoietic progenitors are functionally less complex and do not follow the same genetic program as Gata2-dependent HPCs.
Materials and methods
Gata2Venus ES cells and mice
Generation of the Gata2-Venus mouse model is described in the supplemental Methods, available on the Blood Web site. In short, an IRES-Venus fragment and a loxP-PGK-Puro-loxP fragment were inserted in the Gata2 3′ untranslated region (UTR). IB10 ES cells were transfected and puromycin selected, and 360 clones were polymerase chain reaction (PCR) screened for Gata2Venus (right arm junction, 2292 bp). Correct integration was verified by Southern blot (left arm) for 2 clones with normal karyotype. Founders were identified by Venus PCR. First-generation G2V offspring were crossed with CAG-Cre mice22 and backcrossed (>10 generations) with C57BL/6.
Mice and embryo production
Gata2+/− mice,5 Ly5.1 (6-8 weeks) and C57BL/6 mice were obtained/maintained (Harlan or locally) and genotyped by PCR (supplemental Methods). Day of plug discovery is E0. Embryos were staged by somite pair (sp): E9.5 = 16 to 28 sp, E10 = 28 to 40 sp, early E10 = 28 to 34 sp, E10.5 = 35 to 40 sp, and E11 = 40 to 50 sp. All mouse experimentation was performed under the UK Animals Scientific Procedures Act 1986 Project License 70/8076 and NL Ethics Committee approval and performed in compliance with Standards for Care and Use of Laboratory Animals.
Immunostaining
Whole-mount conceptuses were stained and imaged23; cryosections and cells for flow cytometry were stained8 using anti-CD34-biotin (1:50; BD), anti-Gata3 (1:10 KT122, 111207H09; Absea), anti-Gata4 (1:50 H-112, sc-9053; SantaCruz), and anti-green fluorescent protein antibodies. For flow cytometry,8 cells were stained with anti-CD31 (390; BD), anti-CD34 (RAM34; BD), anti-cKit (2B8; BD), anti-CD41 (MWReg3; SantaCruz), anti-Sca1 (D7; Ebiosciences), and anti-CD16/32 (2.4G2; BD) antibodies and Hoechst 33258 (BD) and analyzed (FACSARIAIII; SORP).
Hematopoietic assays
Venus-sorted E9 to E11 AGM, vitelline+umbilical arteries (VA+UA), PL, and YS and E10 FL or Gata2-deficient E9 to E10 AGM, VA+UA, and PL were seeded in 3.6 mL methylcellulose (1 mL per dish; M3434; Stem Cell Tech) for 10 to 12 days.24 Colonies were counted, isolated, and washed, and Venus expression was examined (FACSARIAIII; Fortessa). Sorted G2V E11 AGM (Ly5.1/Ly5.2) cells were transplanted24 into 9.5-Gy irradiated (C57BL/6X129) F1 recipients (Ly5.2/Ly5.2) together with 2 × 105 spleen cells from the recipient strain. Peripheral blood (PB) donor chimerism was determined by Venus PCR and/or fluorescence-activated cell sorter (FACS) at 4 months after transplantation and scored positive if PB donor chimerism was >10%.
RNA analyses
Detailed RNA procedures are provided in supplemental Methods. The Gene Expression Omnibus data accession number is GSE76254. Briefly, RNA was isolated from E10.5 AGM CD31 and cKit sorted cells with the mirVana miRNA Kit (Ambion), and quality/quantity was assessed using the 2100 Bioanalyzer (Agilent; RNA Nano/Pico chip). For RNA sequencing, samples were prepared with the SMARTER protocol and analyzed on an Illumina HiSeq200 system.25-27 For quantitative reverse transcription-PCR (qRT-PCR), SuperScriptII/III ReverseTranscriptase (LifeTechnologies) was used for first-strand cDNA synthesis. Primers are specified in supplemental Table 1.
Results
Generation and validation of a novel Gata2 reporter mouse model
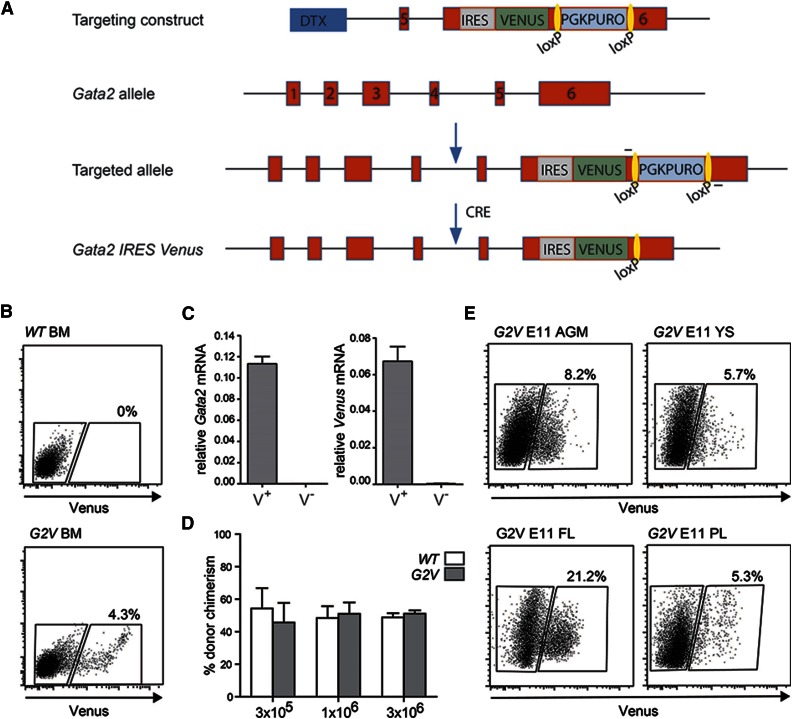
Previously, analysis of Gata2-expressing cells has been limited to a reporter mouse model that results in Gata2 haploinsufficiency.16 Our approach allows for the expression of the reporter within the Gata2 genomic locus without affecting the levels of Gata2 expression or protein function. This is particularly important because Gata2 haploinsufficiency greatly reduces the number of HS/PCs generated during development.7,8,19,20,28 Briefly, an internal ribosome entry site sequence (IRES) followed by the Venus fluorochrome gene was recombined into the Gata2 3′UTR (Figure 1A) in ES cells. The resulting Gata2Venus (G2V) mice bred normally and showed no overt growth or hematopoietic defects.
Figure 1.
Gata2 Venus reporter construction and validation. (A) Schematic diagram of the IRES Venus reporter selection cassette insertion in the 3′UTR of the mouse Gata2 locus and Cre-mediated removal of lox PGK-Puro lox. Primers used for detection of the targeted and recombined alleles are indicated flanking the loxP sites (yellow). (B) Representative flow cytometric analysis and sorting plot of Venus-expressing cells in the BM of adult Gata2 Venus (G2V) mice. Gated regions show percentage positive and negative viable cells. (C) Relative levels of Gata2 and Venus mRNA in sorted Venus+ and Venus− Gata2V/+ BM cells as determined by qRT-PCR. Gata2 transcripts in Venus+ cells = 0.11337 ± 0.00681 and Venus− cells = 0.00012 ± 0.00003 (P = .000076). Venus transcripts in Venus+ cells = 0.06722 ± 0.00799 and Venus− cells = 0.00036 ± 0.00010 (P = .00112). Mean ± standard error of the mean (SEM), n = 3. (D) Competitive limiting dilution transplantation strategy used to test the quantity and robustness of Gata2V/V BM HSCs compared with wild type. Percentage of donor cell chimerism in adult irradiated recipients cotransplanted with the same number of wild-type (WT) Ly5.1/5.2 and Gata2 Venus (G2V/V) Ly5.2 BM cells. Varying numbers (1 × 105, 3 × 105, 3 × 106) of BM cells of each genotype were injected, and peripheral blood of recipients was analyzed for donor cell engraftment by FACS at 1 and 4 months after transplantation. n = 2 (5 mice per group). (E) Representative FACS plots demonstrating frequency of Venus expressing cells in E11 AGM, YS, PL, and FL. Gates indicate Venus− and Venus+ cell fractions. Percentages represent the frequency of Venus+ cells within the viable cell fraction (Table 1).
To determine whether Venus reporter expression parallels that of Gata2, G2V bone marrow (BM) cells were sorted into Venus-expressing (Venus+) and nonexpressing (Venus−) fractions (Figure 1B). qRT-PCR for Gata2 and Venus transcripts (Gata2V/+ BM) demonstrated that only Venus+ cells express Venus and Gata2 mRNA (Figure 1C). Western blot analysis revealed that equivalent amounts of Gata2 protein were present in Gata2+/+ and Gata2V/V adult BM cells (data not shown). FACS analysis showed that Gata2v/v BM LSK frequency (388/105 cells) are comparable to wild-type (WT) BM LSK frequency (378/105 cells). Importantly, the results of competitive limiting dilution transplantation analyses of Gata2V/V and WT BM cells demonstrate that HSCs are qualitatively and quantitatively normal in this mouse model (Figure 1D). Thus, Venus expression correctly reports Gata2 expression without interfering with its normal expression levels or function.
Gata2 is expressed in emerging aortic hematopoietic cluster cells and other embryonic hematopoietic tissues
Venus expression was examined in midgestation G2V hematopoietic tissues. Flow cytometry revealed that E9 to E11 AGM, YS, PL, and FL contained Venus+ cells (Figure 1E). At E9, 6.28 ± 0.47% of viable YS cells and 1.82 ± 0.31% of AGM cells are Venus+. At E10.5 (when the first HSCs are generated), 3.27 ± 0.52% of AGM cells are Venus+, and this increases to 7.86 ± 1.1% at E11. Table 1 shows the frequencies of Gata2-expressing cells.29,30
Table 1.
Frequency of Venus+ cells in embryonic tissues of G2V embryos
| Tissue | Stage | Number of experiments, embryos analyzed | % Venus+ cells/tissue |
|---|---|---|---|
| AGM | E9, 16-25 sp | n = 4, 22 | 1.82 ± 0.31 |
| E10, 28-36 sp | n = 4, 29 | 3.27 ± 0.52 | |
| E11, 43-49 sp | n = 4, 25 | 7.86 ± 1.1 | |
| FL | E9 | nd | nd |
| E10, 28-36 sp | n = 4, 10 | 13.89 ± 0.7 | |
| E11, 43-49 sp | n = 4, 19 | 19.27 ± 2.14 | |
| YS | E9, 16-25 sp | n = 5, 8 | 6.28 ± 0.47 |
| E10, 28-36 sp | n = 6, 9 | 6.17 ± 0.86 | |
| E11, 42-46 sp | n = 1, 3 | 5.25 ± 0.59 | |
| PL | E9, 17-23 sp | n = 3, 4 | 10.91 ± 0.49 |
| E10, 28-35 sp | n = 3, 6 | 10.8 ± 1.92 | |
| E11, 42-46 sp | n = 1, 3 | 10.01 ± 4.64 |
The frequency of Venus+ cells within the viable cell fraction of embryonic tissues per embryo is presented. FACS analysis of single cell suspensions of dissected embryonic tissues was performed to define the percentage of cells expressing Venus. AGM contains part of the vitelline and umbilical arteries. Our data for the total number of cells in each tissue (data not shown) correlated with published data for YS and E9.5 AGM29 and for FL.30 The data represent mean ± SEM of 3 to 6 independent experiments, with the exception of E11 YS and PL data (data represent the mean ± standard deviation of 1 experiment. E, embryonic day; n, number of independent experiments, number of individual embryos analyzed; nd, not done; sp, somite pairs.
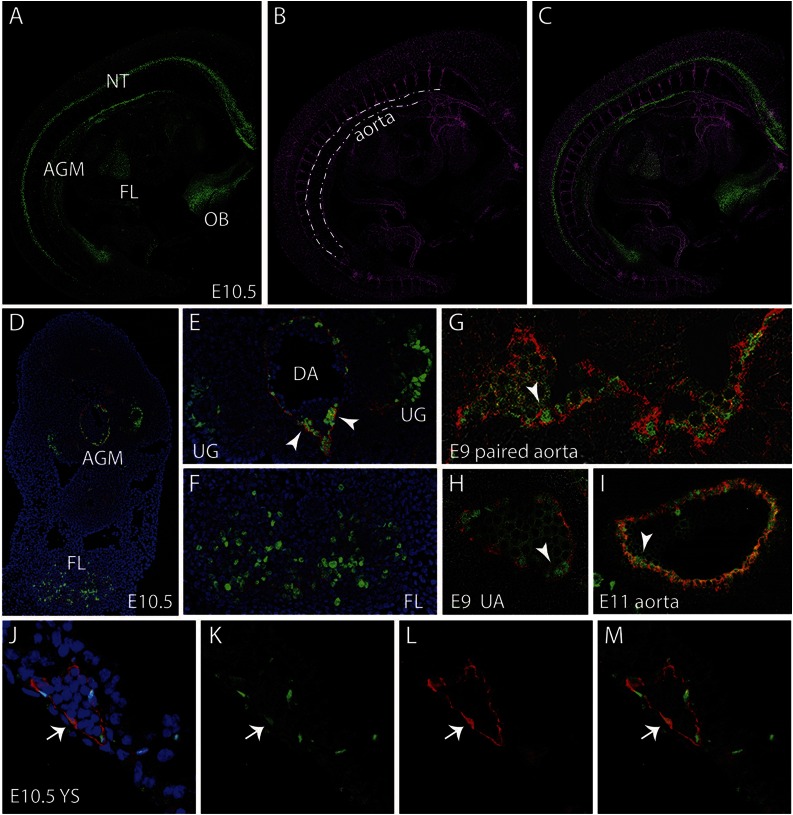
Whole-mount images of E10 and E11 G2V embryos immunostained with anti-CD31 antibody (marks all endothelial cells and hematopoietic cluster cells) shows Venus+ cells along the aorta (DA). Venus+ cells are also observed in cells of the neural tube (NT), olfactory bulb (OB), and FL (Figure 2A-C). In the E10.5 AGM region (4,6 diamidino-2-phenylindole [DAPI] and CD31 stained [blue and red, respectively]), Venus expression is found in endothelial and hematopoietic cluster cells mainly on the ventral side of the DA and in the urogenital (UG) region (Figure 2D-E). In the FL, Venus-expressing cells are found in a punctate distribution pattern (Figure 2D,F). At E9, Venus is expressed in some of the CD34+ (red) endothelial cells of the paired aorta (Figure 2G, arrowhead) and also in some of the endothelial and hematopoietic cluster cells of the UA (Figure 2H). Venus continues to be expressed at E11 in some aortic endothelial cells and emerging/other hematopoietic cluster cells (Figure 2I, arrowhead). The E10.5 YS shows Venus expression in some of the CD31+ (red) endothelial cells. Overall, Venus expression is similar to what has been previously documented for Gata2 in situ hybridization analysis.31,32 Thus, our model allows for the prospective identification, isolation, and characterization of Gata2-expressing cells during normal development.
Figure 2.
Localization of Gata2Venus-expressing cells in embryonic hematopoietic sites. Confocal images of a whole mount immunostained E10.5 Gata2Venus embryo showing (A) Venus (green), (B) CD31 (magenta), and (C) merged expression. Venus-expressing cells are detected in the AGM along the wall of the dorsal aorta (dotted lines), the FL, NT, and OB. (D) Confocal image of a transverse section through the E10.5 AGM. DAPI staining (blue), CD31 (red), and Venus fluorescence (green) revealed Gata2-expressing aortic endothelial and hematopoietic cluster cells and UG and FL cells. Enlarged images of D showing Gata2-expressing cells in (E) AGM (DA, dorsal aorta; UG, urogenital ridges; arrowheads indicate hematopoietic cluster) and (F) FL. Venus (green) and CD34 (red) fluorescence showing endothelial and hematopoietic cluster cells in (G) E9 paired aorta, (H) umbilical artery (UA) at E9, and (I) E11 aorta. Arrowheads indicate hematopoietic cluster. (J-M) Images of E10.5 YS section showing DAPI merged, Venus, CD31, and merged fluorescence. Arrow denotes an endothelial cell expressing Venus and CD31.
All HSCs, but not all HPCs, express Gata2
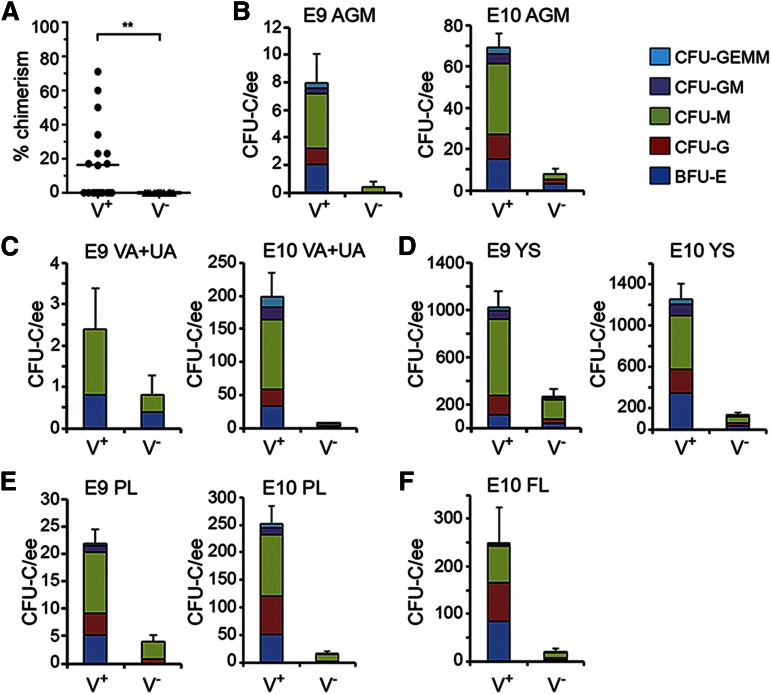
To test for HSC activity, E11 AGM Venus+ and Venus− cells were transplanted into irradiated adult recipients. All long-term repopulating HSCs were found in the Gata2Venus-expressing fraction (Figure 3A). Nine of 19 recipients receiving Venus+ cells were engrafted (15-71%), whereas none of the 14 Venus− recipients showed donor-derived hematopoietic cells. These HSCs were multilineage repopulating (supplemental Figure 1) and self-renewing (8 repopulated of 12 transplanted with 3 × 106 BM cells from primary repopulated mice; n = 4).
Figure 3.
Quantitation of functional HSCs and HPCs in G2V embryonic hematopoietic tissues. HSCs in sorted Venus+ and Venus− cell fractions of E11 AGM were analyzed by transplantation into irradiated adult recipients. (A) Percentage donor cell chimerism was determined by Venus PCR of peripheral blood DNA at 4 months after transplantation. Each dot represents 1 recipient receiving 1.7 to 6.5 embryo equivalent (ee) of AGM cells. n = 7. **P = .0089. (B-F) Hematopoietic progenitor number per tissue in sorted Venus+ and Venus− cell fractions of (B) E9 and E10 AGM, (C) E9 and E10 VA+UA, (D) E9 and E10 YS, (E) E9 and E10 PL, and (F) E10 FL. CFU-C per 1 ee of tissue is shown. Colony types designated by colored bars are CFU-granulocyte, erythroid, monocyte, megakaryocyte (GEMM); CFU-granulocyte, macrophage (GM); CFU-macrophage (M); CFU-granulocyte (G), and burst forming unit-erythroid (BFU-E). SEM of total CFU-C is shown; 2 ee of somite pair–matched tissues were pooled for sorting and yielded 1 ee for colony analysis.
The relationship between Gata2 expression and HPC function was also examined. E9 and E10 AGM Venus+ and Venus− cells were plated in the colony forming unit–culture (CFU-C) assay. High enrichment of HPCs was found in the Venus+ fractions (Table 2). Surprisingly, HPCs were found also in the Venus- fraction, although there were very few. At E9, the Venus+ and Venus− fractions, respectively, yielded 8.0 ± 2.1 and 0.4 ± 0.4 CFU-C per AGM. CFU-C numbers increased in the Venus+ cell fraction at E10 (69.0 ± 7.1) and the Venus− fraction increased to 8.0 ± 2.3 CFU per AGM. However, bi- and multipotent progenitors were found only in the Venus+ fraction (Figure 3B).
Table 2.
CFU-C number in Venus+ and Venus− cell fractions of G2V embryonic tissues
| Tissue | Stage | Number of experiments, embryos analyzed | CFU-C/tissue/sorted cell fraction | |
|---|---|---|---|---|
| Venus− | Venus+ | |||
| AGM | E9, 20-23sp | n = 2, 5 | 0.4 ± 0.4 | 8.0 ± 2.1 |
| E10, 32-35sp | n = 2, 6 | 8.0 ± 2.3 | 69.0 ± 7.1 | |
| VA+UA | E9, 20-23sp | n = 2, 5 | 0.8 ± 0.5 | 2.4 ± 1.0 |
| E10, 32-35sp | n = 2, 6 | 7.0 ± 1.5 | 197.7 ± 36.9 | |
| YS | E9, 20-23sp | n = 2, 5 | 270.0 ± 69.8 | 1020 ± 137.3 |
| E10, 32-35sp | n = 2, 5 | 130.0 ± 25.5 | 1252.0 ± 156.0 | |
| PL | E9, 20-23sp | n = 2, 5 | 4.0 ± 1.3 | 22.0 ± 2.5 |
| E10, 32-35sp | n = 2, 6 | 14.5 ± 5.3 | 251.7 ± 32.4 | |
| FL | E10, 32-35sp | n = 2, 6 | 20.5 ± 7.6 | 248.7 ± 75.2 |
Number of total CFU-C (mean ± SEM) per tissue per G2V embryo for the sorted Venus− and Venus+ cell fractions at E9 and E10.
HPC activity was also examined in the Venus+ and Venus− fractions of other hematopoietic tissues (Table 2). E9 and E10 VA+UA (Figure 3C), YS (Figure 3D), and PL (Figure 3E), and E10 FL (Figure 3F) contained progenitors in both fractions. Most HPCs were Venus+. The greatest number of CFU-C arising from Venus− cells was found in the E9 YS (270.0 ± 69.8 CFU-C/YS). These data indicate that some HPCs are not expressing Gata2. BFU-E, CFU-G, and CFU-M were the predominant colony types in both fractions, and in contrast to the Venus+ fractions, the Venus− fractions of VA+UA, YS, FL, and PL yielded few or no CFU-GEMM. Thus, all AGM HSCs express Gata2, Gata2 expression is associated with immature HPCs, but not all HPCs are Gata2 expressing.
Some HPCs and vascular cluster cells are formed in the absence of Gata2
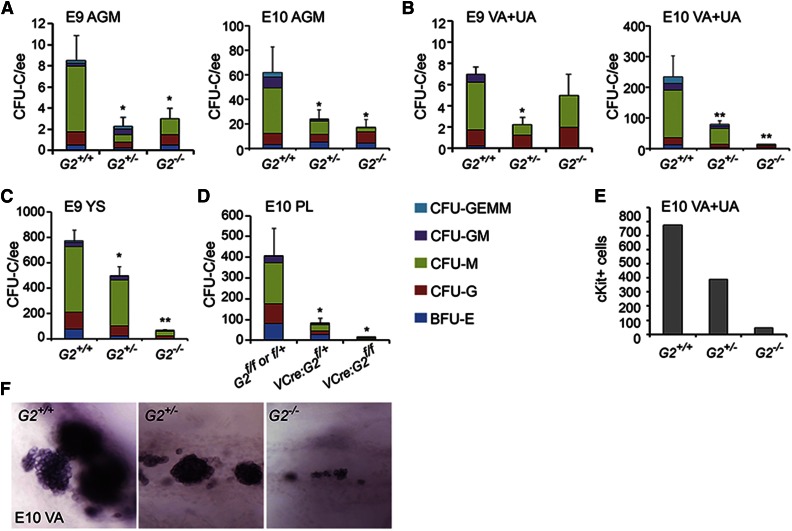
Because the Venus− fractions of midgestation G2V hematopoietic tissues contain CFU-C, we tested whether such hematopoietic progenitors are present in Gata2-deleted embryos. CFU-Cs were detected in the Gata2−/− E9 AGM, E10 AGM (Figure 4A), and E10 VA+UA (Figure 4B), although significantly fewer compared with WT (Table 3). Gata2+/− tissues also contained fewer CFU-Cs compared with WT. The E9 Gata2−/− YS contained the most CFU-C (64.4 ± 12.2; Figure 4C). In VEC-Cre:Gataf/f embryos, E10 PL showed significantly decreased CFU-C numbers (Figure 4D), as did E10 AGM and YS7 compared with WT. The CFU-C remaining in Gata2−/− embryos are predominantly CFU-G and CFU-M. Very few Gata2−/− CFU-GM and no CFU-GEMM were observed. These data support and validate our findings in G2V embryos that not all HPCs are Gata2-expressing, Gata2-independent progenitors exist in each of the early hematopoietic tissues, and the Gata2-expressing cell fraction is more enriched in multipotent progenitors.
Figure 4.
CFU-C numbers and vascular hematopoietic clusters in Gata2−/− embryos. CFU-C numbers per ee found in (A) E9 and E10 AGM, (B) E9 and E10 VA+UA, (C) E9 and E10 YS, and (D) E10 placenta. *P < .05; **P < .01. (E) Quantitation of cKit+ hematopoietic cluster cells in VA+UA of E10 Gata2+/+, Gata2+/−, and Gata2−/− embryos. (F) Representative whole mount images of hematopoietic cluster cells in the VA of E10 Gata2+/+ (30 sp), Gata2+/− (31 sp), and Gata2−/− (30 sp) embryos stained for cKit expression.
Table 3.
CFU-C number per E9-E10 Gata2-deleted hematopoietic tissues
| Tissue | Stage | Genotype | |||||
|---|---|---|---|---|---|---|---|
| WT | Gata2+/− | Gata2−/− | |||||
| AGM | E9, 20-23sp | 8.5 ± 2.3 | n = 1, 4 | 2.25 ± 0.9* | n = 1, 4 | 3.0 ± 1.0 | n = 1, 2 |
| E10, 28-34sp | 62.4 ± 20.8 | n = 3, 7 | 24.0 ± 7.4* | n = 3, 13 | 16.7 ± 6.8* | n = 3, 9 | |
| VA+UA | E9, 20-23sp | 7.0 ± 0.6 | n = 1, 4 | 2.25 ± 0.6* | n = 1, 4 | 5.0 ± 2.0 | n = 1, 2 |
| E10, 28-34sp | 241.1 ± 67.8 | n = 3, 7 | 78.7 ± 12.3** | n = 3, 13 | 12.0 ± 2.5** | n = 3, 9 | |
| YS | E9, 20-23sp | 772.5 ± 85.3 | n = 1, 4 | 500.0 ± 71.5* | n = 1, 4 | 64.4 ± 12.2** | n = 1, 2 |
| E10, 28-34sp | 918.6 ± 147.9 | n = 3, 7 | 584.6 ± 89.0** | n = 3, 9 | 25.7 ± 8.0** | n = 2, 3 | |
| Gata2f/+ or Gata2f/f | VEC-Cre:Gata2f/+ | VEC-Cre:Gata2−/− | |||||
| FL | E10, 30-34sp | 115.8 ± 35.0 | n = 2, 5 | 83.0 ± 20.8 | n = 2, 6 | 22.2 ± 6.7* | n = 2, 6 |
| PL | E10, 30-34sp | 406.0 ± 134.0 | n = 2, 5 | 81.0 ± 25.0* | n = 2, 6 | 12.0 ± 4.0* | n = 2, 6 |
Number of total CFU-C (mean ± SEM) per tissue shown for WT, Gata2 germline, and conditional knockout embryos at E9 and E10.
P < .05.
P < .01.
Because hematopoietic clusters appear in the VA and UA prior to appearance in the aorta, and are larger than in the AGM,33 we further examined these vessels. Whole-mount microscopic analysis demonstrates that clusters form in the absence of Gata2. The number and size of cKit+ hematopoietic clusters in early E10 Gata2+/− and Gata2−/− VA+UA are decreased compared with WT (Figure 4E). The number of cKit+ cells decreases 20-fold in the E10 Gata2−/− VA+UA (Figure 4F) in correspondence to the decrease in VA+UA CFU-C (Figure 4B), suggesting that these emerging cKit+ hematopoietic cluster cells are part of the cohort of Gata2-independent HPCs.
Alternative genetic program is expressed in Venus− hematopoietic cells
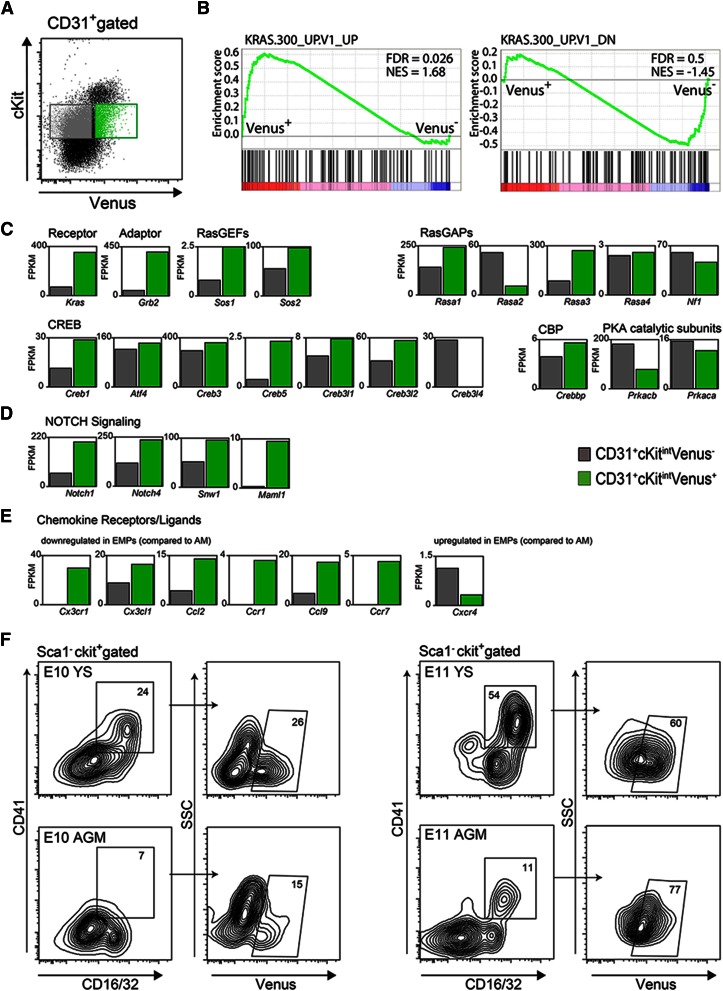
The molecular basis for the functional differences observed in Gata2-dependent and -independent HPCs was examined by RNA sequencing. As most CD31+Venus− HPCs showed cKit intermediate expression, we compared this population to CD31+Venus+cKitint HPCs (Figure 5A). Gene set enrichment analysis on genes sorted by log ratio of Venus+ vs Venus− FPKMs revealed that genes in the Ras signaling pathway were significantly enriched in the Venus+ compared with the Venus− fraction (Figure 5B). Genes upregulated by Ras were enriched in the Venus+ fraction, and highly upregulated genes included Kras, Grb2 (Ras adaptor), and Sos1 and Sos2 (RasGEF activators) (Figure 5C). Genes downregulated by Ras were enriched in the Venus− fraction. RasGAP gene (renders Ras inactive) Rasa2 was highly upregulated in the Venus− fraction, whereas Rasa1 and Rasa3 were highly upregulated in the Venus+ fraction. Rasa4 and NF1 were expressed to similar levels. Also, Venus+ HPCs showed increased levels of CREB and CBP expression compared with Venus− HPCs and express protein kinase A catalytic subunit genes, suggesting that Venus+ HPCs have the potential to activate CREB target genes. Gata2 has CREB response element consensus sites (−3 kb, −300 bp upstream transcription start site), suggesting that it is a downstream target.34,35 As Gata2 is a Notch target,18 a two- to fourfold higher expression of Notch1 and Notch4 was found in the Venus+ fraction (Figure 5D). Moreover, Snw1 and Maml1 (transcriptional coactivators in the Notch pathway that interact with Notch) were upregulated (2- and 30-fold, respectively) in Gata2-expressing HPCs.
Figure 5.
Differential expression of signaling pathway modulators in Gata2-dependent and -independent HPCs. (A) Flow cytometric sorting gates for isolation of E10.5 AGM G2V CD31+cKitintVenus− (gray) and CD31+cKitintVenus− (green) HPCs used for RNA sequence analysis. Gene Expression Omnibus data accession number is GSE76254. (B) Gene enrichment analysis for Ras signaling pathway genes. Bar graphs of fragments per kilobase million (FPKM) values obtained from RNA sequence analysis of CD31+cKitintVenus− (gray bar) and CD31+cKitintVenus+ (green bar) AGM cells for (C) Ras pathway and cyclic AMP response element-binding protein (CREB) and CREB-binding protein (CBP) transcription factor genes and (D) Notch pathway genes. (E) Bar graphs of FPKM values obtained from RNA sequence analysis for a selection of chemokine receptor/ligand genes (see Kierdorf et al37; these genes were down-/upregulated in YS EMPs compared with adult microglia [AM]). (F) Representative FACS plots demonstrating frequency of EMPs in the Venus+ fraction, as defined as Sca1−cKit+CD41+CD16/32+, in YS and AGM of E10 (left) and E11 (right) G2V embryos. Numbers indicate the percentages of gated cells within the parental cell population.
Because Venus− HPCs are mainly restricted in their differentiation potential to the macrophage and granulocytic lineages, we evaluated their similarity to YS-derived erythromyeloid progenitors (EMPs) that give rise to tissue-resident macrophages. Flow cytometric analysis for EMP markers36 showed that 3.89% of E10 YS and 0.72% of E10 AGM cells were EMPs (Sca1−cKit+CD41+CD16/32+). The majority of EMPs were Venus− (74% in YS, 85% in AGM; Figure 5F). At E11, the frequency of EMPs in the E11 YS and AGM decreased to 2.11% and 0.11%, respectively (Figure 5F), with 60% of YS and 77% of AGM EMPs now being Venus+. Published transcriptome data on mouse YS EMPs show the low expression of several chemokine receptors/ligands (Cx3cr1, Cx3cl1, Ccl2, Ccr1, Ccl9, and Ccr7).37 The expression of these genes was low or absent in Venus− AGM cells compared with Venus+ cells (Figure 5F). Also, Cxcr4 (highly expressed in EMPs) was highly expressed in Venus− AGM cells compared with Venus+ cells. These results suggest that the Venus− population shares similarities to EMPs at the transcription level.
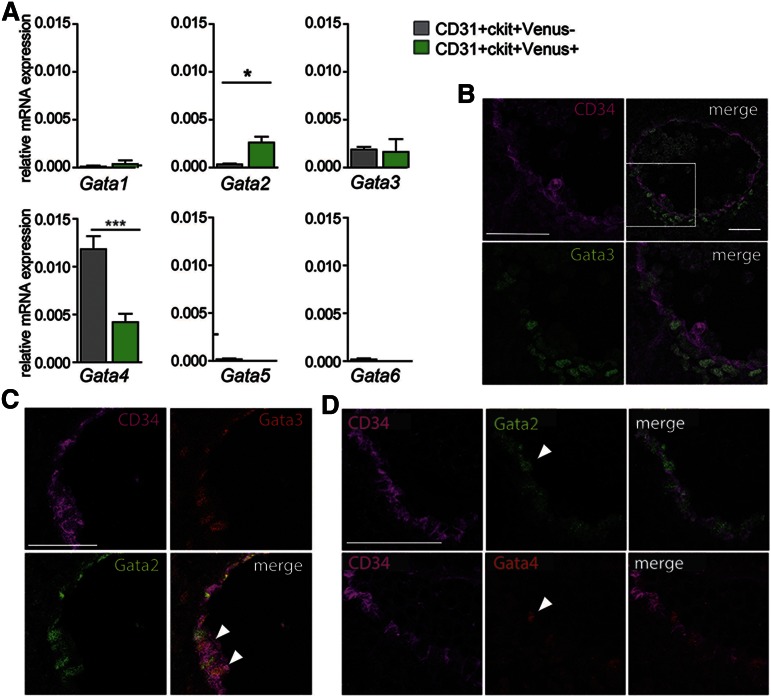
Analysis of FPKMs for heptad transcription factors previously described as expressed in AGM HSCs and HPCs1-3 showed expression in both the Venus+ and Venus− AGM fractions (data not shown). Also, other Gata factors were expressed in both fractions. In the mouse, Gata1, 2, and 3 are hematopoietic transcription factors, whereas the Gata4, 5, and 6 factors are not directly related to hematopoiesis. qRT-PCR performed on E10.5 AGM CD31+cKit+ cells (Figure 6A) confirmed Gata3 expression by both the Venus+ and Venus− fractions, and Gata4 was significantly higher in the Venus− fraction. Gata1, 5, and 6 transcripts were low/not detected. Immunostaining of E10.5 AGM (WT) showed Gata3-expressing cells in the mesenchyme underlying the ventral aspect of the aorta, aortic endothelial cells, and some cells emerging from the aortic wall (Figure 6B) in agreement with Fitch.38 G2V E10.5 AGM confirmed that some aortic endothelial cells coexpress Gata3 and Gata2 (Figure 6C). Gata4 expression was also found in aortic endothelial (CD34+) cells, but it did not overlap with Gata2 expression (Figure 6D). Together, these results suggest that Gata3 and/or Gata4 may provide some function in Gata2-independent hematopoietic cells.
Figure 6.
Gata family gene expression in AGM Gata2-dependent and -independent HPCs. (A) qRT-PCR for expression of Gata1, 2, 3, 4, 5, and 6 transcription factors (normalization with Gapdh) in E11 AGM CD31+cKit+Venus+ and CD31+cKit+Venus− cells. n = 3. SEM shown with *P = .05 and ***P = .001. (B) Transverse section of WT E10.5 AGM immunostained for CD34 (magenta) and Gata3 (green) showing expression of Gata3 in the aortic endothelial cells and some emerging hematopoietic cells and ventral mesenchymal cells directly under the aorta. (C) Transverse section of G2V E10.5 AGM immunostained for CD34 (magenta), Gata2 (green), and Gata3 (red) showing some overlapping expression of Gata2 and Gata3 in aortic endothelial cells (arrowheads). (D) Transverse consecutive sections of E11 G2V AGM immunostained for CD34 (magenta) and Venus (green) in the top panels and for CD34 (magenta) and Gata4 (red) in the bottom panels. Gata4 expression is observed in some ventral aortic endothelial cells and emerging hematopoietic cells (arrow).
Discussion
In this study, we prospectively enriched and characterized Gata2-dependent and -independent HPC subsets from our novel Gata2Venus reporter mouse. Molecular analyses, together with the fact that some vascular hematopoietic cluster cells and HPCs persist in the absence of Gata2 expression, suggest that an alternative genetic program exists for the production of HPCs. The transcriptome differences observed between Venus+ and Venus− HPCs may offer possibilities for pathway modifications to achieve the programming complexities necessary for the generation/function of normal definitive HPCs and provide insights into the factors involved in myeloid leukemogenesis.
Gata2 expression in the developing hematopoietic system
We showed the temporal and quantitatively coordinate transcription of Venus and Gata2 in our G2V mouse model. The strategy used39 eliminates expression level and protein alterations that affect HP/SC development. In G2V embryos, we showed that the cells with the most robust and complex hematopoietic potential (all HSCs and most HPCs) are Gata2 expressing. Imaging and FACS analyses of G2V embryos confirm that Gata2 is expressed in all hematopoietic sites during midgestation and that the numbers of Gata2-expressing cells reflect the developmental and temporal hematopoietic changes occurring in each site. At E9, Gata2-expressing cells are found predominantly in the YS, which at this time produces the highest numbers of the hematopoietic progenitors (EMP) in the conceptus. Slightly later as hematopoiesis begins in the AGM and FL, the numbers of Gata2-expressing cells also increase. The highest numbers of CD31+cKit+ cluster cells are found in the aorta, VA, and UA at E10.5, as quantitated by whole-mount embryo imaging.33 Most, but not all, hematopoietic cluster cells express Gata2, and Gata2 expression may be downregulated as HPCs differentiate. However, we found some hematopoietic cluster cells and HPCs in the E10 Gata2−/− vasculature, confirming the existence of Gata2-independent HPCs.5
Importantly, Gata2 is expressed in the endothelial cells of the DA. Already at E8.5, endothelial cells lining the paired dorsal aortae express Gata2, and it continues to be expressed in the E10.5 aorta when HSCs are generated, thus highlighting an involvement of Gata2 in the hemogenic program of endothelial cells. Data in VE-cadherin conditional Gata2-deficient mice and other models7,19,28,40 strongly support the notion that Gata2 is required in hemogenic endothelium for the emergence of HSCs, as does the morpholino knockdown of Gata2b in zebrafish.41
Gata2 and the relationship with hematopoietic function
Prospective isolation and in vivo transplantation showed that all HSCs are Gata2 expressing. In contrast, some HPCs are present in the Venus− cell fractions of G2V hematopoietic tissues and Gata2−/− hematopoietic tissues. In both cases, the HPCs are restricted in their differentiation potential to predominantly the macrophage and granulocytic lineages. Currently, the EMP population is of high interest as a novel hematopoietic cell subset providing tissue resident macrophages.37,42-44 Our FACS data revealed that EMPs are mainly in the Venus− cell population of E10 YS and AGM and increased in the Venus+ population at E11. Chemokine receptor/ligand gene sets obtained from a study on EMP/microglia transcriptome comparisons allowed us to find similarities in chemokine receptor/ligand expression between EMPs and the Venus− HPC fraction.
Despite prevalence of EMPs in the Venus− cell population in E10 YS and AGM, definitive progenitors are largely Venus+. The coexistence of these HPC subsets highlights the fact that there is more diversity in the types of progenitors generated in the embryo than was previously appreciated. In support of this are recent data from ES cell hematopoietic differentiation cultures suggesting that there are 2 different hemogenic endothelial cell subsets45 and the fact that, in vivo, the AGM, VA/UA, YS, PL, and head are all hemogenic tissues.36,46-49
The highest number of Venus− HPCs was found in E9 and E10 YS (270.0 ± 69.8 and 130.0 ± 25.5 CFU-C, respectively) compared with other tissues (PL, AGM, VA+UA). It is clear that Gata2 has an important role in EHT in the hemogenic endothelial cell compartment before or during the generation/emergence of hematopoietic cells, as evidenced by the decrease (but not absence) in the hematopoietic cluster cells in Gata2−/− aorta, VA, and UA. However, it is as yet unclear at what frequency EHT occurs in the YS, thus raising the possibility that Gata2-independent HPCs arise differently than Gata2-dependent HPCs (perhaps directly from hemangioblasts50).
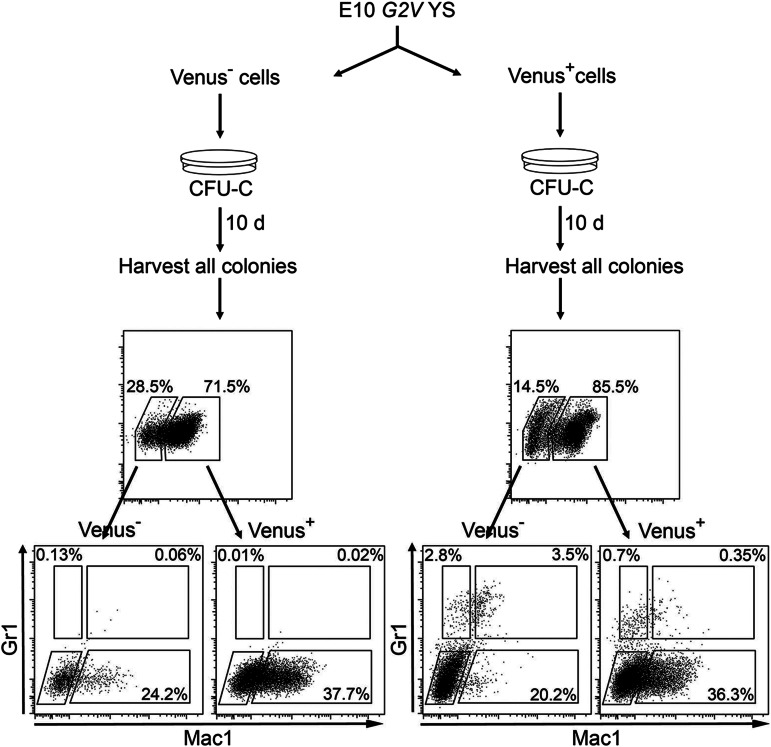
We found differences in the number of CFUs from E9 YS Venus- cells (270.0 ± 69) and Gata2−/− cells (64.4 ± 12.2; Figures 3D and 4C; Tables 2 and 3). The fourfold lower CFU number is likely related to observations (ours and others) that colonies from Gata2−/− embryos, YS explants, and ES cell differentiations were smaller/less proliferative than WT colonies, due to the complete absence of Gata2.6,7 Venus− cells are not defective for Gata2, and the resulting colonies are normal in size. Whereas at the time of sorting they did not express Gata2, Gata2 expression could initiate after seeding Venus− HPCs in methylcellulose, and cells thus undergo normal proliferation/differentiation. To test whether Venus− HPCs can convert to Venus+ cells, we analyzed Venus expression in colonies derived from sorted YS fractions after 10 days of differentiation (Figure 7). Venus expression was found in colonies derived from both fractions, indicating that a portion of Venus− cells start to express Gata2 during formation of a hematopoietic colony. Interestingly, colonies derived from Venus+ cells showed a Gr1+ and Mac1+ phenotype, whereas Venus−-derived colony cells were only Mac1+. This demonstrates that Gata2 is not necessary for a subset of HPCs and that Gata2 promotes more complex hematopoietic function in other progenitor subsets.
Figure 7.
Gata2 is expressed by Venus− cells after culture. Schematic diagram showing method and FACS analysis by which Gata2 expression was found in the progeny of sorted Venus− HPCs. G2V YS tissue was FACS sorted into Venus− and Venus+ fractions. Cells were subsequently seeded in methylcellulose, and colonies were analyzed after 10 days of culture. Colonies were harvested from the dish, cells were washed and stained (with anti-Gr1 and anti-Mac1 antibodies), and Venus, Gr1, and Mac1 expression was analyzed by FACS. FACS plots (top) indicate Venus expression in cells harvested from Venus− (left) and Venus+ (right) CFU-C experiments. Note that both FACS analyses indicate Venus expression in both cultures. FACS plots (bottom) show Gr1 and Mac1 expression in Venus− and Venus+ populations in both cultures and that cells harvested from the Venus+ culture show a more immature phenotype.
Gata3/Gata4 redundancy in Gata2-independent progenitors
The expression of Gata3 and Gata4 in Venus− AGM HPCs and aortic endothelial cells is intriguing and highlights the potential redundancy of Gata transcription factors in hematopoietic cell generation. Gata2 and Gata3 can partially rescue the erythroid phenotype in Gata1-deficient mice,51,52 and although recently it was suggested that Gata3 is redundant in HSCs,53 others clearly show that it regulates HSC cell cycle entry54 and self-renewal.55 The fact that Gata3 and Gata4 are expressed in Gata2-nonexpressing enriched HPCs suggests that they may function in this early progenitor subset. Gata3-deficient embryos show decreased numbers of FL HP/SCs.56 Gata3 affects HSC development non-cell autonomously by activating the expression of Th (tyrosine hydroxylase) and hence, catecholamine production in the ventro-lateral cells of the sympathetic nervous system underlying the embryonic aorta.38 HSC production was rescued when catecholamines were administered to the pregnant dams. These investigators also found that some aortic endothelial cells express a Gata3LacZ reporter, leaving open the possibility of a direct and overlapping role for Gata3 in some HPCs.
Much less is known concerning Gata4 in hematopoietic development. In zebrafish, there is a close relationship between anterior hemangioblasts and cardiac precursors.57 Together with Gata5 and Gata6, Gata4 specifies these 2 anterior mesoderm derivatives. In mice, Gata4 is a key component of the cardiac developmental program, with close associations between cardiac, vascular, and hematopoietic lineages.58-60 Moreover, a subset of mouse endocardial and YS endothelial cells express cardiac markers, possess hemogenic potential, and give rise to transient definitive erythroid/myeloid progenitors.61 Our results suggest the Gata4 aortic endothelial cells and Venus− Gata4-expressing HPCs may be derivatives of mesodermal cells with a genetic program that retains cardiac-vascular-hematopoietic potential and can produce HPCs. Further examination of double reporter and deficient mice should reveal the overlapping and/or redundant roles of these Gata factors.
Gata2 as a pivotal regulator of complex hematopoietic function
RNA sequence comparisons of the functionally distinct Venus+ and Venus− HPC subsets revealed a strong upregulation of Kras and Ras pathway genes in Venus+ HPCs. This pathway is particularly important in cell differentiation, acting as a molecular switch to relay extracellular growth signals.62 Kras mutations confer a competitive-repopulating advantage to BM HSCs in transplantations and initiate leukemia in mice.63 In humans, Kras mutations (together with other cooperating gene mutations) are prevalent in patients with various forms of myelomonocytic and myeloid leukemia.64 Interactions between oncogenic Ras and Gata2 have been proposed.65 The normal function of Kras has not yet been explored fully. However, conditional deletion of Kras by Vav-Cre or Mx1-Cre does not affect HSCs or the adult hematopoietic system.66 However, chimeric mice, produced by Kras−/− ES cell blastocyst injection, show no contribution of Kras−/− cells to the hematopoietic system, suggesting that Kras may be important during the embryonic development of the hematopoietic system but not after its generation.
The low expression of Notch1, Notch4, and coactivators in the Venus− compared with the Venus+ HPC fraction supports the fact that early hematopoietic cells are generated independent of this signaling pathway or implies that these are differentiated cells that have turned off Notch signaling.67,68 Others have shown that Notch1 deletion impairs the development of HSCs and angiogenesis,69 but not YS primitive or definitive hematopoiesis. Moreover, Gata2 expression in the aortic endothelium is lost when Jagged1 (ligand) is deleted.70 Our data demonstrate a direct relationship for Notch and Gata2 expression, strongly supporting a pivotal role for this pathway in the generation of functionally complex hematopoietic cells. In the absence of Notch signaling, less complex HPCs emerge in the AGM or are immigrants from the YS.18,69,70 In addition, our observed upregulated expression of some CREB genes in Venus+ HPCs supports the involvement of these regulators in definitive hematopoietic cell generation.71 Our Gata2Venus model, in combination with recently reported Gata2 distal enhancer-Evi1 mouse model, will allow for a direct examination of the cells relevant to leukemogenesis.12
In conclusion, we enriched, localized, and characterized Gata2-dependent and -independent subsets of hematopoietic progenitors in Gata2Venus embryos. The combination of this reporter with other reporter and knockout models will lead to a better understanding of the role of Gata2 (and other factors) in the development and function of multipotential HP/SCs in health, leukemogenesis, and reprogramming.
Acknowledgments
The authors thank Prof Jim Palis for critical comments on this manuscript; Dr Dorota Kurek for providing anti-Gata3 and Gata4 antibodies; Drs Dorota Kurek and Mihaela Crisan for immunostaining support; Dr Siska Driegen for ES cell culture support; and Dr Derk ten Berge for providing Wnt for ES cell cultures.
This work was supported by ZonMw (Dutch Medical Research Council Grant 911-09-036), FES Netherlands Institute for Regenerative Medicine (101675), the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (RO37 DK54077), ZonMw TOP (91211068), European Research Council Advanced Grant (341096), and Landsteiner Foundation for Blood Transfusion Research (1344).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.K., E.d.P., C.E., M.-L.K., M.J., C.S.V., and T.Y. performed research; P.S.K. analyzed RNAseq data; R.v.d.L. performed/analyzed flow cytometric data; D.M. provided reagents; P.K., E.d.P., C.E., D.M., and E.D. designed experiments and analyzed and interpreted the data; and P.K., E.D., E.d.P., and C.E. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elaine Dzierzak, University of Edinburgh, Centre for Inflammation Research, Queens Medical Research Institute, 47 Little France Crescent, Edinburgh EH16 4TJ, United Kingdom; e-mail: e.dzierzak@erasmusmc.nl or elaine.dzierzak@ed.ac.uk.
References
- 1.Wilson NK, Foster SD, Wang X, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7(4):532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 2.Solaimani Kartalaei P, Yamada-Inagawa T, Vink CS, et al. Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J Exp Med. 2015;212(1):93–106. doi: 10.1084/jem.20140767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swiers G, Baumann C, O’Rourke J, et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat Commun. 2013;4:2924. doi: 10.1038/ncomms3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujiwara Y, Chang AN, Williams AM, Orkin SH. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103(2):583–585. doi: 10.1182/blood-2003-08-2870. [DOI] [PubMed] [Google Scholar]
- 5.Tsai FY, Keller G, Kuo FC, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371(6494):221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 6.Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89(10):3636–3643. [PubMed] [Google Scholar]
- 7.de Pater E, Kaimakis P, Vink CS, et al. Gata2 is required for HSC generation and survival. J Exp Med. 2013;210(13):2843–2850. doi: 10.1084/jem.20130751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling KW, Ottersbach K, van Hamburg JP, et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200(7):871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodrigues NP, Janzen V, Forkert R, et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106(2):477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- 10.Hsu AP, Johnson KD, Falcone EL, et al. GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood. 2013;121(19):S3830-S3837. [DOI] [PMC free article] [PubMed]
- 11.Gröschel S, Sanders MA, Hoogenboezem R, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157(2):369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Yamazaki H, Suzuki M, Otsuki A, et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell. 2014;25(4):415–427. doi: 10.1016/j.ccr.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyworth C, Gale K, Dexter M, May G, Enver T. A GATA-2/estrogen receptor chimera functions as a ligand-dependent negative regulator of self-renewal. Genes Dev. 1999;13(14):1847–1860. doi: 10.1101/gad.13.14.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persons DA, Allay JA, Allay ER, et al. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood. 1999;93(2):488–499. [PubMed] [Google Scholar]
- 15.Nandakumar SK, Johnson K, Throm SL, Pestina TI, Neale G, Persons DA. Low-level GATA2 overexpression promotes myeloid progenitor self-renewal and blocks lymphoid differentiation in mice. Exp Hematol. 2015;43(7):565-577. [DOI] [PubMed]
- 16.Minegishi N, Suzuki N, Yokomizo T, et al. Expression and domain-specific function of GATA-2 during differentiation of the hematopoietic precursor cells in midgestation mouse embryos. Blood. 2003;102(3):896–905. doi: 10.1182/blood-2002-12-3809. [DOI] [PubMed] [Google Scholar]
- 17.Ottersbach K, Dzierzak E. Analysis of the mouse placenta as a hematopoietic stem cell niche. Methods Mol Biol. 2009;538:335–346. doi: 10.1007/978-1-59745-418-6_17. [DOI] [PubMed] [Google Scholar]
- 18.Robert-Moreno A, Espinosa L, de la Pompa JL, Bigas A. RBPjkappa-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development. 2005;132(5):1117–1126. doi: 10.1242/dev.01660. [DOI] [PubMed] [Google Scholar]
- 19.Gao X, Johnson KD, Chang YI, et al. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J Exp Med. 2013;210(13):2833–2842. doi: 10.1084/jem.20130733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim KC, Hosoya T, Brandt W, et al. Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J Clin Invest. 2012;122(10):3705–3717. doi: 10.1172/JCI61619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orlic D, Anderson S, Biesecker LG, Sorrentino BP, Bodine DM. Pluripotent hematopoietic stem cells contain high levels of mRNA for c-kit, GATA-2, p45 NF-E2, and c-myb and low levels or no mRNA for c-fms and the receptors for granulocyte colony-stimulating factor and interleukins 5 and 7. Proc Natl Acad Sci USA. 1995;92(10):4601–4605. doi: 10.1073/pnas.92.10.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237(2):318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 23.Yokomizo T, Yamada-Inagawa T, Yzaguirre AD, Chen MJ, Speck NA, Dzierzak E. Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nat Protoc. 2012;7(3):421–431. doi: 10.1038/nprot.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medvinsky A, Taoudi S, Mendes S, Dzierzak E. Analysis and manipulation of hematopoietic progenitor and stem cells from murine embryonic tissues. Curr Protoc Stem Cell Biol. 2008;Chapter 2:Unit 2A.6. [DOI] [PubMed] [Google Scholar]
- 25.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha VK, Bunkenborg J, Olsen JV, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115(5):629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson KD, Hsu AP, Ryu MJ, et al. Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J Clin Invest. 2012;122(10):3692–3704. doi: 10.1172/JCI61623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dzierzak E, de Bruijn M. Isolation and analysis of hematopoietic stem cells from mouse embryos. Methods Mol Med. 2002;63:1–14. doi: 10.1385/1-59259-140-X:001. [DOI] [PubMed] [Google Scholar]
- 30.Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK. The placenta is a niche for hematopoietic stem cells. Dev Cell. 2005;8(3):365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 31.Nardelli J, Thiesson D, Fujiwara Y, Tsai FY, Orkin SH. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev Biol. 1999;210(2):305–321. doi: 10.1006/dbio.1999.9278. [DOI] [PubMed] [Google Scholar]
- 32.Pimanda JE, Ottersbach K, Knezevic K, et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci USA. 2007;104(45):17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokomizo T, Dzierzak E. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 2010;137(21):3651–3661. doi: 10.1242/dev.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Impey S, McCorkle SR, Cha-Molstad H, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119(7):1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Odom DT, Koo SH, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGrath KE, Frame JM, Fegan KH, et al. Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Reports. 2015;11(12):1892–1904. doi: 10.1016/j.celrep.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kierdorf K, Erny D, Goldmann T, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16(3):273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 38.Fitch SR, Kimber GM, Wilson NK, et al. Signaling from the sympathetic nervous system regulates hematopoietic stem cell emergence during embryogenesis. Cell Stem Cell. 2012;11(4):554–566. doi: 10.1016/j.stem.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201(2):221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khandekar M, Brandt W, Zhou Y, et al. A Gata2 intronic enhancer confers its pan-endothelia-specific regulation. Development. 2007;134(9):1703–1712. doi: 10.1242/dev.001297. [DOI] [PubMed] [Google Scholar]
- 41.Butko E, Distel M, Pouget C, et al. Gata2b is a restricted early regulator of hemogenic endothelium in the zebrafish embryo. Development. 2015;142(6):1050–1061. doi: 10.1242/dev.119180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ginhoux F, Greter M, Leboeuf M, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330(6005):841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez Perdiguero E, Klapproth K, Schulz C, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518(7540):547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238(2):274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- 45.Ditadi A, Sturgeon CM, Tober J, et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat Cell Biol. 2015;17(5):580–591. doi: 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palis J, Yoder MC. Yolk-sac hematopoiesis: the first blood cells of mouse and man. Exp Hematol. 2001;29(8):927–936. doi: 10.1016/s0301-472x(01)00669-5. [DOI] [PubMed] [Google Scholar]
- 47.Frame JM, McGrath KE, Palis J. Erythro-myeloid progenitors: “definitive” hematopoiesis in the conceptus prior to the emergence of hematopoietic stem cells. Blood Cells Mol Dis. 2013;51(4):220–225. doi: 10.1016/j.bcmd.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lux CT, Yoshimoto M, McGrath K, Conway SJ, Palis J, Yoder MC. All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood. 2008;111(7):3435–3438. doi: 10.1182/blood-2007-08-107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhodes KE, Gekas C, Wang Y, et al. The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell. 2008;2(3):252–263. doi: 10.1016/j.stem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jaffredo T, Nottingham W, Liddiard K, Bollerot K, Pouget C, de Bruijn M. From hemangioblast to hematopoietic stem cell: an endothelial connection? Exp Hematol. 2005;33(9):1029–1040. doi: 10.1016/j.exphem.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi S, Shimizu R, Suwabe N, et al. GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood. 2000;96(3):910–916. [PubMed] [Google Scholar]
- 52.Tsai FY, Browne CP, Orkin SH. Knock-in mutation of transcription factor GATA-3 into the GATA-1 locus: partial rescue of GATA-1 loss of function in erythroid cells. Dev Biol. 1998;196(2):218–227. doi: 10.1006/dbio.1997.8842. [DOI] [PubMed] [Google Scholar]
- 53.Buza-Vidas N, Duarte S, Luc S, Bouriez-Jones T, Woll PS, Jacobsen SE. GATA3 is redundant for maintenance and self-renewal of hematopoietic stem cells. Blood. 2011;118(5):1291–1293. doi: 10.1182/blood-2011-02-338046. [DOI] [PubMed] [Google Scholar]
- 54.Ku CJ, Hosoya T, Maillard I, Engel JD. GATA-3 regulates hematopoietic stem cell maintenance and cell-cycle entry. Blood. 2012;119(10):2242–2251. doi: 10.1182/blood-2011-07-366070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frelin C, Herrington R, Janmohamed S, et al. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nat Immunol. 2013;14(10):1037–1044. doi: 10.1038/ni.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pandolfi PP, Roth ME, Karis A, et al. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11(1):40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 57.Peterkin T, Gibson A, Patient R. Common genetic control of haemangioblast and cardiac development in zebrafish. Development. 2009;136(9):1465–1474. doi: 10.1242/dev.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fehling HJ, Lacaud G, Kubo A, et al. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130(17):4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- 59.Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432(7017):625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- 60.Van Handel B, Montel-Hagen A, Sasidharan R, et al. Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell. 2012;150(3):590–605. doi: 10.1016/j.cell.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakano H, Liu X, Arshi A, et al. Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat Commun. 2013;4:1564. doi: 10.1038/ncomms2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pierre S, Bats AS, Chevallier A, et al. Induction of the Ras activator Son of Sevenless 1 by environmental pollutants mediates their effects on cellular proliferation. Biochem Pharmacol. 2011;81(2):304–313. doi: 10.1016/j.bcp.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 63.Sabnis AJ, Cheung LS, Dail M, et al. Oncogenic Kras initiates leukemia in hematopoietic stem cells. PLoS Biol. 2009;7(3):e59. doi: 10.1371/journal.pbio.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang YI, You X, Kong G, et al. Loss of Dnmt3a and endogenous Kras(G12D/+) cooperate to regulate hematopoietic stem and progenitor cell functions in leukemogenesis. Leukemia. 2015;29(9):1847–1856. doi: 10.1038/leu.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katsumura KR, Yang C, Boyer ME, Li L, Bresnick EH. Molecular basis of crosstalk between oncogenic Ras and the master regulator of hematopoiesis GATA-2. EMBO Rep. 2014;15(9):938–947. doi: 10.15252/embr.201438808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Damnernsawad A, Kong G, Liu Y, Chang Y-I, et al. Kras plays an important role in generating differentiated blood cells [abstract]. Blood. 2013;122(21) Abstract 2451. [Google Scholar]
- 67.Richard C, Drevon C, Canto PY, et al. Endothelio-mesenchymal interaction controls runx1 expression and modulates the notch pathway to initiate aortic hematopoiesis. Dev Cell. 2013;24(6):600–611. doi: 10.1016/j.devcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bertrand JY, Cisson JL, Stachura DL, Traver D. Notch signaling distinguishes 2 waves of definitive hematopoiesis in the zebrafish embryo. Blood. 2010;115(14):2777–2783. doi: 10.1182/blood-2009-09-244590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumano K, Chiba S, Kunisato A, et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18(5):699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- 70.Robert-Moreno A, Guiu J, Ruiz-Herguido C, et al. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 2008;27(13):1886–1895. doi: 10.1038/emboj.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim PG, Nakano H, Das PP, et al. Flow-induced protein kinase A-CREB pathway acts via BMP signaling to promote HSC emergence. J Exp Med. 2015;212(5):633–648. doi: 10.1084/jem.20141514. [DOI] [PMC free article] [PubMed] [Google Scholar]