Abstract
Rapsyn is a synapse-specific protein that is required for clustering acetylcholine receptors at the neuromuscular junction. Analysis of the rapsyn promoter revealed a consensus site for the transcription factor Kaiso within a region that is mutated in a subset of patients with congenital myasthenic syndrome. Kaiso is a POZ-zinc finger family transcription factor which recognizes the specific core consensus sequence CTGCNA (where N is any nucleotide). Previously, the only known binding partner for Kaiso was the cell adhesion cofactor, p120 catenin. Here we show that δ-catenin, a brain-specific member of the p120 catenin subfamily, forms a complex with Kaiso. Antibodies against Kaiso and δ-catenin recognize proteins in the nuclei of C2C12 myocytes and at the postsynaptic domain of the mouse neuromuscular junction. Endogenous Kaiso in C2C12 cells coprecipitates with the rapsyn promoter in vivo as shown by chromatin immunoprecipitation assay. Minimal promoter assays demonstrated that the rapsyn promoter can be activated by Kaiso and δ-catenin; this activation is apparently muscle specific. These results provide the first experimental evidence that rapsyn is a direct sequence-specific target of Kaiso and δ-catenin. We propose a new model of synapse-specific transcription that involves the interaction of Kaiso, δ-catenin, and myogenic transcription factors at the neuromuscular junction.
Rapsyn (receptor associated protein of the synapse) is a 43-kDa postsynaptic protein that is essential in the proper functioning of the neuromuscular junction. It is a critical effector of acetylcholine receptor (AChR) clustering upon neural agrin signaling; no AChR clusters form in muscles of rapsyn-deficient mutant mice even following treatment with agrin (17). Messenger RNAs that encode rapsyn are highly concentrated in the subsynaptic region of skeletal muscle (28). These results strongly suggest that there is a mechanism to control rapsyn expression in subsynaptic nuclei.
According to one model of synapse-specific transcription, the six-base-pair element CCGGAA, termed the N box, is required for regulating transcription in subsynaptic nuclei. This motif confers synapse-specific transcription of AChR γ and AChR ɛ subunits, utrophin, and acetylcholine esterase genes (11, 25). N-box-dependent synaptic expression is stimulated by agrin and neuregulin, which triggers the mitogen-activated protein kinase and Jun N-terminal kinase signaling pathways to ultimately allow activation by the N-box binding Ets transcription factor, GABP (reviewed in reference 41). However, the level of some synaptic genes, including rapsyn, was not perturbed in the muscles of mutant mice expressing a skeletal muscle-targeted, general Ets dominant-negative mutant (9). This suggests that rapsyn expression is controlled by a mechanism that does not involve the Ets transcription factor and N box and that other synapse-specific mechanisms are likely to control the expression of rapsyn.
Recent observations of congenital myasthenic syndromes (CMS) that result from genetic defects in endplate-specific presynaptic, synaptic, or postsynaptic proteins revealed the significance of rapsyn gene regulation. Rapsyn mutations were identified in a subset of patients with endplate AChR deficiencies (12, 29, 32, 33, 38). Furthermore, two novel E-box mutations in the rapsyn promoter region have been recently reported in eight patients with CMS (33). These results focused our attention to a specific region of the rapsyn promoter. Sequence analysis of this region revealed two consensus Kaiso binding sites (8). One site partially overlaps with a previously identified E-box motif, and interestingly a mutation within this E-box-Kaiso site was identified in a subset of patients with CMS (32, 33). Collectively, these observations implicate Kaiso as a key regulator of rapsyn transcription.
Kaiso is a ubiquitously expressed new member of the POZ-zinc finger family of transcription factors and was identified as a specific binding partner for p120 catenin (6). Kaiso has been shown to mediate transcriptional repression at methylated loci (36, 45). In addition, Daniel et al. (8) have shown Kaiso to be a dual-specificity DNA-binding protein that recognizes the minimal core sequence CTGCNA (where N is any nucleotide) in addition to the methyl-CpG dinucleotides. However, in electrophoretic mobility shift assays (EMSAs), Kaiso has a higher affinity for the consensus binding site than for the methyl-CpG sites (8). In addition, Kaiso target gene recognition is apparently regulated by interactions with members of the p120 catenin subfamily. To support this notion, the interaction of Kaiso with either the sequence-specific binding site or the methyl-CpG sites, as well as Kaiso-mediated transcriptional repression via the Kaiso binding site, was indeed inhibited by p120 catenin (8, 21).
Notably, the p120 subfamily member δ-catenin (or neural plakophilin-related arm-repeat protein) is specifically expressed in the nervous system (26, 31), where it is thought to partake in neuronal signaling pathways (20, 22, 26). Since the neuromuscular junction is a model synapse and since many of the mechanisms that function at the neuromuscular junction are similar to those in the central nervous system (CNS), it is feasible that δ-catenin also functions at the neuromuscular junction. The possibility therefore exists that Kaiso and δ-catenin partake in a signaling pathway at the neuromuscular junction.
Here we report that the rapsyn promoter is a transcriptional target of Kaiso and δ-catenin in mouse C2C12 myotubes and chicken primary myotubes. Minimal promoter assays showed that the rapsyn promoter can be activated by Kaiso and δ-catenin and that this activation is muscle specific. Site-specific mutation of the rapsyn promoter, which mimics the mutation found in human CMS, resulted in a significant reduction in Kaiso-mediated activation of the rapsyn promoter. This result may explain the reduced levels of rapsyn expression at the endplates of CMS patients who carry this mutation. We demonstrate via the chromatin immunoprecipitation (ChIP) assay that endogenous Kaiso in C2C12 cells binds the rapsyn promoter. Furthermore, antibodies to δ-catenin and Kaiso recognize proteins at the mouse neuromuscular junction and in the nuclei of C2C12 myotubes. We also provide the first experimental evidence that δ-catenin, a brain-specific member of the p120 catenin subfamily, forms a complex with Kaiso in vivo. Collectively, these results demonstrate that Kaiso, δ-catenin, and myogenic transcription factors are components of a novel pathway that regulates synapse-specific transcription of rapsyn.
MATERIALS AND METHODS
Constructs.
The human and mouse rapsyn promoter-reporter constructs were generated by cloning promoters in the forward orientation into BglII/HindIII sites of promoterless pGL3 Basic vector (Promega). Both promoters were amplified by PCR from human genomic DNA; appropriate restriction sites were incorporated in the primers. The primer pair to amplify human promoter consists of (forward) 5′-CCCAGATCTTAGTGGCAATTAATGTGCATC and (reverse) 5′-CCCAAGCTTGCCCTGTGTCCCACGTGG; the primers for the mouse promoter are (forward) 5′-CCCAGATCTGGGGGAATGGGGTGGAAT and (reverse) 5′-CCCAAGCTTATCTCTTTGTAGCGGCCCATC. Three nested deletion constructs of human rapsyn promoter were also generated by PCR by using primers with an incorporated BglII site as follows: 5′-CCCAGATCTGGAAGAAGCAGGGCTGGGCG for deletion 1, 5′-CCCAGATCTGACGGGCTGAACCAGCTTTG for deletion 2, and 5′-CCCAGATCTCTCGCGGGTGGGTGCAGCAGAG for deletion 3. Two more deletion constructs were made by using unique KpnI and ApaI restriction sites. All the constructs were verified by sequencing. Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene).
Full-length mouse δ-catenin in plasmid pcDNA4/His was kindly supplied by K. S. Kosik, Harvard Medical School, Boston, Mass. The pcDNA3-Kaiso expression plasmids are as described by Daniel and Reynolds (6).
Cell lines and transient transfections.
Human embryonic kidney 293T cells and mouse C2C12 myoblasts and myotubes were obtained from the American Type Culture Collection. The 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 4.5 g of glucose per liter and 10% heat-inactivated fetal bovine serum (FBS).
Mouse C2C12 cells were cultured in DMEM supplemented with 10% FBS. Myoblasts were allowed to fuse into multinucleated cells for 2 days, and then FBS was replaced with 10% horse serum to promote myotube formation. The myoblasts were transfected at about 70% of confluence, and luciferase activity was measured in 48 h.
Chicken myotube cultures were prepared from the hind limb of White Leghorn chicken embryos on day 11 by the method of Fischbach (16) with minor modifications (42). Myotubes were cultured in DMEM supplemented with 10% (vol/vol) horse serum, 2% chicken embryo extract (42), 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 1 mM l-glutamine. Cultures were treated on day 3 with 10−5 M cytosine arabinoside for 24 h to reduce the number of fibroblasts. Transient transfections were performed on 4- to 6-day-old cultures.
All transient transfections are performed by the calcium phosphate method as described by Rodova et al. (39). A β-galactosidase expression plasmid is included in each transfection to monitor transfection efficiency. After 48 h the cells are lysed, and activities are determined by using enzyme assay kits for luciferase (Promega) and β-galactosidase (Promega). Relative light units are the luciferase units normalized to β-galactosidase. All experiments are carried out in triplicate and repeated at least three times.
Immunoprecipitation and immunoblotting.
C2C12 myotubes were scraped in M-PER mammalian protein extraction reagent (Pierce) supplemented with protease inhibitor mixture (Roche Applied Science). Cell lysates were centrifuged (at 15,000 × g for 20 min at 4°C), and the resulting clarified supernatants were collected. For the mouse brain samples, mice were killed and brains removed and rinsed in ice-cold phosphate-buffered saline (PBS). The washed brain tissues were homogenized in M-PER with protease inhibitors and spun at 15,000 × g for 20 min at 4°C, and the clarified lysates were used. Immunoprecipitations were performed with G-protein beads (Pierce) according to the manufacturer's instructions. After being boiled for 5 min, immunoprecipitates were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and electrotransferred onto nitrocellulose membranes. The detection of individual proteins was carried out by immunoblotting with anti-δ-catenin (C-20; Santa-Cruz) or polyclonal Kaiso (6) antibodies and visualized by enhanced chemiluminescence.
EMSAs.
EMSAs were performed as previously described (39). The following oligonucleotides were synthesized (core consensus Kaiso sites are italicized): wild-type CTGTCGCTGCCAGTTGGGGCC from human promoter at position −45; mutant Kaiso site CTGTCGCTAACAGCTGGGGCC; and wild-type with a mutation of A to G (T to C in reversed strand) CTGTCGCTGCCAGCTGGGGCC. An aliquot of 10 pM double-stranded oligonucleotides was end labeled with polynucleotide kinase (Promega) and [γ-32P]ATP for 45 min at 37°C. The labeled probe was purified by centrifugation through a Clontech Chroma-spin TE-10 column according to the manufacturer's instructions.
pGEX-Kaiso construct-transformed Escherichia coli DH5α cultures were grown to isolate glutathione transferase (GST) fusion proteins by using glutathione-Sepharose beads as described by Daniel et al. (8). The proteins were subjected to SDS-polyacrylamide gel electrophoresis, followed by Coomassie blue staining to check purity and concentration. Protein concentration was determined by the bicinchoninic acid assay (Pierce). Binding reactions containing 50,000 cpm of labeled DNA were incubated for 10 min at room temperature, followed by a 30-min incubation on ice with 300 ng of fusion protein or 5 μg of nuclear extract and 160 ng of poly(dI · dC) in 25 mM HEPES, 100 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 1 mM EDTA, 0.1% NP-40, and 5% (vol/vol) glycerol. The binding buffer for fusion protein GST-zinc finger 2 and 3 (GST-ZF2,3) did not contain NP-40. DNA-protein complexes were electrophoresed in 4% native polyacrylamide gels and visualized by autoradiography.
ChIP.
ChIP assays were performed as previously described (43). In brief, C2C12 murine myoblast cells were washed in PBS solution and incubated for 1 h with the cross-linking solution containing 1% formaldehyde. The cross-linking reaction was then stopped with glycine at a concentration of 0.125 M. The cells were washed twice with cold 1× PBS and harvested into tubes in PBS-phenylmethylsulfonyl fluoride. The cells were pelleted at 3,000 to 4,000 rpm at 4°C for 5 min and resuspended in cell lysis solution [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 8), 85 mM KCl, 0.5% NP-40]. Cross-linked material was sonicated, and the chromatin was broken into fragments of an average length of ∼500 to 600 bp. The sonicated samples were centrifuged at 14,000 rpm (Eppendorf) for 10 min at 4°C, and the supernatants were precleared with equilibrated protein A-Sepharose. Lysates were incubated overnight with 4 μg of each antibody at 4°C. The immune complexes were captured by rabbit anti-mouse antibody-conjugated protein A-Sepharose beads and subsequently washed three times in immunoprecipitation wash buffer (100 mM Tris-Cl [pH 8], 500 mM LiCl, 1% NP-40, 1% deoxycholic acid). The antibody-antigen-promoter complexes were eluted by adding 35 μl of immunoprecipitation elution buffer (50 mM NaHCO3, 1% SDS) to each sample, followed by light vortexing for 30 min. Each sample was then treated with RNase A at 67°C for 5 h to remove RNA and to reverse formaldehyde cross-links, followed by ethanol precipitation. The precipitated products were treated with proteinase K at 45°C for 2 h, followed by two rounds of phenol-chloroform extraction and ethanol precipitation. The DNA pellets were resuspended in 10 to 20 μl of distilled H2O, and one-fifth of this volume was used for PCR analysis. Ten percent input C2C12 genomic DNA (untreated with antibody) was used as a positive control in the PCR. The following primers specific for a region flanking the two Kaiso binding sites within the murine rapsyn promoter were used for PCR: 5′-GCCTTTGACAAGGCTTCCAAAGAGCAT-3′ (forward) and 5′-GGCTGGTTCAGCCCCTGTCTGCAGCTG-3′ (reverse).
Immunofluorescence and antibodies.
The cells prepared for fluorescence staining were grown on 35-mm dishes. They were rinsed twice with PBS and fixed with ice-cold 100% methanol for 15 min at −20°C. Immunostaining was performed as described previously (39). Confocal microscopy was performed with a Zeiss LSM 510 microscope equipped with Plan-Neofluar oil objectives (either 25×, 0.8 numerical aperture, or 40×, 1.3 numerical aperture). The images were obtained by using 1,024 by 1,024 pixel density, excitation at 488 nm, and emission filter BP 505-550.
Frozen mouse gastrocnemius muscle sections were fixed with 1% paraformaldehyde. The neuromuscular junctions were stained with 6 × 10 −8 M rhodamine-conjugated α-bungarotoxin (Molecular Probes) and immunostained with either δ-catenin C-20 antibodies or with affinity-purified rabbit polyclonal antibodies to Kaiso, followed by a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (Jackson Laboratories). The sections were examined with a Zeiss Axioskop photomicroscope equipped with a 63× oil objective, 1.4 numerical aperture, in antifade medium (80% glycerol, 20% 0.5 M phosphate buffer [pH 8.0], and 0.2% N-propyl gallate). Images were captured at room temperature by using an Optronics Magnifier CCD camera with the Magnifier image acquisition software. The images were processed by using Adobe Photoshop software.
The following antibodies were used: two monoclonal (6F and 11D) and rabbit polyclonal antibodies against Kaiso (described in reference 7), two polyclonal antibodies against δ-catenin (K-20 and C-20; Santa Cruz), and polyclonal antibodies against p120 (m-19; Santa Cruz). Secondary FITC-conjugated anti-rabbit immunoglobulin G (IgG), anti-mouse IgG, and anti-goat IgG were purchased from Jackson Laboratories.
RESULTS
The core rapsyn promoter contains a conserved Kaiso site and a cluster of E boxes.
Comparison of the 5′ ends of genes of evolutionarily distant organisms may reveal potential regulatory promoter elements that are conserved, implying a functional significance. Figure 1A shows a sequence comparison between mouse and human rapsyn promoters, highlighting regions of strong similarity. The transcriptional start site was determined by Ohno et al. (33) for the human rapsyn promoter (Fig. 1A). Sequence analysis revealed the absence of TATA and CAAT motifs. However, promoter activity can rely on a conserved consensus initiator element which overlaps the transcription start site (19). An E-box element is defined by the nucleotides CANNTG and is involved in regulating a number of muscle-specific genes (44). A cluster of three consensus E boxes is conserved between the mouse and human rapsyn promoters. Interestingly, one of these E boxes overlaps with a consensus binding site for the newly discovered transcription factor Kaiso, which encodes an N-terminal protein-protein interaction POZ domain and three C-terminal Kruppel-like C2H2 zinc fingers (6). DNA-binding analysis of Kaiso revealed that the specific core sequence, CTGCNA, is sufficient for Kaiso-DNA interactions (8). We identified two conservative Kaiso core consensus sites in the rapsyn promoter (hereafter called Kaiso 1 and Kaiso 2). Kaiso 1 carries a reverse complementary sequence of CTGCNA and partially overlaps with the second conserved E box at the −36 position (Fig. 1A). These conserved Kaiso sites are in close proximity to the E boxes, which led us to hypothesize that Kaiso is involved in rapsyn transcriptional regulation.
FIG. 1.
Rapsyn promoter has high activity in myotubes and contains a conserved cluster of E boxes and Kaiso binding sites. (A) Sequence comparison of the mouse and human promoter regions. The E boxes and Kaiso core sequences are boxed. The location of the major transcription initiation site in the human promoter is indicated by +1. Asterisks mark identical sequences in both mouse and human promoters. Arrows show the starts of nested deletions in the luciferase fusion constructs of the human promoter. A double mutation (GC to TT) was introduced at the Kaiso 1 site in the −372m and −110m constructs and is shown above the Kaiso 1 site in lowercase font. A mutation of A to G was introduced in the −110 construct to make the −110G construct and is shown above the −40 E box. (B) Deletion mapping of the human rapsyn promoter shows its high level of transcriptional activity in primary chicken myotubes. The bars on the left represent deletion constructs that were used as a reporter in the transient transfection assay shown on the right. The histogram on the right shows the mean of triplicate samples ± standard deviations. Each experiment was repeated at least three separate times with similar results. Reporter activity is plotted as relative light units (RLU); relative light units are the luciferase units normalized to β-galactosidase. The core rapsyn promoter region between positions −17 and −110 maintains a high level of promoter activity in myotubes and contains four E boxes, one of which overlaps with the Kaiso site.
To test rapsyn promoter activity, we first amplified the human rapsyn promoter from human genomic DNA. Nested deletions were made and fused with luciferase in the pGL3Basic vector. The reporters obtained were then tested for promoter activity in transient transfection assays in human embryonic kidney 293T cells, chicken primary myotubes, and mouse C2C12 myotubes. In HEK 293T cells, all the reporters had negligible promoter activity (data not shown). In contrast, high levels of transcriptional activity of all constructs were found in chicken primary myotubes, as shown in Fig. 1B, as well as in mouse C2C12 myotubes (data not shown). These data indicate a strong muscle-specific component in rapsyn transcription regulation. The minimal promoter sequence which retained a high level of activity was the construct beginning at position −110 (−110 construct), and this construct contains a cluster of three conserved E boxes and a proximal Kaiso binding site. The promoter activity of the −17 construct was minimal and similar to that of the promoterless GL3 plasmid. These results delineate a core promoter activity in the −110 to −17 region of the rapsyn promoter which contains four E boxes and a conserved Kaiso site.
Activation of the rapsyn promoter by Kaiso and δ-catenin.
To ascertain whether Kaiso is capable of regulating transcription via the rapsyn promoter, we cotransfected the promoter-reporter constructs and a Kaiso expression vector. Cotransfection of the Kaiso expression vector into primary chicken myotubes resulted in a dramatic increase in the activity of both the −215 and the −110 rapsyn promoter-reporter constructs, while no increase was observed in the −17 construct which lacks any Kaiso consensus binding sites (Fig. 2 A, B, and C). Previously, the only reported Kaiso binding partner was the ubiquitously expressed p120 catenin (6). δ-Catenin is a member of the p120 catenin subfamily and is enriched in the CNS (18). We demonstrate that δ-catenin complexes with Kaiso (see below). To determine if δ-catenin is involved in rapsyn transcription regulation, we cotransfected a δ-catenin expression construct with our rapsyn promoter-reporter constructs into chicken primary myotubes. Similar to the results obtained for Kaiso, δ-catenin induced rapsyn promoter reporter activity and, like Kaiso, δ-catenin had no effect on the −17 construct that lacks any Kaiso binding sites (Fig. 2A, B, and C). This suggests that the proximal Kaiso site which overlaps the E box is functional and is involved in Kaiso- and δ-catenin-mediated activation of the rapsyn promoter. Transfection of primary chicken myotubes with a mouse rapsyn promoter-reporter construct which contained the Kaiso sites was also activated by cotransfection with Kaiso or δ-catenin expression constructs (Fig. 2D). Similar results were obtained with mouse C2C12 myotubes (data not shown). However, we failed to detect any induction when these constructs were cotransfected into nonmuscle 293T cells (data not shown). Since myogenic transcription factors are highly expressed in myotubes, these results suggest the involvement of such factors in Kaiso- and δ-catenin-mediated activation of the rapsyn promoter.
FIG. 2.
Minimal promoter experiments show that the rapsyn promoter can be induced by Kaiso and δ-catenin in primary chicken myotubes. Three micrograms of effector expression plasmid were cotransfected with promoter-reporter constructs; this was replaced by an equal amount of pcDNA3 in the controls. Each bar represents the average of triplicate samples ± standard deviations. Each experiment was repeated at least three separate times with similar results. (A) The −215 construct which contains two Kaiso sites and five E boxes can be induced with the indicated effectors. (B) The −110 construct which contains only one Kaiso site and three conservative E boxes still can be induced with the same effectors. This indicates that the proximal Kaiso site in the −110 construct is functional. (C) There is no induction of the −17 construct which lacks Kaiso and E-box sites. (D) The 1,024-bp mouse rapsyn promoter contains two conserved Kaiso sites and several E boxes. Similar to the human promoter, it can be induced by Kaiso and δ-catenin. (E) The Kaiso 1 site at position −36 was mutated (TGGCAG to TGTTAG) in the −372m construct (white bars) and in the −110m construct (gray bars). The E box which overlaps the Kaiso 1 site at position −40 was also mutated (CAACTG to CAGCTG). This is the same mutation of A to G at the −38 position which is reported in patients with myasthenic syndrome (34). Mutation at either location affects Kaiso and δ-catenin induction of the rapsyn promoter.
To determine whether the response to Kaiso and δ-catenin depends specifically on the Kaiso binding site, we tested the activities of the constructs beginning at positions −372 and −110 and containing a mutation (CTGCCA to CTAACA) in the Kaiso binding site (−372m construct and −110m construct, respectively). Constructs that contain this mutation could not be induced by any Kaiso or δ-catenin effector plasmids (Fig. 2E). This confirmed the involvement of the Kaiso binding site in the induction by Kaiso and δ-catenin. We also tested a construct with a mutation of A to G at position −38 (−38A to G) in the overlapping Kaiso-E-box site; this is the same rapsyn promoter mutation reported in a subset of CMS patients (33). This reporter construct was not induced by Kaiso or δ-catenin effector expression (Fig. 2E). Altogether, these results support the hypothesis that Kaiso and δ-catenin are involved in rapsyn promoter regulation.
Rapsyn-derived promoter sequences bind Kaiso in vitro and in vivo.
To determine whether the rapsyn-derived probe harboring the Kaiso binding site is capable of binding Kaiso in vitro, we performed EMSAs with a synthetic rapsyn-derived double-stranded oligonucleotide (CTGTCGCTGCCAGTTGGGGCC). This oligonucleotide contains the core consensus Kaiso binding motif (italicized). Since Kaiso contains a POZ domain that inhibits but does not abolish DNA binding of POZ-zinc finger transcription factors (3), we used a truncated Kaiso-GST fusion protein containing only Kaiso's three zinc fingers (GST-ZF1,2,3) (Fig. 3A). Previously, Kaiso zinc fingers 2 and 3 were shown to be necessary and sufficient for binding to human and murine matrilysin promoter-derived probes (8). We therefore also tested GST fusion proteins containing only Kaiso zinc fingers 2 and 3 (GST-ZF2,3) (Fig. 3B). The rapsyn-derived probe was able to form a DNA-protein complex with both proteins, although GST-ZF1,2,3 bound much more efficiently than GST-ZF2,3. Both wild-type rapsyn and matrilysin-derived cold competitors (100-fold excess) out-competed the probe, confirming the specificity of DNA binding. A probe containing a mutation within the Kaiso binding site, previously shown to inhibit Kaiso-DNA binding (8) and shown in this study to be crucial for Kaiso and δ-catenin activation of the rapsyn promoter (Fig. 2E), displayed significantly reduced binding to GST-ZF1,2,3 and did not bind GST-ZF2,3. A probe that contains the mutation −38A to G in the E-box site had slightly reduced GST-ZF1,2,3 binding and no binding to GST-ZF2,3. These results suggest that Kaiso can bind the rapsyn promoter and that rapsyn may be a direct Kaiso target in C2C12 myotubes.
FIG. 3.
Kaiso specifically binds a rapsyn-derived probe with a Kaiso core binding site by EMSA. Radiolabeled CTGTCGCTGCCAGTTGGGGCC oligonucleotide (Kaiso binding motif in italics) was used as a wild-type (wt) probe. The following mutated probes carrying the same mutations that abolish Kaiso induction of the rapsyn promoter in transient transfection assays were also used: CTGTCGCTAACAGTTGGGGCC (mut) and CTGTCGCTGCCAGCTGGGGCC (G) (Kaiso binding motif in italics). A 100-fold excess of wild-type unlabeled oligonucleotide (wt comp) as well as human matrilysin promoter-derived Kaiso binding unlabeled oligonucleotide GTGCTTCCTGCCAATAACG (matr comp; Kaiso binding site in italics) were used as competitors to show specificity of the binding. (A) A total of 30 ng of GST-Kaiso zinc fingers 1, 2, and 3 fusion protein (GST-ZF1,2,3) was incubated with the indicated probes. Wild-type probe and the probe with a mutation of A to G bind GST-ZF1,2,3 fusion protein, whereas the probe with a mutation in the Kaiso site has greatly reduced binding. (B) Thirty nanograms of GST-Kaiso zinc fingers 2 and 3 fusion protein (GST-ZF2,3) was incubated with the indicated probes. The wild-type probe was able to shift the specific band only in the absence of 0.1% NP-40, which is present in the incubation buffer for the binding reactions shown in panel A.
To investigate whether endogenous Kaiso in C2C12 cells binds to the rapsyn promoter in vivo, we performed a ChIP assay. Rapsyn-specific primers were designed to amplify a promoter region harboring both Kaiso binding sites. A rapsyn promoter fragment of ∼230 bp was successfully amplified by PCR from C2C12 genomic DNA (Fig. 4, Input). PCR analyses with chromatin from Kaiso immunoprecipitates amplified the ∼230-bp rapsyn promoter fragment (Fig. 4, 6F Kaiso), while neither antihemagglutinin (12CA5) immunoprecipitates nor Kaiso immunoprecipitations lacking input genomic DNA amplified this region (Fig. 4). Taken together, these results demonstrate definitively that Kaiso interacts with the rapsyn promoter both in vitro and in vivo.
FIG. 4.
Kaiso binds the rapsyn promoter in vivo. Murine C2C12 genomic DNA was isolated and fragmented by sonication, and endogenous Kaiso was immunoprecipitated by using the 6F monoclonal antibody. A 237-bp fragment of the murine rapsyn promoter was amplified by PCR from Kaiso immunoprecipitates (6F Kaiso), while negligible amounts of rapsyn promoter were amplified from the irrelevant antihemagglutinin antibody immunoprecipitates (12CA5). PCRs from Kaiso immunoprecipitates lacking input chromatin (No Input) or a PCR lacking template (No Template) are presented as controls. Lane 1 (Input) represents the PCR amplification of the rapsyn promoter directly from C2C12 genomic DNA, which was purified from 1/10 of the amount of lysate that was used for each Kaiso immunoprecipitation. IP, immunoprecipitate.
FIG. 6.
Immunohistochemistry of Kaiso and δ-catenin in C2C12 cells. Images were obtained by laser-scanning confocal microscopy. (A and C) δ-Catenin immunostaining following treatment with 10 ng of LMB per ml for 3 h shows that LMB inhibits nuclear export of δ-catenin in C2C12 cells. (B) Nuclear Kaiso immunostaining with 6F monoclonal antibodies. (D) LMB treatment induces the rapsyn promoter transfected into C2C12 cells. C2C12 myoblasts were transfected overnight with the indicated promoter-reporters; the luciferase assay was performed 18 h after transfection. Cells were treated with 10 ng of LMB per ml for 3 h before harvesting. LMB treatment induces rapsyn promoter-reporter (−215 construct), whereas LMB failed to induce the −110m construct with a Kaiso site mutation. However, the −110 construct with the mutation of A to G (−110G) in the overlapping Kaiso site E box still can be induced by LMB. Data are presented as the means of triplicate samples ± standard deviations. Each experiment was repeated at least three separate times with similar results. Scale bar, 20 μm.
Coimmunoprecipitation of Kaiso with antibodies to δ-catenin.
Our hypothesis that both Kaiso and δ-catenin are involved in rapsyn regulation suggests that these proteins may form a complex at the neuromuscular junction. Coimmunoprecipitation experiments were performed by using whole-cell lysates of C2C12 myotubes. Because δ-catenin is specifically expressed in the nervous system (46), mouse brain was also used for immunoprecipitation analysis as a positive control. As shown in the right panel of Fig. 5B, Kaiso was detected in anti-δ-catenin immunoprecipitates from C2C12 myotubes (lane 1) but not in immunoprecipitates of preimmune sera captured with protein A- and protein G-Sepharose, respectively (lanes 3 and 4). The specificity of this experiment was verified by probing the same δ-catenin immunoprecipitates with irrelevant Sp1 antibodies, which yielded no signal (data not shown). These results reveal that Kaiso forms a complex not only with p120 catenin, as has been shown by Daniel and Reynolds (6), but also with the highly related p120 subfamily member, δ-catenin.
FIG. 5.
Kaiso and δ-catenin are expressed and form a complex in C2C12 myotubes. (A) Antibodies to δ-catenin (C-20; Santa Cruz) recognize a band of approximately 160 kDa in total cell lysate of C2C12 myotubes (left panel). Immunoprecipitations with antibodies to δ-catenin (C-20; Santa Cruz) precipitated the same proteins that migrate at 160 kDa from both C2C12 myotubes and mouse brain (right panel). Mouse brain is included in the immunoprecipitation experiment to verify the size of δ-catenin (160 kDa); C2C12 lysate (1/50 of the lysate used for immunoprecipitation) represents the input. Thus, the anti-δ-catenin antibodies can be used to immunoprecipitate δ-catenin from C2C12 myotubes. (B) Polyclonal antibodies to Kaiso recognize a band of approximately 110 kDa in lysate from C2C12 myotubes (left panel). Immunoprecipitation with monoclonal antibodies to Kaiso (6F or 12G) precipitated these same proteins from C2C12 myotubes (lane 2, right panel). In the right panel, lane 1 demonstrates specific coprecipitation of Kaiso by δ-catenin antibodies (C20; Santa Cruz) from C2C12 myotube lysates, whereas Kaiso is not precipitated by the preimmune sera in the context of protein G- or protein A-Sepharose (lanes 3 and 4, respectively). This indicates that Kaiso is a binding partner of δ-catenin. IP, immunoprecipitate.
Leptomycin B sensitivity of δ-catenin in C2C12 myotubes: evidence for nucleocytoplasmic shuttling of δ-catenin.
To further test our hypothesis that Kaiso and δ-catenin regulate rapsyn transcription, we examined the subcellular localization of these proteins by using immunofluorescence staining of methanol-fixed C2C12 myotubes. While at steady state δ-catenin localizes predominantly to the cytosol, the possibility remained that, like p120 (21), δ-catenin shuttles between nuclear and cytoplasmic subcellular compartments. To test this hypothesis, we utilized a specific inhibitor of CRM-1-mediated nuclear export called leptomycin B (LMB) to assess the nucleocytoplasmic shuttling activity of δ-catenin in these cells. Prior to fixation and immunofluorescence staining, C2C12 cells were treated with 10 ng of LMB per ml for 3 h. This treatment significantly increased nuclear levels of δ-catenin, indicating that it is subject to nucleocytoplasmic shuttling in C2C12 cells (Fig. 6A and C). In contrast, this treatment led to the cytoplasmic perinuclear localization of p120 catenin in these cells (data not shown). Immunofluorescence staining with Kaiso monoclonal antibodies (Fig. 6B) revealed a diffuse nuclear localization, consistent with previous reports (6). These data suggest that in C2C12 cells, δ-catenin, rather than p120, undergoes rapid nucleocytoplasmic shuttling.
To ascertain whether the activity of rapsyn promoter-reporters in myoblasts is sensitive to LMB, C2C12 cells were treated with 10 ng of LMB per ml for 3 h before harvesting (16 h after transfection) (Fig. 6D). The −215 construct containing two Kaiso binding sites was induced up to threefold upon the treatment. The −110 reporter construct containing a mutation in the core Kaiso binding site did not respond to LMB treatment whereas the −110 construct with the mutation −38A to G could be induced, although to a lesser extent than the −215 reporter construct. These results imply that rapsyn promoter activity is mediated by a CRM-1-dependent factor, possibly δ-catenin.
Kaiso and δ-catenin are present at the neuromuscular junction.
Our hypothesis that Kaiso and δ-catenin are involved in rapsyn promoter regulation requires both proteins to be present at the neuromuscular junction. To address this issue we performed immunohistochemistry of mouse muscle sections with antibodies to Kaiso and δ-catenin, in addition to α-bungarotoxin, to identify the location of neuromuscular junctions. Both Kaiso and δ-catenin were localized at the neuromuscular junction (Fig. 7). The identical pattern of colocalization was observed at the chicken neuromuscular junction (data not shown). These data are consistent with our hypothesis of rapsyn promoter regulation at the neuromuscular junction and suggest a new pathway of synapse-specific transcription involving the POZ-zinc finger transcription factor Kaiso.
FIG. 7.
Antibodies to Kaiso and δ-catenin recognize proteins concentrated at the mouse neuromuscular junction. Confocal micrographs of sections of mouse thigh muscle stained with antibodies to Kaiso (upper left panel) and δ-catenin (upper right panel), followed by FITC-conjugated secondary antibodies. The locations of neuromuscular junctions are indicated by the concentrations of rhodamine-conjugated α-bungarotoxin which binds to the AChR (second row). Combined images of the antibody staining (green) and AChR staining (red) are presented in the bottom row to show the extent of the synaptic localization. Anti-δ-catenin and anti-Kaiso staining are found in the region of the muscle directly under the AChRs, but there is also staining that extends beyond the limits of the AChR staining. Scale bar, 20 μm.
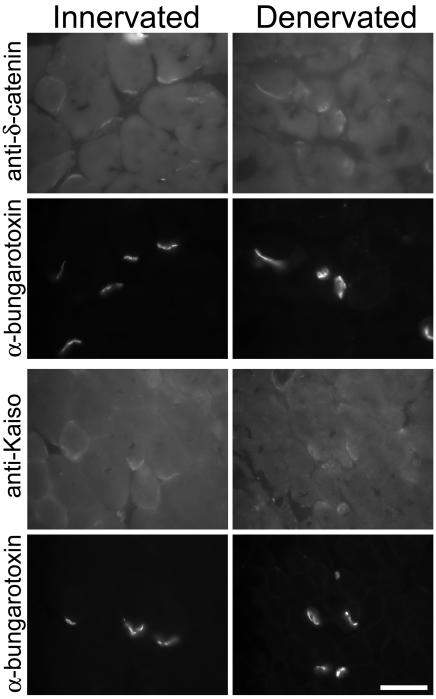
To determine whether Kaiso and δ-catenin have a postsynaptic localization that is consistent with such a function, we denervated mouse gastrocnemius muscle by sciatic nerve resection and compared their localization in innervated and denervated tissue. Two weeks after nerve resection, postsynaptic AChR clusters still remained. Kaiso and δ-catenin staining persisted after denervation and colocalized with postsynaptic AChR clusters (Fig. 8). Thus, Kaiso and δ-catenin at the neuromuscular junction concentrate postsynaptically, consistent with a role in rapsyn regulation.
FIG. 8.
Antibodies to Kaiso and δ-catenin recognize molecules in the postsynaptic apparatus of mouse gastrocnemius muscle. Denervation of the muscles was performed by sectioning the sciatic nerve at midthigh 2 weeks previously. Images from denervated muscles are shown in the right column and images from the contralateral innervated muscles are shown in the left column. Neuromuscular junctions are identified by rhodamine-conjugated α-bungarotoxin. Antibodies to δ-catenin (top row) and Kaiso (third row) recognize molecules concentrated at the synaptic regions in both the innervated and denervated muscles. The presence of staining in the denervated muscles reveals that antibodies are recognizing molecules located in the postsynaptic apparatus. Scale bar, 10 μm.
DISCUSSION
Here we demonstrate that the rapsyn promoter has functional Kaiso binding sites and that rapsyn promoter activity is regulated by Kaiso and δ-catenin in mouse and chicken myotubes. Immunofluorescence analyses verified that Kaiso and δ-catenin are concentrated at the postsynaptic region of mouse and chicken skeletal muscles. Based on these observations, we hypothesize that there is a pathway involving Kaiso and δ-catenin that regulates synapse-specific transcription at the neuromuscular junction. Since Kaiso and δ-catenin specifically induced the rapsyn promoter in muscle cells, and not in nonmuscle 293T cells, we further hypothesize that myogenic factors are also involved in this pathway. Current experiments are directed at determining the exact interactions of these components both in vitro and in vivo. We have shown that Kaiso and δ-catenin induce rapsyn promoter activity, but we have yet to show that Kaiso and/or δ-catenin are required for rapsyn transcription in vivo. However, our results strongly support the existence of a pathway that would explain the synapse-specific transcription of rapsyn and other synapse-specific proteins at the neuromuscular junction and also at other synapses throughout the body.
The neuromuscular junction has long served as a model synapse (reviewed in reference 40). The fact that we find antibody staining of Kaiso and δ-catenin at the neuromuscular junction is not surprising since many proteins found concentrated at synapses in the CNS are also found at the neuromuscular junction. Recently, another newly discovered member of the POZ-zinc finger family, myoneurin, has been reported to localize preferentially to subsynaptic nuclei at the neuromuscular junction (5). Kaiso is ubiquitously expressed; however, Xenopus Kaiso expression was detected predominantly in neural tissues (23), providing the first clue that Kaiso may function in neural tissues. The Kaiso binding partner, p120 catenin, is also ubiquitously expressed (37). However, the p120 family member δ-catenin has an expression pattern reported to be limited to the nervous system (18, 34). The observed colocalization between Kaiso and δ-catenin at the neuromuscular junction is consistent with our hypothesis that the Kaiso and δ-catenin interaction is integral in regulating synapse-specific transcription at the neuromuscular junction and raises the strong possibility that these molecules are involved with the regulation of synapse-specific transcription throughout the nervous system.
δ-Catenin was discovered in a two-hybrid assay as a bona fide interactor with presenilin-1 (46), a protein which carries mutations that cause familial Alzheimer's disease. Presenilin-1 is expressed in the brain as well as at the postsynaptic domain of the neuromuscular junction (2). δ-Catenin is a potent substrate and forms a stable complex with Abl kinase (27). Abl kinase has been found in the postsynaptic domain of the neuromuscular junction and is a critical component of the tyrosine kinase signaling cascade downstream of agrin (15). These results raise the possibility that δ-catenin can be phosphorylated by Abl kinase at the neuromuscular junction and this phosphorylation can modulate its activity. Experiments directed at testing this hypothesis are currently under way.
We found Kaiso and δ-catenin localized to nuclei of C2C12 myotubes. We additionally demonstrated for the first time that Kaiso either directly binds or forms an indirect complex with δ-catenin. Our data show that LMB treatment, which is a specific inhibitor of CRM-1-mediated nuclear export, leads to nuclear δ-catenin accumulation in C2C12 cells. Intriguingly, the rapsyn promoter is induced by LMB treatment in chicken myotubes. This induction is inhibited by mutations of the Kaiso binding site. Altogether, this supports the hypothesis that nuclearly localized δ-catenin is involved in transcriptional regulation in C2C12 myotubes and at the neuromuscular junction.
Mutations in the promoter regions of genes can often lead to disease. The mutation in the N box of the AChR ɛ-subunit promoter causes AChR deficiency at the neuromuscular junction and, consequently, CMS (1, 30, 31). This strongly implies a functional significance of the N box for transcription regulation of the AChR at the neuromuscular junction (41). Similarly, two point mutations in the E box of the rapsyn promoter have been reported in a subset of patients with CMS (33). One of these mutations (−38A to G) is located in the E box that partially overlaps the functional Kaiso binding site that we describe here. However, this mutation does not impact the core consensus Kaiso sequence as defined by Daniel et al. (8). In contrast, it changes the nonconserved dinucleotide of the E-box consensus (CANNTG) and does not disrupt the binding of DNA with myogenic factors. Consistent with this view, Ohno et al. (33) did not find the expected induction by MyoD of the simian virus 40 promoter when fused with several copies of this E box or the E box carrying the mutation −38A to G. In our study, however, this mutation reduces the activation of the rapsyn promoter by Kaiso and δ-catenin and implicates the influence of bases adjacent to the consensus Kaiso binding site. The possibility therefore exists that CMS associated with this mutation may result from disruption of the Kaiso binding site and not the adjacent E box.
Kaiso is a sequence-specific DNA-binding protein, and CTGCNA (where N is any nucleotide) was identified as the core consensus sequence (8). Two copies of the optimal consensus were found in both the human and mouse matrilysin promoters, and Kaiso was shown to demonstrate sequence-specific binding to matrilysin-derived oligonucleotides (8). These and other data implicate matrilysin as a Kaiso target gene (C. M. Spring, K. F. Kelly, I. O'Kelly, M. Graham, H. C. Crawford, and J. M. Daniel, submitted for publication). While most POZ-zinc finger proteins are transcriptional repressors (10, 14), some are activators (24, 35), and some repress as well as activate transcription (4, 13). In our system, Kaiso behaves as a transcriptional activator, suggesting that the transcriptional properties of Kaiso may be context dependent. Consistent with this, Kaiso fails to activate the rapsyn promoter-reporter in nonmuscle human embryonic kidney 293T cells which do not express δ-catenin or myogenic factors. We hypothesize that the nuclear targeting of δ-catenin induces rapsyn transcription in muscle cells. We speculate that there is a balance between the inducing activity of the E-box-binding myogenic factors and Kaiso activity; this balance is likely regulated by δ-catenin nuclear trafficking. The mutation −38A to G decreases the induction by Kaiso and δ-catenin and apparently shifts the balance between myogenic factors and Kaiso and δ-catenin. This may cause the reduction or loss of rapsyn protein at the neuromuscular junction and the consequent endplate AChR deficiency and may explain the phenotype observed in CMS patients with a mutation in this region of the rapsyn promoter.
In conclusion, these results strongly suggest that Kaiso and δ-catenin are components of a mechanism that regulates synapse-specific transcription. We therefore propose the following novel model of synapse-specific transcription: activation of relevant target genes, including rapsyn, requires the binding of Kaiso and δ-catenin in addition to myogenic transcription factors. Promoter activation is modulated by the expression levels of Kaiso and δ-catenin as well as the translocation of δ-catenin from the subsynaptic cytoplasm to the subsynaptic nuclei. The proposed Kaiso/δ-catenin complex regulates the affinity of Kaiso-DNA binding and modulates the transcriptional activity of Kaiso target genes at synapses throughout the body.
Acknowledgments
We thank K. S. Kosik (Department of Neurology, Harvard Medical School) for the δ-catenin expression construct. We also thank Douglas Wright, Robin L. Maser, and Gregory Vanden Heuvel (University of Kansas Medical Center) for critical readings of the manuscript.
This work was supported by grants from the NIH (NS33320) to M.J.W. and from CIHR (MOP-42405) to J.M.D.
REFERENCES
- 1.Abicht, A., R. Stucka, C. Schmidt, A. Briguet, S. Hopfner, I. H. Song, D. Pongratz, W. Muller-Felber, M. A. Ruegg, and H. Lochmuller. 2002. A newly identified chromosomal microdeletion and an N-box mutation of the AChR epsilon gene cause a congenital myasthenic syndrome. Brain 125:1005-1013. [DOI] [PubMed] [Google Scholar]
- 2.Askanas, V., W. K. Engel, C. C. Yang, R. B. Alvarez, V. M. Lee, and T. Wisniewski. 1998. Light and electron microscopic immunolocalization of presenilin 1 in abnormal muscle fibers of patients with sporadic inclusion-body myositis and autosomal-recessive inclusion-body myopathy. Am. J. Pathol. 152:889-895. [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, V. J., and R. Treisman. 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 8:1664-1677. [DOI] [PubMed] [Google Scholar]
- 4.Bereshchenko, O. R., W. Gu, and R. Dalla-Favera. 2002. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 32:606-613. [DOI] [PubMed] [Google Scholar]
- 5.Cifuentes-Diaz, C., M. Bitoun, D. Goudou, N. Seddiqi, N. Romero, F. Rieger, J. P. Perin, and P. M. Alliel. 2004. Neuromuscular expression of the BTB/POZ and zinc finger protein myoneurin. Muscle Nerve 29:59-65. [DOI] [PubMed] [Google Scholar]
- 6.Daniel, J. M., and A. B. Reynolds. 1999. The catenin p120ctn interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell. Biol. 19:3614-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel, J. M., R. C. Ireton, and A. B. Reynolds. 2001. Monoclonal antibodies to Kaiso: a novel transcription factor and p120ctn-binding protein. Hybridoma 20:159-166. [DOI] [PubMed] [Google Scholar]
- 8.Daniel, J. M., C. M. Spring, H. C. Crawford, A. B. Reynolds, and A. Baig. 2002. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 30:2911-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Kerchove D'Exaerde, A., J. R. Cartaud, and L. Schaeffer. 2002. Expression of mutant Ets protein at the neuromuscular synapse causes alterations in morphology and gene expression. EMBO Rep. 3:1075-1081. (First published 22 October 2002; 10.1093/embo-reports/kvf220.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deltour, S., S. Pinte, C. Guerardel, B.Wasylyk, and D. Leprince. 2002. The human candidate tumor suppressor gene HIC1 recruits CtBP through a degenerate GLDLSKK motif. Mol. Cell. Biol. 22:4890-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duclert, A., N. Savatier, L. Schaeffer, and J. P. Changeux. 1996. Identification of an element crucial for the sub-synaptic expression of the acetylcholine receptor epsilon-subunit gene. J. Biol. Chem. 271:17433-17438. [DOI] [PubMed] [Google Scholar]
- 12.Dunne, V., and R. A. Maselli. 2003. Identification of pathogenic mutations in the human rapsyn gene. J. Hum. Genet. 48:204-207. [DOI] [PubMed] [Google Scholar]
- 13.Faucheux, M., J. Y. Roignant, S. Netter, J. Charollais, C. Antoniewski, and L. Theodore. 2003. batman interacts with Polycomb and trithorax group genes and encodes a BTB/POZ protein that is included in a complex containing GAGA factor. Mol. Cell. Biol. 23:1181-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedele, M., G. Benvenuto, R. Pero, B. Majello, S. Battista, F. Lembo, E. Vollono, P. M. Day, M. Santoro, L. Lania, C. B. Bruni, A. Fusco, and L. Chiariotti. 2000. A novel member of the BTB/POZ family, PATZ, associates with the RNF4 RING finger protein and acts as a transcriptional repressor. J. Biol. Chem. 275:7894-7901. [DOI] [PubMed] [Google Scholar]
- 15.Finn, A. J., G. Feng, and A. M. Pendergast. 2003. Postsynaptic requirement for Abl kinases in assembly of the neuromuscular junction. Nat. Neurosci. 6:717-723. [DOI] [PubMed] [Google Scholar]
- 16.Fischbach, G. D. 1972. Synapse formation between dissociated nerve and muscle cells in low density cell cultures. Dev. Biol. 28:407-429. [DOI] [PubMed] [Google Scholar]
- 17.Gautam, M., P. G. Noakes, J. Mudd, M. Nichol, G. C. Chu, J. R. Sanes, and J. P. Merlie. 1995. Failure of postsynaptic specialization to develop at neuromuscular junctions of rapsyn-deficient mice. Nature 377:232-236. [DOI] [PubMed] [Google Scholar]
- 18.Ho, C., J. Zhou, M. Medina, T. Goto, M. Jacobson, P. G. Bhide, and K. S. Kosik. 2000. δ-Catenin is a nervous system-specific adherens junction protein which undergoes dynamic relocalization during development. J. Comp. Neurol. 420:261-276. [PubMed] [Google Scholar]
- 19.Javahery, R., A. Khachi, K. Lo, B. Zenzie-Gregory, and S. T. Smale. 1994. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol. 14:116-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, S. B., G. W. Lanford, Y. H. Chen, M. Moribito, K. Kim, and Q. Lu. 2002. Glutamate-induced delta-catenin redistribution and dissociation from postsynaptic receptor complexes. Neuroscience 115:1009-1021. [DOI] [PubMed] [Google Scholar]
- 21.Kelly, K. F., C. M. Spring, A. A. Otchere, and J. M. Daniel. 2004. NLS-dependant nuclear localization of p120 ctn is necessary to relieve Kaiso-mediated transcriptional repression. J. Cell Sci. 117:2675-2686. [DOI] [PubMed]
- 22.Kim, K., A. Sirota, Y. H. Chen, S. B. Jones, R. Dudek, G. W. Lanford, C. Thakore, and Q. Lu. 2002. Dendrite-like process formation and cytoskeletal remodeling regulated by delta-catenin expression. Exp. Cell Res. 275:171-184. [DOI] [PubMed] [Google Scholar]
- 23.Kim, S. W., X. Fang, H. Ji, A. F. Paulson, J. M. Daniel, M. Ciesiolka, F. van Roy, and P. D. McCrea. 2002. Isolation and characterization of XKaiso, a transcriptional repressor that associates with the catenin Xp120(ctn) in Xenopus laevis. J. Biol. Chem. 277:8202-8208. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, A., H. Yamagiwa, H. Hoshino, A. Muto, K. Sato, M. Morita, N. Hayashi, M. Yamamoto, and K. Igarashi. 2000. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol. Cell. Biol. 20:1733-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koike, S., L. Schaeffer, and J. P. Changeux. 1995. Identification of a DNA element determining synaptic expression of the mouse acetylcholine receptor delta-subunit gene. Proc. Natl. Acad. Sci. USA 92:10624-10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, Q., M. Paredes, M. Medina, J. Zhou, R. Cavallo, M. Peifer, L. Orecchio, and K. S. Kosik. 1999. δ-Catenin, an adhesive junction-associated protein which promotes cell scattering. J. Cell Biol. 144:519-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, Q., N. K. Mukhopadhyay, J. D. Griffin, M. Paredes, M. Medina, and K. S. Kosik. 2002. Brain armadillo protein delta-catenin interacts with Abl tyrosine kinase and modulates cellular morphogenesis in response to growth factors. J. Neurosci. Res. 67:618-624. [DOI] [PubMed] [Google Scholar]
- 28.Moscoso, L. M., J. P. Merlie, and J. R. Sanes. 1995. N-CAM, 43K-rapsyn, and S-laminin mRNAs are concentrated at synaptic sites in muscle fibers. Mol. Cell. Neurosci. 6:80-89. [DOI] [PubMed] [Google Scholar]
- 29.Muller, J. S., G. Mildner, W. Muller-Felber, U. Schara, K. Krampfl, B. Petersen, S. Petrova, R. Stucka, W. Mortier, J. Bufler, G. Kurlemann, A. Huebner, L. Merlini, H. Lochmuller, and A. Abicht. 2003. Rapsyn N88K is a frequent cause of congenital myasthenic syndromes in European patients. Neurology 60:1805-1810. [DOI] [PubMed] [Google Scholar]
- 30.Nichols, P., R. Croxen, A. Vincent, R. Rutter, M. Hutchinson, J. Newsom-Davis, and D. Beeson. 1999. Mutation of the acetylcholine receptor epsilon-subunit promoter in congenital myasthenic syndrome. Ann. Neurol. 45:439-443. [PubMed] [Google Scholar]
- 31.Ohno, K., B. Anlar, and A. G. Engel. 1999. Congenital myasthenic syndrome caused by a mutation in the Ets-binding site of the promoter region of the acetylcholine receptor epsilon subunit gene. Neuromuscul. Disord. 9:131-135. [DOI] [PubMed] [Google Scholar]
- 32.Ohno, K., A. G. Engel, X. M. Shen, D. Selcen, J. Brengman, C. M. Harper, A. Tsujino, and M. Milone. 2002. Rapsyn mutations in humans cause endplate acetylcholine-receptor deficiency and myasthenic syndrome. Am. J. Hum. Genet. 70:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohno, K., M. Sadeh, I. Blatt, J. M. Brengman, and A. G. Engel. 2003. E-box mutations in the RAPSN promoter region in eight cases with congenital myasthenic syndrome. Hum. Mol. Genet. 12:739-748. [DOI] [PubMed] [Google Scholar]
- 34.Paffenholz, R., and W. W. Franke. 1997. Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation 61:293-304. [DOI] [PubMed] [Google Scholar]
- 35.Peukert, K., P. Staller, A. Schneider, G. Carmichael, F. Hänel, and M. Eilers. 1997. An alternative pathway for gene regulation by Myc. EMBO J. 16:5672-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prokhortchouk, A., B. Hendrich, H. Jorgensen, A. Ruzov, M. Wilm, G. Georgiev, A. Bird, and E. Prokhortchouk. 2001. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds, A. B., J. Daniel, P. D. McCrea, M. J. Wheelock, J. Wu, and Z. Zhang. 1994. Identification of a new catenin: the tyrosine kinase substrate p120cas associates with E-cadherin complexes. Mol. Cell. Biol. 14:8333-8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richard, P., K. Gaudon, F. Andreux, E. Yasaki, C. Prioleau, S. Bauche, A. Barois, C. Ioos, M. Mayer, M. C. Routon, M. Mokhtari, J. P. Leroy, E. Fournier, B. Hainque, J. Koenig, M. Fardeau, B. Eymard, and D. Hantai. 2003. Possible founder effect of rapsyn N88K mutation and identification of novel rapsyn mutations in congenital myasthenic syndromes. J. Med. Genet. 40:e81. [Online.] [DOI] [PMC free article] [PubMed]
- 39.Rodova, M., M. R. Islam, R. L. Maser, and J. P. Calvet. 2002. The polycystic kidney disease-1 promoter is a target of the beta-catenin/T-cell factor pathway. J. Biol. Chem. 277:29577-29583. [DOI] [PubMed] [Google Scholar]
- 40.Sanes, J. R., and J. W. Lichtman. 1999. Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci. 22:389-442. [DOI] [PubMed] [Google Scholar]
- 41.Schaeffer, L., A. de Kerchove D'Exaerde, and J. P. Changeux. 2001. Targeting transcription to the neuromuscular synapse. Neuron 31:15-22. [DOI] [PubMed] [Google Scholar]
- 42.Wallace, B. G. 1986. Aggregating factor from Torpedo electric organ induces patches containing acetylcholine receptors, acetylcholinesterase, and butyrylcholinesterase on cultured myotubes. J. Cell Biol. 102:783-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinmann, A. S., and P. J. Farnham. 2002. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 26:37-47. [DOI] [PubMed] [Google Scholar]
- 44.Weintraub, H., V. J. Dwarki, I. Verma, R. Davis, S. Hollenberg, L. Snider, A. Lassar, and S. J. Tapscott. 1991. Muscle-specific transcriptional activation by MyoD. Genes Dev. 5:1377-1386. [DOI] [PubMed] [Google Scholar]
- 45.Yoon, H. G., D. W. Chan, A. B. Reynolds, J. Qin, and J. Wong. 2003. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 12:723-734. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, J., U. Liyanage, M. Medina, C. Ho, A. D. Simmons, M. Lovett, and K. S. Kosik. 1997. Presenilin 1 interaction in the brain with a novel member of the Armadillo family. Neuroreport 8:2085-2090. [DOI] [PubMed] [Google Scholar]