Abstract
Background
Parasitic nematodes threaten the health of humans and livestock and cause a major financial and socioeconomic burden to modern society. Given the widespread distribution of diseases caused by parasitic nematodes there is an urgent need to develop tools that will elucidate the genetic complexity of host-parasite interactions. Heterorhabditis bacteriophora is a parasitic nematode that allows simultaneous monitoring of nematode infection processes and host immune function, and offers potential as a tractable model for parasitic nematode infections. However, molecular tools to investigate these processes are required prior to its widespread acceptance as a robust model organism. In this paper we describe microinjection in adult H. bacteriophora as a suitable means of dsRNA delivery to knockdown gene transcripts.
Methods
RNA interference was used to knockdown four genes by injecting dsRNA directly into the gonad of adult hermaphrodite nematodes. RNAi phenotypes were scored in the F1 progeny on the fifth day post-injection, and knockdown of gene-specific transcripts was quantified with real-time quantitative RT-PCR (qRT-PCR).
Results
RNAi injection in adult hermaphrodites significantly decreased the level of target transcripts to varying degrees when compared with controls. The genes targeted by RNAi via injection included cct-2, nol-5, dpy-7, and dpy-13. In each case, RNAi knockdown was confirmed phenotypically by examining the progeny of injected animals, and also confirmed at the transcriptional level by real-time qRT-PCR.
Conclusions
Here we describe for the first time the successful use of microinjection to knockdown gene transcripts in H. bacteriophora. This technique can be used widely to study the molecular basis of parasitism.
Keywords: Heterorhabditis bacteriophora, Microinjection, RNAi, Gene knockdown, qRT-PCR
Background
Diseases caused by parasitic nematodes are a major concern, resulting in human health and socioeconomic consequences [1, 2]. It is estimated that more than half of the human population is infected with gastrointestinal nematodes alone [3, 4], and around 20 species routinely cause disease [5]. Parasitic nematodes are also a concern to the livestock industry, as they cause diseases and financial loss estimated in the tens of millions of dollars per year [6–8]. Parasitic nematode control in humans and livestock is limited to periodic treatment with anthelmintics, but development of resistance to the commonly used drugs is an ongoing problem, limiting efficacy and requiring continual development of new anthelmintics [9, 10]. The search for new drugs, as well as alternate novel control methods such as vaccines, requires comprehensive knowledge of the host-parasite interaction.
An important interaction between the parasite and its potential host is the active avoidance of the host immune system by the parasite [11]. Understanding this interaction at the molecular level provides useful insight into pathogen virulence and host immunity [12, 13]. Advances in understanding these mechanisms require powerful molecular tools to investigate gene function, which are unavailable for most parasitic nematodes. Discovery of RNA interference (RNAi) in Caenorhabditis elegans was a major step forward in the analysis of gene function [14]. RNAi is the RNA-induced silencing of gene-specific mRNA targets [15]. In C. elegans RNAi is performed either by soaking in double-stranded RNA (dsRNA), feeding the nematodes with bacteria expressing dsRNA or by injecting the dsRNA into the gonad of mature adult hermaphrodite nematodes [16]. In C. elegans, RNAi has been successfully used to knockdown almost all the genes in the genome [17]. Several studies have adapted this technique to parasitic nematodes in attempts to knockdown genes in order to understand host-pathogen interactions [18, 19]. RNAi based gene knockdown has been tested, with mixed success, in both plant and animal parasitic nematodes. Animal parasitic nematodes in which RNAi has been tested include Nippostrongylus brasiliensis [20, 21], Brugia malayi [22, 23], Onchocerca volvulus [24], Litomosoides sigmodontis [25], Ascaris suum [26, 27], Trichostrongylus colubriformis [28], Haemonchus contortus [29–31], Ostertagia ostertagi [32], Teladorsagia circumcincta [33], Trichinella spiralis [34], Heligmosomoides polygyrus [35] and Heterorhabditis bacteriophora [36, 37].
RNAi experiments conducted in parasitic nematodes have been performed by soaking or feeding the dsRNA. However, significant variability in both detecting the desired phenotype and reduction in target gene transcripts is common [38, 39]. RNAi in Heligmosomoides polygyrus has failed to produce any observable knockdown [35]. Considerable variation was observed between genes knocked down in Haemonchus contortus; in one study, gene knockdown occurred only in genes that are expressed in tissues that come in direct contact with dsRNA, such as the intestine [40]. Another study found that only administration of dsRNA by feeding resulted in expected phenotypes [31]. Nearly complete knockdown of several target genes has also been reported in H. bacteriophora when dsRNA is administered by soaking [36]. This disparity in observed phenotypes cannot be explained solely by differences in the RNAi machinery existing in parasitic nematode species. Analysis of the RNAi machinery has revealed that most parasites contain at least the minimal requirements for a functional RNAi pathway. Noticeably absent are the genes involved in amplification, uptake and spread of dsRNA, including SID-2, RSD-2 and RSD-6 [41]. This variation in RNAi efficacy has hampered investigations of parasitic gene function. Hence, there is a need to increase the efficacy and reliability of gene knockdown by RNAi.
Heterorhabditis bacteriophora is an entomopathogenic nematode, a member of the family Heterorhabditidae in which all nematodes are obligate parasites [42]. It is grouped in the Eurhabditis clade that includes the hookworms Ancylostoma ceylanicum, A. duodenale and Necator americanus that infect humans and other vertebrates. The Eurhabditis clade also includes the well-studied model nematode C. elegans [43]. The divergence time between H. bacteriophora and C. elegans is estimated to be between 86 and 331 MYA [44]. H. bacteriophora has several important similarities with C. elegans. These include small size, transparency, ease of in vitro culture, short generation time, both hermaphroditic and gonochoristic reproduction, and an annotated genome with about the same number of genes [44]. Moreover, the infective stage of H. bacteriophora and strongylid nematodes is an obligate arrested third larval stage (L3) that is similar to the facultative arrested dauer stage of C. elegans. These similarities, as well as its close phylogenetic relationship with C. elegans and other important parasites of humans and animals, make H. bacteriophora a potentially excellent model for molecular studies of nematode infection mechanisms. The only free living stage of H. bacteriophora is a non-feeding, developmentally arrested infective juvenile (IJ) stage. The other stages (i.e. L1, L2, L3, L4 and adult) develop inside the host. The developmentally arrested IJs can survive for months in the soil while seeking a host. After entry into the insect host, IJs regurgitate their endosymbiotic bacterium Photorhabdus luminescens into the insect haemocoel, where it replicates and contributes to the inevitable death of the host within 24–72 h [42, 45]. IJs resume their development in the host, developing only into phenotypically female hermaphrodites. Males, females and hermaphrodites are produced in approximately equal numbers in subsequent generations in the presence of abundant food [46]. The nematodes reproduce for another 2–3 generations feeding on the insect cadaver. However, nutrient limitation and accumulation of density limiting pheromones [47] impedes further progression of the life-cycle and causes mass production of IJs. IJs colonized with P. luminescens then exit the cadaver in large numbers into the external environment. These IJs search for and infect other insect hosts to continue the cycle [48].
Two previous studies in H. bacteriophora have demonstrated that delivery of dsRNA by soaking can successfully knockdown gene transcripts [36, 37]. However, dsRNA delivery either by feeding or by microinjection in H. bacteriophora has not been reported. Very few studies have successfully demonstrated the use of microinjection as a method of introducing dsRNA in parasitic nematodes [26, 49], yet RNAi via microinjection represents the most reliable and least variable method of gene silencing in C. elegans for most targets [16]. In the present study we describe RNAi in H. bacteriophora by injecting dsRNA into the gonads of the adult hermaphrodite. We show that gene function can be successfully knocked down in the progeny of injected nematodes by this technique. RNAi by microinjection provides an alternative approach that can be widely used to study the function of genes involved in parasitism of H. bacteriophora.
Methods
Nematodes
H. bacteriophora strain TT01 was kindly provided by Dr. David Clarke (University College Cork, Ireland). Nematode stocks were maintained in the lab by infecting the Greater Wax Moth larvae (Galleria mellonella) with IJs [50]. IJs emerging from white traps [51] were propagated on lawns of P. luminescens [36] to raise young hermaphrodites for injection.
Primer design
Primers for dsRNA were designed to target ~500 base pair exonic regions of H. bacteriophora DNA. In order to identify possible exons, protein BLASTs were performed to identify regions of similarity between H. bacteriophora and C. elegans for dpy-7 and dpy-13 genes. Primers for regions of interest were determined using Primer3 [52], selecting for an optimum product length of 500 base pairs, Tm of 60 °C, and primer length of 22 nucleotides. The primer pair for Green Fluorescent Protein (GFP) was designed using the same settings in Primer3. T7 sites (TAATACGACTCACTATAGGG) were added to the 5′ ends of each primer to allow for in vitro transcription.
dsRNA synthesis
Genomic DNA was isolated from frozen pellets of ~50,000 H. bacteriophora infective juveniles. An IJ pellet was resuspended in 50 μl of lysis buffer (50 mMKCl, 0.05 % (w/v) gelatin, 10mMTris-HCl pH 8.2, 0.45 % Tween 20, 60 μg/ml Proteinase K, 2.5 mM MgCl2) and placed at −80 °C for 30 min. The solution was then warmed to room temperature and incubated at 60 °C for 2 h, with vortexing every 15 min. Proteinase K was denatured by incubating the homogenized tissue for 15 min at 95 °C. The sample was then cooled to 4 °C and centrifuged at 3,400 × g for 1 min. The resulting supernatant was used as template for subsequent PCR. A 50 μl PCR reaction was carried out using ChoiceTaq Mastermix (Denville Scientific, South Plainfield, NJ, USA) with 200 ng of template DNA, 0.2 μM of each primer, and the manufacturer’s suggested cycling conditions. PCR reactions were analyzed on a 1.2 % agarose gel to verify that the reactions produced single bands of the predicted size.
Five μl of the PCR reaction was used for in vitro transcription using the Ambion Megascript T7 Kit (Thermo Fisher Scientific, Waltham, MA, USA). Reactions were carried out following the manufacturer’s instructions and incubated for 16 h at 37 °C. In vitro transcription reactions were cleaned up using the AmbionMegaclear Kit (Thermo Fisher, Waltham, MA, USA) followed by ammonium acetate/ethanol precipitation to concentrate the dsRNA. Pelleted dsRNA was suspended in 10 μl RNase-free water, quantified using a NanoDrop spectrophotometer, and the quality assessed by separating the dsRNA on a 1.2 % agarose gel.
RNAi by injection
dsRNA was injected into the gonad of adult hermaphrodite nematodes as described for C. elegans [53]. Young adults were obtained by placing IJs collected from white traps on aNA + chol agar plate (3 g yeast extract, 5 g peptone, 12 g agarose per liter with 2 ml of 5 mg/ml cholesterol added after autoclaving) growing P. luminescens bacteria for 68 h at 27 °C. dsRNA was injected at a concentration of 6 μg/μl for cct-2 and 4 μg/μl fornol-5, dpy-13 and dpy-7. The injected nematodes were removed from the injection pad with a pipette using 1xPBS and placed on a rescue plate. After 1 h the nematodes were picked to a new plate seeded with fresh P. luminescens and maintained at 27 °C. Three batches of nematodes were used, representing three different biological replicates each with 15 nematodes. The progeny of the injected hermaphrodite were screened on the fifth day after injection. Images were taken on a stereomicroscope (LeicaS6 D, Leica, Germany).
RNA extraction and qRT-PCR
Total RNA was extracted in TRIzol (Thermo Fisher). Briefly, 20 worms for Hb-cct-2 and Hb-nol-5 and 2 worms for Hb-dpy-7 and Hb-dpy-13 were picked from the P. luminescens plate into a 1.5 ml tube with M9 buffer (3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 ml 1 M MgSO4, H2O to 1 l). Nematodes were washed with M9 two more times after which 250 μl of Trizol was added. The tubes were stored at −80 °C. On the day of RNA extraction, the tubes were thawed and vortexed at maximum speed for 30 min at 4 °C. RNA was extracted according to the manufacturer’s instructions. Total RNA was treated with DNase I, Amplification Grade (Thermo Fisher). RNA was then converted to cDNA with Verso™ cDNA Kit (Thermo Fisher). Real-time quantitative reverse transcription PCR (qRT-PCR) was performed in a 96-well Bio-Rad CFX96 RealTime PCR System (Bio-Rad, Inc., Hercules, CA, USA). PCR reactions were done in 96-well optical reaction plates (Bio-Rad, Hercules, CA, USA) using Agilent Brilliant II SYBR® Green QPCR Master Mix. A 20 μL PCR reaction was set up in each well with 10 μL Brilliant II SYBR Green QRT-PCR master mix, 1/20th of the converted cDNA and 25 μM primers. To quantify the efficiency of the primers, a standard curve was constructed with serial dilutions of gene-specific PCR products that were obtained by amplifying cDNA from adults collected 72 h after plating as IJs. The amplification efficacy (E = 101/-slope-1) for each primer was calculated from the slope generated by the standard curve. The primer sets used had efficiency between 90–100 %. Relative quantification of the amplified gene was done by delta-delta Ct method [54]. Large ribosomal subunit L32 protein (rpl-32) was used as an internal control. Expression of Hba-rpl-32 was shown to be stable in three different life stages of H. bacteriophora (IJs and 48 h and 72 h developing nematodes derived from IJs) before it was used as an internal reference gene. For every gene at least three independent biological samples were tested, each with three technical replicates. Primers used in this study are listed in Table 1.
Table 1.
Primer sequences used for qRT-PCR
| Primer name | Sequence |
|---|---|
| dpy-13Fwd | AGCCCGGAGCTAAAGGTAAC |
| dpy-13Rev | TACGAGTCATCAATGGCACA |
| dpy-7-Fwd | GGTAGACCAGGTCGTCCAGT |
| dpy-7-Rev | ACCAGGCAAACCAGGACTT |
| rpl-32-Fwd | ATCGGATAGATACCACCGCC |
| rpl-32-Rev | TTGTGGGCATAGCACGC |
Results and discussion
We tested the efficacy of dsRNA delivery by microinjection on four H. bacteriophora genes with known RNAi phenotypes. The C. elegans cct-2 (T21B10.1) gene encodes a component of eukaryotic T-complex chaperonin and is expressed in most tissues. It is required for proper folding of proteins including actin, tubulin and cyclin [55–57]. In C. elegans, RNAi of cct-2 causes sterility, embryonic lethality and protruding vulva [17, 58]. The C. elegans nol-5 is an ortholog of human NOP58 (NOP58 ribonucleoprotein) and is involved in reproduction, embryo development, and larval development. Knockdown of nol-5 in C. elegans causes larval arrest, multivulva phenotype, sterility and slow growth [58, 59]. Ciche et al. [36] showed previously that knockdown of Hba-cct-2 and Hba-nol-5by soaking eggs in dsRNA also causes sterility in H. bacteriophora, as evidenced by the absence of visible germline and defective gonad.
In this study, injecting dsRNA of Hba-cct-2 and Hba-nol-5 in the H. bacteriophora hermaphrodites resulted in sterile progeny, with a noticeable lack of any germline and eggs in the adult nematodes. The observed phenotype was similar to the post-embryonic phenotype described by Ciche et al. [36], although the percentage of the sterile progeny was lower. We observed that 40 to 50 % of the progeny of cct-2 dsRNA injected nematodes (Fig. 1b, Table 2), and 20 to 60 % of the progeny of nol-5 dsRNA injected nematodes were sterile in three different biological replicates (Fig. 2b, Table 2). This difference in penetrance may be due to our examination of the phenotype of F1 progeny of injected nematodes, and rather than in adults derived from eggs soaked in dsRNA. For both Hba-cct-2 and Hba-nol-5, dsRNA was generated using primers previously described [36]. Microinjection as the means of dsRNA delivery has never been reported in H. bacteriophora. Knockdown of gene transcripts resulting in visible phenotypes in progeny for both genes in three different trials confirmed that this technique can be used to reliably knockdown gene transcripts in H. bacteriophora.
Fig. 1.
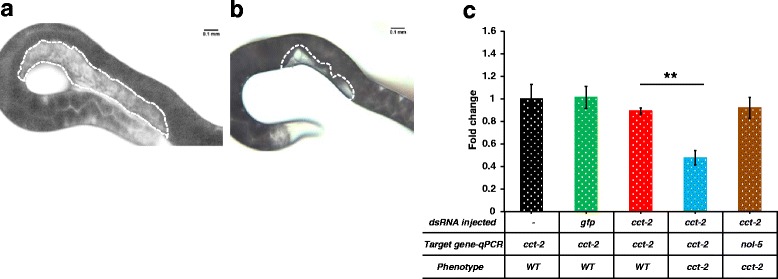
RNAi mediated phenotype and transcript changes in H. bacteriophora injected with cct-2 dsRNA. Adult H. bacteriophora hermaphrodites that were injected with cct-2 dsRNA produced progeny with no germline and empty gonad. a. Progeny of non-injected H. bacteriophora. The area with white dotted lines indicates the position of the gonads. b. Progeny of H. bacteriophora injected with cct-2 dsRNA. c. Expression of cct-2 gene in the progeny of injected worms. The y-axis represents the fold change in mRNA expression in the progeny of H. bacteriophora hermaphrodites. The mRNA levels are normalized to cct-2 expression in the progeny of non-injected hermaphrodites (black bar). The green bar represents cct-2 levels in progeny of gfp injected worms. The red bar represents cct-2 levels in a phenotypically wild type sibling, and the blue bar represents the cct-2 levels in worms with empty gonads. To control for off-target effects, expression of an unrelated gene (nol-5) in worms with empty gonads was determined (brown bar). The graph was obtained by combining data from at least three independent biological replicates. Error bars indicate the standard error of the mean. Asterisks depict the statistical significance of the observed differences in unpaired, two-tailed t-tests with P-values < 0.001 (**)
Table 2.
RNAi phenotypes observed in the progeny of H. bacteriophora worms injected with dsRNA
| Trial-1 | Trial-2 | Trial-3 | Average | |
|---|---|---|---|---|
| GFP | 0 (0/53) | 0 (0/95) | 0 (0/127) | 0 (0/127) |
| nol-5 (ste) | 20 (35/173) | 25 (29/113) | 60 (92/152) | 36 (156/438) |
| cct-2 (ste) | 44 (74/165) | 50 (91/179) | 51 (102/198) | 50 (267/542) |
| dpy-13 (dpy) | 7 (3/42) | 8 (2/25) | 6 (3/43) | 7 (8/110) |
| dpy-7 (dpy) | 3 (1/26) | 5 (3/53) | 2 (2/98) | 3 (6/177) |
For each trial, percentage of observed phenotype is followed in parenthesis by total number of progeny that had the predicted phenotype over total number of progeny from the injection. Last column is the average of all the trials. Ste (Sterile) and dpy (dumpy)
Fig. 2.
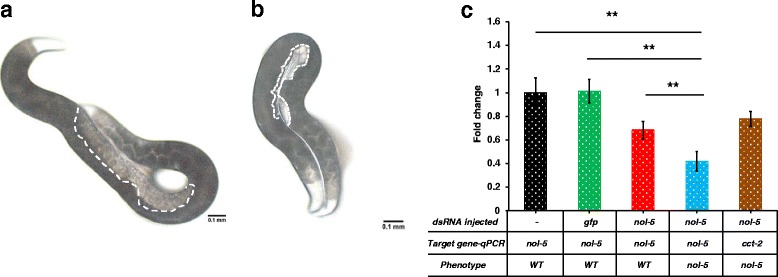
RNAi mediated phenotype and transcript changes in H. bacteriophora injected with nol-5 dsRNA. Adult H. bacteriophora hermaphrodites that were injected with nol-5 dsRNA produced progeny with no germline and empty gonad. a. Progeny of non-injected H. bacteriophora. The area with white dotted lines indicates the position of the gonads. b. Progeny of H. bacteriophora injected with nol-5dsRNA. c. Expression of nol-5 gene in the progeny of nol-5 dsRNA injected worms. The y-axis represents the fold change in mRNA expression in the progeny of H. bacteriophora hermaphrodites. The mRNA levels are normalized to nol-5 expression in the progeny of non-injected hermaphrodites (black bar). The green bar represents nol-5 levels in progeny of gfp injected worms. The red bar represents nol-5 levels in phenotypically wild type siblings, and the blue bar represents the nol-5 levels in worms with empty gonads. To control for off-target effects, expression of an unrelated gene (cct-2) in worms with empty gonads was determined (brown bar). The graph was obtained by combining data from at least three independent biological replicates. Error bars indicate the standard error of the mean. Asterisks depict the statistical significance of the observed differences in unpaired, two-tailed t-tests with P-values < 0.001 (**)
We also tested two other genes, Hba-dpy-13 and Hba-dpy-7 that were previously described to cause a dumpy phenotype in C. elegans. Dumpy mutant nematodes are almost half the length of the wild type nematode. Cel-dpy-13 and Cel-dpy-7 encode two different collagen proteins required for proper cuticular morphology and normal body length [60, 61]. Hba-dpy-13 was also used as a positive control for RNAi by soaking in H. bacteriophora [37]. Injecting Hba-dpy-13 dsRNA resulted in dumpy-like progeny with short, chunky morphology similar to that seen in C. elegans dpy mutants (Fig. 3b, Table 2). Injecting Hba-dpy-7 dsRNA also caused the progeny to be phenotypically similar to Hba-dpy-13RNAi (Fig. 4b, Table 2). However, the numbers of dumpy nematodes observed in both Hba-dpy-13 and Hba-dpy-7 dsRNA injections were significantly lower than observed with nol-5 and cct-2 dsRNA injections. We observed that 6 to 8 % of the progeny of dpy-13 dsRNA injected nematodes and 2 to 5 % of the progeny of dpy-7 dsRNA injected nematodes exhibited the dumpy phenotype across three different biological replicates (Table 2).
Fig. 3.
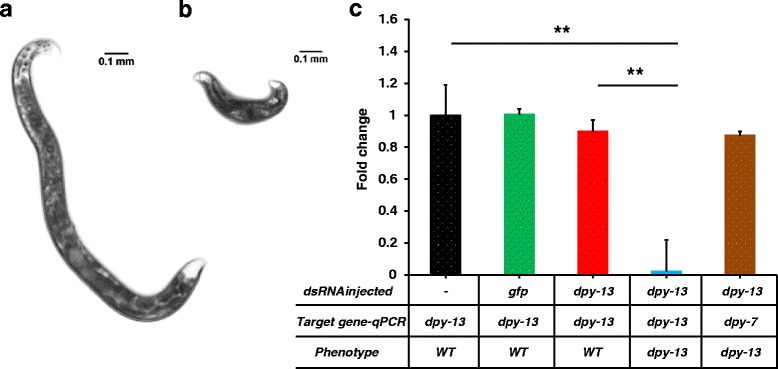
RNAi mediated phenotype and transcript changes in H. bacteriophora injected with dpy-13 dsRNA. Adult H. bacteriophora hermaphrodites that were injected with dpy-13 dsRNA produced progeny with dumpy phenotype. a. Phenotypically wild type progeny of H. bacteriophora worms injected with dpy-13 dsRNA. b. Progeny of H. bacteriophora worms injected with dpy-13 dsRNA exhibiting the dumpy phenotype. c. Expression of dpy-13 gene in the progeny of dpy-13 dsRNA injected worms. The y-axis represents the fold change in mRNA expression in the progeny of H. bacteriophora hermaphrodites. The mRNA levels are normalized to dpy-13 expression in the progeny of non-injected hermaphrodites (black bar). The green bar represents dpy-13 levels in progeny of gfp injected worms. The red bar represents dpy-13 levels in phenotypically wild type siblings, and the blue bar represents the dpy-13 transcript levels in phenotypically dumpy worms. To control for off-target effects, expression of an unrelated gene (dpy-7) in phenotypically dumpy worms was determined (brown bar). The graph was produced by combining data from at least three independent biological replicates. Error bars indicate the standard error of the mean. Asterisks depict the statistical significance of the observed differences in unpaired, two-tailed t-tests with P-values < 0.001 (**)
Fig. 4.
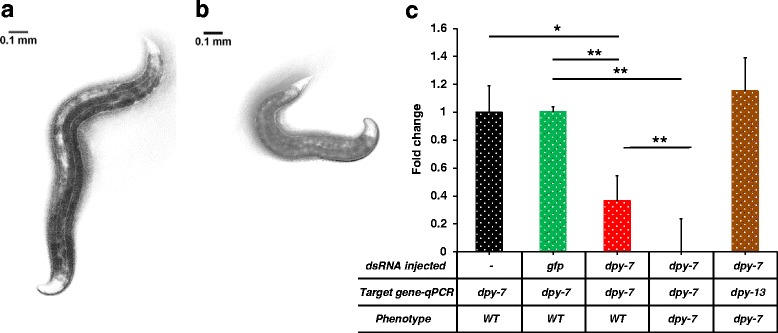
RNAi mediated phenotype and transcript changes in H. bacteriophora injected with dpy-7 dsRNA. Adult H. bacteriophora hermaphrodites that were injected with dpy-7 dsRNA produced progeny with dumpy phenotype. a. Phenotypically wild type progeny of H. bacteriophora worms injected with dpy-7 dsRNA. b. Progeny of H. bacteriophora worms injected with dpy-7 dsRNA exhibiting the dumpy phenotype. c. Expression of dpy-7 gene in the progeny of dpy-7 dsRNA injected worms. The y-axis represents the fold change in mRNA expression in the progeny of H. bacteriophora hermaphrodites. The mRNA levels are normalized to dpy-7 expression in the progeny of non-injected hermaphrodites (black bar). The green bar represents dpy-7 levels in progeny of gfp injected worms. The red bar represents dpy-7 levels in phenotypically wild type siblings, and the blue bar represents the dpy-7 transcript levels in phenotypically dumpy worms. To control for off-target effects, expression of an unrelated gene (dpy-13) in dumpy worms was determined (brown bar). The graph was produced by combining data from at least three independent biological replicates. Error bars indicate the standard error of the mean. Asterisks depict the statistical significance of the observed differences in unpaired, two-tailed t-tests with P-values < 0.01 (*) and < 0.001 (**)
We observed significant variability in phenotypic expression between the genes tested. The number of nematodes with the expected phenotype ranged from 2 to 60 %. Variation in the RNAi efficacy of knockdown has been documented previously in parasitic nematodes [38, 39, 62, 63], with generally greater success in plant pathogenic nematodes than in animal pathogenic nematodes. Since the discovery of RNAi, the technique has been successfully used to understand the function and interaction of most genes in C. elegans. The inability to reliably knockdown gene expression by RNAi in parasitic nematodes is perplexing as many of the genes involved are conserved, with few exceptions [41]. Sequencing of the genome revealed that all the major classes of genes required for RNAi are present in H. bacteriophora [44]. Further analysis on the molecular interaction of RNAi effectors in parasitic nematodes is essential to understand the variability in RNAi efficacy.
RNAi mediated knockdown was confirmed by examining the transcript levels of the targeted genes by qRT-PCR. Knockdown of the target gene transcripts was determined by comparing transcript levels in progeny of non-injected nematodes, nematodes injected with gfp dsRNA, and to siblings of injected worms that were phenotypically wild type. Off-target effects of RNAi were checked by quantifying the transcript level of a non-targeted gene in nematodes with RNAi phenotype. To normalize the transcription of the genes relative to other nematodes, ribosomal protein 32 (Hba-rpl-32) was used as an internal control. The efficacy of all the primers used was tested before checking the transcript levels in the nematodes.
In all four RNAi knockdowns, there was a significant reduction in the mRNA levels of targeted genes when compared to both wild type and gfp dsRNA transcript levels. Hba-cct-2 mRNA transcripts decreased 52 % compared to the wild type (unpaired two-tailed t-test, t(4) = 8.82976, P < 0.001,), and 46 % compared to the sibling of the affected nematode (unpaired two-tailed t-test, t(4) = 6.34638, P < 0.01) (Fig. 1c). We observed 58 % reduction in Hba-nol-5 transcripts when compared to wild type nematodes (unpaired two-tailed t-test, t(4) = 8.76645, P < 0.001), and 38 % reduction in the transcripts when compared to the siblings (unpaired two-tailed t-test, t(4) = 11.25743, P < 0.001) (Fig. 2c). There was a reduction in the Hba-nol-5 transcripts in the unaffected siblings compared to wild type but the reduction was not significant (unpaired two-tailed t-test, t(4) = 0.25366, P = 0.812). Contrary to cct-2 and nol-5, there was almost complete knockdown of the Hba-dpy-13 and Hb-dpy-7 transcripts in these nematodes. Hba-dpy-13 had 97 % reduction in transcript compared to wild type animals and also sibling animals (unpaired two-tailed t-test, t(4) = 11.37752, P < 0.001) (Fig. 3c), and Hba-dpy-7 had 99 % reduction in transcript compared to wild type animals and siblings (unpaired two-tailed t-test, t(4) = 20.28947, P < 0.001) (Fig. 4c). However, in the non-dumpy siblings of the dumpy nematodes there was also a 63 % reduction in Hba-dpy-7 transcripts (unpaired two-tailed t-test, t(4) = 4.7676, P < 0.01) (Fig. 4c). The variation in the reduction of mRNA transcripts by dsRNA knockdown was surprising. Considering the clear RNAi phenotypes observed for cct-2 and nol-5 targeted animals, we predicted concomitant decreases at the transcriptional level as measured by qRT-PCR. This discordance suggests that there is a threshold transcript level above which the nematodes appear phenotypically wild type, and that this level may vary significantly across different genes, thereby highlighting the importance of evaluating phenotype in addition to transcript levels when determining successful gene silencing by RNAi.
Conclusions
In this study we demonstrate that gonadal microinjection is a viable method of delivering dsRNA into H. bacteriophora to cause gene-specific silencing in the F1 generation. Injecting H. bacteriophora with dsRNA provides another strategy that researchers can deploy to generate F1 IJs with silenced genes. This provides a feasible approach to study different aspects of nematode parasitism including IJ recovery during infection, virulence factors used during infection, and the host response to parasitic infection.
Acknowledgements
The authors thank Dr. David Clarke of University College Cork, Ireland for providing the TT01 strain of H. bacteriophora. The project was supported by grant 5R21AI109517 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- dsRNA
double stranded RNA
- GFP
green fluorescent protein
- IJ
infective juvenile
- L3
third stage larva
- qRT-PCR
quantitative reverse transcriptase-polymerase chain reaction
- RNAi
RNA interference
Footnotes
Ioannis Eleftherianos Damien O’Halloran and John M. Hawdon co-senior authors.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RR participated in the study design, performed microinjections and qRT-PCR, and drafted the manuscript. JV designed primers for RNAi, synthesized ds RNA, and maintained the parasites. MK assisted with parasite husbandry. JMH, IE, and DO conceived of, designed, and coordinated the study, and edited the manuscript. All authors read and approved the final manuscript.
Contributor Information
Ramesh Ratnappan, rameshratnappan@email.gwu.edu.
Jonathan Vadnal, Email: jvadnal@gwu.edu.
Melissa Keaney, Email: mkeaney@gwu.edu.
Ioannis Eleftherianos, Email: ioannise@gwu.edu.
Damien O’Halloran, Email: damienoh@gwu.edu.
John M. Hawdon, Email: jhawdon@gwu.edu
References
- 1.Hotez PJ, Molyneux DH, Fenwick A, Ottesen E, Ehrlich Sachs S, Sachs JD. Incorporating a rapid-impact package for neglected tropical diseases with programs for HIV/AIDS, tuberculosis, and malaria. PLoS Med. 2006;3(5):e102. doi: 10.1371/journal.pmed.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotez PJ, Fenwick A, Savioli L, Molyneux DH. Rescuing the bottom billion through control of neglected tropical diseases. Lancet. 2009;373(9674):1570–5. doi: 10.1016/S0140-6736(09)60233-6. [DOI] [PubMed] [Google Scholar]
- 3.Stepek G, Buttle DJ, Duce IR, Behnke JM. Human gastrointestinal nematode infections: are new control methods required? Int J Exp Pathol. 2006;87(5):325–41. doi: 10.1111/j.1365-2613.2006.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pullan RL, Smith JL, Jasrasaria R, Brooker SJ. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasit Vectors. 2014;7:37. doi: 10.1186/1756-3305-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton J. Human gastrointestinal helminth infections: are they now neglected diseases? Trends Parasitol. 2003;19(11):527–31. doi: 10.1016/j.pt.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Bennett R, Ijpelaar J. Updated estimates of the costs associated with thirty four endemic livestock diseases in Great Britain: A note. J Agr Econ. 2005;56(1):135–44. doi: 10.1111/j.1477-9552.2005.tb00126.x. [DOI] [Google Scholar]
- 7.Sackett D, Holmes P, Abbott K, Jephcott S, Barber M. Assessing the economic cost of endemic disease on the profitability of Australian beef cattle and sheep producers. Meat Livestock Australia. 2006;364:1–33. [Google Scholar]
- 8.McLeod RS. Costs of major parasites to the Australian livestock industries. Int J Parasitol. 1995;25(11):1363–7. doi: 10.1016/0020-7519(95)00071-9. [DOI] [PubMed] [Google Scholar]
- 9.Olliaro P, Seiler J, Kuesel A, Horton J, Clark JN, Don R, Keiser J. Potential drug development candidates for human soil-transmitted helminthiases. PLoS Negl Trop Dis. 2011;5(6):e1138. doi: 10.1371/journal.pntd.0001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levecke B, Montresor A, Albonico M, Ame SM, Behnke JM, Bethony JM, Noumedem CD, Engels D, Guillard B, Kotze AC, et al. Assessment of anthelmintic efficacy of mebendazole in school children in six countries where soil-transmitted helminths are endemic. PLoS Negl Trop Dis. 2014;8(10):e3204. doi: 10.1371/journal.pntd.0003204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capron A, Dessaint JP. Molecular basis of host-parasite relationship: towards the definition of protective antigens. Immunol Rev. 1989;112:27–48. doi: 10.1111/j.1600-065X.1989.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 12.Eleftherianos I, ffrench-Constant RH, Clarke DJ, Dowling AJ, Reynolds SE. Dissecting the immune response to the entomopathogen Photorhabdus. Trends Microbiol. 2010;18(12):552–60. doi: 10.1016/j.tim.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Castillo JC, Reynolds SE, Eleftherianos I. Insect immune responses to nematode parasites. Trends Parasitol. 2011;27(12):537–47. doi: 10.1016/j.pt.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 15.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahringer J, ed. Reverse genetics. The C. elegans Research Community, WormBook. WormBook; 2006. doi:10.1895/wormbook.1.47.1, http://www.wormbook.org.
- 17.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–21. doi: 10.1016/S1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 18.Lilley CJ, Davies LJ, Urwin PE. RNA interference in plant parasitic nematodes: a summary of the current status. Parasitology. 2012;139(5):630–40. doi: 10.1017/S0031182011002071. [DOI] [PubMed] [Google Scholar]
- 19.Rosso MN, Jones JT, Abad P. RNAi and functional genomics in plant parasitic nematodes. Annu Rev Phytopathol. 2009;47:207–32. doi: 10.1146/annurev.phyto.112408.132605. [DOI] [PubMed] [Google Scholar]
- 20.Selkirk ME, Huang SC, Knox DP, Britton C. The development of RNA interference (RNAi) in gastrointestinal nematodes. Parasitology. 2012;139(5):605–12. doi: 10.1017/S0031182011002332. [DOI] [PubMed] [Google Scholar]
- 21.Hussein AS, Kichenin K, Selkirk ME. Suppression of secreted acetylcholinesterase expression in Nippostrongylus brasiliensis by RNA interference. Mol Biochem Parasitol. 2002;122(1):91–4. doi: 10.1016/S0166-6851(02)00068-3. [DOI] [PubMed] [Google Scholar]
- 22.Winter AD, McCormack G, Myllyharju J, Page AP. Prolyl 4-hydroxlase activity is essential for development and cuticle formation in the human infective parasitic nematode Brugia malayi. J Biol Chem. 2013;288(3):1750–61. doi: 10.1074/jbc.M112.397604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song C, Gallup JM, Day TA, Bartholomay LC, Kimber MJ. Development of an in vivo RNAi protocol to investigate gene function in the filarial nematode, Brugia malayi. PLoS Pathog. 2010;6(12):e1001239. doi: 10.1371/journal.ppat.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lustigman S, Zhang J, Liu J, Oksov Y, Hashmi S. RNA interference targeting cathepsin L and Z-like cysteine proteases of Onchocerca volvulus confirmed their essential function during L3 molting. Mol Biochem Parasitol. 2004;138(2):165–70. doi: 10.1016/j.molbiopara.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Pfarr K, Heider U, Hoerauf A. RNAi mediated silencing of actin expression in adult Litomosoides sigmodontis is specific, persistent and results in a phenotype. Int J Parasitol. 2006;36(6):661–9. doi: 10.1016/j.ijpara.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 26.McCoy CJ, Warnock ND, Atkinson LE, Atcheson E, Martin RJ, Robertson AP, Maule AG, Marks NJ, Mousley A. RNA interference in adult Ascaris suum - an opportunity for the development of a functional genomics platform that supports organism-, tissue- and cell-based biology in a nematode parasite. Int J Parasitol. 2015;45(11):673–8. doi: 10.1016/j.ijpara.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen N, Xu MJ, Nisbet AJ, Huang CQ, Lin RQ, Yuan ZG, Song HQ, Zhu XQ. Ascaris suum: RNAi mediated silencing of enolase gene expression in infective larvae. Exp Parasitol. 2011;127(1):142–6. doi: 10.1016/j.exppara.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 28.Issa Z, Grant WN, Stasiuk S, Shoemaker CB. Development of methods for RNA interference in the sheep gastrointestinal parasite, Trichostrongylus colubriformis. Int J Parasitol. 2005;35(9):935–40. doi: 10.1016/j.ijpara.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Geldhof P, Murray L, Couthier A, Gilleard JS, McLauchlan G, Knox DP, Britton C. Testing the efficacy of RNA interference in Haemonchus contortus. Int J Parasitol. 2006;36(7):801–10. doi: 10.1016/j.ijpara.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Kotze AC, Bagnall NH. RNA interference in Haemonchus contortus: Suppression of beta-tubulin gene expression in L3, L4 and adult worms in vitro. Mol Biochem Parasitol. 2006;145(1):101–10. doi: 10.1016/j.molbiopara.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Zawadzki JL, Kotze AC, Fritz JA, Johnson NM, Hemsworth JE, Hines BM, Behm CA. Silencing of essential genes by RNA interference in Haemonchus contortus. Parasitology. 2012;139(5):613–29. doi: 10.1017/S0031182012000121. [DOI] [PubMed] [Google Scholar]
- 32.Visser A, Geldhof P, de Maere V, Knox DP, Vercruysse J, Claerebout E. Efficacy and specificity of RNA interference in larval life-stages of Ostertagia ostertagi. Parasitol. 2006;133(Pt 6):777–83. doi: 10.1017/S0031182006001004. [DOI] [PubMed] [Google Scholar]
- 33.Tzelos T, Matthews JB, Whitelaw B, Knox DP. Marker genes for activation of the RNA interference (RNAi) pathway in the free-living nematode Caenorhabditis elegans and RNAi development in the ovine nematode Teladorsagia circumcincta. J Helminthol. 2015;89(2):208–16. doi: 10.1017/S0022149X13000801. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Yang Y, Yang J, Zhang Z, Zhu X. RNAi-mediated silencing of paramyosin expression in Trichinella spiralis results in impaired viability of the parasite. PLoS One. 2012;7(11):e49913. doi: 10.1371/journal.pone.0049913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lendner M, Doligalska M, Lucius R, Hartmann S. Attempts to establish RNA interference in the parasitic nematode Heligmosomoides polygyrus. Mol Biochem Parasitol. 2008;161(1):21–31. doi: 10.1016/j.molbiopara.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Ciche TA, Sternberg PW. Postembryonic RNAi in Heterorhabditis bacteriophora: a nematode insect parasite and host for insect pathogenic symbionts. BMC Dev Biol. 2007;7:101. doi: 10.1186/1471-213X-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moshayov A, Koltai H, Glazer I. Molecular characterisation of the recovery process in the entomopathogenic nematode Heterorhabditis bacteriophora. Int J Parasitol. 2013;43(10):843–52. doi: 10.1016/j.ijpara.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Britton C, Samarasinghe B, Knox DP. Ups and downs of RNA interference in parasitic nematodes. Exp Parasitol. 2012;132(1):56–61. doi: 10.1016/j.exppara.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Dalzell JJ, Warnock ND, McVeigh P, Marks NJ, Mousley A, Atkinson L, Maule AG. Considering RNAi experimental design in parasitic helminths. Parasitology. 2012;139(5):589–604. doi: 10.1017/S0031182011001946. [DOI] [PubMed] [Google Scholar]
- 40.Samarasinghe B, Knox DP, Britton C. Factors affecting susceptibility to RNA interference in Haemonchus contortus and in vivo silencing of an H11 aminopeptidase gene. Int J Parasitol. 2011;41(1):51–9. doi: 10.1016/j.ijpara.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 41.Dalzell JJ, McVeigh P, Warnock ND, Mitreva M, Bird DM, Abad P, Fleming CC, Day TA, Mousley A, Marks NJ, et al. RNAi effector diversity in nematodes. PLoS Negl Trop Dis. 2011;5(6):e1176. doi: 10.1371/journal.pntd.0001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poinar GO. Description and biology of a new insect parasitic Rhabditoid, Heterorhabditis bacteriophora N. Gen., N. Sp. (Rhabditida; Heterorhabditidae N. Fam.) Nematologica. 1975;21(4):463–70. doi: 10.1163/187529275X00239. [DOI] [Google Scholar]
- 43.Kiontke K, Fitch DHA. The Phylogenetic relationships of Caenorhabditis and other rhabditids. The C. elegans Research Community, WormBook. WormBook; 2005. doi:10.1895/wormbook.1.11.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 44.Bai X, Adams BJ, Ciche TA, Clifton S, Gaugler R, Kim KS, Spieth J, Sternberg PW, Wilson RK, Grewal PS. A lover and a fighter: the genome sequence of an entomopathogenic nematode Heterorhabditis bacteriophora. PLoS One. 2013;8(7):e69618. doi: 10.1371/journal.pone.0069618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciche TA. Ensign JC: “For the insect pathogen Photorhabdus luminescens, which end of a nematode is out?Appl”. Environ Microbiol. 2003;69(4):1890–7. doi: 10.1128/AEM.69.4.1890-1897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strauch O, Stoessel S, Ehlers RU. Culture conditions define automictic or amphimictic reproduction in entomopathogenic rhabditid nematodes of the genus Heterorhabditis. Fundam Applied Nematol. 1994;17(6):575–82. [Google Scholar]
- 47.Noguez JH, Conner ES, Zhou Y, Ciche TA, Ragains JR, Butcher RA. A novel ascaroside controls the parasitic life cycle of the entomopathogenic nematode Heterorhabditis bacteriophora. ACS Chem Biol. 2012;7(6):961–6. doi: 10.1021/cb300056q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ciche T. The biology and genome of Heterorhabditis bacteriophora. The C. elegans Research Community, WormBook. WormBook; 2007. doi:10.1895/wormbook.1.47.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 49.Cinkornpumin JK, Hong RL. RNAi mediated gene knockdown and transgenesis by microinjection in the necromenic Nematode Pristionchus pacificus. J Vis Exp. 2011;56:e3270. doi: 10.3791/3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White GF. A Method for obtaining infective nematode larvae from cultures. Science. 1927;66(1709):302–3. doi: 10.1126/science.66.1709.302-a. [DOI] [PubMed] [Google Scholar]
- 51.Lindegren JE, Valero KA, Mackey BE. Simple in vivo production and storage methods for Steinernema carpocapsae infective juveniles. J Nematol. 1993;25(2):193–7. [PMC free article] [PubMed] [Google Scholar]
- 52.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 53.Evans TC, ed. Transformation and microinjection. The C. elegans Research Community, WormBook. WormBook; 2006. doi:10.1895/wormbook.1.108.1, http://www.wormbook.org.
- 54.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 55.Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69(6):1043–50. doi: 10.1016/0092-8674(92)90622-J. [DOI] [PubMed] [Google Scholar]
- 56.Yaffe MB, Farr GW, Miklos D, Horwich AL, Sternlicht ML, Sternlicht H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358(6383):245–8. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]
- 57.Won KA, Schumacher RJ, Farr GW, Horwich AL, Reed SI. Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol Cell Biol. 1998;18(12):7584–9. doi: 10.1128/MCB.18.12.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1(1):E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piano F, Schetter AJ, Morton DG, Gunsalus KC, Reinke V, Kim SK, Kemphues KJ. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr Biol. 2002;12(22):1959–64. doi: 10.1016/S0960-9822(02)01301-5. [DOI] [PubMed] [Google Scholar]
- 60.von Mende N, Bird DM, Albert PS, Riddle DL. dpy-13: a nematode collagen gene that affects body shape. Cell. 1988;55(4):567–76. doi: 10.1016/0092-8674(88)90215-2. [DOI] [PubMed] [Google Scholar]
- 61.Johnstone IL, Shafi Y, Barry JD. Molecular analysis of mutations in the Caenorhabditis elegans collagen gene dpy-7. Embo J. 1992;11(11):3857–63. doi: 10.1002/j.1460-2075.1992.tb05478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maule AG, McVeigh P, Dalzell JJ, Atkinson L, Mousley A, Marks NJ. An eye on RNAi in nematode parasites. Trends Parasitol. 2011;27(11):505–13. doi: 10.1016/j.pt.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 63.Geldhof P, Visser A, Clark D, Saunders G, Britton C, Gilleard J, Berriman M, Knox D. RNA interference in parasitic helminths: current situation, potential pitfalls and future prospects. Parasitology. 2007;134(Pt 5):609–19. doi: 10.1017/S0031182006002071. [DOI] [PubMed] [Google Scholar]


