Abstract
Self-association of β-amyloid (Aβ) into oligomers and fibrils is associated with Alzheimer’s disease (AD), motivating the search for compounds that bind to and inhibit Aβ oligomerization and/or neurotoxicity. Peptides are an attractive class of such compounds, with potential advantages over small molecules in affinity and specificity. Self-complementation and peptide library screening are two strategies that have been employed in the search for peptides that bind to Aβ. Alternatively, one could design Aβ-binding peptides based on knowledge of complementary binding proteins. One candidate protein, transthyretin (TTR), binds Aβ, inhibits aggregation, and reduces its toxicity. Previously, strand G of TTR was identified as part of a specific Aβ binding domain, and G16, a 16-mer peptide with a sequence that spans strands G and H of TTR, was synthesized and tested. Although both TTR and G16 bound to Aβ, they differed significantly in their effect on Aβ aggregation, and G16 was less effective than TTR at protecting neurons from Aβ toxicity. G16 lacks the β-strand/loop/β-strand structure of TTR’s Aβ binding domain. To enforce proper residue alignment, we transplanted the G16 sequence onto a β-hairpin template. Two peptides with 18 and 22 amino acids were synthesized using an orthogonally protected glutamic acid derivative, and an N-to-C cyclization reaction was carried out to further restrict conformational flexibility. The cyclized 22-mer (but not the noncyclized 22-mer nor the 18-mer) strongly suppressed Aβ aggregation into fibrils, and protected neurons against Aβ toxicity. The imposition of structural constraints generated a much-improved peptidomimetic of the Aβ binding epitope on TTR.
Keywords: Beta-amyloid, transthyretin, cyclic peptide, Alzheimer’s disease, peptidomimetics, amyloid fibrils
Graphical Abstract
Alzheimer’s disease (AD) is the most common form of dementia, affecting more than 35 million people worldwide. Intracellular neurofibrillary tangles and extracellular senile plaques, primarily in the hippocampus and cerebral cortex, are two characteristic features of the disease. The main component of the extracellular plaques is β-amyloid (Aβ), a proteolytic product of amyloid precursor protein (APP). When cleaved from APP, Aβ spontaneously self-assembles into soluble oligomers that progress into insoluble fibrillar aggregates. Although the precise mechanism of AD pathogenesis is unknown, several studies have suggested that the aggregation of Aβ plays a key causal role, and that the soluble oligomers are the most toxic Aβ species.1–3
Numerous small molecules that can alter Aβ production and/or aggregation have been explored as therapeutic agents against AD, although to date none have been clinically effective.4 As an alternative approach, the use of peptides and peptidomimetics that bind to Aβ and alter Aβ aggregation was introduced, since peptides have potential advantages over small molecules in terms of better target affinity and specificity.5 In general, peptides that bind to Aβ have been either designed based on self-complementary domains, or found by screening random-peptide libraries.6–9 Alternatively, one could design a peptide by mimicking the binding epitope of a complementary binding protein. Molecular recognition involving proteins is often mediated by relatively large interfaces with secondary structural motifs, and thus has the potential for higher specificity than small molecules. Peptides mimicking the three-dimensional structure of protein binding sites have been recognized as a promising alternative to large biologics such as antibodies.10,11 This approach is particularly useful when one has precise structural information about the protein–protein interaction site.
Transthyretin (TTR), a stably folded homotetrameric transport protein that circulates in blood and cerebrospinal fluid, is neuroprotective in AD mouse models.12–14 We and others have shown that TTR binds to Aβ and inhibits its toxicity in vitro.15,16 Inhibition of toxicity is mediated at 1:10 to 1:100 TTR:Aβ molar ratio, implying that TTR is selective for toxic Aβ oligomers.17 Therefore, we speculated that a peptide mimicking the epitope of Aβ-TTR interaction would have great therapeutic potential.
Previously, we identified strand G in the inner hydrophobic pocket of TTR as a putative Aβ-binding site.17,18 We tested peptides of variable length with sequences derived from the binding domain on strand G, and we conducted alanine scanning to determine the specific residues required for binding.19 Based on these screening studies, we synthesized a 16-mer linear peptide, G16 (Table 1), corresponding to residues 102–117 of TTR, along with a scrambled-sequence peptide. G16 bound Aβ and was protective against Aβ toxicity in vitro, whereas the scrambled-sequence peptide was inactive. However, G16 required a much higher concentration than TTR to afford the same level of protection against Aβ toxicity. In addition, G16 differed substantially from TTR in its effect on Aβ aggregation.19
Table 1.
Sequences of TTR-Derived Peptides
| peptide name |
sequence | TTR residue numbers |
mol wt (expected/ measured) |
modification from native TTR sequence |
|---|---|---|---|---|
| G16 | PRRYTIAALLSPYSWS | 102–117 | 1922.2/1922.0 | Y116W, N- and C-capped |
| GF | PRRYTIAALLSPYSWSDYKDDDDK | 102–117 | 2917.2/2916.3 | Y116W, FLAG tag added to C-terminus |
| LG2 | SKVDPPTIAALLSPYSYSQ | 106–121 | 1921.2/1921.2 | T118Q, T119S, A120K, Grafted onto DPP template |
| CG2 | SKVDPPTIAALLSPYSYSQ | 106–121 | 1903.2/1904.0 | T118Q, T119S, A120K, Grafted onto DPP template, cyclized |
| LG3 | SKVVTDPPRYTIAALLSPYSYSQ | 104–123 | 2441.8/2441.2 | T118Q, T119S, A120K, Grafted onto DPP template |
| CG3 | SKVVTDPPRYTIAALLSPYSYSQ | 104–123 | 2423.8/2423.2 | T118Q, T119S, A120K, Grafted onto DPP template, cyclized |
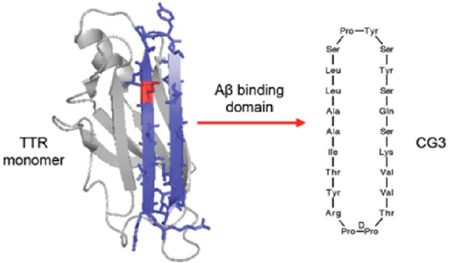
We hypothesized that the gap in efficacy between native TTR and G16 could be attributed to the fact that G16 was a sequence but not structural mimic of the Aβ binding domain on TTR. G16 spans two β-strands on TTR (strand G and part of H) plus a short connecting loop. However, G16 lacks the β-strand/loop/β-strand structure of the binding domain in TTR. Here, we report the synthesis and characterization of peptides that mimic the structural features of the Aβ -binding domain on TTR. The register of a β-hairpin determines which pairs of cross-strand amino acids participate in hydrogen bonding and this in turn defines which side chains are displayed on the same face. Various templates can be been used to mimic the alignment of antiparallel strands connected by a loop; one approach is to transplant antiparallel β-strands onto a semirigid hairpin-stabilizing template. We therefore used the G16 sequence but enforced a hairpin via DPro-LPro. We also examined whether increasing conformational rigidity via cyclization would further improve peptide performance. We tested peptides for their ability to bind Aβ, alter Aβ aggregation, and inhibit Aβ toxicity. Our work demonstrates that conformational constraint improves the anti-Aβ activity of TTR-derived peptides.
RESULTS AND DISCUSSION
Design and Synthesis of Cyclic Peptides
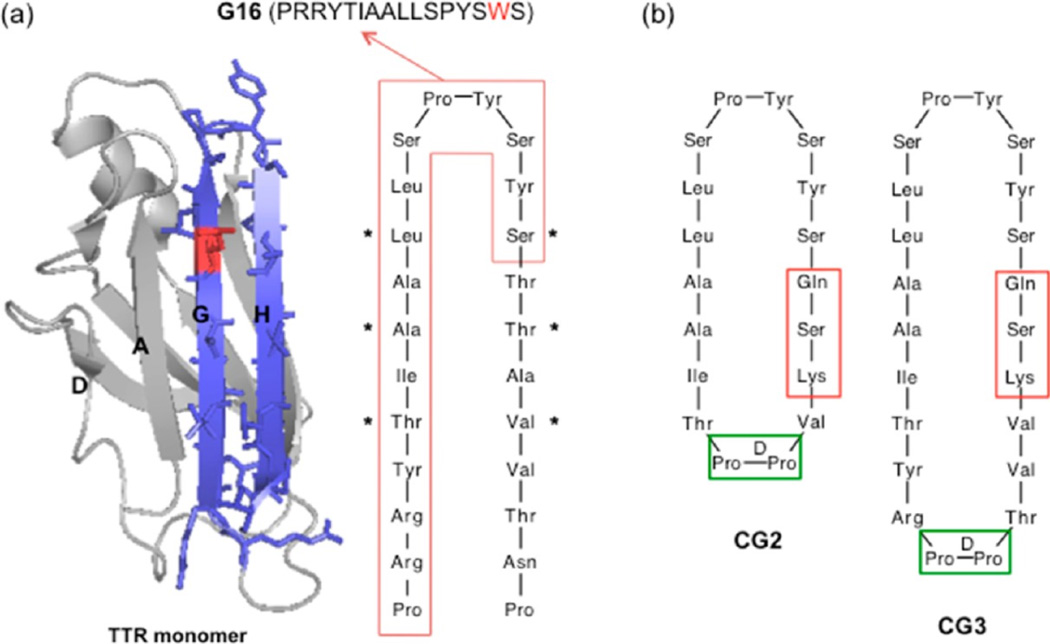
Strand G of TTR contains Leu110, a critical residue for binding of TTR to Aβ. Other residues in strand G as well as those in the adjacent antiparallel strand H and the connecting G-H loop also contribute to the Aβ binding domain. A linear peptide, G16, designed from the sequence of the binding domain (Figure 1a), showed moderate neuroprotective activity against Aβ toxicity.19
Figure 1.
Cyclic peptide design strategy (a) Ribbon structure of transthyretin (PDB entry 1DVQ) monomer, showing strands G and H in blue. Sidechains from residues 102 to 125 are shown explicitly. Leu110, a critical residue for interaction, is shown in red. Sequence of strands G and H is shown, with the boxed region corresponding to the sequence of G16. (b) Design of cyclic peptides CG2 and CG3, mimicking the structure of strand G/loop/strand H using a dipeptide DPro-LPro template (green box). Residues in red boxes are changed from the native sequence in order to facilitate cyclization.
We hypothesized that constraining the conformation of G16 would better mimic the structure of native TTR, leading to improved interaction with Aβ. Several different strategies can be employed to mimic antiparallel β-strands connected by a loop, including disulfide bridge formation, turn stabilization by strong β-hairpin promoters, or cyclization. Since it was important to preserve the residues critical for interaction with Aβ, we decided to make a cyclic structure by connecting the N-terminus of strand G and C-terminus of strand H using a β-hairpin stabilizing template, without modifying residues Thr106 to Ser117. Successful synthesis of short cyclic peptides that mimic the hairpin-like structure of a native protein have been previously reported.10,20 Each aspect of the synthesis of this cyclic binding-site mimic required careful consideration. A dipeptide containing DPro at position i+1 prefers to adopt β-turns with a right-handed twist,21,22 so dipeptides such as DPro-Gly and DPro-LPro are commonly used as β-turn inducing templates.23 When transplanting a hairpin loop structure from the native protein onto a dipeptide template, it is important to insert it at a non-hydrogen bonding position to maintain the hydrogen-bonding pattern along the antiparallel β-strands. In addition, a regular hairpin structure is only seen in cyclic peptides that contain in total (4n+2) residues, where n is an integer.24 Given these considerations, we initially decided to transplant Arg104 to Thr123 onto a DPro-Gly turn-inducing template, as mimics of the G and H strands of TTR.
On-resin cyclization was chosen to prevent the formation of peptide oligomeric byproducts. Trifunctional amino acids (such as glutamic or aspartic acid), where the amino acid can be anchored to the resin through the side chain, are incorporated into the sequence to allow on-resin cyclization. Although it is common practice to place the turn-inducing template in the middle of the linear sequence to enhance cyclization efficiency, we wanted to preserve all the residues near the loop; therefore the glutamic acid was placed adjacent to Ser117, replacing the native Thr118. It was thus anticipated that the cyclization efficiency would be lower than what has been reported by others using a template in the middle. After cleavage from the amide-resin, the glutamic acid is converted to glutamine. The end coupling reaction occurs between the C-terminal Gln and N-terminal residue, which would be Thr119 in the native sequence. To enhance the coupling efficiency, the branched residue Thr was changed to Ser. The residue corresponding to Ala120 was changed to Lys for two reasons: (1) to increase the solubility of the cyclic peptide without interfering with Aβ binding, because it is pointing toward the opposite site of the interacting domain in the native TTR structure and (2) to increase the stability of β-hairpin via interaction with Tyr105.25
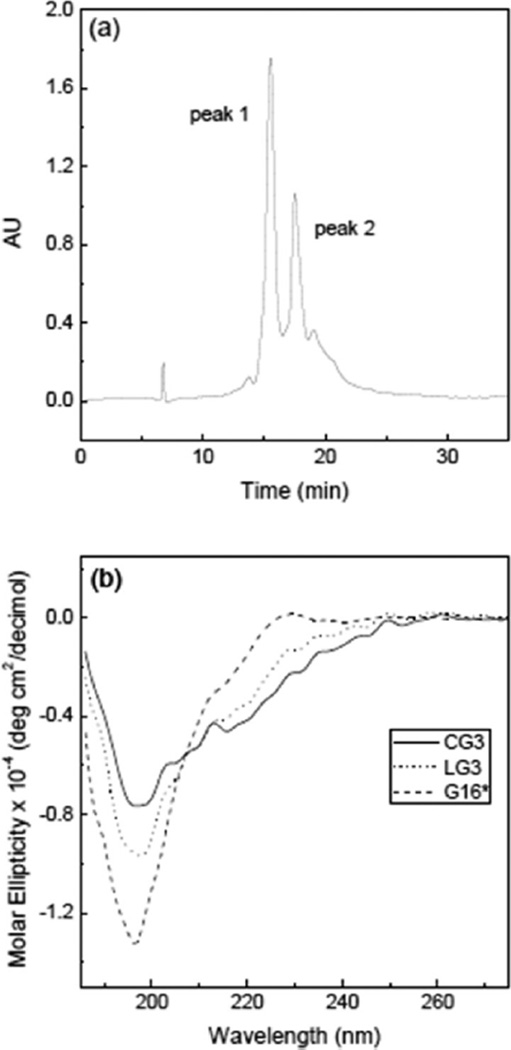
In the first attempt at producing a structural mimic, SKVVTDPGRYTIAALLSPYSYSQ (CG1) was successfully synthesized as the linear peptide by standard Fmoc-solid phase methods. The reagent PyBOP was added for the end-coupling (cyclization) reaction. Mass spectra analysis of the reaction product showed that cyclization failed, and a side product with mass 53 amu higher than the starting peptide was produced instead. Formation of this side product was traced to the presence of small amounts of pyrrolidine contaminant in PyBOP, and to the difficulty of the desired coupling.26 Therefore, we designed a second cyclic peptide (CG2) where the DPro-Gly template was changed to the more rigid DPro-LPro and the length was shortened to 18 amino acids (Thr106 to Val121) to preorganize reactive ends in closer proximity (Figure 1b). The diproline peptide has been reported to strongly prefer a type II′ β-turn backbone and to stabilize a second opposite turn.27 Freshly crystallized PyBOP, confirmed free of pyrrolidine by mass spectrometry, was used. After 18 h reaction with PyBOP, we obtained a mixture of cyclized (CG2) and noncyclized (LG2) peptides confirmed by mass spectrometry (not shown). With the success of CG2 synthesis, we attempted a longer peptide, and successfully synthesized a 22-residue peptide containing the DPro-LPro template, and performed on-resin cyclization (Figure 1b). The mixture of CG3 and LG3 was separated by RP-HPLC (Figure 2a), and the identity of the peptides in each peak was confirmed by mass spectrometry. From the HPLC peak area, we estimated the ratio of cyclized to noncyclized peptide at ~40:60. Table 1 lists the sequences of TTR-derived peptides used in this study.
Figure 2.
Purification and characterization of β-hairpin peptides LG3 and CG3. (a) RP-HPLC analysis of crude peptide after on-resin cyclization and cleavage. Mass spectrometry analysis of the purified fractions confirmed the identity of peak 1 (LG3) and peak 2 (CG3). (b) CD spectra of G16, LG3, and CG3. *For this experiment, G16 was not capped.
Characterization of Peptides
The effect of the hairpin template and cyclization on secondary structure was confirmed by circular dichroism (Figure 2b). CD spectra for G16 was consistent with a disordered peptide, as expected. The reduction in the absolute molar ellipticity at ~197 nm as well as the appearance of negative ellipticity at 215–220 nm for LG3 may indicate a decrease in disorder and adoption of a β-turn conformation attributable to templating. N-to-C cyclization led to a further dampening of the negative band at 197 nm and greater (negative) ellipticity at 215 to 240 nm. These changes in CD spectra are very similar to those reported by others upon increase of β-sheet/β-turn structure.28
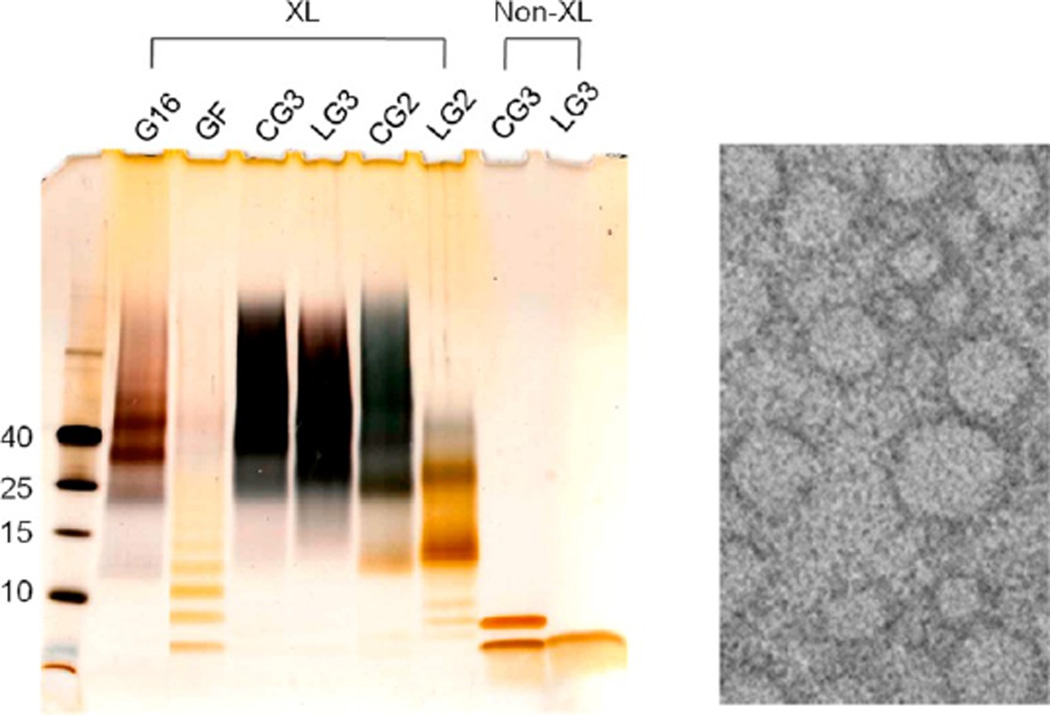
Peptide self-assembly was probed by photoinduced cross-linking (PICUP) followed by gel electrophoresis. We observed discrete oligomers ranging from ~24 to 45 kDa (apparent) for G16. CG2 and LG2 contained bands ~13 kDa and ~25 kDa, but CG2 also contained larger cross-linked oligomers, demonstrating that cyclization strongly promoted self-association for this peptide. Both LG3 and CG3 contained cross-linked species covering a broad molecular weight range starting from 25 kDa (Figure 3a), indicative of the strong tendency of these peptides to self-associate in aqueous buffers. We also synthesized a linear peptide (TTR residues 102–117) with a C-terminal FLAG tag (GF). Cross-linking of GF produced a ladder of lower-molecular-weight multimers but none of the larger multimers observed for G16, CG2 or CG3 (Figure 3a), demonstrating that addition of a highly charged tail reduces self-assembly. (A ladder of multimers in which the concentration declines exponentially with size can be formed due to diffusional collisions occurring during exposure rather than the existence of stable pre-existing oligomers29). TEM images reveal some micelle-like aggregates in solutions of CG3 (Figure 3b), similar in appearance to what we previously observed in G16.19 In the absence of cross-linker, for LG3, a clean single band was observed at ~5 kDa. Although the apparent molecular weight based on the standard is larger than the actual molecular weight of the LG3, we believe that the band is the peptide monomer.30 Two bands were observed for CG3 in the absence of cross-linker, suggesting that a fraction of CG3 forms a SDS-stable dimer, or possibly that there are two conformers that migrate differently.
Figure 3.
(a) Gel electrophoresis analysis of photoinduced cross-linked (XL) and un-cross-linked (Non-XL) peptides. Peptide concentration was 36 µM, and peptides were visualized by silver staining. Non-cross-linked CG3 and LG3 were boiled and separated on the right two lanes for comparison. (b) TEM image of CG3, illustrating micelle-like aggregates of ~10–40 nm diameter. Vertical height of the image =200 nm.
To further probe for self-association, G16, LG3, and CG3 were incubated at 80 µM at 37 °C and analyzed by light scattering. After 1h of incubation, LG3 did not scatter light above background, while both G16 and CG3 scattered light above background, with about 5 times higher intensity for CG3 (data not shown). Since scattering is attributed to larger particle formation, these results indicate that CG3 is the most susceptible to self-association, followed by G16 and then LG3.
Solution-Phase Binding of TTR-Derived Peptides to Aβ
Two methods were used to probe for solution-phase interaction between Aβ and the cyclic peptides CG2 and CG3 and noncyclized peptides LG2 and LG3: photoinduced cross-linking (PICUP) and proteolytic fragmentation. For comparison G16 and GF were also tested.
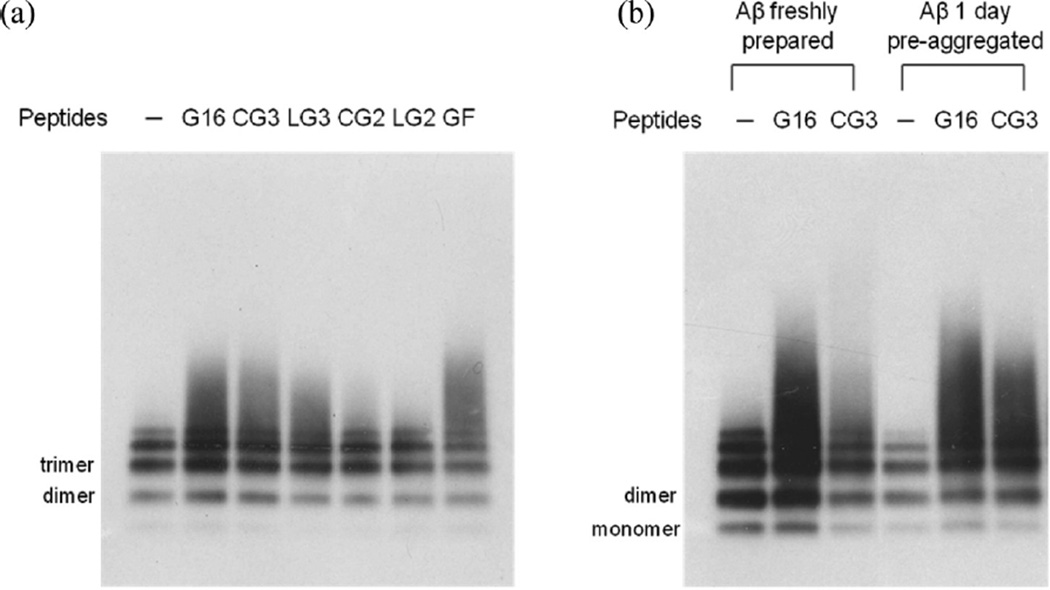
Preaggregated Aβ (30 µM Aβ incubated for 24 h at 37 °C) was mixed with peptides at 10:1 molar excess of Aβ, cross-linked, separated by gel electrophoresis, and detected by anti-Aβ antibody 4G8 (Figure 4a). Aβ incubated for 1 day under these conditions spontaneously aggregates into a heterogeneous mixture of small multimers and soluble oligomers. We observed a ladder of monomer, dimer, trimer, tetramer and pentamer for Aβ alone, consistent with previous reports.19,31 When Aβ was mixed with G16, a smear of oligomers larger than Aβ pentamers was observed, as previously reported.19 Similarly, Aβ with CG3, LG3 or GF produced a broad band of larger oligomers. In contrast, neither CG2 nor LG2 caused any shift in Aβ size distribution. We interpret these data to indicate that G16, CG3, LG3 and GF all interact with Aβ, while CG2 and LG2 do not show signs of interaction with Aβ at these conditions. Since CG2 self-associates but does not bind Aβ, whereas GF binds Aβ but is much less prone to self-association, we conclude that self-association of the peptide is neither necessary nor sufficient for binding to Aβ.
Figure 4.
Evidence of interaction between Aβ and peptides investigated by photoinduced cross-linking (PICUP). (a) Aβ was incubated without (−) or with peptides G16, CG3, LG3, CG2, LG2 and GF where Aβ concentration is in 10-fold excess. Samples were cross-linked prior to application to gels. Aβ was detected via Western blotting with 4G8. (b) Aβ at different aggregation state (freshly prepared or preaggregated) was incubated without (−) or with peptides G16 and CG3 prior to cross-linking and electrophoresis. Aβ was detected via Western blotting with 4G8.
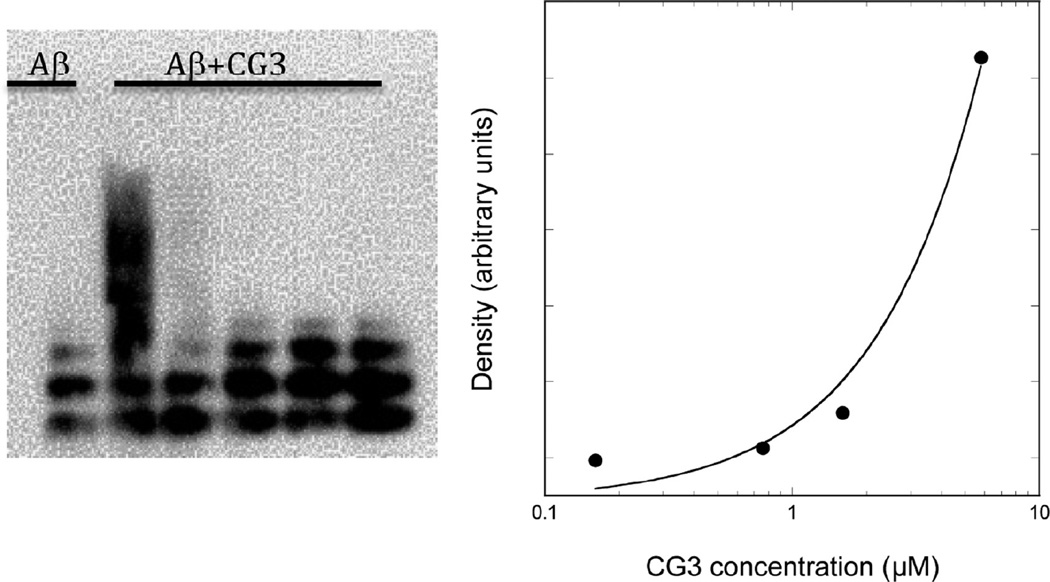
The cross-linking experiment was repeated but with freshly prepared as well as preaggregated Aβ (Figure 4b). With G16 added, a shift toward higher molecular weight oligomers was observed for either fresh or preaggregated Aβ. In contrast, CG3 interacted much less with freshly prepared Aβ than with preaggregated Aβ. This result suggests that CG3 is selective for preaggregated over freshly prepared Aβ, whereas G16 showed no such selectivity. We hypothesize that the conformational constraint imposed by templating and cyclization results in a peptide that is less adaptable and more rigid in its geometric requirements for binding, and therefore more selective. We previously reported that TTR preferentially interacts with oligomers of Aβ rather than monomers;17 these data suggest that CG3, by being a better structural mimic of TTR, retains the higher selectivity for Aβ oligomers of the native folded protein. Finally, we examined the effect of CG3 concentration on binding. Briefly, CG3 at a final concentration of 0.16, 0.8, 1.6, or 5.8 µM was incubated with 24 µM preaggregated Aβ, cross-linked, and analyzed by gel electrophoresis. The density of the broad oligomer band increased 9-fold as the CG3 concentration increased from 1.6 to 5.8 µM (Figure 5). No oligomer band was detected at 0.8 or 0.16 µM. These results indicate that, within the sensitivity of this assay, the apparent binding affinity of CG3 to Aβ is in the ~1 µM range. This is an apparent affinity only, as it does not consider the reduction in molar concentration due to oligomerization, or the heterogeneity and multivalency of the CG3- Aβ interaction.
Figure 5.
Concentration dependence of the binding interaction between Aβ and CG3 investigated by photoinduced cross-linking (PICUP). Preaggregated Aβ (24 µM) was incubated without or with CG3 at varying concentrations. Samples were cross-linked prior to application to gels. Aβ was detected via Western blotting with 4G8. Density of the broad oligomer band at each concentration was measured using ImageJ and density of background was subtracted.
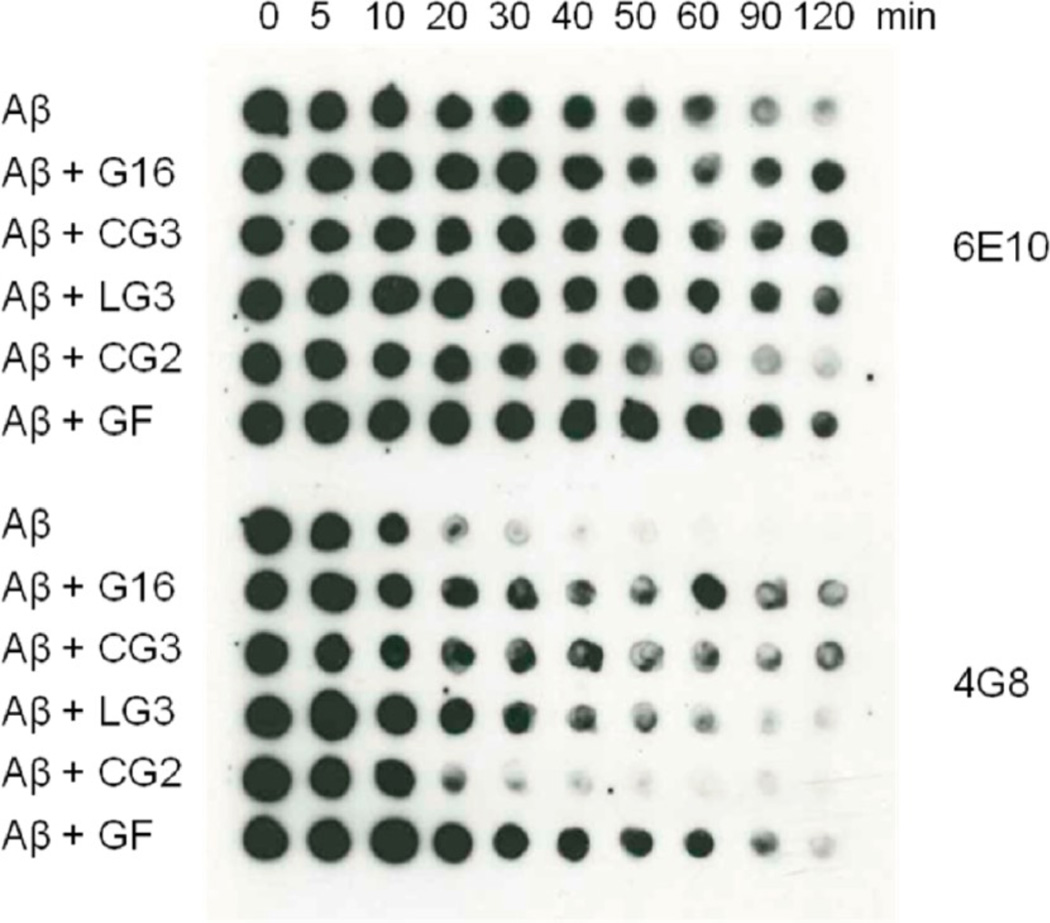
Evidence for interaction between Aβ and peptides was also sought using a proteolytic fragmentation assay. Peptides were mixed with Aβ at 3:1 molar excess of peptides, and then protease was added. Aliquots were removed at defined time points and degradation of Aβ was assessed by probing with anti-Aβ antibody. We anticipated that binding of peptides to Aβ would inhibit access of proteases and delay degradation. Two monoclonal antibodies were used, which bind to either N-terminal (6E10) or central (4G8) domains of Aβ. As shown in Figure 6, Aβ degradation was detected within 30 min by 4G8. When incubated with G16, CG3, LG3 or GF, Aβ degradation was delayed. G16 and CG3 were most effective, followed by GF and LG3. CG2 did not have any effect on the time course of degradation. The N-terminal domain (6E10) was protected from protease for longer times overall, but the same effects of peptides were shown after 120 min. These results are consistent with the cross-linking studies (Figure 4a), indicating that G16, CG3, LG3, and GF interact with Aβ while CG2 does not. Why does CG2 not interact with Aβ while CG3 does? The critical sequence, TIAALLSPYSYS,19 is present in both cases. One possibility is that the β-hairpin turn in CG2 interferes sterically with binding of Aβ to T106. Or, shortening the β-strands compared to the native sequence may impose a non-native twist.32
Figure 6.
Evidence of interaction between Aβ and peptides investigated by proteolytic fragmentation assay. Aβ was incubated with or without G16, CG3, LG3, CG2 and GF for 24 h where peptide is in 3-fold excess. The relative rate of proteolytic fragmentation of Aβ was measured by addition of Proteinase K followed by dotting onto nitrocellulose membrane at different time points. Unfragmented species were detected by 6E10 and 4G8 antibodies.
Effect of Peptides on Aβ Aggregation Kinetics
We next evaluated whether any of the peptides influenced Aβ aggregation. Several orthogonal methods were used: dynamic light scattering (DLS) to measure hydrodynamic size of soluble aggregates, sedimentation/filtration to measure mass fraction of large dense particles, thioflavin T (ThT) fluorescence to measure relative mass of specifically fibrillar aggregates, and transmission electron microscopy (TEM) to visualize morphology, size, and relative number of aggregates.
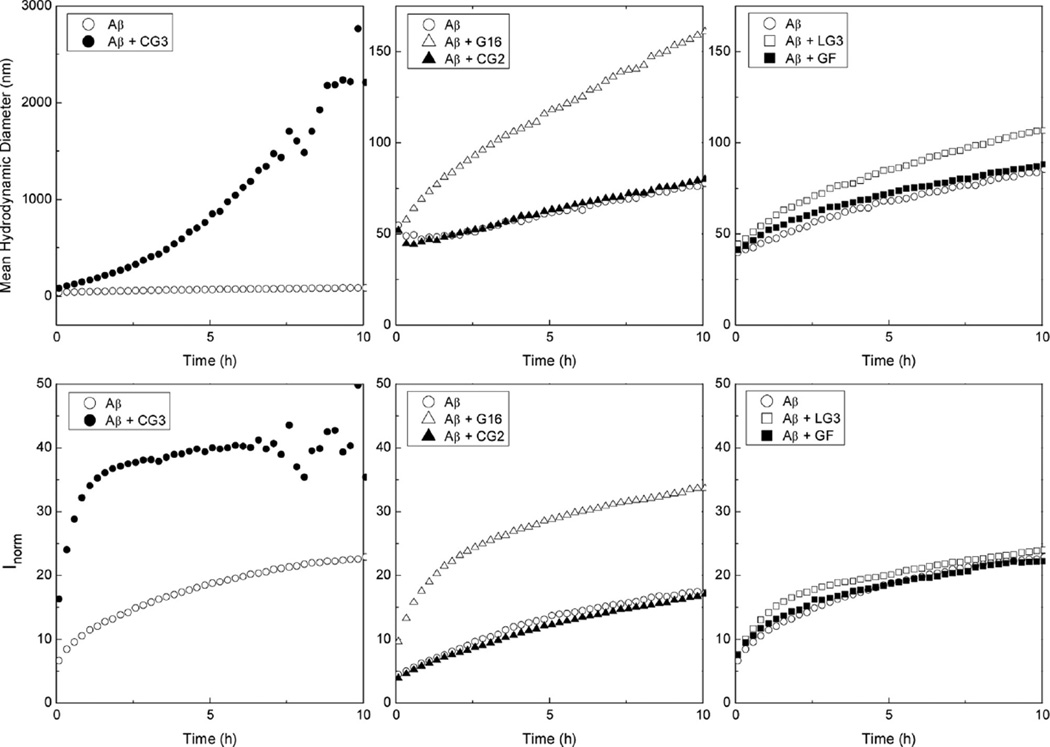
For light scattering analysis, Aβ (140 µM) was prepared either alone or with added peptides, at a 10:1 molar excess of Aβ. Aβ alone aggregated steadily over the course of 10 h, with both the mean aggregate size and total scattered intensity increasing (Figure 7). At the low peptide concentration (14 µM) used for this test, there was no detectable scattering above background from the peptides in the absence of Aβ (data not shown). Both G16 and CG3 increased the size of aggregates of Aβ, but the effect of CG3 was much more pronounced. For example, after 5 h, the mean hydrodynamic size was ~1000 nm for Aβ-CG3 mixture, while that of Aβ-G16 mixture was about 120 nm. The total intensity of scattered light was increased with addition of either G16 or CG3 by about 2-fold. To confirm the presence of large aggregates, we examined the solutions using nanoparticle tracking (NTA). This technique detects scattering from individual particles as they diffuse in the optical chamber. Solutions were prepared at 28 µM Aβ and at 10:1 Aβ:peptide molar ratio, and observed 5 or 24 h after preparation. Large particles (~micron) were detected with both Aβ-CG3 mixtures and Aβ-G16 mixtures. Aβ alone had much smaller (typically ~200–300 nm particles, data not shown).The large size of the particles precluded quantitative analysis of NTA data.
Figure 7.
Effect of peptides on Aβ aggregate growth kinetics measured by light scattering. The mean hydrodynamic diameter (top) and normalized scattering intensity (bottom) were measured for samples containing 140 µM Aβ without or with 14 µM CG3, LG3, CG2, G16 or GF. At the conditions of these experiments, concentrations of the peptides (in the absence of Aβ) were too low to contribute to the scattering signal.
Consistent with the lack of binding interaction in PICUP and protease degradation assays, CG2 did not affect Aβ aggregation as measured by either hydrodynamic diameter or total scattered intensity (Figure 7). Although LG3 and GF showed a binding interaction with Aβ in the PICUP and protease degradation assays, they had little or no effect on Aβ aggregation (Figure 7). The distinction between binding and effect on aggregation might be attributable to the different self-assembly characteristics of the peptides. Both G16 and CG3 readily self-assemble, as shown by PICUP (Figure 3) and light scattering. Although cross-linking of LG3 produced oligomers similar to CG3 (Figure 3), LG3 did not scatter light above background at the condition (80 µM at 37 °C) where scattered light from CG3 and G16 was detectable, as previously noted. GF, presumably due to its tail of charged residues, self-assembles much less than the other peptides (Figure 3). We hypothesize that preorganization of G16 and CG3 into larger oligomers provides a mechanism for their interaction with multiple Aβ oligomers, leading to formation of large clusters of Aβ-peptide aggregates. Weaker binding affinity for GF and LG3 compared to G16 and CG3 (Figure 6, bottom) may also contribute to the reduced influence of GF and LG3 on Aβ aggregation.
As a different measure of aggregation, we prepared Aβ alone (30 µM) or with CG3 (10:1 Aβ excess), incubated them together for 0 or 24 h at 37 °C, then filtered through 0.45 µm filter and measured concentrations using the BCA assay. With Aβ alone, 85% of the material was filterable initially, but only 31% after 24 h. In contrast, about 90% of the CG3-Aβ mixture was filterable at either 0 or 24 h. Similar observations were made by sedimentation. Briefly, samples were centrifuged for 30 min and the concentration of the supernatant (top 80%) was measured. After 24 h, only 15% of Aβ remained in the supernatant while 91% of the Aβ-CG3 mixture remained in solution. As previously reported, G16 also significantly retained Aβ in the supernatant.19 Taken together with the DLS and NTA data, these results suggest one of two possibilities: that in the presence of CG3 (or G16), only a small fraction of Aβ is incorporated into very large aggregates while most of it remains unaggregated, or that the Aβ aggregates are too loosely associated and/or too highly hydrated to be removed by centrifugation or filtration.
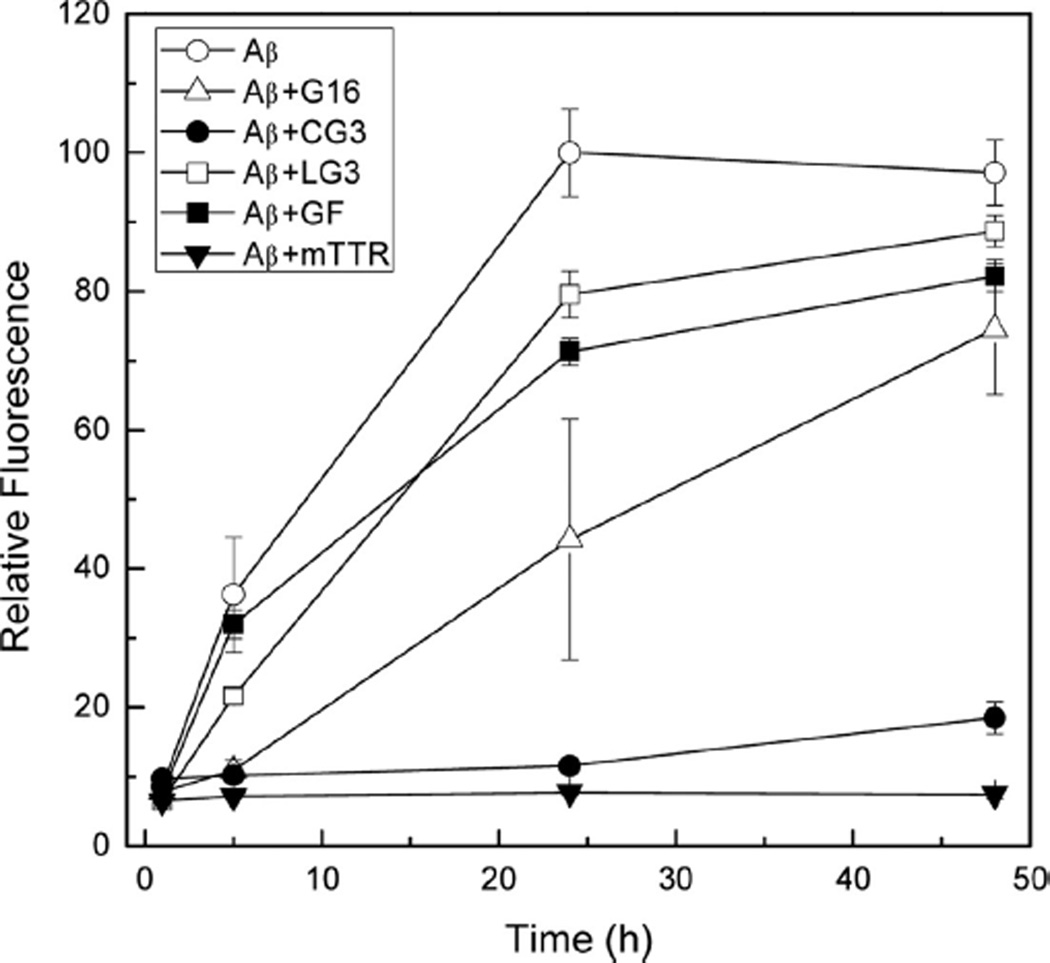
We used ThT fluorescence intensity as another measure of the effect of peptides on Aβ aggregation. ThT binds to fibrillar aggregates with a β-sheet structure but does not bind to amorphous aggregates; binding causes a sharp increase in fluorescence intensity that is presumed proportional to the mass of fibrillar aggregates.33 Samples were prepared at 10:1 Aβ molar excess. After 1 h, fluorescence intensity of all samples was low, as expected given the well-known lag time in appearance of Aβ fibrils (Figure 8). After 5 h, Aβ alone showed the highest ThT fluorescence intensity while that of Aβ with G16 or with CG3 remained low. A difference between G16 and CG3 appeared after 24 h: while ThT-positive species were detected in the Aβ-G16 mixture, the fluorescence still remained low for Aβ-CG3. LG3 and GF had at most a minor inhibitory effect on ThT fluorescence increase (Figure 8), consistent with their minor effect on Aβ aggregation as measured by light scattering (Figure 7). For comparison, ThT fluorescence of Aβ mixed with mTTR was measured. mTTR is a mutant TTR protein that is stable as a monomer and is even more effective than wild-type TTR at inhibiting Aβ aggregation.34 There was no increase in fluorescence observed for Aβ-mTTR even after 48 h, and it remained the sample with the lowest ThT intensity (Figure 8).
Figure 8.
Effect of peptides on Aβ aggregation measured by ThT fluorescence intensity. Samples containing 28 µM Aβ without or with 2.8 µM of the indicated peptide or protein were incubated at 37 °C for up to 48 h. After 1, 5, 24, and 48 h of incubation, samples were diluted 14-fold into a ThT-containing solution, and fluorescence emission intensity was measured immediately. Three replicates were averaged for each sample and the data for Aβ alone, Aβ+G16 and Aβ+CG3 represent the average of two independent experiments.
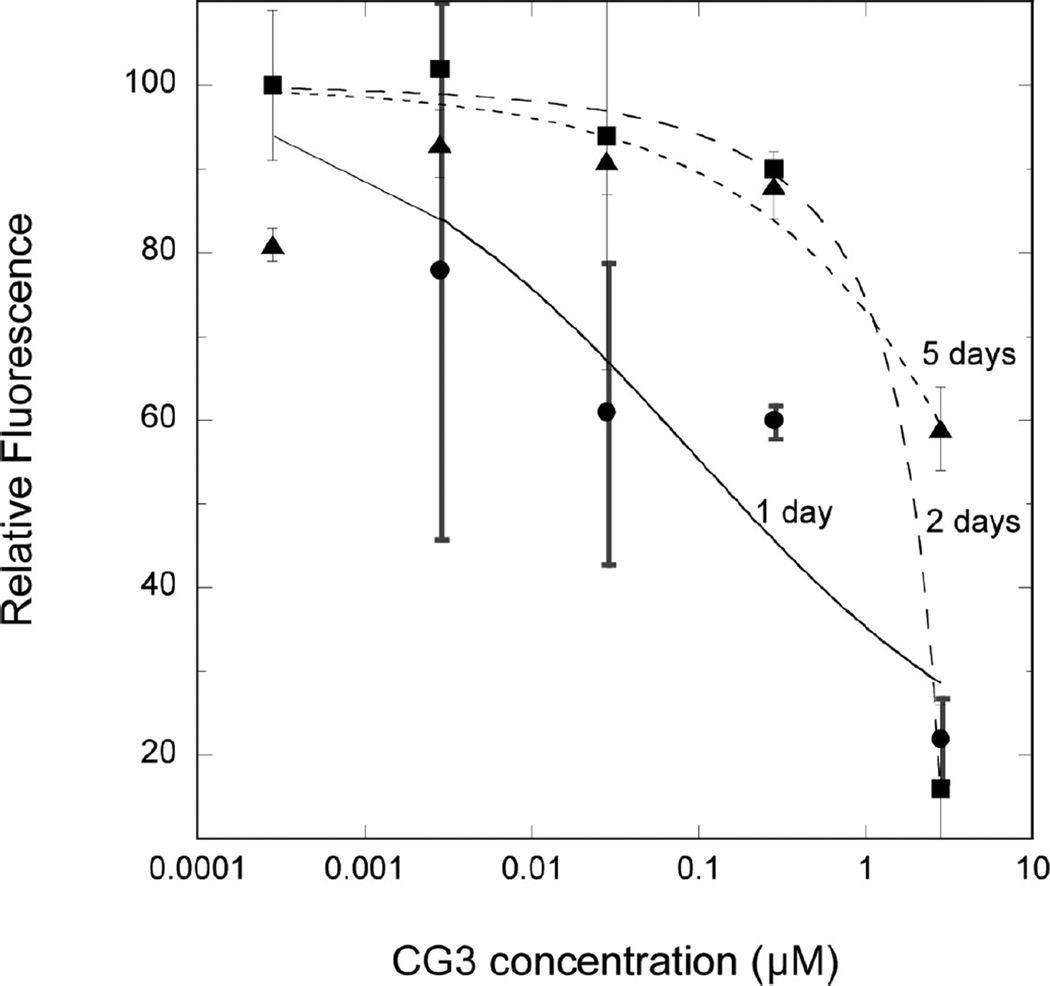
We next measured the effect of CG3 concentration and of coaggregation time on ThT intensity (Figure 9). After one-day coincubation of CG3 with Aβ, we observed an increase in ThT intensity with decreasing CG3 concentration, demonstrating a dose-dependent decrease in the efficacy of CG3 at inhibiting Aβ fibrillogenesis. The concentration dependence was fairly weak, as we observed partial inhibitory activity even at ~10 nM CG3. The relatively weak concentration dependence likely reflects the heterogeneous multivalency of self-assembly as well as coassembly of both Aβ and CG3. With 2 days coincubation, inhibition of Aβ fibrillogenesis was observed only at the highest concentration tested (2.8 µM). Longer (5 day) coincubation led to a partial loss of inhibitory activity even at the highest concentration. The time-dependent loss of CG3 activity could be due to several causes, including: (a) continued aggregation of Aβ eventually overwhelms CG3 because Aβ is present at 10-fold higher concentration, (b) the mechanism of action of CG3 is such that it delays Aβ aggregation but does not totally prevent it–in other words it acts kinetically rather than thermodynamically, or (c) CG3 undergoes some chemical or physical degradation with longer incubation at 37 °C.
Figure 9.
Effect of CG3 concentration and coincubation time on Aβ aggregation, measured by ThT fluorescence intensity. Samples containing 28 µM Aβ with the indicated concentration of CG3 were incubated at 37 °C for 1, 2, or 5 days. Samples were diluted 14-fold into a ThT-containing solution, fluorescence emission intensity was measured immediately and compared to a similarly prepared sample containing Aβ alone. Two to three measurements were averaged for each sample, and the data are the mean ± SD of two independent experiments. The curves are the nonlinear regression fit to the logistic equation but do not indicate any mechanistic interpretation.
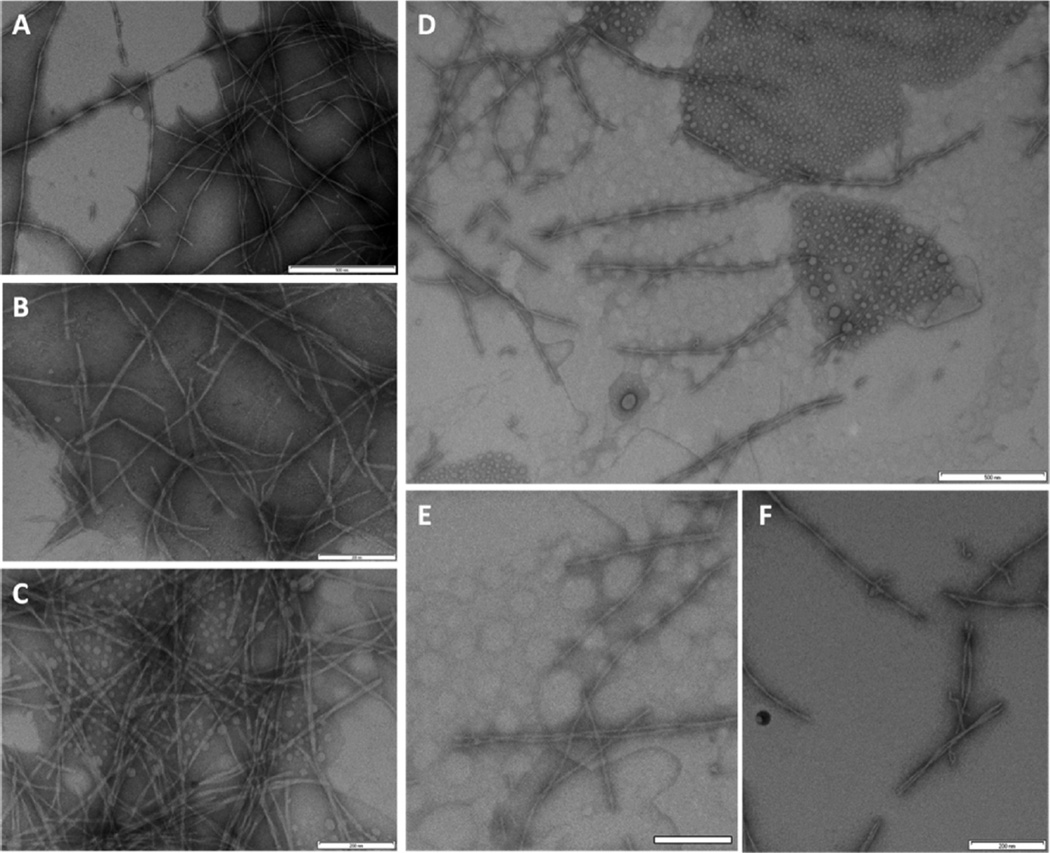
Finally, we examined samples of Aβ alone or with G16 or CG3 by TEM (Figure 10). As expected, Aβ alone aggregated into well-defined fibrils of characteristic diameter and length (Figure 10a and b). With G16, there were still a large number of fibrils, many of which appeared clumped, along with numerous spherical globules (Figure 10c), as has been previously reported.19 The CG3+Aβ sample was characterized by a strikingly lower density of aggregates (Figure 10d). Those aggregates that were present were fibrillar but much shorter than fibrils of Aβ alone (Figure 10e and f). This result is in line with previously obtained TEM images of mTTR+Aβ, where addition of mTTR to Aβ inhibited fibril initiation and suppressed aggregate growth, resulting in shorter and fewer fibrils.33 We examined many fields and did not observe any large (>micron) clusters. With CG3+Aβ, lightly stained spherical globules were also observed (Figure 10d), similar to those observed in TEM images of CG3 alone (Figure 3b).
Figure 10.
TEM images, taken after 48 h incubation at 37 °C. (A and B) 28 µM Aβ, (C) 28 µM Aβ + 2.8 µM G16, and (D, E and F) 28 µM Aβ + 2.8 µM CG3. Scale bar for A and D is 500 nm and for B, C, E and F is 200 nm.
We propose the following hypothesis to reconcile all of the data on CG3′s effect on Aβ aggregation, and to compare CG3 with mTTR and G16. Previously we observed that mTTR greatly inhibited Aβ aggregation, by both suppressing fibril initiation and arresting fibril growth, as measured by DLS, NTA, ThT, and TEM.17,34 Binding of G16 to Aβ decreased fibril formation and/or growth (as measured by ThT or filtration), but caused an increase in mean aggregate size and a general shift toward clustered aggregates (DLS, NTA, and TEM data).19 CG3 resembles the mode of action of mTTR, in that it inhibits Aβ fibrillar growth (as measured by ThT and TEM), and is selective for Aβ oligomers. However, CG3 (like G16) has a propensity to self-associate. As a consequence, some CG3-Aβ complexes may form large aggregate clusters, which are nonfibrillar and weakly associating. These clusters are detected in solution by light scattering methods (DLS and NTA), but are unstable and break apart easily when subjected to sedimentation or filtration.
Effect of Peptides on Aβ Toxicity
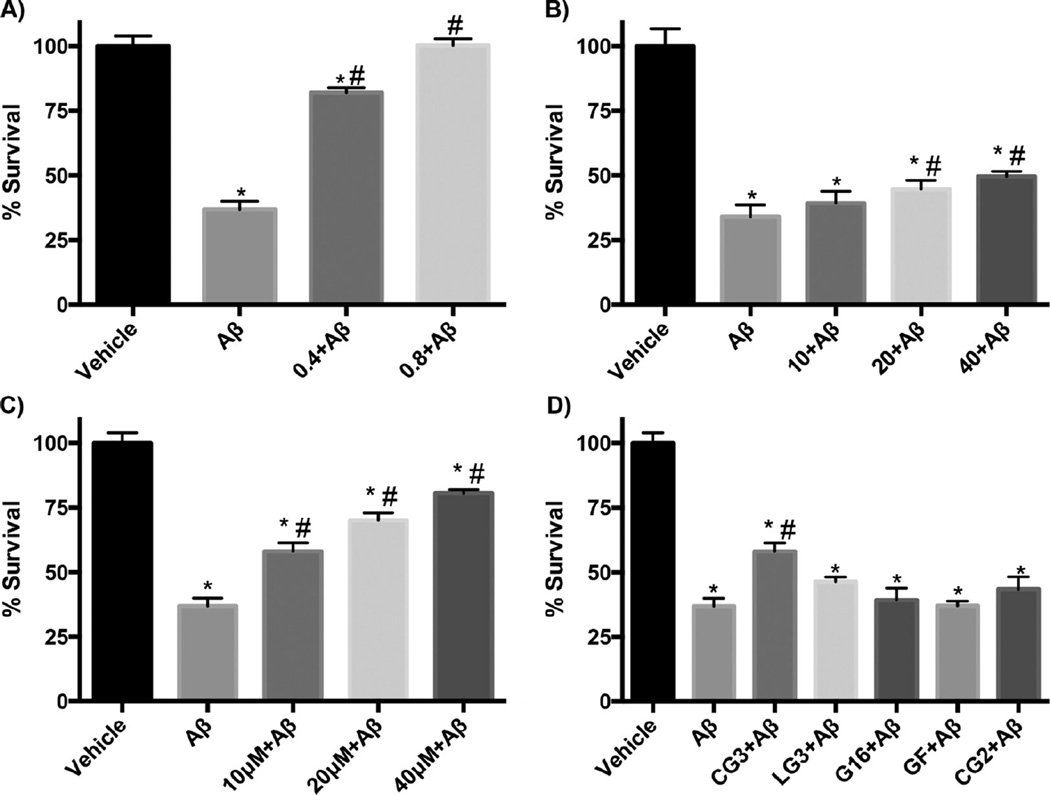
We tested whether TTR-derived peptides were effective at preventing Aβ toxicity, using primary mouse neuronal cultures and the MTS assay. None of the peptides by themselves had any effect on neuronal viability (data not shown). The Aβ preparation alone was toxic, with ~35% cell viability (Figure 11). Consistent with previous observations, mTTR conferred complete protection from Aβ-mediated toxicity at substoichiometric concentrations (0.8 µM, Figure 11a). G16 conferred a small but significant protection at the highest dose tested (Figure 11b). CG3 was much more effective than G16, showing significant protection at 10 µM and greater recovery of cell viability at higher doses (Figure 11c,d). At 10 µM, CG2, LG3 and GF had no protective effect (Figure 11d). GF did show modest protection at the highest dose (40 µM) tested (data not shown).
Figure 11.
Effect of peptides on Aβ induced toxicity measured using the MTS assay with primary neuronal cultures. Dose-dependent protection afforded by (A) mTTR (B) G16, (C) CG3. In each graph, Aβ concentration was 10 µM and the molar concentration of the added protein or peptide is shown. (D) Comparison of efficacy of several peptides, all at 10 µM. *statistically different from vehicle (p < 0.05). #statistically different from Aβ (p < 0.05).
Summary
Building on prior identification of the Aβ binding site domain in TTR,18,35 we synthesized several conformationally constrained peptides, and tested them for binding to Aβ, interference with Aβ aggregation, and inhibition of Aβ toxicity. Despite containing all the required residues for linear sequences to bind Aβ, neither LG2 nor CG2 bound Aβ, nor did they affect Aβ aggregation or inhibit Aβ toxicity. We speculate that lack of binding is due to either steric restriction (particularly to Thr106 near the hairpin) or to unwanted adoption of a non-native-like conformation. The longer LG3 bound to Aβ, but had only minor effects on aggregation and was ineffective as a toxicity inhibitor. Cyclization to CG3 greatly enhanced the effect on both Aβ aggregation and toxicity, presumably due to the additional conformational rigidity afforded.
Presumably because of its structural mimicry, CG3′s effect on Aβ aggregation is overall more like mTTR than is G16. CG3 effectively suppressed formation of ThT-positive species, much like mTTR, and with both mTTR and CG3 we observed fewer and much shorter Aβ fibrils by TEM. This is in counter-distinction to G16, which delayed but did not prevent the formation of ThT-positive Aβ aggregates, and caused morphological changes in the appearance of Aβ aggregates but did not shorten fibrils. CG3 differs from mTTR, though, when interaction with Aβ is probed with light scattering -a technique that is particularly sensitive to large particles. Specifically, CG3, like G16 and unlike TTR or mTTR, facilitates the formation and rapid growth of a few, very large, Aβ aggregates. However, both CG3 (this paper) and G1619 reduce the fraction of Aβ aggregates that can be removed by filtration or sedimentation. These results may seem at first contradictory, but can be explained once it is recognized that DLS reports on the size of aggregates whereas the filtration/sedimentation assays report on the mass fraction of insoluble/filterable aggregates. Taken together, we interpret the data as follows: CG3′s primary mechanism of interaction with Aβ is similar to mTTR, in that it inhibits formation of Aβ fibrillar aggregates and arrests growth of those fibrils that do form. However, due to its self-assembly characteristics, CG3 facilitates clustering of some Aβ, resulting in large, weakly associating aggregates. G16, presumably because of its greater conformational flexibility, has lower selectivity to Aβ oligomers, and does not suppress Aβ fibril growth as effectively as CG3, nor is it as effective at inhibiting Aβ toxicity.
Even though the protective activity of CG3 was greatly increased compared to G16, CG3 was not as effective at inhibiting Aβ toxicity as mTTR. It is likely that CG3 does not fully mimic TTR’s structure at the Aβ-binding interface, resulting in overall reduced binding affinity for CG3 relative to mTTR. Characterization of the solution structure(s) of CG3 by NMR would be useful in ascertaining the extent to which the peptide is a true structural mimic of TTR’s G and H strands. It is possible that CG3 adopts two or more conformational and/or oligomeric states, as hinted by the presence of two bands on the denaturing gel (Figure 3), and that only certain conformers of CG3 are active at protecting against Aβ toxicity. Additionally, the self-assembly of CG3 might reduce its effective concentration. Further improvement in activity might be achieved by enforcing tighter conformational constraints and/or controlling self-assembly.
Our work suggests a new strategy for designing peptide-based drugs against Aβ by mimicking the binding epitope of native proteins that exhibit natural protective action. Small molecules still dominate the pharmaceutical industry because of their small size, relatively low cost of manufacture, oral bioavailability and membrane permeability. Decades of advances in medicinal chemistry have paved the way for rational design and optimization of leads. On the other hand, small molecules may have poorer selectivity and affinity, especially when the targeted binding domain encompasses a large surface area.5,11 Antibody-based compounds and other large protein biologics possess potential advantages of very high affinity and exquisite selectivity. Their disadvantages include the need for injection or intranasal delivery, and their high cost. Peptides occupy a mid range between small molecules and protein biologics, and potentially could have the advantages of both (or, more pessimistically, the disadvantages of both!). Since peptides are chemically synthesized, they can be more easily engineered than proteins to incorporate desired features such as stability or membrane permeability. For example, cyclization, incorporation of D-amino acids or other non-natural amino acids, methylation, or biotinylation can greatly improve resistance to proteolysis and oral availability.5,36 To date, only a few peptides have been developed into clinically successful drugs. It remains a daunting challenge to tailor a peptide-based drug to possess desirable pharmacokinetic properties without loss of selectivity and affinity. Particularly challenging is the treatment of central nervous system disorders, if the drug must cross the blood-brain barrier (BBB). Peptides may need to be conjugated to vectors that target BBB-specific receptors, or packaged in liposomes or other nanocarriers, for effective delivery.36 Finally, more cost-effective methods are needed for manufacture of peptide-based drugs.5
METHODS
Peptide Synthesis and Purification
Linear peptides (G16, GF) were synthesized using standard Fmoc solid-phase method on the Symphony peptide synthesizer (Protein Technologies, In., Tuscon, AZ). The resin used was Fmoc–PAL-PEG-PS from Applied Biosystems (Foster City, CA). Extended cycles and double couplings were used to improve yield and peptides were modified with N-terminal acetylation and C-terminal amidation. Peptides were cleaved and purified as previously described.19
Hairpin-templated peptides (LG2, CG2, LG3, CG3) were synthesized using an orthogonally protected glutamic acid derivative and standard Fmoc solid-phase method. Fmoc-d-Glu-ODmab (Novabiochem, La Jolla, CA) was coupled to Fmoc–PAL-PEG-PS resin through the unprotected side chain. Assembly of the peptide backbone was done using standard Fmoc solid-phase method and the final Fmoc group was removed. The resin bound peptides were then treated with 2% hydrazine monohydrate in dimethylformamide (DMF) to remove Dmab protecting group and reacted with 10% diisopropylethylamine (DIPEA) in DMF. For peptide cyclization, an equimolar amount of PyBOP was added to the resin bound peptides, and the mixture was incubated for 18 h. Peptides were cleaved from the resin and purified by reverse-phase HPLC. Fractions corresponding to the two major peaks were collected, lyophilized, and analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
Purified peptides were dissolved in 0.22 µm filtered water, and the concentration of the peptides were determined by the absorbance at 205 nm with molar absorptivity calculated for each peptides based on their sequence.37 Peptides were then diluted to 0.7 mM, aliquoted, snap-frozen, and stored at −80 °C.
Aβ Sample Preparation
Aβ(1–40) was purchased from Anaspec, Inc. (Fremont, CA) as lyophilized powder. For dynamic light scattering, ThT fluorescence assay, TEM, NTA measurement and filtration/sedimentation assay, Aβ was dissolved in 50% ACN, relyophilized, and reconstituted in urea/glycine buffer at 12 mg/mL as described.17 Aβ stock was then aliquoted, snap-frozen in ethanol with dry ice, and stored at −80 °C. For PICUP and proteolytic fragmentation analysis, lyophilized Aβ was dissolved in prechilled hexafluoroisopropanol (HFIP, Acros Organics, Geel, Belgium), aliquoted, dried and stored at −20 °C as previously described.19 Right before use, dried peptide was dissolved in 50 mM NaOH and diluted into phosphate-buffered saline with azide (PBSA) [10 mM Na2HPO4/NaH2PO4, 150 mM NaCL and 0.02% NaN3 (pH 7.4)] to 30 µM as previously described.19
Circular Dichroism
CG3 and LG3 stock solutions in water at 0.7 mM were diluted into phosphate/NaF buffer [10 mM K2HPO4/KH2PO4 and 140 mM NaF (pH 7.4)] to 45 µM. Samples were filtered through 0.45 µm filter and transferred into a 1 mm cell. Circular dichroism (CD) spectra were collected on an Aviv 202SF CD spectrophotometer from Aviv Biomedical (Lakewood, NJ) at room temperature. Blank solvent spectra was collected and subtracted.
Photoinduced Cross-Linking (PICUP) of Aβ and Peptides
Aβ (30 µM in PBSA) was used immediately (freshly prepared) or after incubation for 24 h at 37 °C (1 day preaggregated). Aβ was mixed with peptides to a final concentration of 24 µM Aβ and 2.4 µM peptides. Samples were incubated for 1 h at 37 °C and cross-linked by photoinduced method (PICUP) as previously described.19,38 Cross-linked samples were separated on a 10–20% Tris-Tricine gradient gel (Invitrogen, Carlsbad, CA), transferred onto a 0.2 µm poly(vinylidene difluoride) (PVDF) membrane, and detected by monoclonal mouse anti-Aβ antibody 4G8 (Covance, Princeton, NJ) as previously described.19
To examine peptides for self-association, 20 µL of peptides at 36 µM in PBSA was incubated for 1 h at 37 °C and cross-linked using the PICUP method. Cross-linked samples were then heated at 95 °C for 5 min and separated on a 10–20% Tris-Tricine gradient gel. Peptides were visualized by silver staining (SilverStain Kit, Pierce, Rockford, IL).
Proteolytic Fragmentation Assay
The proteolytic fragmentation assay of Aβ with peptides was performed as previously described in detail.19 Briefly, Aβ alone (20 µM) or with peptide (60 µM) was incubated for 24 h at 37 °C. Proteinase K (Promega, Madison, WI) was added to the samples at a final concentration of 0.5 µg/mL and samples were dotted onto a 0.45 µm nitrocellulose membrane (Pierce) after incubation for 0, 5, 10, 20, 30, 40, 50, 60, 90, and 120 min. Membranes were then reacted with monoclonal antibodies against Aβ (6E10, Covance, against Aβ(3–8) and 4G8 against Aβ(18–22)) and visualized.
Dynamic Light Scattering (DLS)
Aβ stock in 8 M urea was thawed and sonicated for 2 min before being diluted into PBSA. Aβ alone (140 µM) or mixed with peptides (14 µM) was filtered through 0.45 µm filter directly into a light scattering cuvette and placed into a bath of the index-matching solvent decahydronaphthalene with temperature controlled to 37 °C. Light scattering data were collected using a Brookhaven BI-200SM system (Brookhaven Instruments Corp., Holtsville, NY) and an Innova 90C-5 argon laser (Coherent, Santa Clara, CA) operating at 488 nm and 150 mW. The average scattering intensity at 90° was measured over 10 h and data were normalized to the total mass concentration. The z-averaged hydrodynamic diameter was determined from the autocorrelation function using the method of cumulants.
Thioflavin T (ThT) Fluorescence Assay
ThT stock solutions were prepared in PBSA and filtered through 0.22 µm filter. The concentration of ThT was measured using an extinction coefficient of 26,620 M−1 cm−1 at 416 nm and the stock solution was diluted to 10 µM in PBSA. Aβ alone (28 µM) and with peptides (2.8 µM) or mTTR (2.8 µM) were prepared in PBSA and incubated at 37 °C. After 1, 5, 24, and 48 h of incubation, 10 µL of each sample was mixed with 130 µL of 10 µM ThT and analyzed. ThT fluorescence emission was measured using a QuantaMaster spectrofluometer (PTI, Birmingham, NJ), with excitation at 440 nm and emission spectra recorded from 460 to 500 nm. Three serial spectra were averaged for each sample and the background signal of ThT in PBSA was subtracted from the averaged data. Fluorescence intensity at a wavelength of 480 nm was compared. Using the same protocol, additional experiments were conducted in which the CG3 concentration and aggregation time were varied.
Transmission Electron Microscopy (TEM)
Aβ alone (28 µM) or with peptides (G16 or CG3 at 2.8 µM) were prepared in PBSA and incubated for 48 h at 37 °C. A drop of sample was placed on a pioloform-coated grid and stained with methylamine tungstate stain. Images were taken with a Philips CM120 scanning transmission electron microscope (FEI Corp., Eindhoven, The Netherlands).
Nanoparticle Tracking (NTA)
NTA measurements were taken with a Nanosight LM10 (Nanosight, Amesbury, U.K.) equipped with a 405 nm laser as previously described.17 Aβ alone (28 µM) or with CG3 (2.8 µM) were prepared in PBSA and incubated for 5 h at 37 °C. Three 60 s videos were taken for each sample.
Filtration/Sedimentation Assay
Aβ alone (30 µM) or with CG3 (3 µM) was prepared in PBSA and incubated for 24 h at 37 °C. For filtration, 175 µL of sample was filtered through 0.45 µm filter. For sedimentation, 100 µL of sample was transferred onto a separate tube and centrifuged for 30 min at 19,500 rcf. The supernatant (80%) was removed. Concentration of untreated, filtered, and centrifuged samples were measured by BCA assay (Pierce).
Expression and Purification of mTTR
Recombinant human transthyretin mutant F87M/L110M (mTTR) was produced and purified as previously described in detail.33
In Vitro Cellular Toxicity
Aβ toxicity was assessed as described in detail previously.19 Briefly, primary cortical neuronal cells were cultured from embryonic15.5 day mice and were maintained in neurobasal media until they matured as previously described.17 Aβ(1–42) (American Peptide, Sunnyvale, CA) at 1 mg/mL in PBS was diluted to 10 µM without or with peptides as previously described.19 At 6 days in vitro (DIV), the cells were treated with either Aβ alone or Aβ incubated with different doses of mTTR and peptides. Appropriate vehicle and controls were run along with the treatments. At 8 DIV, the neurons were incubated with MTS reagent (Promega) as per manufacturer protocol for 2–3 h and absorbance was measured at 490 nm.
Acknowledgments
Funding
This work was supported by National Institutes of Health Grant R01AG033493.
Stacy Maynard provided valuable insight into the design of the cyclic peptide. Dennis Yang collected CD data on G16*. Claire Brickson assisted with data collection.
ABBREVIATIONS
- Aβ
beta-amyloid
- AD
Alzheimer’s disease
- APP
amyloid precursor protein
- ACN
acetonitrile
- CD
circular dichroism
- DLS
dynamic light scattering
- DIV
days in vitro
- HFIP
hexafluoroisopropanol
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl) −2 (4-sulfophenyl)-2H–tetrazolium salt
- NTA
nanoparticle tracking analysis
- PBS
phosphate-buffered saline
- PICUP
photoinduced cross-linking of unmodified proteins
- PyBOP
benzotriazol-1-yl-N-oxy-tris(pyrrolidino)-phosphonium hexafluorophosphate
- TEM
transmission electron microscopy
- TTR
transthyretin
Footnotes
Author Contributions
PYC and MDB developed the cyclic peptide synthesis strategy, and synthesized all peptides. PYC carried out most of the biochemical and biophysical characterization experiments. GJ conducted the toxicity inhibition experiments. JAJ and RMM developed the overall strategy and implementation, and interpreted the data. PYC and RMM wrote the manuscript.
The authors declare the following competing financial interest(s): We have submitted a provisional patent application covering the synthesis and use of the peptide described herein.
REFERENCES
- 1.Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. Amyloid beta-protein assembly and Alzheimer disease. J. Biol. Chem. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 3.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 4.Karran E, Mercken M, De Strooper B. The amyloid cascade hypothesis for Alzheimer’s disease: an appraisal for the development of therapeutics. Nat. Rev. Drug Discovery. 2011;10:698–712. doi: 10.1038/nrd3505. [DOI] [PubMed] [Google Scholar]
- 5.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 6.Tjernberg LO, Naslund J, Lindqvist F, Johansson J, Karlstrom AR, Thyberg J, Terenius L, Nordstedt C. Arrest of beta-amyloid fibril formation by a pentapeptide ligand. J. Biol. Chem. 1996;271:8545–8548. doi: 10.1074/jbc.271.15.8545. [DOI] [PubMed] [Google Scholar]
- 7.Taylor M, Moore S, Mayes J, Parkin E, Beeg M, Canovi M, Gobbi M, Mann DM, Allsop D. Development of a proteolytically stable retro-inverso peptide inhibitor of beta-amyloid oligomerization as a potential novel treatment for Alzheimer’s disease. Biochemistry. 2010;49:3261–3272. doi: 10.1021/bi100144m. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi T, Mihara H. Peptide and protein mimetics inhibiting amyloid beta-peptide aggregation. Acc. Chem. Res. 2008;41:1309–1318. doi: 10.1021/ar8000475. [DOI] [PubMed] [Google Scholar]
- 9.Soto C, Sigurdsson EM, Morelli L, Kumar RA, Castano EM, Frangione B. Beta-sheet breaker peptides inhibit fibrillogenesis in a rat brain model of amyloidosis: Implications for Alzheimer’s therapy. Nat. Med. 1998;4:822–826. doi: 10.1038/nm0798-822. [DOI] [PubMed] [Google Scholar]
- 10.Robinson JA. Beta-hairpin peptidomimetics: design, structures and biological activities. Acc. Chem. Res. 2008;41:1278–1288. doi: 10.1021/ar700259k. [DOI] [PubMed] [Google Scholar]
- 11.Eichler J. Peptides as protein binding site mimetics. Curr. Opin. Chem. Biol. 2008;12:707–713. doi: 10.1016/j.cbpa.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 12.Stein TD, Johnson JA. Lack of neurodegeneration in transgenic mice overexpressing mutant amyloid precursor protein is associated with increased levels of transthyretin and the activation of cell survival pathways. J. Neurosci. 2002;22:7380–7388. doi: 10.1523/JNEUROSCI.22-17-07380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buxbaum JN, Ye Z, Reixach N, Friske L, Levy C, Das P, Golde T, Masliah E, Roberts AR, Bartfai T. Transthyretin protects Alzheimer’s mice from the behavioral and biochemical effects of Abeta toxicity. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2681–2686. doi: 10.1073/pnas.0712197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein TD, Anders NJ, DeCarli C, Chan SL, Mattson MP, Johnson JA. Neutralization of transthyretin reverses the neuroprotective effects of secreted amyloid precursor protein (APP) in APPSW mice resulting in tau phosphorylation and loss of hippocampal neurons: Support for the amyloid hypothesis. J. Neurosci. 2004;24:7707–7717. doi: 10.1523/JNEUROSCI.2211-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Masliah E, Reixach N, Buxbaum JN. Neuronal production of transthyretin in human and murine Alzheimer’s disease: Is it protective? J. Neurosci. 2011;31:12483–12490. doi: 10.1523/JNEUROSCI.2417-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giunta S, Valli MB, Galeazzi R, Fattoretti P, Corder EH, Galeazzi L. Transthyretin inhibition of myloid beta aggregation and toxicity. Clin. Biochem. 2005;38:1112–1119. doi: 10.1016/j.clinbiochem.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Yang DT, Joshi G, Cho PY, Johnson JA, Murphy RM. Transthyretin as both a sensor and a scavenger of beta-amyloid oligomers. Biochemistry. 2013;52:2849–2861. doi: 10.1021/bi4001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du J, Cho PY, Yang DT, Murphy RM. Identification of beta-amyloid-binding sites on transthyretin. Protein Eng., Des. Sel. 2012;25:337–345. doi: 10.1093/protein/gzs026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho PY, Joshi G, Johnson JA, Murphy RM. Transthyretin-derived peptides as beta-amyloid inhibitors, ACS Chem. Neurosci. 2014;5:542–551. doi: 10.1021/cn500014u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milroy LG, Grossmann TN, Hennig S, Brunsveld L, Ottmann C. Modulators of protein-protein interactions. Chem. Rev. 2014;114:4695–4748. doi: 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- 21.Haque TS, Gellman SH. Insights on beta-hairpin stability in aqueous solution from peptides with enforced type I′ and type II′ beta-turns. J. Am. Chem. Soc. 1997;119:2303–2304. [Google Scholar]
- 22.Gellman SH. Minimal model systems for beta sheet secondary structure in proteins. Curr. Opin. Chem. Biol. 1998;2:717–725. doi: 10.1016/s1367-5931(98)80109-9. [DOI] [PubMed] [Google Scholar]
- 23.Favre M, Moehle K, Jiang LY, Pfeiffer B, Robinson JA. Structural mimicry of canonical conformations in antibody hypervariable loops using cyclic peptides containing a heterochiral diproline template. J. Am. Chem. Soc. 1999;121:2679–2685. [Google Scholar]
- 24.Gibbs AC, Kondejewski LH, Gronwald W, Nip AM, Hodges RS, Sykes BD, Wishart DS. Unusual beta-sheet periodicity in small cyclic peptides. Nat. Struct. Biol. 1998;5:284–288. doi: 10.1038/nsb0498-284. [DOI] [PubMed] [Google Scholar]
- 25.Syud FA, Stanger HE, Gellman SH. Interstrand side chain–side chain interactions in a designed beta-hairpin: Significance of both lateral and diagonal pairings. J. Am. Chem. Soc. 2001;123:8667–8677. doi: 10.1021/ja0109803. [DOI] [PubMed] [Google Scholar]
- 26.Alsina J, Barany G, Albericio F, Kates SA. Pyrrolidide formation as a side reaction during activation of carboxylic acids by phosphonium salt coupling reagents. Lett. Pept. Sci. 1999;6:243–245. [Google Scholar]
- 27.Bean JW, Kopple KD, Peishoff CE. Conformational analysis of cyclic hexapeptides containing the d-Pro-l-Pro sequence to fix beta-turn positions. J. Am. Chem. Soc. 1992;114:5328–5334. [Google Scholar]
- 28.Blanco FJ, Jimenez MA, Pineda A, Rico M, Santoro J, Nieto JL. NMR solution structure of the isolated N-terminal fragment of Protein-G B-1 domain. Biochemistry. 1994;33:6004–6014. doi: 10.1021/bi00185a041. [DOI] [PubMed] [Google Scholar]
- 29.Bitan G. Structural study of metastable amyloidogenic protein oligomers by photo-induced cross-linking of unmodified proteins. Methods Enzymol. 2006;413:217–236. doi: 10.1016/S0076-6879(06)13012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X, Murphy RM. Synthesis and disaggregation of asparagine repeat-containing peptides. J. Pept. Sci. 2014;20:860–867. doi: 10.1002/psc.2677. [DOI] [PubMed] [Google Scholar]
- 31.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid beta -protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. U. S. A. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dias RL, Fasan R, Moehle K, Renard A, Obrecht D, Robinson JA. Protein ligand design: from phage display to synthetic protein epitope mimetics in human antibody Fc-binding peptidomimetics. J. Am. Chem. Soc. 2006;128:2726–2732. doi: 10.1021/ja057513w. [DOI] [PubMed] [Google Scholar]
- 33.Biancalana M, Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta. 2010;1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du J, Murphy RM. Characterization of the interaction of beta-amyloid with transthyretin monomers and tetramers. Biochemistry. 2010;49:8276–8289. doi: 10.1021/bi101280t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Zhang X, Ladiwala AR, Du D, Yadav JK, Tessier PM, Wright PE, Kelly JW, Buxbaum JN. Mechanisms of transthyretin inhibition of beta-amyloid aggregation in vitro. J. Neurosci. 2013;33:19423–19433. doi: 10.1523/JNEUROSCI.2561-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlieghe P, Khrestchatisky M. Medicinal chemistry based approaches and nanotechnology-based systems to improve CNS drug targeting and delivery. Med. Res. Rev. 2013;33:457–516. doi: 10.1002/med.21252. [DOI] [PubMed] [Google Scholar]
- 37.Anthis NJ, Clore GM. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 2013;22:851–858. doi: 10.1002/pro.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bitan G, Lomakin A, Teplow DB. Amyloid beta-protein oligomerization: prenucleation interactions revealed by photoinduced cross-linking of unmodified proteins. J. Biol. Chem. 2001;276:35176–35184. doi: 10.1074/jbc.M102223200. [DOI] [PubMed] [Google Scholar]