Abstract
To investigate the role of promoters in regulating variable gene rearrangement and allelic exclusion, we constructed mutant mice in which a 1.2-kb region of the Vβ13 promoter was either deleted (P13−/−) or replaced with the simian virus 40 minimal promoter plus five copies of Gal4 DNA sequences (P13R/R). In P13−/− mice, cleavage, rearrangement, and transcription of Vβ13, but not the flanking Vβ gene segments, were significantly inhibited. In P13R/R mice, inhibition of Vβ13 rearrangement was less severe and was not associated with any apparent reduction in Vβ13 cleavage. Expression of a T-cell receptor (TCR) transgene blocked cleavages at the normal Vβ13-recombination signal sequence junction and Vβ13 coding joint formation of both wild-type and mutant Vβ13 alleles. However, a low level of aberrant Vβ13 cleavage was consistently detected, especially in TCR transgenic P13R/R mice. These findings suggest that the variable gene promoter is required for promoting local recombination accessibility of the associated Vβ gene segment. Although the promoter is dispensable for allelic exclusion, it appears to suppress aberrant Vβ cleavages during allelic exclusion.
The variable regions of immunoglobulin (Ig) and T cell receptor (TCR) genes are assembled from variable (V), diversity (D), and joining (J) gene segments. V(D)J recombination is tightly regulated in the context of lymphocyte development, exhibiting lineage, developmental stage, and allele specificity (1, 13, 24, 32). Although recombination at different TCR and Ig loci is mediated by the same recombinase complex and conserved recombination signal sequences (RSS), complete rearrangements of TCR genes are limited to T cells, while complete rearrangements of Ig genes are limited to B cells. Within the appropriate cell lineage, recombination is regulated temporally and in a stage-specific manner. In addition, in a given lymphocyte, only one of two alleles of antigen receptor loci usually undergoes functional rearrangement, a process known as allelic exclusion.
Studies have shown that transcriptional regulatory elements, such as promoters and enhancers, play an important role in targeting specific gene segments for recombination. Deletion of the enhancer at any of the TCRβ, TCRδ, TCRα, IgH, and Igκ loci results in a severe reduction in the level of rearrangement at the respective loci (4, 5, 7, 11, 14, 27, 31, 41, 42). Similarly, deletion of the PDβ1 promoter immediately upstream of the Dβ1 gene segment at the TCRβ locus significantly impairs Dβ1 rearrangement (40, 41). To date, most evidence suggests that enhancers and promoters regulate V(D)J recombination by modulating chromatin structures and rendering gene segments accessible to RAG cleavage (13, 24, 32).
Among the various levels of control, the most complex regulation is probably allelic exclusion. Like lineage- and stage-specific regulations, allelic exclusion is apparently also achieved through modulating access of gene segments to RAG cleavage (13, 15, 24, 32). For example, TCRβ allelic exclusion is regulated at the Vβ-to-DβJβ rearrangement step. In CD4− CD8− (DN) thymocytes, where TCRβ rearrangement normally occurs, Vβ gene segments are transcribed, sensitive to nuclease, and associated with acetylated histones (6, 17, 35). In CD4+ CD8+ (DP) thymocytes, where TCRβ allelic exclusion is in effect, Vβ transcription, nuclease sensitivity, and association with acetylated histones are markedly reduced. When a Vβ gene segment together with 3.6-kb 5′ sequences, encompassing the promoter, were inserted 6.8 kb upstream of the Dβ1 gene segment, the inserted Vβ gene segment rearranged at the same frequency as the natural copy but was no longer subject to allelic exclusion control (29), indicating distinct controls of the frequency and allelic exclusion of Vβ gene rearrangement. However, beyond these preliminary observations, little is known about the specific cis elements that modulate variable gene accessibility for rearrangement initially and then for allelic exclusion.
Another cardinal feature of V(D)J recombination is site specificity, which is achieved by precise cleavage of DNA at the junction of coding sequences and RSS (9, 10). The precise cleavages occur probably because RAG proteins interact directly with two RSS to form an enzymatically active complex, known as a synapse (8, 34, 38). Within this synapse, RAG proteins cleave the DNA at the junctions of RSS and coding sequences. Studies have shown that the presence of two intact RSS is required for synapse formation and coupled cleavages (8, 34, 38). However, RAG proteins have also been shown to cleave DNA that contains only a single RSS or RSS-like sequence (19, 20, 36, 37), indicating that RAG proteins are capable of uncoupled and imprecise cleavages. At the endogenous antigen receptor loci, gene segments are packed into chromatin and can be separated by hundreds of kilobase pairs on linear DNA. It is unclear whether RSS alone provide a sufficient cis signal for synapse formation and therefore precise and coupled cleavages. Nor is it known how aberrant cleavages are suppressed at the endogenous antigen receptor loci.
In mice, the TCRβ locus spans approximately 600 kb (22). All Vβ gene segments, except Vβ14, are clustered together at the 5′ end of the locus and separated from Dβ, Jβ, and Cβ by at least 330 kb. The two cis elements, PDβ1 promoter and Eβ enhancer, which have been shown to regulate Dβ-to-Jβ rearrangements, do not appear to play a significant role in regulating Vβ accessibility and allelic exclusion (17, 18, 40, 41). We have now investigated the role of a variable gene promoter in regulating Vβ rearrangement and allelic exclusion by targeted deletion or replacement in the endogenous TCRβ locus. By examining the effects of the promoter mutations on specific as well as flanking Vβ gene cleavage, joining, allelic exclusion, and transcription, our results show that the normal variable gene promoter is required for promoting local recombination accessibility. Although the promoter is not required for mediating allelic exclusion per se, it appears to suppress aberrant Vβ cleavages during allelic exclusion.
MATERIALS AND METHODS
Targeting vectors and mice.
The targeting vector used for electroporation into J1 embryonic stem cells consisted of a floxed phosphoglycerate kinase (PGK) promoter-driven neomycin resistance gene (neo) flanked upstream by a 2.4-kb PstI-NdeI fragment and downstream by a 7.6-kb NcoI-ClaI fragment (Fig. 1A). The targeting vector for the replacement mutation contained an additional 113-bp concatemer of five copies of Gal4 sequences and the 111-bp simian virus 40(SV40) minimal promoter downstream of neo. A PGK promoter-driven thymidine kinase gene (tk) was inserted downstream of the 7.6-kb fragment. Embryonic stem cell clones with proper deletion of the Vβ13 promoter were identified by Southern blotting and were injected into C57BL/6 blastocysts to generate chimeric mice, which were bred with deleter mice to remove the floxed neo (25). Heterozygous mutant mice were bred with each other to generate homozygous mutant mice. A single loxP site was left in place of the Vβ13 promoter in the final P13−/− mutant mice, while a single loxP site, Gal4 sequences, and the SV40 minimal promoter were left in place of the Vβ13 promoter in the final P13R/R mutant mice. Mutant mice were analyzed in mixed 129 × C57BL/6 backgrounds and maintained under specific-pathogen-free conditions. Routine genotype analyses of the Vβ13 promoter mutations were performed by PCR of tail DNA using primers 5′-GGTCAAGCATCTACTTATTGTTC-3′ and 5′-AGCCAAGAAGCCTGGTGCCCAT-3′. All promoter mutationswere confirmed by another round of PCR using the wild-type Vβ13 promoter-specific primers 5′-GGACTGTGCTAAGACTGATTC-3′ and 5′-GGACTGCATATCTGGGAGACTG-3′. TCR transgenic mice expressing the 2C TCR (28) were assayed by flow cytometry using an antibody specific for the 2C TCR.
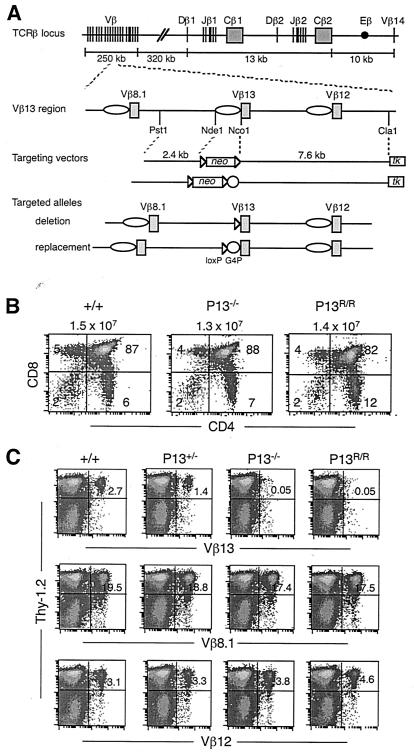
FIG. 1.
Inactivation of the normal Vβ13 promoter specifically blocks development of Vβ13-expressing T cells. (A) Schematic diagrams of the TCRβ locus, Vβ13 region, targeting vectors, and targeted alleles. Gene segments are shown as vertical lines or filled boxes. Vβ promoters are shown as open ovals. Triangles indicate loxP sequences. Open circles represent five copies of Gal4 sequences plus the SV40 minimal promoter. Selectable neo marker was deleted from the targeted alleles in mutant mice by Cre/loxP-mediated recombination. As a result, the 1.2-kb Nde1-Nco1 fragment, encompassing the Vβ13 promoter plus transcriptional initiation sites, but not the start codon, was replaced by one copy of the loxP sequence in deletion mutant mice (P13−/−). In replacement mutant mice (P13R/R), the promoter was replaced by loxP followed by Gal4 sequences and the SV40 minimal promoter. (B) CD4 versus CD8 staining profiles of thymocytes from 4- to 6-week-old wild-type (+/+), P13−/−, and P13R/R mice. Numbers outside the plots indicate the average numbers of thymocytes from four to six mice. Numbers inside the plots indicate percentages of cells in each quadrant. (C) Flow cytometry analysis of Vβ13-expressing T cells. Cells from lymph nodes of +/+, P13+/−, P13−/−, and P13R/R mice were stained with antibodies specific for the pan-T-cell marker Thy-1.2 plus Vβ13, Vβ8.1, or Vβ12. Expression of Thy1.2 versus Vβ is shown for live cells. Numbers indicate the percentages of Thy-1.2+ cells that are also Vβ+. Representative data from one mouse of each genotype are shown.
Flow cytometry.
Antibodies specific for CD4, CD8, Thy-1.2, Vβ13, Vβ8.1, and Vβ12 were direct conjugates from PharMingen (San Diego, Calif.). Flow cytometry was performed on a FACSCalibur apparatus (Becton Dickinson), and data were analyzed using CellQuest software. CD4− CD8− thymocytes were purified by complement-mediated lysis of CD4+ and/or CD8+ cells followed by cell sorting. The sorted cell populations were more than 97% pure.
PCR assays for Vβ CJ.
The semiquantitative nested PCR assays for measuring Vβ13-to-DβJβ1.1 coding joints (CJ) were performed in a 50-μl reaction mixture containing 0.2 μg of thymocyte DNA, 100 ng of each primer (primer no. 1 and 2), a 0.2 μM concentration of each deoxynucleoside triphosphate, 3.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 2.5 U of Taq polymerase. Primary reactions were run for 12 cycles of 30 s at 95°C, 30 s at 61°C, and 2 min at 72°C. Two-microliter reaction mixtures were transferred from the primary reactions to new tubes for secondary PCRs that were performed under identical conditions, except with nested primers (no. 3 and 4) and 18 cycles of amplification. Quantitative titrations of DNA templates were performed by serially diluting wild-type thymic DNA into RAG2-deficient kidney DNA such that the final amount of DNA remained at 200 ng per reaction mixture. Twenty-five-microliter aliquots of secondary PCR mixtures were electrophoresed on a 1.5% agarose gel, transferred to Zeta-probe membranes (Bio-Rad), and hybridized with 32P-labeled Vβ13 cDNA probe or 32P-end-labeled oligonucleotides corresponding to the sequence downstream of Jβ1.1 (primer no. 5). Filters were washed at 50°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and subjected to autoradiography. Vβ13-to-DβJβ2.1 rearrangement was done as described above, except with primers 1 and 6 for the primary reaction and primers 3 and 7 for the secondary reaction. Southern blotting was done with 32P-end-labeled oligonucleotides corresponding to sequence downstream of Jβ2.1 (primer no. 8). Vβ8.1-to-DβJβ1.1 rearrangement was done with primers 9 and 2 for the primary reaction and primers 9 and 4 for the secondary reaction. Vβ12-to-DβJβ1.1 rearrangement was done with primers 10 and 2 for the primary reaction and primers 10 and 4 for the secondary reaction. Southern blotting for Vβ8.1 to DβJβ1.1 or Vβ12 to DβJβ1.1 was done using specific Vβ cDNA probes or an oligonucleotide probe corresponding to sequence downstream of Jβ1.1 (no. 5). Rearrangement products were quantified by using PhosphorImager analysis software (Fujifilm). PCR products were cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.) for sequencing. Semiquantitative JAK3 PCR was done as previously described (40). PCR primer sequences were as follows: primer 1, 5′-CTGCCATGGGCACCAGGCTTCTTG-3′; primer 2, 5′-AGATACTCGAATATGGACACGGAG-3′; primer 3, 5′-GGCACCAGGCTTCTTGGCTGGGCAG-3′; primer 4, 5′-TGGACACGGAGGACATGCTTTGTC-3′; primer 5, 5′-AGAGAGACCTGGAAATTTACCTG-3′; primer 6, 5′-GGTTTTCTGCTCCGGGGGTCTTTG-3′; primer 7, 5′-GGTCTTTGTGGCTGACTGTCCTAC-3′; primer 8, 5′-TCTCTCCCACCTGTATGGCCTCTG-3′; primer 9, 5′-ACTCTTCTTTGTGGTTTTGATT-3′; primer 10, 5′-GCTGGAGTTACCCAGACACCC-3′.
PCR assay for SE.
Linker-mediated PCR (LM-PCR) assays for Vβ13, Vβ12, Vβ8.1, 5′ Dβ1, or 5′ Dβ2 signal ends (SE) were performed as described elsewhere (23, 40) with slight modification. For LM-PCR, 150 ng of ligated DNA was used as template in a 50-μl reaction mixture. In the primary reaction for Vβ13, Vβ12, and Vβ8.1 SE, PCR was carried out for 15 cycles of 45 s at 95°C, 30 s at 63°C, and 45 s at 72°C, followed by a 7-min extension at 72°C. Five microliters was transferred to fresh tubes and amplified in a secondary reaction for 27 cycles with a nested primer and BW-1H using the same conditions. PCR for 5′ Dβ2 SE was performed using the same conditions as for Vβ13 SE, except for 20 cycles in the secondary reaction. For 5′ Dβ1 SE, the primary reaction was 15 cycles of 30 s at 95°C, 30 s at 62°C, and 30 s at 72°C, followed by a 7-min extension at 72°C, and the secondary reaction was 20 cycles using the same condition as the primary reaction. Twenty microliters of the secondary reaction mixtures was electrophoresed on 1.6% agarose gels, transferred to nylon N+ membranes (Amersham), and probed with end-labeled oligonucleotide probes. The rest of the PCR products were cloned into pCR2.1-TOPO (Invitrogen) for sequencing. The specific primers and probes used were as follows: 3′Vβ13 distal primer, 5′-GTCGCTTTCAGTTTGGGGTTCTTG-3′; 3′Vβ13 proximal primer, 5′-AAAAAATTACTTGGAGTCCCTGAG-3′; 3′Vβ13 probe, 5′-CAGAGACCTGGGACTATT-3′; 3′Vβ12 distal primer, 5′-AAATCTCTGAACTACCTTCAAGGTC-3′; 3′Vβ12 proximal primer, 5′-TTTCTTAATACTCGATTATCTTCTG-3′; 3′Vβ12 probe, 5′-AGGATGCCCTGCCTGTGC-3′; 3′Vβ8.1 distal primer, 5′-AAATCTGTCAGAATGACCTTAGTA-3′; 3′Vβ8.1 proximal primer, 5′-GTAAGGATGAGACTCATGCTGTGT-3′; 3′Vβ8.1 probe, 5′-TGGCTTCCTTCACTCTGC-3′; 5′Dβ1 distal primer, 5′-GGTAGACCTATGGGAGGGTC-3′; 5′Dβ1 proximal primer, 5′-ACCTATGGGAGGGTCCTTTTTTGTATAAAG-3′; 5′Dβ1 probe, 5′-TGTAACATTGTGGAATTC-3′; 5′Dβ2 distal primer, 5′-GATTTACCCAGCTTGAGACTTTTTCC-3′; 5′Dβ2 proximal primer, 5′-CAGCCCCTCTCAGTCAGACAAACC-3′; 5′Dβ2 probe, 5′-TGCCACCTGGTCTCCCTGCCCCTG-3′.
PCR assays for SJ.
The PCRs and ApaLI digestion used to measure Vβ13 signal joints (SJ) were performed as described previously with slight modification (40). The primers for Vβ13-to-Dβ2 SJ were the same as primers for Vβ13-to-Dβ2 SE. The same upstream primers and two downstream primers, 5Dβ1A (5′-GAACAGGGGGTAAAGAGGAAACCC-3′) and 5Dβ1B (5′-CATTAGCTCGCATCTTACCACCAC-3′), were used to assay Vβ13-to-Dβ1 SJ. Two micrograms of thymocyte DNA was used in all PCR mixtures. Both undigested and ApaLI-digested products were detected by Southern hybridization with the same end-labeled Vβ13 probe that was used to detect Vβ13 SE.
PCR assays for Vβ GT.
Total RNA was isolated using TRIzol (Invitrogen) from thymocytes of various mice between 6 and 10 weeks of age. To remove residual genomic DNA, 10 μg of the RNA sample was treated with 2 U of amplification-grade DNase I (Invitrogen) for 15 min at 25°C. The reaction was inactivated by the addition of 2 μl of 25 mM EDTA (Invitrogen) and incubation at 65°C for 15 min. First-strand cDNA was synthesized from 50 ng of starting RNA (and 5- and 25-fold dilutions of each of these RNAs) using the Titan One-Tube RT-PCR System (Roche, Indianapolis, Ind.), following the manufacturer's instructions. PCR cycling parameters were 30 min at 50°C, 30 s at 94°C, 45 s at temperatures specific for each primer pair (Tables 1 and 2), and 1 min at 72°C for 15 (Vβ genes) or 24 (β-actin) cycles. Twenty microliters of β-actin primary PCR mixture was directly loaded onto an agarose gel without further amplification. To amplify Vβ germ line transcripts (GT), 5 μl of cDNA from the primary reverse transcription-PCR (RT-PCR) mixture was used as template for seminested PCR. PCR amplification was performed for 30 s at 94°C, 45 s at temperatures specific for each pair of primers, and 1 min at 72°C. Twenty-two to 30 cycles were normally used. PCR products were run on 1% agarose gels and visualized by ethidium bromide staining. Quantification was performed using the software IQ Mac version 1.2. Signals were first normalized to β-actin and expressed relative to that of the wild type, which was given the value of 1.0.
TABLE 1.
PCR primers for assaying Vβ GT
| GT | Primer | Sequence |
|---|---|---|
| Vβ8.1 | F | TCC TGT GTG CAA AAC ACA TGG AGG T |
| R1 | TCT TAC AGA GGC AGA CAC TTC CTG G | |
| R2 | TCC TGG GGT ACA CAG AGA G | |
| Vβ12 | F1 | TTC TGA TAG CAA ATC ACA CAG ATG T |
| F2 | CAG ACA CCC AGA CAT GAG | |
| R | AGG AGC ACA GAA AAG TTC AGA ACT G | |
| Vβ13 | F | TTC TTG ACA CAG TAC TGT CTG AAG C |
| R1 | CTC TGG ATA CAC GCA GCA TGG CCT | |
| R2 | GTC CCA GGT CTC TGC TGA AAG CTG G | |
| β-actin | B-F | CCT AAG GCC AAC CGT GAA AAG |
| B-R | CAT GGT GCT AGG AGC CA |
TABLE 2.
PCR conditions for assaying Vβ GT
| TCR Vβ segment | RT-PCR conditions | Nested PCR conditions |
|---|---|---|
| Vβ8.1 | Primers Vβ8.1F and Vβ8.1R1; 60°C; 15× | Primers Vβ8.1F and Vβ8.1R2; 62°C; 26× |
| Vβ12 | Primers Vβ12F1 and Vβ12R; 56°C; 15× | Primers Vβ12F2 and Vβ12R; 58°C; 27× |
| Vβ13 | Primers Vβ13F and Vβ13R1; 60°C; 15× | Primers Vβ13F and Vβ13R2; 62°C; 28× |
| β-actin | Primers B-F and B-R; 60°C; 24× |
Rapid amplification of 5′ cDNA ends (5′ RACE).
One microgram of DNase I-treated total thymic RNA from various mice was reverse transcribed with random hexamers by using the SMART RACE cDNA amplification kit (Clontech, Palo Alto, Calif.), according to the manufacturer's instructions. cDNA was amplified according to the instructions for primary PCRs, with 2.5 μl of cDNA, UPM, and V13-5 (5′-CTGCTGGCACAGAGATAGGTGGCTGTGTC-3′) primers for 20 cycles. Five microliters of the primary reaction mixture (diluted 1:50 in Tris-EDTA buffer) was used in secondary PCRs with NUP and V13-4 (5′-CTGCTTTGCAGACTGGATCTTGAGAGTGGA-3′) primers at 94°C for 5 s, 68°C for 10 s, and 72°C for 3 min for 20 cycles. Twenty microliters of PCR product was electrophoresed on a 1% agarose gel, followed by hybridization with a Vβ13-specific probe.
RESULTS
Effect of promoter mutations on Vβ13-expressing T-cell development.
We constructed mutant mice in which a 1.2-kb region, encompassing the known promoter and transcription initiation site, but not the translation start codon, of the Vβ13 gene segment, was deleted (P13−/−) (Fig. 1A). The same promoter region was also replaced with the SV40 minimal promoter plus five copies of the Gal4 DNA sequences (P13R/R) in order to inactivate the promoter and potentially afford future opportunities to target heterologous proteins to the site. The effects of the promoter mutations on Vβ13 rearrangement and expression were examined, using the flanking Vβ8.1 and Vβ12 gene segments, each of which has its own independent promoter, as controls.
In both homozygous P13−/− and P13R/R mice, cell numbers in the thymi and CD4 and CD8 staining profiles of thymocytes were indistinguishable from those of wild-type mice (Fig. 1B). In peripheral lymphoid organs, the number of T cells, the relative ratio of CD4+ versus CD8+ T cells, and frequencies of Vβ8.1- or Vβ12-expressing T cells were also similar between wild-type and promoter-mutant mice (Fig. 1C and data not shown). However, in contrast to wild-type mice, which had an average of 2.7% Vβ13-expressing T cells, very few Vβ13+ T cells were detected in homozygous P13−/− or P13R/R mice (Fig. 1C). In heterozygous P13+/− and P13+/R mice, the percentage of Vβ13-positive T cells was reduced by half compared to that in wild-type mice (Fig. 1C and data not shown). Thus, the promoter mutations specifically impaired development of Vβ13-expressing T cells.
Effect of promoter mutations on Vβ13 rearrangement.
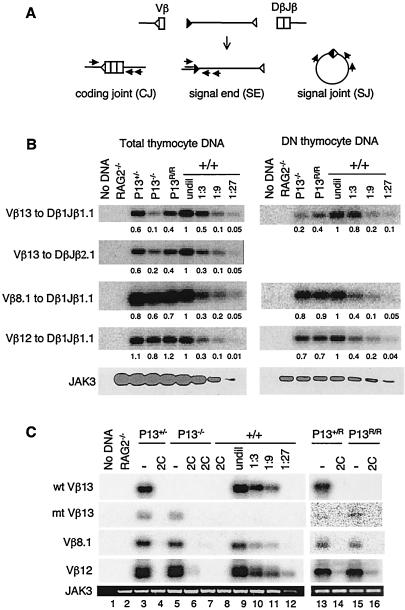
To investigate the mechanisms underlying the diminished Vβ13+ T-cell development, the effect of the promoter mutations on Vβ13 rearrangement was examined by semiquantitative PCR assays (Fig. 2A). Compared to levels in wild-type mice, CJ of Vβ13 to Dβ1Jβ1.1 or to DβJβ2.1 in total thymocyte DNA were decreased approximately 2-fold in P13+/− mice and 5- to 10-fold in P13−/− mice (Fig. 2B, left panel). In P13R/R mice, the levels of Vβ13 CJ were reduced approximately twofold. Similar results were also obtained with DNA from purified DN thymocytes (Fig. 2B, right panel), in which TCRβ rearrangement usually occurs and TCRβ-expressing thymocytes have not yet undergone β-selection (16). In contrast, there was no significant difference in the levels of Vβ8.1 and Vβ12 CJ between wild-type and mutant mice. Sequence analysis of the CJ products revealed that the frequencies and lengths of nucleotide deletions and additions at the Vβ13-Dβ1Jβ1 junction were indistinguishable among wild-type, P13−/−, and P13R/R mice (unpublished data). Despite the substantial Vβ13 rearrangements, both P13−/− and P13R/R mice had virtually no Vβ13-expressing T cells (Fig. 1C), indicating that the variable gene promoter is required for efficient rearrangement as well as expression of the rearranged products of specific variable genes.
FIG. 2.
Promoter mutations diminish Vβ13 rearrangement but not allelic exclusion. (A) Schematic diagram of recombination reaction and assays for CJ, SE, and SJ. Gene segments are shown as open boxes. RSS are shown as open or filled triangles. Arrows indicate the position and direction of PCR primers. (B) Vβ13 CJ formation is inhibited in promoter-mutant mice. Vβ13, Vβ8.1, and Vβ12 CJ were amplified by PCR in DNA from total or purified DN thymocytes of +/+, P13+/−, P13−/−, and P13R/R mice. PCR products were separated on agarose gels and then transferred and hybridized with specific Vβ probes. DNA from total or DN thymocytes of wild-type mice was serially diluted into RAG2−/− kidney DNA and then amplified to determine the linear range of the PCR assay. JAK3 was amplified to verify DNA quality and relative amount. Numbers indicate the band intensities normalized to that of JAK3. The level of rearrangement in wild-type DNA was arbitrarily defined as 1. PCR assays were performed five times with two independently isolated total thymocyte DNA samples and three times with DN thymocyte DNA. Representative results from one set of the PCR assays are shown. (C) Vβ13 CJ formation is inhibited by expression of a TCR transgene. Vβ13-to-Dβ1Jβ1 CJ of the wild-type (wt) or the mutant (mt) Vβ13 allele were assayed separately using primers specific for the promoter sequences in the wild-type allele or the loxP sequences in the mutant allele, respectively. PCR assays for Vβ8.1 and Vβ12 rearrangements and for JAK3 were the same as in panel B. Representative results from three independent assays are shown.
Effect of promoter mutations on Vβ13 allelic exclusion.
To investigate the effect of the promoter mutations on allelic exclusion, functionally assembled TCR transgenes, encoding both the α and β chains of a TCR called 2C (28), were introduced into the promoter-mutant mice. As shown for other TCRs, expression of the 2C TCR reduced the fraction of DP thymocytes slightly and promoted T-cell development into either the CD4 or CD8 lineage (28). However, the number and CD4 and CD8 staining profiles of thymocytes were similar in 2C TCR transgenic mice on either the wild-type or the promoter-mutant background (data not shown). As expected, expression of the TCR transgene blocked Vβ8.1 and Vβ12 CJ formation in wild-type, P13+/−, P13−/−, P13+/R, and P13R/R mice (Fig. 2C). Similarly, expression of the TCR transgene also blocked the CJ formation of both the wild-type and the mutant Vβ13 alleles in wild-type, P13+/−, P13−/−, P13+/R, and P13R/R mice. When a TCRβ transgene alone was introduced into P13+/− mice, rearrangement of both the wild-type and the mutant Vβ13 allele was also inhibited (data not shown). Thus, in the absence of the normal variable gene promoter, the Vβ13 gene segment appears to undergo allelic exclusion.
Effect of promoter mutations on Vβ13 SJ formation.
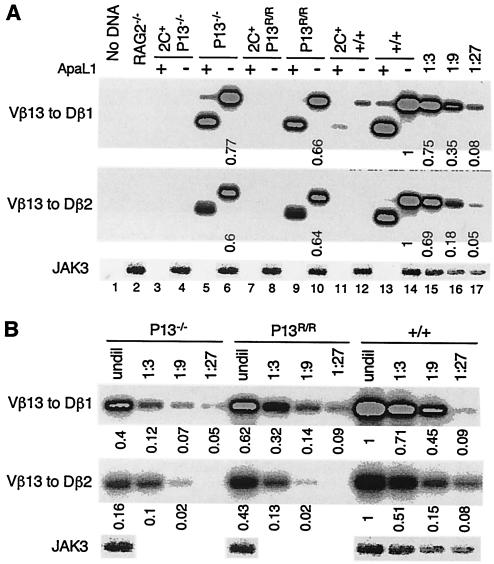
During V(D)J recombination, SE are joined to form SJ (Fig. 2A). The diminished Vβ13 rearrangement in promoter-mutant mice is expected to be accompanied by a corresponding decrease in Vβ13 SJ. In wild-type mice, SJ resulting from Vβ13-to-Dβ1 or -to-Dβ2 rearrangements were readily detected (Fig. 3A, lanes 14 to 17). Most of these SJ were cleaved by ApaL1 (lane 13), indicating precise joining of two SE. In the presence of the TCR transgene, only a low level of Vβ13-to-Dβ1 SJ was detected (lanes 11 and 12), consistent with inhibition of Vβ13 rearrangement under allelic exclusion. In both P13−/− and P13R/R mice, Vβ13 (to Dβ1 or to Dβ2) SJ were detected, but the levels were reduced approximately nine- and threefold, respectively, compared to those in wild-type mice (Fig. 3). As in wild-type mice, the majority of these SJ were cleaved by ApaL1. In the presence of the TCR transgene, no Vβ13 SJ were detected in the promoter-mutant mice (Fig. 3A, lanes 3, 4, 7, and 8). Thus, diminished Vβ13 rearrangement in promoter-mutant mice is associated with a corresponding reduction in the level of Vβ13 SJ, in either the presence or the absence of a TCR transgene.
FIG. 3.
Expression of a TCR transgene abolishes Vβ13 SJ formation in promoter-mutant mice. (A) Analysis of Vβ13 SJ in various types of mice. Vβ13-to-Dβ1 or -to-Dβ2 SJ were assayed by PCR in thymocyte DNA from RAG2−/− mice or +/+, P13−/−, and P13R/R mice in the presence or absence of the 2C TCR transgene. Half of the PCR products were digested with ApaLI before separation on agarose gels. PCR products were hybridized with a Vβ13-specific oligonucleotide probe. Shown are representative data from three experiments. (B) Comparison of the levels of Vβ13 SJ in wild-type and promoter-mutant mice. DNA samples were serially diluted and then used to assay for Vβ13-to-Dβ1 or -to-Dβ2 SJ. Numbers indicate the band intensities normalized to that of JAK3. The level of Vβ13 SJ in wild-type DNA was arbitrarily defined as 1. Shown are representative data from three experiments.
Effect of promoter mutations on Vβ13 cleavages.
To examine whether reduced Vβ13 CJ and SJ formation in promoter-mutant mice is associated with diminished Vβ13 cleavage, we measured the levels of Vβ13 SE in thymocyte DNA using LM-PCR (Fig. 2A). Compared to that in wild-type mice, a similar level of Vβ13 SE was detected in P13R/R mice, whereas the level of Vβ13 SE in P13−/− mice was reduced approximately threefold (Fig. 4A). In general, the same relative levels of Vβ13 SE were detected in wild-type, P13−/−, and P13R/R mice using independently isolated DNA samples, although there were variations in specific amounts of SE detected in different experiments. The smaller-than-expected reduction in SE in P13−/− and in P13R/R mice could be because the LM-PCR assay for SE is not as sensitive as PCR assays for CJ and SJ. As a result, the assay may not be sufficiently sensitive to detect the difference in Vβ13 SE corresponding to a twofold difference in Vβ13 rearrangement in P13R/R mice. It is also possible that in P13R/R mice the introduced Gal4 sequences and SV40 minimal promoter may have impaired Vβ13 CJ formation without significantly affecting Vβ13 cleavage. Nevertheless, in P13−/− mice, the reduced Vβ13 rearrangement is correlated in large part with a reduced level of Vβ13 cleavage (accessibility).
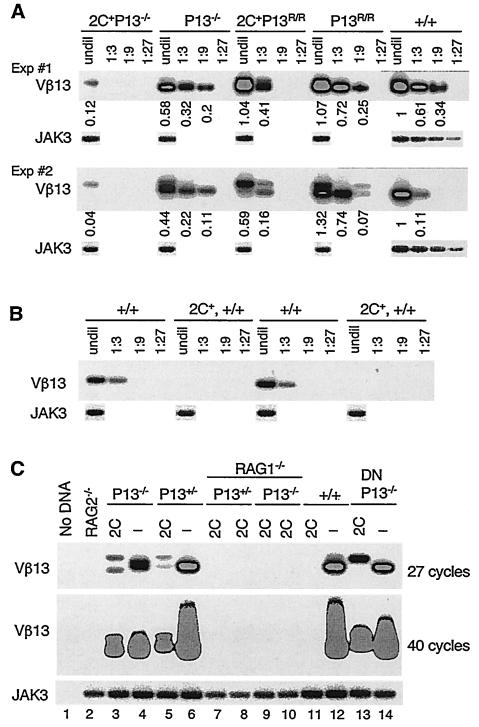
FIG. 4.
Expression of a TCR transgene fails to abolish Vβ13 cleavages in promoter-mutant mice. (A and B) Comparison of the levels of Vβ13 SE in wild-type and promoter-mutant mice. The levels of Vβ13 SE were assayed by LM-PCR in thymocyte DNA from +/+, P13−/−, and P13R/R mice with or without the TCR transgene. DNA was either undiluted (undil) or serially diluted (every threefold) and then amplified. PCR products were separated on agarose gels and hybridized with an oligonucleotide probe. JAK3 was amplified as in Fig. 2B and then hybridized with an oligonucleotide probe. Numbers indicate the band intensities normalized to that of JAK3. The level of Vβ13 SE in wild-type DNA was arbitrarily defined as 1. Shown are representative data using two sets of independently isolated DNA samples of each genotype. (C) Vβ13 cleavages in TCR transgenic promoter-mutant mice are RAG1 dependent. Vβ13 SE were assayed by LM-PCR in thymocyte DNA of RAG2−/− mice, +/+, P13+/−, and P13−/− mice with or without the 2C TCR transgene and 2C+ P13−/− RAG1−/− and 2C+ P13+/− RAG1−/− mice. Lanes 13 and 14 are DNA from purified DN thymocytes of 2C+ P13−/− and P13−/− mice, respectively. One set of PCR assays was carried out for 27 cycles, and the other set was carried out for 40 cycles. Shown are representative data from three experiments.
Vβ13 cleavages under allelic exclusion.
Allelic exclusion is associated with an inhibition of variable gene cleavage (33). Corresponding to the block of CJ formation, no Vβ13 SE was detected in thymocyte DNA of TCR transgenic mice on the wild-type background (Fig. 4B and C). In 2C+ P13−/− mice, although a Vβ13 cleavage product was consistently detected, the level was reduced approximately 15- to 20-fold and another 3-fold compared to those in P13−/− and wild-type mice, respectively. Noticeably, the size of the cleavage products was altered compared to that in P13−/− mice (Fig. 4A and C). Similarly, Vβ13 cleavage products were detected in 2C+ P13R/R mice, but the levels varied considerably in independent experiments using thymocyte DNA from different mice, despite our best efforts (Fig. 4A and data not shown). Because the sizes of PCR products varied in some of the assays with different DNA, the PCR product may represent amplification of rare cleavage products. Nevertheless, in all 2C+ P13−/− and 2C+ P13R/R samples analyzed (>5 for each), Vβ13, but never Vβ8.1 or Vβ12, cleavage products were always detected (Fig. 4 and unpublished data). When a different TCR transgene was expressed in P13−/− mice, a low level of Vβ13 cleavage products was also detected (data not shown).
Vβ13 cleavage products were cloned and sequenced. In wild-type mice, among 31 Vβ13 SE products sequenced, 26 started with the heptamer (CACAGTA) at the 5′ end (Table 3), 3 started with ACTCAGA 8 nucleotides downstream of the heptamer (the first C of the heptamer is counted as nucleotide −1), and 2 started within the Vβ13 coding sequences. These results indicate that the majority of Vβ13 cleavages in wild-type thymocytes occur precisely at the junction between RSS and coding sequences and only a small fraction of the cleavages is imprecise (13%). As in wild-type mice, most of Vβ13 SE in P13−/− mice were derived from precise cleavages at the Vβ13-RSS junction, and a similar fraction of the cleavages was aberrant (18%). In contrast, in the presence of the TCR transgene, all 30 Vβ13 cleavage products in P13−/− mice were derived from imprecise cleavages either within the Vβ13 coding sequences or the RSS. Similarly, in 2C+ P13R/R mice, all 24 Vβ13 cleavage products sequenced were derived from imprecise cleavages within either the Vβ13 coding sequences or the RSS. Furthermore, in P13R/R mice in the absence of the TCR transgene, among 19 Vβ13 SE products sequenced 9 were derived from aberrant cleavages (47%).
TABLE 3.
Comparison of Vβ13 cleavage sites in wild-type and promoter-mutant mice
| Genotype | Cleavage sitea | No. sequenced | % Aberrant cleavages |
|---|---|---|---|
| +/+ | CACAGTA (Hep) | 26 | 13 |
| CTCCACT (+68) | 1 | ||
| CACCTAT (+26) | 1 | ||
| ACTCAGA (−8) | 2 | ||
| P13−/− | CACAGTA (Hep) | 23 | 18 |
| ACTAACT (+73) | 3 | ||
| ACTCAGA (−8) | 2 | ||
| 2C+ P13−/− | ACTAACT (+73) | 5 | 100 |
| CACTCTC (+65) | 19 | ||
| ACTCCTC (+69) | 3 | ||
| CACCTAT (+26) | 2 | ||
| ACTCAGA (−8) | 1 | ||
| P13R/R | CACAGTA (Hep) | 10 | 47 |
| ACTAACT (+73) | 2 | ||
| ACTCCAC (+69) | 3 | ||
| ACACAGC (+33) | 3 | ||
| ACTCAGA (−8) | 1 | ||
| 2C+ P13R/R | ACTAACT (+73) | 2 | 100 |
| CACTCTC (+65) | 8 | ||
| ACACAGC (+33) | 2 | ||
| ACTCAGA (−8) | 10 |
Boldface indicates the normal heptamer (Hep) of RSS.
Although cleavage products that have ends starting within the RSS might have been generated by nuclease processing of SE derived from cleavage at the normal Vβ13-RSS junction, the postcleavage processing of normal SE does not generate products that have ends starting within Vβ13 coding sequences. The aberrant Vβ13 cleavages observed in TCR transgenic promoter-mutant mice are RAG dependent, because in the absence of RAG1 no Vβ13 cleavage products were detected in thymocytes of 2C+ P13−/− or 2C+ P13+/− mice, even when PCRs were carried out for 40 cycles (Fig. 4C, lanes 7 to 11). In addition, the aberrant Vβ13 cleavage products were detected in both DN and DP thymocytes of 2C+ P13−/− and 2C+ P13R/R mice (Fig. 4C, lanes 13 and 14, and data not shown). Thus, the observed Vβ13 SE in TCR transgenic promoter-mutant mice most likely reflects rare aberrant cleavages of Vβ13, consistent with a lack of Vβ13 CJ and SJ formation.
Effect of promoter mutations on Vβ13 GT.
To examine the effect of the promoter mutations on Vβ13 transcription, we measured the levels of Vβ13 GT in thymocytes by RT-PCR. Vβ13 GT was readily detected in wild-type thymocytes (Fig. 5A, lanes 17 to 19). In the presence of the 2C TCR transgene, the level was decreased approximately 10- to 20-fold (lanes 2 to 4), consistent with previous observations (6, 26). In P13−/− thymocytes, the levels of Vβ13 GT were about fivefold lower than in wild-type mice (lanes 5 to 7). In the presence of the TCR transgene, the level was further reduced two- to threefold (lanes 8 to 10). Similarly, the level of Vβ13 GT was about fivefold lower in P13R/R mice than in wild-type mice (lanes 11 to 13). However, expression of the TCR transgene in P13R/R mice did not result in any further reduction in the levels of Vβ13 GT (lanes 14 to 16). As controls, similar levels of Vβ12 and Vβ8.1 GT were detected in thymocytes of wild-type, P13−/−, and P13R/R mice. Expression of the TCR transgene resulted in a 10- to 20-fold decrease in the levels of Vβ12 and Vβ8.1 GT in wild-type and P13−/− mice, but somewhat less in P13R/R mice. Thus, germ line Vβ13 is transcribed in the absence of the normal promoter, and most of the residual transcription is inhibited by the expression of a TCR transgene in P13−/−, but not in P13R/R, mice.
FIG. 5.
Initiation sites of Vβ13 germ line transcription are altered by the expression of a TCR transgene. (A) Vβ13 germ line transcription occurs in promoter-mutant mice. GT of Vβ13, Vβ12, and Vβ8.1 were assayed by nested RT-PCR in thymocyte RNA from +/+, P13−/−, and P13R/R mice with or without the 2C TCR transgene. cDNA was serially diluted before the PCR assay. The levels of β-actin transcript were assayed to verify RNA quality and relative amounts. Numbers indicate the band intensities normalized to that of β-actin. The level of Vβ13 GT in wild-type thymocytes was arbitrarily defined as 1. Data from two separate experiments are shown. (B) Initiation sites of Vβ13 germ line transcription are altered by the expression of a TCR transgene. RNA from +/+, P13−/−, P13R/R, and RAG2−/− mice with or without the 2C TCR transgene was reverse transcribed into cDNA. The initiation sites were then determined by RACE-PCR using the cDNA samples. PCR products were separated on agarose gels and hybridized with a Vβ13-specific probe. The sizes of the marker DNA are indicated. Shown are representative data from three experiments. (C) Schematic diagrams of initiation sites of Vβ13 GT in various mice. PCR products from the experiment shown in panel B were cloned and sequenced (results not shown). The initiation sites were determined based on sequencing and the sizes of the PCR products in panel B. The leader and Vβ13 exon are depicted. Triangles indicate loxP sequences. Open circles represent five copies of Gal4 sequences plus the SV40 minimal promoter. The higher arrows indicate the major initiation sites.
In promoter-mutant mice, a 1.2-kb region including the known promoter and transcription initiation site was deleted or replaced. Vβ13 GT in P13R/R mice might have been initiated from the SV40 minimal promoter that was used to replace the endogenous promoter. However, Vβ13 GT in P13−/− mice must have been initiated from previously unknown sites upstream of the Vβ13 coding sequence. To map the transcription initiation sites in the absence of the normal promoter, we carried out RACE assays. In RAG2−/− thymocytes, most Vβ13 transcription was initiated from one major site (Fig. 5B, lane 8), which corresponds to the correct promoter and is consistent with a previous report (2). In P13−/− and P13R/R thymocytes, however, two major RACE products were detected (lanes 4 and 6), suggesting that Vβ13 transcription is initiated from two major sites. Noticeably, in the presence of the TCR transgene, the sizes of the RACE products were dramatically altered in RAG2−/−, P13−/−, and P13R/R mice. Based on the sizes and sequences of the RACE products (data not shown), most Vβ13 transcription initiation sites were mapped to the normal promoter region in RAG2−/− and wild-type mice (Fig. 5C) and to a 1.5-kb region immediately upstream of the normal promoter in P13−/− and P13R/R mice.
DISCUSSION
The variable gene promoter is required for efficient Vβ cleavage, rearrangement, and transcription.
In mice, the Vβ13 gene segment resides approximately in the middle of the Vβ cluster and is closely flanked by Vβ8.1 and Vβ12 gene segments (22). In the promoter-mutant mice, Vβ8.1 and Vβ12 rearrangements were not significantly affected, as indicated by their normal cleavage, joining, allelic exclusion, and expression. In contrast, deletion of the Vβ13 promoter resulted in a significant reduction of Vβ13 CJ formation (5- to 10-fold) (Table 4). Correspondingly, the levels of Vβ13 SJ, SE, and GT were reduced ∼9-, ∼3-, and ∼5-fold, respectively. The smaller-than-expected reduction in the levels of Vβ13 SE could be due to the limitations of semiquantitative LM-PCR assays for steady-state levels of SE. Furthermore, the residual Vβ13 rearrangements in P13−/− mice had normal levels of nucleotide deletions and additions, indicating that the promoter deletion did not affect the recombination reaction per se. Together, these findings suggest that the variable gene promoter regulates Vβ rearrangement by promoting access of its associated Vβ gene segment to RAG-mediated cleavage.
TABLE 4.
Reduction of Vβ13 CJ, SJ, SE, and GT in P13−/− and P13R/R mice compared to wild-type mice
| Segment | Fold reduction
|
|||
|---|---|---|---|---|
| P13−/− | 2C+ P13−/− | P13R/R | 2C+ P13R/R | |
| CJ | 5-10 | NDa | 2 | ND |
| SJ | 9 | ND | 3 | ND |
| SE | 3 | 45-60 | No change | 2-10 |
| GT | 5 | 10-15 | 5 | 5 |
ND, not detectable.
Studies have shown that the Vβ gene segments themselves as well as the sequences between gene segments are associated with acetylated histones in DN thymocytes (35). Based on these data, the presence of a global regulator has been proposed to control the accessibility of the entire Vβ cluster (35). While the putative global regulator might have contributed to the residual Vβ13 accessibility and rearrangement observed in P13−/− mice, our results strongly suggest that the individual promoter also plays a critical role in targeting specific Vβ gene for rearrangement. Previously, our investigators have shown that when the Vβ13 gene segment together with the promoter were inserted upstream of the Dβ1 gene segment, the inserted Vβ13 gene segment rearranged at the same frequency as the natural copy (29). Together, these findings strongly suggest that the variable gene promoter plays an essential role in regulating local access of gene segments for rearrangement.
Regulation of local recombination accessibility by the variable gene promoter is analogous to the PDβ1 promoter that regulates access and rearrangement of the proximal Dβ1 and Jβ1, but not the distal Dβ2 and Jβ2, gene segments (22, 40, 41). Thus, a general mechanism of promoter control of V(D)J recombination is probably by regulating local accessibility. The local control of accessibility may also contribute to the observed differences in usages and recombination efficiencies among different Vβ gene segments (21). Different Vβ promoters share conserved sequence motifs but also exhibit significant sequence differences (2, 12). The conserved cis elements may contribute to the overall regulation, and the different cis elements may underlie the differences among gene segments. As in the TCRβ locus, each variable gene segment in other antigen receptor loci is associated with its own promoter. It is possible that these promoters also regulate their associated variable gene rearrangement by a similar mechanism (3).
A critical role of the variable gene promoter in mediating Vβ cleavage, joining, and allelic exclusion is further supported by the differences observed between the deletion and replacement mutations of the Vβ13 promoter. Higher levels of Vβ13 CJ and SE were observed in P13R/R mice than in P13−/− mice (Table 4), indicating that the inserted SV40 minimal promoter and/or Gal4 sequences can partly compensate for the loss of the normal promoter. However, no significant difference in the level of Vβ13 SE was detected between P13R/R and wild-type mice. Several nonexclusive mechanisms could account for this discrepancy. First, the LM-PCR assay may not be sufficiently sensitive to detect differences in SE corresponding to twofold differences in Vβ13 rearrangement. Consistent with this possibility, reduction of Vβ13 SE in P13−/− mice was also less than expected from the reduction in the levels of Vβ13 CJ and SJ. Second, Vβ13 SJ could be recleaved, resulting in an elevated level of SE. Third, some of the Vβ13 cleavages may not lead to CJ formation either because the normal variable gene promoter is required for efficient CJ formation or because the cleavages were aberrant. In support of the latter possibility, a significantly higher fraction of Vβ13 cleavages in P13R/R mice was imprecise, and these cleavages did not contribute to CJ formation as indicated by PCR amplification and sequencing (unpublished data). Regardless of the precise mechanisms, the observed differences in Vβ13 cleavage and rearrangement in P13−/− and P13R/R mice suggest a critical influence of the sequences in the promoter region on Vβ accessibility and cleavage.
The variable gene promoter is not required for allelic exclusion.
One of the remarkable regulations of antigen receptor gene assembly is allelic exclusion. TCRβ allelic exclusion is controlled at the step of Vβ gene rearrangement and is initiated by expression of the pre-TCR complex, consisting of TCRβ, pTα, and CD3 proteins (39). Studies have shown that TCRβ allelic exclusion is associated with changes in Vβ chromatin structures and accessibility to nuclease (6, 17, 26, 35). However, cis elements that mediate allelic exclusion are not known. When the Vβ13 gene segment together with the promoter were inserted upstream of the Dβ1 gene segment, the inserted Vβ13 gene segment continued to rearrange in the presence of a TCR transgene expression (29), indicating that the promoter alone is not sufficient to mediate allelic exclusion. Complementary to this observation, we have now shown that in the absence of the normal promoter in both P13−/− and P13R/R mice, Vβ13 rearrangement was excluded by the expression of a TCR transgene. These findings suggest that the normal variable gene promoter is dispensable for allelic exclusion.
Unexpectedly, Vβ13 cleavage products were detected in both 2C+ P13−/− and 2C+ P13R/R mice (Fig. 4). Although the levels of cleavage were variable and low, especially in 2C+ P13−/− mice, Vβ8.1 and Vβ12 cleavage products were never detected in the same DNA samples. The more-abundant cleavages in 2C+ P13R/R mice seem to correlate with a higher level of germ line Vβ13 transcription and a lack of inhibition of this transcription by the TCR transgene. Although RAG dependent, all Vβ13 cleavages in 2C+ P13−/− and 2C+ P13R/R mice took place within Vβ13 coding sequence and RSS and did not result in CJ or SJ formation, suggesting aberrant cleavage of the Vβ13 gene segment alone. In addition, we found that initiation sites of Vβ13 germ line transcription were altered by the expression of the TCR transgene in RAG2−/− mice as well as in P13−/− and P13R/R mice. Because of the promoter mutations, the initiation sites and their alterations following TCR transgene expression were different between wild-type and mutant mice. These differences might underlie the occurrence of aberrant Vβ13 cleavages in promoter-mutant mice. Consistent with this hypothesis, a recent study showed that Dβ1 accessibility in a recombination substrate is determined by position and orientation of the PDβ1 promoter but not by histone acetylation (30).
Because of the low frequency and variability, the implication of the observed aberrant Vβ cleavages on the role of the variable gene promoter in V(D)J recombination is unclear at the present time. Findings reported here suggest a need to further investigate whether the variable gene promoter suppresses aberrant Vβ cleavage during allelic exclusion. Importantly, our findings, for the first time, demonstrate that the normal variable gene promoter is required for efficient cleavage, rearrangement, and transcription of its associated Vβ gene segment, but not for allelic exclusion.
Acknowledgments
We thank Tara Schmidt and Maria Fragoso for technical assistance, Herman N. Eisen, Tyler Jacks, Tania Baker, and Phillip Sharp for critical review of the manuscript, and members of the Chen laboratory for helpful discussions.
This work was supported in part by Public Health Service grants CA100875 and AI40146 (to J.C.) and a grant from the Ministry of Health & Welfare, Republic of Korea (02-PJ1-PG3-20908-0002 to C.J.R.). B.B.H. was partly supported by a Postdoctoral Fellowship from the American Cancer Society.
REFERENCES
- 1.Alt, F., T. K. Blackwell, and G. Yancopoulos. 1987. Development of the primary antibody repertoire. Science 238:1079-1087. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, S. J., H. S. Chou, and D. Y. Loh. 1988. A conserved sequence in the T-cell receptor β-chain promoter region. Proc. Natl. Acad. Sci. USA 85:3551-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, J. E., D. Cado, and D. H. Raulet. 1998. Developmentally programmed rearrangement of T cell receptor Vγ genes is controlled by sequences immediately upstream of the Vγ genes. Immunity 9:159-168. [DOI] [PubMed] [Google Scholar]
- 4.Bories, J.-C., J. Demengeot, L. Davidson, and F. W. Alt. 1996. Gene-targeted deletion and replacement mutations of the T-cell receptor β-chain enhancer: the role of enhancer elements in controlling V(D)J recombination accessibility. Proc. Natl. Acad. Sci. USA 93:7871-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvier, G., F. Watrin, M. Naspetti, C. Verthuy, P. Naquet, and P. Ferrier. 1996. Deletion of the mouse T-cell receptor β gene enhancer blocks αβ T-cell development. Proc. Natl. Acad. Sci. USA 93:7877-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chattopadhyay, S., C. Whitehurst, F. Schwenk, and J. Chen. 1998. Biochemical and functional analyses of chromatin changes at the TCRβ locus during CD4− CD8− to CD4+ CD8+ thymocyte differentiation. J. Immunol. 160:1256-1267. [PubMed] [Google Scholar]
- 7.Chen, J., F. Young, A. Bottaro, V. Stewart, R. Smith, and F. Alt. 1993. Mutations of the intronic IgH enhancer and its flanking sequences differentially affect accessibility of the JH locus. EMBO J. 12:4635-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eastman, Q. M., T. M. J. Leu, and D. G. Schatz. 1996. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature 380:85-88. [DOI] [PubMed] [Google Scholar]
- 9.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 10.Gellert, M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 11.Gorman, J. R., N. van der Stoep, R. Monroe, M. Cogne, L. Davidson, and F. W. Alt. 1996. The Igκ 3′ enhancer influences the ration of Igκ versus Igλ B lymphocytes. Immunity 5:241-252. [DOI] [PubMed] [Google Scholar]
- 12.Halle, J.-P., P.-H. Seuffert, C. Woltering, G. Stelzer, and M. Meisterernst. 1997. A conserved tissue-specific structure at a human T-cell receptor β-chain core promoter. Mol. Cell. Biol. 17:4220-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hempel, W. M., I. Leduc, N. Mathieu, R. K. Tripathi, and P. Ferrier. 1998. Accessibility control of V(D)J recombination: lessons from gene targeting. Adv. Immunol. 69:309-352. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez-Munain, C., B. P. Sleckman, and M. S. Krangel. 1999. A developmental switch from TCR delta enhancer to TCR alpha enhancer function during thymocyte maturation. Immunity 10:723-733. [DOI] [PubMed] [Google Scholar]
- 15.Krangel, M. S. 2001. V(D)J recombination becomes accessible. J. Exp. Med. 193:F27-F30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levelt, C. N., and K. Eichmann. 1995. Receptors and signals in early thymic selection. Immunity 3:667-672. [DOI] [PubMed] [Google Scholar]
- 17.Mathieu, N., W. M. Hempel, S. Spicuglia, C. Verthuy, and P. Ferrier. 2000. Chromatin remodeling by the T cell receptor (TCR)-beta gene enhancer during early T cell development: implications for the control of TCR-beta locus recombination. J. Exp. Med. 192:625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathieu, N., S. Spicuglai, S. Gorbatch, O. Cabaud, C. Fernex, C. Verthuy, W. M. Hempel, A.-O. Huber, and P. Ferrier. 2003. Assessing the role of the T cell receptor β gene enhancer in regulating coding joint formation during V(D)J recombination. J. Biol. Chem. 278:18101-18109. [DOI] [PubMed] [Google Scholar]
- 19.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 20.Neiditch, M. B., G. S. Lee, L. E. Huye, V. L. Brandt, and D. B. Roth. 2002. The V(D)J recombinase efficiently cleaves and transposes signal joints. Mol. Cell 9:871-878. [DOI] [PubMed] [Google Scholar]
- 21.Okada, C. Y., and I. L. Weissman. 1989. Relative Vβ transcript levels in thymus and peripheral lymphoid tissues from various mouse strains. J. Exp. Med. 169:1703-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowen, L., B. F. Koop, and L. Hood. 1996. The complete 685-kilobase DNA sequence of the human β T cell receptor locus. Science 272:1755-1762. [DOI] [PubMed] [Google Scholar]
- 23.Schlissel, M., A. Constantinescu, T. Morrow, M. Baxter, and A. Peng. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent and cell cycle regulated. Genes Dev. 7:2520-2532. [DOI] [PubMed] [Google Scholar]
- 24.Schlissel, M. S., and P. Stanhope-Baker. 1997. Accessibility and the developmental regulation of V(D)J recombination. Semin. Immunol. 9:161-170. [DOI] [PubMed] [Google Scholar]
- 25.Schwenk, F., U. Baron, and K. Rajewsky. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senoo, M., and Y. Shinkai. 1998. Regulation of Vβ germline transcription in RAG-deficient mice by the CD3ɛ-mediated signals: implication of Vβ transcriptional regulation in TCRβ allelic exclusion. Int. Immunol. 10:553-560. [DOI] [PubMed] [Google Scholar]
- 27.Serwe, M., and F. Sablitzky. 1993. V(D)J recombination in B cells is impaired but not blocked by targeted deletion of the immunoglobulin heavy chain intron enhancer. EMBO J. 12:2321-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sha, W. C., C. A. Nelson, R. D. Newberry, D. M. Kranz, J. H. Russell, and D. Y. Loh. 1988. Selective expression of an antigen receptor on CD8-bearing T lymphocytes in transgenic mice. Nature 335:271-274. [DOI] [PubMed] [Google Scholar]
- 29.Sieh, P., and J. Chen. 2001. Distinct control of the frequency and allelic exclusion of the Vβ gene rearrangement at the TCRβ locus. J. Immunol. 167:2121-2129. [DOI] [PubMed] [Google Scholar]
- 30.Sikes, M. L., A. Meade, R. Tripathi, M. S. Krangel, and E. M. Oltz. 2002. Regulation of V(D)J recombination: a dominant role for promoter positioning in gene segment accessibility. Proc. Natl. Acad. Sci. USA 99:12309-12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sleckman, B. P., C. G. Bardon, R. Ferrini, L. Davidson, and F. W. Alt. 1997. Function of the TCRα enhancer in αβ and γδ T cells. Immunity 7:505-515. [DOI] [PubMed] [Google Scholar]
- 32.Sleckman, B. P., J. R. Gorman, and F. W. Alt. 1996. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu. Rev. Immunol. 14:459-481. [DOI] [PubMed] [Google Scholar]
- 33.Stanhope-Baker, P., K. M. Hudson, A. L. Shaffer, A. Constantinescu, and M. S. Schlissel. 1996. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell 85:887-897. [DOI] [PubMed] [Google Scholar]
- 34.Steen, S. B., L. Gomelsky, and D. B. Roth. 1996. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells 1:543-553. [DOI] [PubMed] [Google Scholar]
- 35.Tripathi, R., A. Jackson, and M. S. Krangel. 2002. A change in the structure of Vβ chromatin associated with TCR beta allelic exclusion. J. Immunol. 168:2316-2324. [DOI] [PubMed] [Google Scholar]
- 36.Tycko, B., and J. Sklar. 1990. Chromosomal translocations in lymphoid neoplasia: a reappraisal of the recombinase model. Cancer Cells 2:1-8. [PubMed] [Google Scholar]
- 37.van Gent, D. C., J. F. McBlane, D. A. Ransden, M. J. Sadofsky, J. E. Hesse, and M. Gellert. 1995. Initiation of V(D)J recombination in a cell-free system. Cell 81:925-934. [DOI] [PubMed] [Google Scholar]
- 38.van Gent, D. C., D. A. Ramsden, and M. Gellert. 1996. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell 85:107-113. [DOI] [PubMed] [Google Scholar]
- 39.von Boehmer, H., and H. J. Fehling. 1997. Structure and function of the pre-T cell receptor. Annu. Rev. Immunol. 15:433-452. [DOI] [PubMed] [Google Scholar]
- 40.Whitehurst, C., S. Chattopadhyay, and J. Chen. 1999. Control of V(D)J recombination accessibility of the Dβ1 gene segment at the TCRβ locus by a germline promoter. Immunity 10:313-322. [DOI] [PubMed] [Google Scholar]
- 41.Whitehurst, C. E., M. S. Schlissel, and J. Chen. 2000. Deletion of germline promoter PDβ1 from the TCRβ locus causes hypermethylation that impairs Dβ1 recombination by multiple mechanisms. Immunity 13:703-714. [DOI] [PubMed] [Google Scholar]
- 42.Xu, Y., L. Davidson, F. W. Alt, and D. Baltimore. 1996. Deletion of the Igκ light chain intronic enhancer/matrix attachment region impairs but does not abolish VκJκ rearrangement. Immunity 4:377-385. [DOI] [PubMed] [Google Scholar]