Abstract
Gene silencing in the budding yeast Saccharomyces cerevisiae requires the enzymatic activity of the Sir2 protein, a highly conserved NAD-dependent deacetylase. In order to study the activity of native Sir2, we purified and characterized two budding yeast Sir2 complexes: the Sir2/Sir4 complex, which mediates silencing at mating-type loci and at telomeres, and the RENT complex, which mediates silencing at the ribosomal DNA repeats. Analyses of the protein compositions of these complexes confirmed previously described interactions. We show that the assembly of Sir2 into native silencing complexes does not alter its selectivity for acetylated substrates, nor does it allow the deacetylation of nucleosomal histones. The inability of Sir2 complexes to deacetylate nucleosomes suggests that additional factors influence Sir2 activity in vivo. In contrast, Sir2 complexes show significant enhancement in their affinities for acetylated substrates and their sensitivities to the physiological inhibitor nicotinamide relative to recombinant Sir2. Reconstitution experiments showed that, for the Sir2/Sir4 complex, these differences stem from the physical interaction of Sir2 with Sir4. Finally, we provide evidence that the different nicotinamide sensitivities of Sir2/Sir4 and RENT in vitro could contribute to locus-specific differences in how Sir2 activity is regulated in vivo.
Heterochromatin, the highly condensed portion of eukaryotic chromosomes, is essential for proper chromosome structure and function (20, 21). In the budding yeast Saccharomyces cerevisiae, a mechanism akin to heterochromatin (referred to as gene silencing) blocks transcription and recombination at certain regions of the genome (39, 48). At the silent mating-type cassettes, gene silencing ensures the maintenance of haploid cell identity (23). Gene silencing adjacent to telomeres contributes to normal telomere positioning within the nucleus as well as to telomere stability (19, 43). In addition, a form of gene silencing limits recombination within the ribosomal DNA (rDNA) repeats (7, 18, 59). This function also governs the replicative life span of budding yeast mother cells (57).
Silencing at mating-type loci, telomeres, and the rDNA repeats requires histone deacetylation by the Sir2 protein (2, 6, 25, 49, 67). Sir2 is the founding member of a broadly conserved class of histone deacetylase enzymes known as NAD-dependent deacetylases (27, 35, 60). In contrast to other histone deacetylases, which catalyze the simple hydrolysis of the acetyl-lysine linkage on acetylated histone tails, these enzymes employ a more complicated mechanism that requires the participation of NAD. The deacetylation reaction is coupled to cleavage of the N-glycosidic bond at C-1 of NAD, the release of nicotinamide, and the transfer of the acetyl group from the substrate to ADP-ribose (66, 68). The products of this reaction are deacetylated lysine, nicotinamide, and 2′-O-acetyl-ADP-ribose, a novel metabolite (10, 30, 51). Although the biological significance of this mechanism is not understood, it may serve to couple the deacetylation of certain substrates to the metabolic state of the cell (22).
The deacetylase activity of Sir2 operates within the context of two distinct sets of silencing proteins. A complex of Sir2 and Sir4 as well as the Sir3 protein is the primary constituent of silent chromatin at mating-type loci and at telomeres, whereas a complex containing Sir2, Net1 (also known as Cfi1), and Cdc14 mediates silencing at rDNA (40, 55, 63, 72). Whereas the mechanism of the Sir2 reaction has been extensively studied by the use of recombinant Sir2 proteins, there is little information on the activity of native Sir2 complexes, and thus a number of questions regarding Sir2 substrate specificity and regulation remain unanswered. First, which lysine residues on the histone tails are deacetylated by Sir2 in vivo? The recombinant Sir2 enzyme has a marked preference for the deacetylation of lysine 16 of histone H4 in vitro, and mutation of lysine 16 has a much more pronounced effect on silencing than mutation of other lysine residues in the histone tails (27, 33, 68). However, chromatin immunoprecipitation studies demonstrate hypoacetylation of every histone tail lysine in silent chromatin in vivo (65). Second, is Sir2 alone sufficient to deacetylate histone tails in the context of a nucleosome? Although recombinant Sir2 is active on histone peptides and mixtures of histones, its activity on more physiological chromatin substrates has not been tested.
Third, is Sir2 regulated in vivo by its assembly into silencing complexes? With enzymatically inactive mutants of Sir2, it has been shown that activity is required for the proper localization of Sir2, as well as Sir3 and Sir4, to the silent mating-type loci and to telomeres in vivo. These mutant proteins can still localize to rDNA, suggesting that there may be locus-specific differences in the function and regulation of Sir2 activity (25).
In this study, we addressed these questions by purifying and characterizing native Sir2 complexes from budding yeast. Using a combination of affinity purification and mass spectrometry, we provide a complete analysis of the protein compositions of these complexes. We also compare the enzymatic activities of Sir2 complexes to the activity of recombinant Sir2. Our results provide further insight into the physiological targets of Sir2 deacetylase activity. In addition, they implicate protein-protein interactions as an important mechanism for the regulation of Sir2 in vivo.
MATERIALS AND METHODS
Yeast strains and plasmids.
The yeast strains used for this study are listed in Table 1. The SIR2-TAP (DMY1704) and NET1-TAP (DMY1690) strains have been described previously (26). Deletion of the SIR4 gene in DMY1704 by the transformation of a BglII-PvuII fragment from plasmid pSIR4::LEU2 (29) yielded strain DMY1821. A NET1-FLAG allele was engineered in this strain by the transformation of a PCR product from plasmid pDM714 to yield strain DMY2636 (referred to as SIR2-TAP, sir4Δ, and NET1-FLAG in Fig. 1). Plasmid pDM714 was derived from pFA6a-GST-kanMX6 (38), whereby glutathione S-transferase (GST) was replaced with sequences encoding a single FLAG epitope on an AscI-PacI restriction fragment. PCR and transformation with plasmid pDM714 were performed as described previously (38).
TABLE 1.
Yeast strains used for this study
| Strain | Genotype | Source |
|---|---|---|
| SF10 | BJ459 MATaura3-52 trp1 lys2-801 leu2Δ1 pep4Δ::HIS3 prb1Δ1.6R can1 | 24 |
| DMY1704 | SF10 SIR2-TAP::TRP1Kl | 25 |
| DMY2636 | SF10 SIR2-TAP::TRP1Kl sir4Δ::LEU2 NET1-FLAG::kanMX6 | This study |
| DMY1690 | SF10 NET1-TAP::TRP1Kl | 25 |
| DMY2377 | W303-1a pep4Δ::LEU2 | A. Rudner |
| DMY2640 | DMY2377 TAP-SIR4 | This study |
| DMY2843 | W303-1a TEL-VIIL::URA3 | A. Rudner |
| DMY2844 | W303-1a adh4::URA3 | A. Rudner |
| DMY2839 | DMY2843 sir2Δ::kanMX6 | This study |
| DMY2840 | DMY2843 pnc1Δ::kanMX6 | This study |
| DMY2841 | DMY2844 sir2Δ::kanMX6 | This study |
| DMY2842 | DMY2844 pnc1Δ::kanMX6 | This study |
| DMY2798 | W303-1a leu2::mURA3-LEU2 | J. Huang |
| DMY2800 | W303-1a NTS2::mURA3-LEU2 | J. Huang |
| DMY2804 | W303-1a NTS1::mURA3-LEU2 | J. Huang |
| DMY2828 | DMY2798 sir2Δ::kanMX6 | This study |
| DMY2829 | DMY2798 pnc1Δ::kanMX6 | This study |
| DMY2831 | DMY2800 sir2Δ::kanMX6 | This study |
| DMY2833 | DMY2800 pnc1Δ::kanMX6 | This study |
| DMY2835 | DMY2804 sir2Δ::kanMX6 | This study |
| DMY2837 | DMY2804 pnc1Δ::kanMX6 | This study |
FIG. 1.
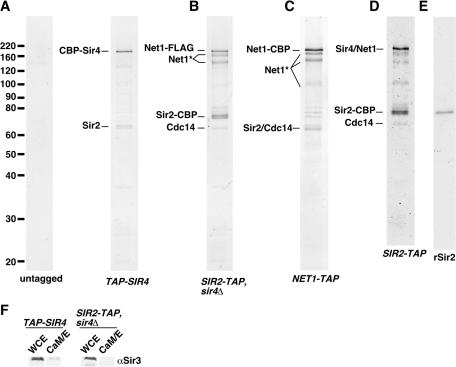
TAP of yeast Sir2. TAP purifications from various strains (the final elutions from the CaM-Sepharose column) were concentrated by TCA precipitation and analyzed by SDS-PAGE on 12.5% gels, followed by colloidal Coomassie blue staining. The positions of size markers (in kilodaltons) are shown on the left. The strains are listed in Table 1 as follows. (A) DMY2377 (untagged) and DMY2640 (TAP-SIR4); (B) DMY2636 (SIR2-TAP sir4Δ); (C) DMY1690 (NET1-TAP); and (D) DMY1704 (SIR2-TAP). (E) Twenty nanograms of recombinant 6His-Sir2 (rSir2) was analyzed in parallel with complexes. (F) Fractions from purifications from the TAP-SIR4 or SIR2-TAP sir4Δ strain were analyzed by Western blotting with a Sir3 antibody. WCE, whole-cell extract; CaM/E, final elution from the CaM-Sepharose column.
The TAP-SIR4 allele in strain DMY2640 was generated in strain DMY2377 in two steps. First, a Candida albicans URA3 gene was inserted immediately upstream of the SIR4 coding region, removing the ATG codon. This marker was transformed on a PCR fragment amplified from plasmid pAG60 (17). The URA3 cassette was then replaced by a markerless PCR fragment containing an N-terminal version of the tandem affinity purification (TAP) tag (from plasmid pBS1761) such that the TAP tag was integrated in frame with the SIR4 coding region (46). Transformants at this step were identified by counterselection on a medium containing 5-fluoroorotic acid (5-FOA). Correct integration in all transformants was verified by PCR. Strain DMY2377 is the untagged strain shown in Fig. 1; equivalent results were obtained for SF10, a strain that is isogenic to DMY2636 (data not shown).
Strains DMY2843 and DMY2844 were kindly provided by A. Rudner. Strains DMY2798, DMY2800, and DMY2804 were generated by the transformation of strain W303-1a, as described previously (26), and were kindly provided by J. Huang. Strains DMY2839, DMY2840, DMY2841, and DMY2842 were generated by the transformation of DMY2843 or DMY2844 with PCR products as described previously (38). Strains DMY2828, DMY2829, DMY2831, DMY2833, DMY2835, and DMY2837 were similarly derived from strain DMY2798, DMY2800, or DMY2804.
To generate a plasmid to overexpress SIR3-TAP, we generated a C-terminal fragment of an integrated SIR3-TAP allele by a PCR using genomic DNA from strain DMY1737 (25). This fragment was digested with XhoI and inserted into XhoI-cut pAR16 (a gift from S. Holmes [24]) to generate pDM598. Plasmid pTrx-Sir4, used to express the Trx-6His-Sir4(745-1172) protein, was a gift from J. Chang and T. Ellenberger. The plasmid used to express the CobB protein was a gift from R. Frye. Plasmids pDM607 (sir2-H364Y) and pDM608 (SIR2) were generated by subcloning an EcoRI-SalI fragment bearing the SIR2 locus from either pSIR2-LEU2 or pH364Y-LEU2 (67) into pRS314 (56).
Purification of TAP-tagged proteins.
For Sir2-TAP and TAP-Sir4 purification, cells were grown to late log phase (optical density at 600 nm [OD600] of ∼4) in a rich medium containing 1% yeast extract, 2% Bacto Peptone, and 4% glucose at 30°C. The cells were harvested, washed once in ice-cold 50 mM HEPES, pH 7.9, and frozen in liquid nitrogen in small chunks. Cell lysis was performed as described previously (25). After lysis, the frozen cell powder was suspended in an equal volume of ice cold buffer L (50 mM HEPES [pH 7.9], 300 mM KCl, 10% glycerol, 10 mM magnesium acetate, 1 mM EGTA, 0.2 mM EDTA, 10 mM β-glycerophosphate, 20 mM β-mercaptoethanol, 0.5% Nonidet P-40 [NP-40], 2 mM phenylmethylsulfonyl fluoride [PMSF], 4 mM benzamidine, and 2 μg each of leupeptin, bestatin, and pepstatin/ml). All subsequent steps were carried out at 4°C.
Extracts were mixed by stirring for 30 min and then were centrifuged at 15,000 × g for 15 min. The supernatant was bound directly to an immunoglobulin G (IgG)-Sepharose resin (Amersham). The IgG-Sepharose resin was prewashed in buffer W (20 mM HEPES [pH 7.9], 150 mM KCl, 5% glycerol, 1 mM EGTA, 0.1 mM EDTA, 10 mM β-mercaptoethanol, 0.1% NP-40, 1 mM PMSF, 2 mM benzamidine, 1 μg each of leupeptin, bestatin, and pepstatin/ml), and 0.5 ml of resin was bound to 30 ml of supernatant. After incubation with constant rocking, the resin was collected by centrifugation and transferred to a small column (Bio-Rad). The resin was washed with 15 to 20 ml of buffer W, followed by 5 ml of buffer TEV/C (20 mM Tris [pH 8.0], 150 mM KCl, 5% glycerol, 1 mM EDTA, 10 mM β-mercaptoethanol, 1 mM PMSF). Bound proteins were eluted by the addition of 1.5 ml of buffer TEV/C containing 100 U of recombinant TEV protease (Invitrogen) and rocking overnight (12 to 15 h). The eluates were collected and the resin was washed with a further 10 ml of buffer TEV/C. This wash was pooled with the eluate, and magnesium acetate, CaCl2, and imidazole were added to 1, 2, and 1 mM, respectively. Purification with a calmodulin (CaM)-Sepharose resin (Amersham) was performed essentially as described previously (46, 47), except that the final elution buffer (CaM/E) contained 300 mM KCl and 10 mM EGTA. Final eluates were used directly in activity assays.
For the purification of Sir3-CBP (Sir3 containing the CaM binding peptide) expressed from plasmid pDM598, cells carrying the plasmid were grown to an OD600 of 0.6 in synthetic complete medium lacking leucine and containing 2% raffinose (54). Galactose was then added to a 2% final concentration, and the cells were grown for another 12 h. Harvesting of the cells and protein purification were done as described above.
Gel filtration chromatography of purified complexes was done with a prepacked 24-ml Superose 6 column (Amersham) equilibrated with running buffer (20 mM Tris [pH 8.0], 300 mM KCl, 5% glycerol, 1 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM PMSF, 10 μg of purified GST protein/ml). Samples were loaded in a volume of 0.5 ml, and 0.5-ml fractions were collected. Chromatography was performed with an AKTA fast-performance liquid chromatography system (Amersham). The fractions were concentrated by precipitation with trichloroacetic acid (TCA) before analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Anti-FLAG immunoprecipitations were carried out in 130-μl mixtures containing TAP-purified material, CaM/E, and 15 μl of prewashed anti-FLAG affinity resin (Sigma). Recombinant insulin (Sigma) was added to 50 μg/ml to reduce nonspecific binding. Control reactions contained 200 μg of FLAG peptide/ml as a competitor; a nonspecific 3× hemagglutinin peptide competitor was included in experimental samples. After binding for 2 h at 4°C, the beads were collected by centrifugation. Supernatants were used directly in activity assays. The beads were washed three times with 100 μl of CaM/E and then suspended in 1.5× SDS-PAGE sample buffer.
SDS-PAGE and Western blotting.
SDS-PAGE was performed by standard techniques (50). For Coomassie blue staining, samples were concentrated by TCA precipitation before being loaded and the gels were stained with Simply Blue Safestain (Invitrogen). For Western blotting, the gels were transferred onto nitrocellulose filters and the filters were blocked with Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat milk. Incubations with primary and secondary antibodies were done in the same solution. The Sir2, Sir3, and Sir4 antibodies have been described previously (40, 67). The anti-FLAG M2 antibody (Sigma); the H3-Ac(K9, K14), H3-AcK9, and H4-AcK12 antibodies (Upstate Biotechnology); and the general H3 antibody (Abcam) were used at a 1:1,000 dilution. The H4-AcK5 and H4-AcK16 antibodies (Serotec) were used at a 1:600 dilution.
Mass spectrometry.
The identification of proteins from Coomassie blue-stained bands and from protein mixtures was done by tandem mass spectrometry (MS/MS) as described previously (25, 71). For mixture analysis, proteins were precipitated with TCA and washed with ice-cold acetone. The proteins were resuspended in 50 mM ammonium bicarbonate-10% acetonitrile and digested for 8 to 12 h at 37°C in the presence of 50 ng of trypsin (Promega, Madison, Wis.). Digested peptides were dried completely and resuspended in 5% acetonitrile-5% formic acid. The peptides were loaded (20 min) by an Endurance autosampler (Michrom Bioresources) onto a hand-pulled fused silica microcapillary column packed with C18 reversed-phase material (75-μm internal diameter, 12 cm long). Once in the column, the peptides were resolved across a 30-min gradient ranging from 12 to 40% acetonitrile in 0.1% formic acid. MS/MS spectra were obtained with an LCQ DecaXP mass spectrometer (ThermoElectron, San Jose, Calif.) and searched against the yeast nonredundant database by the use of SEQUEST software. Protein matches were validated by manual inspection.
Acetate release assays.
Reactions contained 50 mM glycine (pH 9.0), 5 mM Tris (pH 8.0), 150 mM KCl, 2.5% glycerol, 5 mM EGTA, 0.5 mM magnesium acetate, 0.5 mM imidazole, 0.05% NP-40, 5 mM β-mercaptoethanol, 1 mM DTT, 0.1 mg of purified GST/ml, ∼40,000 cpm of [14C]acetyl-H4 N-terminal peptide (amino acids 1 to 20), various amounts of Sir2 protein, and either 0 or 200 μM NAD in a volume of 20 μl. Incubations were done at 30°C for 2 h. The reactions were then quenched by the addition of 80 μl of a mixture of 0.03 M HCl and 0.03 M acetic acid and were extracted with 0.5 ml of ethyl acetate. Acetate release was quantified by scintillation counting of 0.4 ml of the ethyl acetate phase in 2 ml of scintillation fluid.
The H4 peptide was chemically acetylated by mixing 10 μg of peptide with 200 μCi of [14C]acetic acid (50 mCi/mmol) (Amersham) in a reaction containing 12 mM BOP reagent and 10 mM triethylamine (both from Sigma). The reaction was incubated at room temperature with gentle rocking for >12 h. Labeled peptide was purified by the use of Microcon-SCX spin columns (Amicon), vacuum dried, and resuspended in double-distilled H2O at a concentration of ∼80,000 cpm/μl (0.35 mM). The H4 N-terminal peptide and all other peptides used in this study were synthesized and purified by reversed-phase high-pressure liquid chromatography at the Tufts University Core Facility.
Histone purification and nucleosome assembly.
HeLa cells grown in the presence of sodium butyrate were purchased from the National Cell Culture Center. Histone purification was performed as described previously (74). Nucleosomes were assembled by salt dialysis as described previously (73). The DNA fragment used to assemble mononucleosomes has been described elsewhere (28). After assembly, the nucleosomes were loaded onto 5-ml linear 5 to 25% sucrose gradients containing 10 mM HEPES (pH 7.6), 100 mM KCl, 1 mM EDTA, 1 mM DTT, 2 mM benzamidine, 0.1% NP-40, and 2 mM sodium butyrate. The gradients were centrifuged at 100,000 × g for 14 h at 4°C. The peak fractions were pooled, frozen in liquid nitrogen, and stored at −80°C.
Nicotinamide release assays.
Nicotinamide release assays were adapted from the work of Landry et al. (34). For peptide assays, reactions contained 50 mM Tris (pH 7.5), 5 mM Tris (pH 8.0), 150 mM KCl, 2.5% glycerol, 5 mM EGTA, 0.5 mM magnesium acetate, 0.5 mM imidazole, 0.05% NP-40, 5 mM β-mercaptoethanol, 1 mM DTT, 0.1 mg of purified GST/ml, 0.5 μCi of [3H]nicotinamide-NAD (3 Ci/mmol) (Amersham), 105 μM NAD (Sigma), various amounts of histone peptide, and 2.5 ng of Sir2 protein, unless otherwise indicated. The reaction volume was 10 μl. The final concentration of NAD in these reactions was 120 μM.
For histone or nucleosome assays, reactions contained 5 mM HEPES (pH 7.6), 50 mM KCl, 0.05% NP-40, 1 mM sodium butyrate, 1 mM benzamidine, 5% sucrose, and 0.05 mM DTT in addition to the components included in the peptide reactions. Histones or nucleosomes were added to the reactions in sucrose gradient buffer. These reactions contained 0.5 μCi of [3H]NAD and 10 μM NAD, giving a final concentration of 25 μM. The reaction volume was 20 μl.
The reactions were incubated at 30°C for 2 h. The reactions were then quenched by the addition of 73.5 ml of a mixture of 0.37 M sodium borate (pH 8.0) and 50 mM glycine (pH 9.0). Extraction with ethyl acetate and scintillation counting were performed as described above. Calculations of the picomoles of NAD consumed were carried out with the following formula: [(counts released/total input counts) − (counts released without enzyme/total input counts)] × 1,200. Michaelis constants (Km) were determined by fitting the curves shown in Fig. 4A to the Michaelis-Menten formula (64) by using GraphPad Prism software.
FIG. 4.
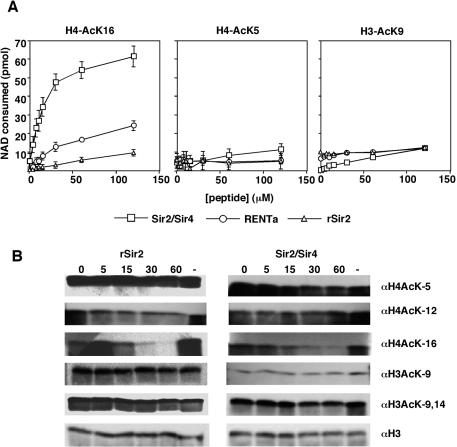
rSir2 and Sir2 complexes both preferentially deacetylate lysine 16 of histone H4 in vitro. (A) rSir2 or Sir2 complexes were incubated with [3H]nicotinamide-NAD and increasing concentrations of one of the following peptides: histone H4 amino acids 1 to 20 acetylated at lysine 16 (left), histone H4 amino acids 1 to 20 acetylated at lysine 5 (middle), or histone H3 amino acids 1 to 20 acetylated at lysine 9 (right). The released nicotinamide was extracted and counted, and the data were converted to picomoles of NAD consumed as described in Materials and Methods. (B) rSir2 (5 ng) or the Sir2/Sir4 complex (1.25 ng) was incubated with NAD and hyperacetylated HeLa core histones (600 ng) for the indicated times (in minutes), after which the reactions were terminated by the addition of SDS sample buffer and analyzed by Western blotting with the indicated antibodies. —, control reactions lacking NAD.
Assay of histone and nucleosome deacetylation by Western blotting.
Reactions were set up as described for nicotinamide release assays, except that a fixed amount of total histones (indicated in figure legends) was included and that only unlabeled NAD was included at a concentration of 200 μM. After incubation at 30°C for the indicated time, the reactions were terminated by the addition of an equal volume of 2× SDS-PAGE loading buffer. The samples were subjected to SDS-PAGE in an 18% acrylamide gel, transferred to nitrocellulose, and then blotted with the indicated antibodies.
Recombinant proteins.
The 6His-Sir2 protein purified from baculovirus-infected insect cells (referred to as rSir2; see Fig. 1) was kindly provided by A. Farrell. The GST-Net1(566-801) protein and the GST-Sir4(1-730) protein were kindly provided by J. Huang. The untagged Sir4(743-1358) fragment was initially purified as a GST fusion protein as described previously (40), and the GST tag was removed by thrombin cleavage. CobB was expressed and purified as described previously (14).
For expression of the Trx-6His-Sir4(745-1172) protein, Escherichia coli BL21 cells transformed with pTrx-Sir4 were grown in Luria-Bertani broth supplemented with 30 μg of kanamycin/ml at 30°C to an OD600 of 0.6 and then were induced with 1 mM isopropyl-thio-β-galactoside for 15 h at 16°C. The cells were harvested, washed once in ice-cold phosphate-buffered saline, and frozen in liquid nitrogen. The cell pellets were thawed on ice and resuspended in an equal volume of buffer A (50 mM Tris [pH 8.0], 200 mM NaCl, 2 mM benzamidine, 1 mM PMSF, 0.2 mg of lysozyme/ml). NaCl was then added to a final concentration of 350 mM, DTT was added to 1 mM, and Triton X-100 was added to 0.5%. All subsequent steps were carried out at 4°C. The suspension was sonicated 3 times for 30 s each at a 40% amplitude, with 2-min rests on ice (Branson sonifier). The extract was centrifuged at 15,000 × g for 10 min, and the supernatant was bound to 1 ml of Ni-nitrilotriacetic acid-agarose resin (Qiagen) that had been prewashed in buffer B (20 mM Tris [pH 8.0], 350 mM NaCl, 2 mM benzamidine, 1 mM PMSF, 1 mM DTT, 0.1% Triton X-100). After incubation for 2h with gentle rocking, beads were collected by centrifugation and transferred to a small column. The beads were washed first with 30 ml of buffer W1 (buffer B plus 20 mM imidazole) and then with 5 ml of buffer W2 (buffer W1 lacking Triton X-100). Bound proteins were eluted with 3 ml of buffer E (buffer W2 plus 250 mM imidazole), and 20 0.2-ml fractions were collected. The peak fractions were pooled and dialyzed extensively against buffer D (20 mM Tris [pH 8.0], 300 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 1 mM PMSF).
Silencing assays.
Cells were grown in nonselective liquid medium to an OD600 of ∼1, and fivefold serial dilutions were spotted onto plates as described previously (19). The plates were incubated for 2 days at 30°C and then photographed.
RESULTS
Affinity purification of native yeast Sir2.
Native Sir2 complexes were purified from yeast extracts by the TAP method (47). Since Sir2 is known to be a component of two distinct complexes, purifications were conducted with extracts from several different yeast strains. In one strain, a TAP tag was engineered at the N terminus of SIR4, generating the TAP-SIR4 allele. It was anticipated that purification from this strain would yield the Sir2/Sir4 complex, which has been purified previously (25). In a second strain, a TAP tag was engineered at the C terminus of SIR2, generating the SIR2-TAP allele. In addition, this strain also carried a deletion of the SIR4 gene, which abolishes silencing at mating-type loci and telomeres and increases silencing at rDNA (61), as well as a FLAG tag at the C terminus of the NET1 gene to allow for detection of the Net1 protein. We expected that TAP from this strain would yield only the rDNA-associated RENT complex. Finally, an alternative purification of the RENT complex employed a strain carrying a NET1-TAP allele. All of the tagged proteins were functional for silencing at the various silenced loci (26; also data not shown).
The protein composition of the material purified from the different strains was visualized by SDS-PAGE followed by Coomassie blue staining (Fig. 1). The identities of the proteins in the indicated gel bands were determined by tandem mass spectrometry. Consistent with previous results, TAP of Sir4 yielded CBP-Sir4 and Sir2 (Fig. 1A) (25). The purification of Sir2-TAP from a sir4Δ background yielded primarily Sir2-CBP, Net1-FLAG, and Cdc14, which have all been shown to be components of the RENT complex (Fig. 1B) (55, 63, 72). Sir2-CBP and Net1-FLAG appeared to be present in equal amounts, while Cdc14 appeared to be substoichiometric. Several smaller fragments of Net1-FLAG were also present, the significance of which is not yet clear. Purification from the NET1-TAP strain yielded primarily Net1-CBP, Sir2, and Cdc14, strongly suggesting that these three proteins form the core of the RENT complex (Fig. 1C). The specificity of all of the observed interactions was confirmed by the absence of these proteins from a parallel purification from an untagged strain (Fig. 1A). TAP of Sir2 from a strain carrying wild-type SIR4 gave a pattern of bands consistent with a mixture of the purifications from the other two strains (as shown by mass spectrometry), indicating that the deletion of SIR4 has no indirect effects on the protein composition of the RENT complex (Fig. 1D).
To ascertain the composition of these complexes more thoroughly, we further analyzed each purification reaction as a mixture, without gel separation. This approach involved trypsin digestion of precipitated protein mixtures followed by liquid chromatography and tandem mass spectrometry (LC-MS/MS) (see Materials and Methods). This analysis confirmed all of the protein identifications assigned by the analysis of individual gel bands (Table 2). In addition, the extra sensitivity gained from mixture analysis allowed the identification of Tof2 in purification reactions from the SIR2-TAP, sir4Δ, and NET1-TAP strains. Tof2 is an 86-kDa protein of unknown function that was originally described as a factor that interacts with topoisomerase I (44). It was present at very low levels in both purification reactions, since it was not detectable in Coomassie blue-stained SDS-PAGE gels (no detectable bands at 86.3 kDa) (Fig. 1B and C).
TABLE 2.
Mass spectrometry analysis of Sir2 complexesa
| Complex or protein | No. of peptides (% coverage) |
|---|---|
| Sir2/Sir4 (TAP-SIR4) | |
| Sir4 | 25 (28.6) |
| Sir2 | 6 (20.1) |
| RENTa (SIR2-TAP sir4Δ) | |
| Net1 | 48 (45.4) |
| Sir2 | 14 (48.2) |
| Cdc14 | 9 (21.1) |
| Tof2 | 2 (4.2) |
| RENTb (NET1-TAP) | |
| Net1 | 61 (50.1) |
| Cdc14 | 13 (32.7) |
| Sir2 | 9 (28.8) |
| Tof2 | 4 (12.7) |
Peptide mixtures from solution digests of Sir2/Sir4, RENTa, and RENTb were resolved by liquid chromatography and directly sequenced by ion-trap mass spectrometry as described in Materials and Methods. Peptides that correspond to proteins present in the various TAP purifications, but not in a parallel purification from an untagged strain, are shown.
Previous reports have observed little or no interaction between the Sir2/Sir4 complex and the Sir3 protein in yeast extracts (16, 25, 40). Although the mixture analysis failed to detect Sir3 in the purification from the TAP-SIR4 strain, a weak association of Sir3 with the Sir2/Sir4 complex could be detected by Western blotting using anti-Sir3 antibodies (Fig. 1F).
We wished to ascertain whether our affinity purification reactions contained a single complex or multiple complexes. Fractionation of the material purified from the TAP-SIR4 strain by gel filtration chromatography showed that all of the affinity-purified CBP-Sir4 was eluted with Sir2 in a single peak that was consistent with a size of >1 MDa (Fig. 2A). This suggests that Sir4 and Sir2 form a large, stable complex in solution. Interestingly, we found that the small amount of Sir3 present in these purifications was not eluted in the peak of Sir2/Sir4, but instead was eluted as a single peak at approximately 450 kDa, suggesting a more dynamic interaction between Sir2/Sir4 and Sir3.
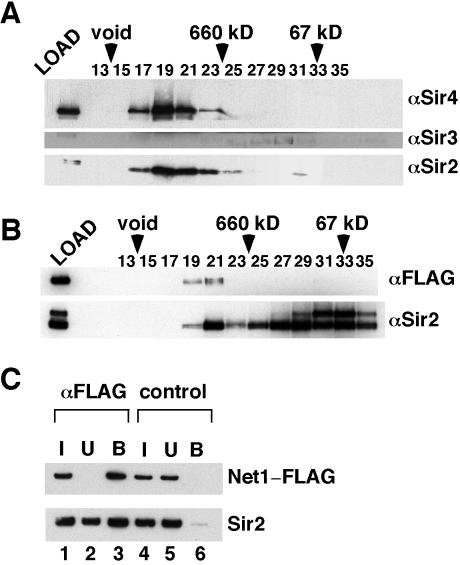
FIG. 2.
Integrity of TAP-purified Sir2 complexes. (A and B) Sir2 complexes were fractionated on a Superose 6 column. The fractions were concentrated by TCA precipitation and analyzed by Western blotting with the indicated antibodies (right). Fraction numbers are indicated above each blot. Elution positions of the size standards are indicated by arrowheads. (A) Sir2/Sir4 complex. (B) RENTa. (C) For RENTa, the purification product was immunoprecipitated with anti-FLAG M2 resin and fractions were analyzed by Western blotting with either an anti-FLAG antibody (top) or an anti-Sir2 antibody (bottom). I, input (10%; lanes 1 and 4); U, unbound (10%; lanes 2 and 5); B, bound (33%; lanes 3 and 6). The reactions in lanes 4 to 6 contained a FLAG peptide to control for nonspecific binding to the M2 resin.
Gel filtration analysis of the material purified from the SIR2-TAP sir4Δ strain revealed that affinity-purified Sir2-CBP was eluted in two distinct peaks (Fig. 2B). The first peak, with a size of >1 MDa, also contained Net1-FLAG, while the second peak was devoid of Net1-FLAG and was eluted at the size predicted for free Sir2 (∼70 to 150 kDa). Most (∼90%) of the Sir2-CBP was eluted in the second peak. This suggests that the majority of the Sir2-CBP purified from this strain is not in a stable complex with Net1-FLAG. It is unclear whether the fraction of Sir2-CBP that was coeluted with Net1-FLAG is tightly bound, with the remainder being free Sir2-CBP, or whether there is a dynamic exchange between Net1-FLAG-bound and free Sir2-CBP during chromatography. As an alternative way to measure the fraction of Sir2-CBP associated with Net1-FLAG in these purifications, we immunoprecipitated Net1-FLAG in the TAP-purified material with an anti-FLAG M2 resin. In parallel, we also carried out immunoprecipitations that included an excess of FLAG peptide in order to control for nonspecific binding to the M2 resin. Net1-FLAG was present entirely in the bound fraction in the absence of the FLAG peptide, whereas the presence of the peptide completely prevented binding (Fig. 2C, compare lanes 3 and 6). In contrast to the gel filtration results, we found that >50% of the input Sir2-CBP coprecipitated with Net1-FLAG in these experiments (Fig. 2C, lanes 1 to 3). This interaction was specific, as it was not detected in the presence of the FLAG peptide (Fig. 2C, lane 6). Thus, the Sir2-CBP/Net1-FLAG complex is partially disassembled during gel filtration chromatography.
Western blots performed on the SIR2-TAP sir4Δ purifications with a polyclonal Sir2 antibody consistently detected a slower-migrating band above Sir2 on the gel (Fig. 2B). This band could also be detected by using a monoclonal Sir2 antibody, suggesting that it may represent a modified form of Sir2 (data not shown). The nature of this potential modification is currently unclear, but we noted that the modified form appears to be excluded from the RENT complex.
For the remainder of this paper, the material purified from the TAP-SIR4 strain will be referred to as the Sir2/Sir4 complex, the material purified from the SIR2-TAP sir4Δ strain will be referred to as RENTa, and the material purified from the NET1-TAP strain will be referred to as RENTb. We emphasize that RENTa and RENTb both refer to the same complex (Table 2); the designations a and b are only meant to distinguish the methods of purification.
Enzymatic activity of yeast Sir2 complexes.
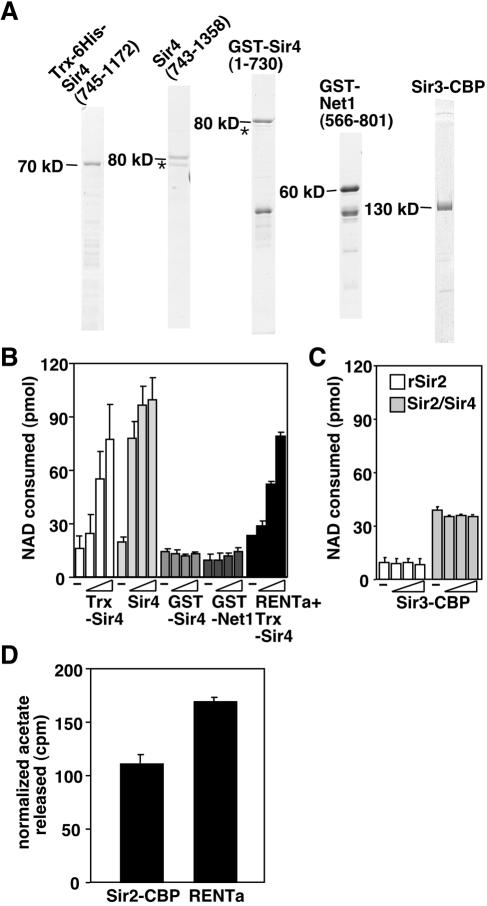
We tested the purified Sir2 complexes for NAD-dependent deacetylase activity by using an acetate release assay. This assay measures the amount of free acetate released from a substrate that has been acetylated with radiolabeled acetyl groups. We used a peptide substrate corresponding to amino acids 1 to 20 of histone H4 that was chemically acetylated with [14C]acetic acid and compared the activities of Sir2/Sir4, RENTa, and RENTb to that of a recombinant, His-tagged Sir2 that was purified from baculovirus-infected insect cells (which we will refer to as rSir2; Fig. 1E).
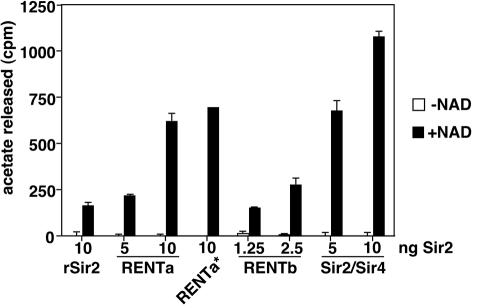
When deacetylation assays were conducted in the absence of NAD, no activity above that of the buffer-only control was observed (Fig. 3). In the presence of NAD, deacetylase activity was stimulated in all cases. Notably, in reactions normalized for the amount of Sir2 added (as assessed by silver staining and Western blotting [data not shown]), both RENTa and Sir2/Sir4 were significantly more active than rSir2. The deacetylase activity of Sir2/Sir4 was particularly robust, showing an enhancement of approximately sevenfold relative to rSir2. The addition of increasing amounts of the native complexes to the reactions enhanced the activity in the presence of NAD but did not change the background counts released in its absence, indicating that all of the deacetylase activity of Sir2/Sir4 and RENT is NAD dependent.
FIG. 3.
NAD-dependent deacetylase activity of Sir2 complexes. Various amounts of rSir2 or Sir2 complexes (indicated at the bottom) were incubated with [14C]acetyl-histone H4 peptide in the presence or absence of 200 μM NAD. The data shown are counts released per minute, averaged from two independent experiments, after the subtraction of background counts extracted from a buffer-only control. Error bars represent standard deviations. RENTa*, deacetylation reaction using a one-step purification from the SIR2-TAP sir4Δ strain.
It has been reported previously that a partially purified fraction that contained Sir2 and Net1 had significant NAD-independent deacetylase activity (16). Since our RENT purification reactions did not contain any detectable NAD-independent activity, we wondered whether such an activity could have been lost during one of the later stages of purification. However, material purified by single-step binding of Sir2-TAP to IgG-Sepharose also showed only NAD-dependent activity (Fig. 3, column RENTa*). Therefore, there is no NAD-independent deacetylase activity associated with Sir2 or the RENT complex in our purification reactions.
To more carefully analyze the differences in activity between rSir2 and the native Sir2 complexes, we assayed the activities of these proteins in reactions that contained a singly acetylated peptide substrate and [3H]nicotinamide-NAD. Since the deacetylation of each acetyl-lysine by Sir2 is accompanied by the removal of nicotinamide from NAD, deacetylation of the peptides in these reactions was measured by quantifying the released nicotinamide. The peptide substrate used for these assays corresponded to the N-terminal 20 amino acids of histone H4 with a single acetyl group at lysine 16 (H4-AcK16), which has been shown to be a preferred substrate for Sir2 in vitro.
Increasing amounts of the H4-AcK16 peptide stimulated nicotinamide release in reactions containing an equal amount of rSir2, RENTa, or Sir2/Sir4 (Fig. 4A, left). Sir2/Sir4 again had the most activity, followed by RENTa and rSir2, respectively. None of the proteins stimulated nicotinamide release in reactions containing an unacetylated version of the same peptide, showing that nicotinamide release is activity dependent (data not shown). We determined the Michaelis constant (Km) for nicotinamide release stimulated by the H4-AcK16 peptide. RENTa, RENTb, and Sir2/Sir4 gave Km values of 56.33 ± 15.43, 26.31 ± 4.51, and 15.16 ± 3.49 μM, respectively (Table 3). In contrast, rSir2 could not be saturated with the peptide substrate under the conditions of the assay. We found the same result in assays using 10-fold higher concentrations of rSir2. These data strongly suggest that the assembly of Sir2 into either the RENT complex or the Sir2/Sir4 complex increases its affinity for the H4-AcK16 peptide.
TABLE 3.
Substrate affinities of rSir2 and Sir2 complexesa
| Complex | KmH4-AcK16 (μM) | KmNAD (μM)b |
|---|---|---|
| rSir2 | >120 | 121.2 ± 11.74 |
| RENTa | 56.33 ± 15.43 | ND |
| RENTb | 26.31 ± 4.51 | ND |
| Sir2/Sir4 | 15.16 ± 3.49 | 65.0 ± 20.68 |
Affinities for the H4-AcK16 peptide were determined by using 120 μM NAD, whereas affinities for NAD were determined by using 120 μM H4-AcK16 peptide (see Materials and Methods).
ND, not determined.
We also compared the NAD affinities of rSir2 and Sir2/Sir4 at a fixed concentration of the H4-AcK16 peptide. Sir2/Sir4 had an enhanced affinity for NAD relative to rSir2 (Km values were 65.0 ± 20.68 μM for Sir2/Sir4 and 121.2 ± 11.74 for rSir2) (Table 3), although the effect was smaller than that seen for the H4-AcK16 peptide.
Substrate specificity of deacetylase activity for recombinant and native Sir2 proteins.
Experiments conducted with recombinant Sir2 proteins have shown that Sir2 preferentially deacetylates lysine 16 on the tail of histone H4 relative to other sites on the H4 tail (27, 68). To compare the substrate specificity of rSir2 to that of the native Sir2 complexes, we conducted nicotinamide release assays using various acetylated peptide substrates. Reactions containing increasing amounts of an H4 N-terminal peptide acetylated at lysine 5 failed to stimulate any nicotinamide release by rSir2 or RENT (Fig. 4A, middle panel). A slight increase in activity was observed for Sir2/Sir4 at the highest peptide concentrations. Similar results were obtained for a histone H3 N-terminal peptide acetylated at lysine 9 (Fig. 4A, right panel). These results indicate that native Sir2 complexes and rSir2 have a similar preference for H4-AcK16 in vitro.
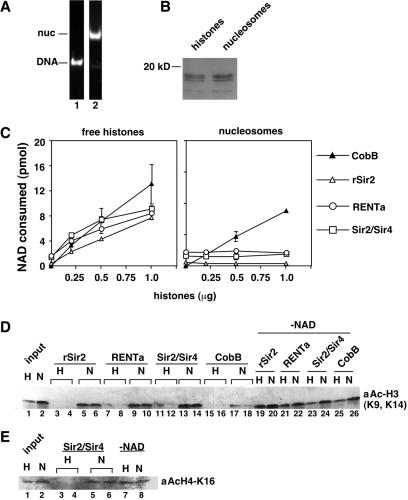
We wished to further investigate the substrate specificity of Sir2 by using mixtures of acetylated histones, rather than singly acetylated peptides, as substrates. We purified hyperacetylated histones from HeLa cells grown in sodium butyrate and tested them as substrates in the nicotinamide release assay. In agreement with the peptide data, we found that the histones stimulated nicotinamide release by the native Sir2 complexes more effectively than that by rSir2 (data not shown). We next examined the site specificity of deacetylation by the different Sir2 proteins by using a Western blot assay with various acetyl-lysine-specific antibodies as well as a control antibody recognizing total histone H3 (Fig. 4B). rSir2 or Sir2/Sir4 was incubated with histones and unlabeled NAD, and aliquots were removed at various time points. An excess amount of rSir2 was used in these reactions relative to Sir2/Sir4 to ensure that deacetylation by both enzymes could be detected. We found that lysine 16 of histone H4 was rapidly deacetylated by both rSir2 and Sir2/Sir4 (Fig. 4B). In contrast, the kinetics of deacetylation at lysines 5 and 12 of histone H4 or lysines 9 and 14 of histone H3 were much slower for both enzymes, with little deacetylation detected in the time scale of the reaction (Fig. 4B). These results further argue that lysine 16 of histone H4 is the preferred site of deacetylation by Sir2. However, we noted that some of the other sites, particularly lysines 9 and/or 14 of histone H3, could be deacetylated efficiently with longer incubation times (Fig. 5D; also data not shown).
FIG. 5.
rSir2 and Sir2 complexes fail to deacetylate nucleosomes in vitro. Mononucleosomes were assembled from hyperacetylated HeLa histones and a 150-bp DNA fragment by salt dialysis and then purified by sedimentation on a sucrose gradient. (A) Representative gradient fractions containing free DNA (lane 1) or nucleosomes (lane 2) were analyzed by agarose gel electrophoresis and ethidium bromide staining. (B) Equal amounts of histones or nucleosomes were analyzed by SDS-PAGE and Coomassie blue staining. (C) rSir2 (25 ng), RENTa (10 ng), Sir2/Sir4 (2.5 ng), or CobB (12.5 ng) was incubated with [3H]nicotinamide-NAD and increasing amounts of either histones (left) or mononucleosomes (right). The released nicotinamide was extracted and counted, and the data were converted to picomoles of NAD consumed as described in Materials and Methods. (D) Deacetylation of histones and nucleosomes assayed by Western blotting. Reactions contained increasing amounts of rSir2 (25 ng in lanes 3 and 5 or 50 ng in lanes 4, 6, 19, and 20), RENTa (5 ng in lanes 7 and 9 or 10 ng in lanes 8, 10, 21, and 22), Sir2/Sir4 (2.5 ng in lanes 11 and 13 or 5 ng in lanes 12, 14, 23, and 24), or CobB (3 ng in lanes 15 and 17 or 6 ng in lanes 16, 18, 25, and 26) and either free histones (H) or histones reconstituted into mononucleosomes (N). Each reaction contained 300 ng of total histones. Lanes 1 and 2, controls that contained buffer instead of enzyme; lanes 19 to 26, controls that lacked NAD. The blot was probed with the anti-H3-Ac(K9, 14) antibody. (E) Data are as for panel D, except that the reactions contained increasing amounts of the Sir2/Sir4 complex (2.5 ng in lanes 3 and 5 or 5 ng in lanes 4, 6, 7, and 8). Lanes 1 and 2, controls that contained buffer instead of enzyme; lanes 7 and 8, controls that contained Sir2/Sir4 and lacked NAD. The blot was probed with the anti-H4-AcK16 antibody.
Nucleosomal histones are poor substrates for Sir2 proteins in vitro.
The deacetylation of histone tails by Sir2 in vivo is likely to occur in the context of chromatin. Therefore, we wished to test the ability of rSir2 and native Sir2 complexes to deacetylate histones that had been assembled into nucleosomes. Hyperacetylated HeLa histones were assembled onto a 150-bp fragment of DNA by salt dialysis, and the resulting mononucleosomes were purified on a sucrose gradient (73) (Fig. 5A). Equal amounts of either free histones or histones assembled into mononucleosomes (Fig. 5B) were included in reactions containing rSir2, RENTa, or Sir2/Sir4. To allow for a direct comparison of the activity of each protein on histones to that on nucleosomes, we normalized the amount of enzyme added such that each reaction contained approximately equal activity as measured on free histones.
As expected, increasing amounts of free histones resulted in a similar amount of deacetylation by rSir2, RENTa, and Sir2/Sir4, as determined by the nicotinamide release assay (Fig. 5C, left panel). However, no stimulation of activity was observed upon the addition of nucleosomes (Fig. 5C, right panel). In contrast, histones and nucleosomes both effectively stimulated nicotinamide release by a similar amount of CobB, the E. coli homologue of Sir2 (Fig. 5C). This indicates that the nucleosomal histones are intact in these experiments. As an additional control, we mixed 1 μg of the nucleosomes into a reaction containing Sir2/Sir4 and the H4-AcK16 peptide used for Fig. 4A. No decrease in the stimulation of nicotinamide release by the H4-AcK16 peptide was observed, which rules out the possibility that the nucleosomes were contaminated with an inhibitor of Sir2 (data not shown). These results indicate that both recombinant and native Sir2 proteins are unable to deacetylate nucleosomal histones in vitro.
As an additional test, we measured the deacetylation of free histones and nucleosomes by using a Western blot assay. For this experiment, increasing amounts of rSir2, RENTa, Sir2/Sir4, or CobB were added to reactions containing a fixed amount of either free histones or nucleosomes, and the reactions were analyzed by Western blotting with either the H3-Ac(K9, K14) antibody or the H4-AcK16 antibody. All of the enzymes deacetylated free histones nearly to completion in these reactions (Fig. 5D and E). In contrast, rSir2, RENT, and Sir2/Sir4 all failed to deacetylate the nucleosomal histones. CobB deacetylated both free histones and nucleosomes effectively, with a slight preference for free histones (Fig. 5D). These results are consistent with the observations shown in Fig. 5C and further support the idea that some aspect of nucleosome structure poses a barrier to histone deacetylation by Sir2.
Enhancement of rSir2 activity by addition of Sir4.
The results in Fig. 3 and 4 show that native Sir2 complexes have a higher specific activity than rSir2. This difference could be due to Sir2 modifications that enhance activity, to interacting proteins, or simply to improper folding of the recombinant protein. We tested the possibility that protein-protein interactions might affect Sir2 activity by adding various protein fragments to deacetylation reactions containing rSir2 and the H4-AcK16 peptide substrate.
We tested fragments of both of the major Sir2-binding partners, Net1 and Sir4, which have been shown to interact directly with Sir2 in vitro. Three different Sir4 fragments were tested in these assays: one consisted of amino acids 745 to 1172 fused to a thioredoxin-six-His tag [Trx-Sir4(745-1172)], one consisted of amino acids 743 to 1358 and lacked a tag [Sir4(743-1358)], and one consisted of amino acids 1 to 730 fused to GST [GST-Sir4(1-730)]. The former two fragments bind directly to Sir2 in vitro, whereas the latter fragment does not (40). In the case of Net1, we used amino acids 566 to 801 fused to GST [GST-Net1(566-801)]. This region of Net1 is within the Sir2-binding domain defined by two-hybrid studies (12) and binds directly to rSir2 in vitro (J. Huang and D. Moazed, unpublished results). All fragments were expressed in E. coli and then purified (Fig. 6A). Strikingly, the addition of either of the Sir2-binding Sir4 fragments caused a dose-dependent stimulation of rSir2 activity (Fig. 6B). The longer Sir4 fragment, which encompasses the C-terminal coiled-coil motif that mediates Sir4 dimerization, afforded the most stimulation. At the highest dose of either of these Sir4 fragments (an ∼12-fold molar excess over rSir2), the activity equaled or surpassed that of the native Sir2/Sir4 protein complex. In contrast, the addition of the N-terminal fragment of Sir4, which lacks a Sir2 interaction domain, failed to stimulate rSir2 activity (Fig. 6B).
FIG. 6.
Interaction of Sir2 with Sir4 enhances its activity. (A) Recombinant protein fragments added to rSir2 reactions were subjected to SDS-PAGE and visualized by Coomassie blue staining. Size markers denote the approximate sizes of the full-length fragments. A contaminating bacterial heat shock protein is denoted by asterisks. (B) Deacetylase activity of rSir2 or RENTa (black bars only) measured by a nicotinamide release assay in the presence of increasing amounts (1.25, 6.25, and 31.25 ng) of recombinant fragments. −, control reactions without any additions. (C) Deacetylase activity of rSir2 (2.5 ng) or Sir2/Sir4 (1.25 ng) measured by a nicotinamide release assay in the presence of increasing amounts (1.25, 6.25, and 31.25 ng) of Sir3-CBP. (D) Deacetylase activity of RENTa complex depleted of Net1-FLAG (Sir2-CBP) or undepleted (RENTa) measured by an acetate release assay. Each reaction contained ∼1.5 ng of Sir2-CBP. The data shown are counts per minute released after the subtraction of background counts from control reactions lacking NAD.
The Sir4 fragments may enhance the activity of rSir2 indirectly by promoting proper protein folding. To test this possibility, we added increasing amounts of Trx-Sir4(745-1172) to reactions containing RENTa instead of rSir2. We reasoned that native Sir2 was less likely to be misfolded than the recombinant protein. We observed that the Trx-Sir4(745-1172) fragment also stimulated the activity of RENTa (Fig. 6B, black bars). This suggests that Trx-Sir4(745-1172) does not enhance activity by assisting the folding of the enzyme, but rather by altering its catalytic properties. Although it is likely that Trx-Sir4(745-1172) stimulates activity by binding to the free Sir2-CBP present in the RENTa preparation, it is also possible that this fragment can exchange with Net1-FLAG during the reaction.
Sir3 has been proposed to act downstream of Sir2's enzymatic activity in the assembly of silent chromatin, based on its ability to interact with deacetylated histone tails in vitro (8). We reasoned that Sir3 may have effects on Sir2 activity in vitro by binding to the deacetylated peptide product or possibly to 2′-O-acetyl-ADP-ribose. However, the addition of full-length Sir3 to rSir2 or Sir2/Sir4 had no effect on activity (Fig. 6C).
The addition of increasing amounts of the GST-Net1 fragment had little effect on rSir2 activity (Fig. 6B). It is possible that although this fragment can interact with Sir2 in vitro, other parts of the Net1 protein act to modulate Sir2 activity. We used a second approach to evaluate the possible contribution of a Net1-Sir2 interaction to the activity of RENT. Since our RENTa purification contained a mixture of both free Sir2-CBP and Sir2-CBP in complex with Net1-FLAG, we tested whether Net1-FLAG contributed to the activity of Sir2-CBP by depleting it from the RENTa purification with an anti-FLAG M2 resin (as described for Fig. 2C). We then tested the activities of equal amounts of Sir2-CBP from depleted samples (which should contain only Sir2-CBP) and control samples (in which a FLAG peptide competed for Net1-FLAG binding to the anti-FLAG resin) by using the acetate release assay. The data showed that the presence of Net1-FLAG increased (by ∼50%) the activity of Sir2-CBP (Fig. 6D).
Native and recombinant Sir2 proteins have different sensitivities to nicotinamide.
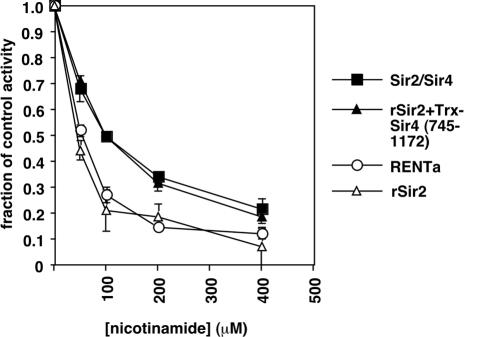
The differences in activity between rSir2 and the native Sir2 complexes led us to test whether nicotinamide, a physiological inhibitor of Sir2 and a product of the Sir2 reaction (5), might impact the various forms of Sir2 in different ways. We added increasing amounts of nicotinamide to reactions containing unlabeled NAD and a [14C]acetyl-H4 peptide and measured deacetylation with the acetate release assay. We found that nicotinamide reduced rSir2 activity by 50% at a concentration of ∼45 μM (Fig. 7). The RENTa activity was similarly inhibited with ∼55 μM nicotinamide. However, the concentration required to reduce Sir2/Sir4 activity to the same extent was ∼100 μM, suggesting that the assembly of Sir2 into a complex with Sir4 confers some resistance to nicotinamide inhibition. This was further supported by the finding that the addition of the Trx-Sir4(745-1172) fragment to reactions containing rSir2 caused the dose-response curve for nicotinamide to resemble that for native Sir2/Sir4.
FIG. 7.
rSir2 and Sir2 complexes are differentially sensitive to nicotinamide inhibition in vitro. The deacetylase activity of rSir2 (10 ng), RENTa (2.5 ng), Sir2/Sir4 (2.5 ng), or rSir2 plus Trx-Sir4(745-1172) (10 ng and 31.25 ng, respectively) was measured by an acetate release assay in the presence of increasing concentrations of nicotinamide. Activities are expressed as fractions of the activity measured in control reactions lacking nicotinamide.
We next tested whether these differences in nicotinamide sensitivity also apply in vivo. The growth of yeast cells in media containing high concentrations of nicotinamide (5 mM) has been shown to disrupt all forms of silencing, presumably by inhibiting the activity of Sir2 (5). Based on the differences in nicotinamide sensitivity observed above (Fig. 7), we hypothesized that silencing at the telomeres, which requires the Sir2/Sir4 complex, might be less sensitive to nicotinamide than silencing in the rDNA, which requires the RENT complex. We tested this idea by assaying the silencing of reporter genes inserted at either telomeres or rDNA in the presence of various concentrations of exogenous nicotinamide (Fig. 8). To improve the sensitivity of our analysis, we also assayed an isogenic set of reporter strains with a deletion of the PNC1 gene. PNC1 encodes a nicotinamidase, which converts nicotinamide into nicotinic acid (1). We reasoned that pnc1Δ cells may show silencing defects at lower concentrations of exogenous nicotinamide due to an increase in intracellular nicotinamide levels, which could be useful for discerning subtle differences in sensitivity between the different silenced loci.
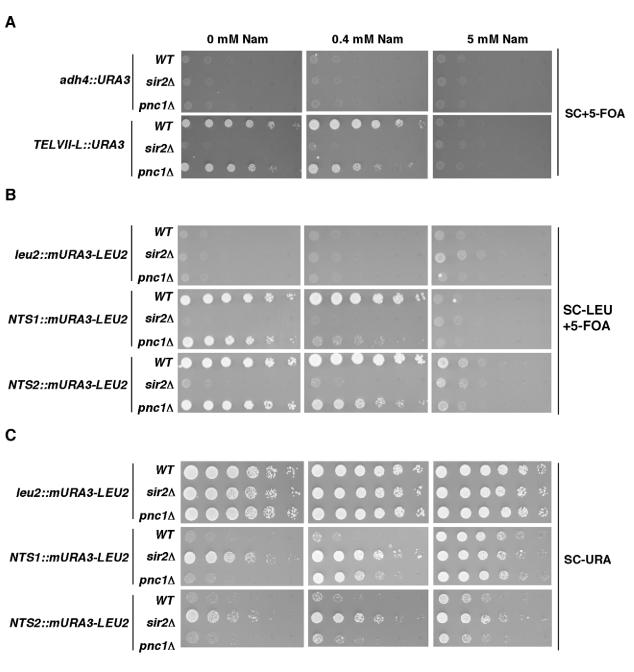
FIG. 8.
Silencing at telomeres and at rDNA is differentially sensitive to nicotinamide inhibition. (A) Fivefold serial dilutions of cultures of the indicated strains were spotted onto complete medium with 5-FOA (SC+5-FOA) to assay telomeric silencing. (B) Fivefold serial dilutions of cultures of the indicated strains were spotted onto complete medium with 5-FOA but lacking leucine (SC-LEU+5-FOA) to assay rDNA silencing. (C) The same as panel B, except that growth was on complete medium lacking uracil (SC-URA). Plates also contained various concentrations of nicotinamide (Nam), as indicated at the top.
The silencing of a telomeric URA3 reporter gene was assayed by monitoring growth on a medium containing 5-FOA, a drug that is toxic to URA3-expressing cells. In the absence of nicotinamide, both the wild-type strain and the pnc1Δ strain were able to grow on 5-FOA medium, whereas the sir2Δ strain, in which silencing fails and URA3 is expressed, did not grow at all (Fig. 8A). At a low nicotinamide concentration (0.4 mM), silencing was unaffected in the wild-type strain, but an ∼25-fold decrease in growth on 5-FOA was observed for the pnc1Δ strain. With 5 mM nicotinamide, silencing was abolished in all three strains, and expression of the telomeric URA3 gene matched that of a nonsilenced URA3 gene (compare with adh4::URA3 panels).
We conducted similar assays with cells carrying a URA3 reporter gene in the nontranscribed spacer (NTS) regions of the rDNA unit (Fig. 8B). At NTS1, we observed robust growth of the wild-type and pnc1Δ strains in the absence of nicotinamide, whereas the sir2Δ strain failed to grow. There was no effect of 0.4 mM nicotinamide on silencing in the wild-type strain, but a dramatic loss of silencing was observed in the pnc1Δ strain, which showed a >125-fold decrease in growth on 5-FOA. As for the experiments with telomeres, 5 mM nicotinamide abolished silencing in all three strains, such that growth was similar to that of control strains carrying the marker at the nonsilenced LEU2 gene. These results revealed an enhanced nicotinamide sensitivity at rDNA relative to telomeres, in agreement with the in vitro results presented in Fig. 7. However, silencing of the same reporter gene inserted at NTS2 displayed a nicotinamide sensitivity similar to that of telomeres (compare the growth of the pnc1Δ strain at 0.4 mM nicotinamide in Fig. 8A and B). This difference between NTS1 and NTS2 suggests that silencing at the two loci has different requirements with respect to the enzymatic activity of the RENT complex.
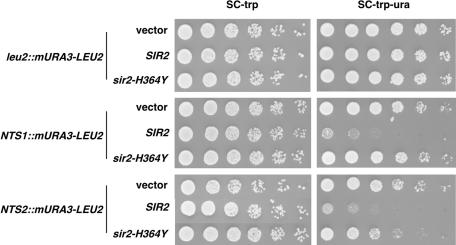
Further insight into the nature of this difference came from silencing assays that measured URA3 expression by monitoring growth on a medium lacking uracil (Fig. 8C). In these assays, we detected a small amount of SIR2-dependent silencing at NTS2 that was resistant even to high concentrations of nicotinamide (Fig. 8C, bottom panel in 5 mM column), suggesting that some of the silencing at NTS2 may not involve Sir2 enzymatic activity. To confirm this possibility, we attempted to rescue the rDNA silencing defects in the sir2Δ strains by introducing low-copy-number plasmids carrying either wild-type SIR2 or an enzymatically inactive mutant (sir2-H364Y) (67). In the NTS1 reporter strain, the sir2Δ defect was completely rescued by the SIR2 plasmid, but not by the sir2-H364Y plasmid (Fig. 9), highlighting the importance of Sir2 activity at this locus. In the NTS2 reporter strain, the sir2-H364Y plasmid caused an approximately fivefold increase in silencing relative to cells transformed with vector alone. Together with the results shown in Fig. 8, these observations indicate the existence of some activity-independent silencing function of Sir2 at this location.
FIG. 9.
Silencing at NTS2 region of rDNA is partially independent of Sir2 activity. Fivefold serial dilutions of cultures of the indicated strains carrying plasmid pRS314 (vector), pDM608 (SIR2), or pDM607 (sir2-H364Y) were spotted onto complete medium lacking tryptophan (SC-trp) to assay growth or onto complete medium lacking tryptophan and uracil (SC-trp-ura) to assay rDNA silencing.
DISCUSSION
NAD-dependent deacetylases are present in organisms ranging from bacteria to humans and participate in diverse biological processes (4, 41, 42, 53, 62). The budding yeast Sir2 protein is the most extensively studied enzyme in this class and thus provides an ideal entry point for a study of the molecular function and regulation of NAD-dependent deacetylation. Most previous studies of the enzymatic activity of Sir2 and its homologues have used recombinant proteins. These studies have provided valuable information regarding the mechanism of the Sir2 reaction, but further insight into how Sir2 activity contributes to silencing in vivo requires study of the activity under more physiological conditions. In addition, although previous studies have identified the major binding partners of Sir2 in the cell, a detailed biochemical study of Sir2 complexes has been lacking. In this paper, we define the composition of native budding yeast Sir2 complexes and characterize their activities in vitro.
Budding yeast Sir2 complexes.
The protein compositions of the Sir2/Sir4 and RENT complexes were simple, and they confirmed previously described interactions. Our analyses also uncovered an association between the topoisomerase I-interacting factor Tof2 and the RENT complex (Table 2). Although we do not know the significance of this interaction, it is interesting that the amino-terminal regions of Tof2 and Net1 (within the first 200 amino acids) share 29% identity. This raises the possibility that Tof2 and Net1 may share some function that contributes to rDNA silencing.
An analysis of the purified complexes by gel filtration chromatography suggested that the Sir2/Sir4 and RENT complexes are both very large (>1 MDa) (Fig. 2A and B). However, accurate estimations of their molecular masses were not possible since neither complex could be sedimented into a glycerol gradient (data not shown). The reason for this aberrant behavior is unclear, but it may have been due to the complexes adopting a highly elongated shape, which would also account for their apparently large gel filtration sizes. In the case of Sir2/Sir4, this would also agree with the structure of the Sir4 C terminus, which is a long parallel coiled-coil dimer (9).
The purification of TAP-Sir4 yielded Sir2 and a small amount of Sir3 (Fig. 1A and F). Interestingly, the Sir3 in the preparation did not cofractionate with the major peak of Sir2 and Sir4 by gel filtration, suggesting that its association with the Sir2/Sir4 complex is dynamic under these conditions (Fig. 2A). This supports previous findings that have suggested that Sir2/Sir4 and Sir3 are distinct biochemical entities that may be brought to chromatin separately (16, 25, 40).
Substrate specificity of Sir2 complexes highlights the importance of H4 lysine 16 for silencing and suggests the involvement of nucleosome remodeling.
The native and recombinant Sir2 proteins had a similar preference for the deacetylation of lysine 16 on the tail of histone H4 (Fig. 4). This observation suggests that this preference results from intrinsic features of the Sir2 active site that are not altered by interaction with other proteins. In contrast, structural studies of the archaeal Sir2-Af2 protein and the budding yeast Hst2 protein in complex with acetylated peptides argue that these enzymes do not effectively discriminate between different acetylated lysine residues (3, 77). Specific substrate recognition may be a property that is unique to budding yeast Sir2. A determination of the structure of the Sir2 active site will be needed to understand the basis for this specificity.
Genetic experiments have clearly indicated the importance of lysine 16 of histone H4 for silencing at the silent mating-type cassettes, telomeres, and rDNA repeats (25, 33, 69). This argues that the substrate specificity of Sir2 that we observed in vitro is physiologically relevant. The fact that all of the other tail lysine residues are also deacetylated in silent chromatin in vivo remains puzzling. It is possible that the deacetylation of lysine 16 by Sir2 can indirectly promote deacetylation of other sites on the tails, either by activating a different deacetylase or by inhibiting an acetyltransferase. An alternative explanation is that the difference in activity measured with the differentially acetylated peptides resulted from kinetic barriers to deacetylation at nonpreferred sites. Even in activity assays containing a large molar excess of peptide over enzyme, it may be impossible to achieve the local concentration of acetyl-lysine that Sir2 is exposed to in vivo when it is recruited to chromatin. These high local concentrations would minimize the kinetic barriers that might limit Sir2 activity in the peptide assay in vitro. By this model, deacetylation at lysine 16 could still promote deacetylation at other sites if (i) lysine 16 were the preferred site for deacetylation and (ii) deacetylation at lysine 16 increased the residence time of Sir2 on chromatin. This model predicts that upon its initial recruitment, Sir2 first deacetylates lysine 16, thereby stabilizing the interaction of silencing complexes with chromatin and increasing the probability of deacetylation at other sites.
Native and recombinant Sir2 proteins all had activity in assays containing a mixture of core histones, but the assembly of the same histones into mononucleosomes completely blocked deacetylation by all three Sir2 proteins (Fig. 5C to E). This was not the case for CobB, the E. coli homologue of Sir2, which effectively deacetylated both histones and nucleosomes (Fig. 5C and D). Thus, the block to deacetylation was a specific property of Sir2 and did not result from a defect in the nucleosomal histones.
The failure to efficiently deacetylate nucleosomal histones in vitro has been noted for other histone deacetylases as well. The NuRD complex, which has both histone deacetylase and chromatin remodeling activities, has less deacetylase activity on nucleosomes than on free histones (70, 75). Further, the deacetylase activities immunoprecipitated with antibodies to various class I deacetylases do not deacetylate nucleosomes (32). The addition of ATP to these assays stimulated the deacetylation of both histones and nucleosomes, suggesting the involvement of either copurifying heat shock proteins or chromatin remodeling factors (see below). In our deacetylase assays, the addition of ATP alone had no effect on the deacetylation of free or assembled histones (data not shown). Finally, a recent report on the Drosophila melanogaster dSir2 protein failed to detect the deacetylation of nucleosome arrays by a recombinant version of the enzyme (45).
The reason for this pronounced difference between histones and nucleosomes is unclear, but it raises the possibility that a factor that is not stably associated with Sir2 complexes is involved in assisting Sir2 to deacetylate nucleosomes. One exciting possibility is that an unidentified nucleosome remodeling factor acts together with Sir2 for the assembly of silent chromatin.
The deacetylase activity measured for Sir2/Sir4 and the RENT complex was entirely NAD dependent (Fig. 3). It was reported previously that a partially purified fraction containing Net1 and Sir2 possessed deacetylase activity that was largely NAD independent (16). Since we did not detect such an activity associated with Sir2, even in single-step pull-down experiments, we attribute this difference to a contaminating deacetylase activity in the partially purified fraction.
Enhancement of Sir2 activity by interaction with Sir4.
In deacetylase reactions normalized for the Sir2 concentration, native Sir2 complexes were more active than rSir2 (Fig. 3). Our data strongly argue that this can be at least partially accounted for by differences in affinity for acetylated peptide substrates. Under conditions that yielded low micromolar Km values for the native complexes with respect to the H4-AcK16 peptide, we were unable to saturate rSir2 (Fig. 4A and Table 3). Interestingly, the affinities of rSir2 and Sir2/Sir4 for NAD were much more comparable (Table 3). This result could be relevant to recent findings that NADH can inhibit Sir2 by acting as an NAD competitor (37). Since the NAD affinities of rSir2 and Sir2/Sir4 are similar, one would predict that the assembly of Sir2 into silencing complexes would not alter its sensitivity to inhibition by NADH.
The differences in activity between rSir2 and native Sir2 complexes could be due to covalent modification of the native Sir2, to a difference in folding of the native proteins, or to interacting factors. Our data indicate that Sir2-binding partners directly enhance Sir2 activity. The addition of recombinant fragments of a Sir4 protein that contains the known Sir2-binding domain potently stimulated the activity of rSir2 in a manner that was sufficient to reconstitute the activity of the native Sir2/Sir4 complex. In contrast, a fragment of Sir4 that lacked the known Sir2-binding domain had no effect on Sir2 activity (Fig. 6B). Although RENTa and RENTb also showed higher affinities for acetylated peptide substrates than rSir2 (Table 2), the rSir2 activity was not stimulated by the addition of a Net1 fragment. However, it seems likely that the Net1-Sir2 interaction also enhances Sir2 activity. Immunodepletion of Net1-FLAG from the RENTa purification resulted in a nearly 50% decrease in the specific activity of the remaining free Sir2-CBP (Fig. 6D). Further, RENTb purified from a NET1-TAP strain contained a higher amount of Net1 relative to Sir2 than did RENTa (compare Fig. 1B and C) and also showed a higher affinity for acetylated substrates (Table 3). Note that these data are also consistent with the participation of Cdc14 in stimulating Sir2 activity, perhaps in combination with Net1. Future experiments will be directed at a more complete reconstitution of the RENT complex in vitro with full-length recombinant components.
The modulation of Sir2 activity by its interaction with other proteins could serve as an important regulatory mechanism in vivo by coupling activity to the assembly of Sir2 into silencing complexes. This would restrict unwanted deacetylation by free Sir2 at normally active regions of the genome. The control of activity of free Sir2 could be particularly important during mitosis, when silencing complexes are known to be disassembled (36, 63). It is intriguing to speculate that the regulation of activity by interacting proteins might be a universal mode of regulation for NAD-dependent deacetylases.
The regions of the Sir2 protein that participate in interactions with Sir4 and Net1 are largely outside of the conserved catalytic domain (11, 12). Thus, the effects of Sir2 complex formation on activity imply that the state of the nonconserved parts of the protein influences the catalytic domain. Although most of the structural studies of NAD-dependent deacetylases to date have focused on the catalytic domain, a recent structure of the budding yeast Hst2 shows that a nonconserved helix can interact in trans with the active site of a neighboring Hst2 molecule, causing a small decrease in activity (76). A more detailed understanding of the mechanism of Sir2 activity stimulation by Sir4 and Net1 binding will require a study of the structure of the entire Sir2 protein, both without ligands and in complex with its binding partners.
Nicotinamide inhibition reveals locus-specific differences in regulation of Sir2 activity.
We found that rSir2 and native complexes had differing sensitivities to inhibition by nicotinamide in vitro. Sir2/Sir4 was approximately twofold less sensitive than rSir2, but rSir2 and RENTa showed similar sensitivities (Fig. 7). Recent studies using recombinant Sir2 enzymes have shown that nicotinamide inhibits the deacetylation reaction by acting as a competing nucleophile that attacks the reactive ADP-ribose-substrate intermediate, thereby promoting the reformation of NAD over deacetylation (31, 52). Recombinant yeast Sir2 was shown to have a catalytic preference for the base exchange reaction (that results in the reformation of NAD) over the deacetylation reaction. The binding of Sir2 to Sir4 may enhance deacetylation relative to base exchange either by occluding the binding of nicotinamide to the active site or by changing the reactivity of the ADP-ribose-substrate intermediate in favor of deacetylation. Further experiments will be required to distinguish between these possibilities.
These data suggest that nicotinamide could differentially regulate Sir2 activity at different silenced loci in yeast. Further in vivo support for this idea came from silencing assays that tested the repression of reporter genes in cells grown on medium containing various amounts of nicotinamide. We found that, in cells impaired in the breakdown of intracellular nicotinamide, a reporter gene in the NTS1 region of rDNA was derepressed at lower concentrations of exogenous nicotinamide than those for a gene that was close to a telomere (Fig. 8). This result agrees with the difference in nicotinamide sensitivity between RENTa and Sir2/Sir4 that was observed in vitro. Our findings contrast with recent data from Gallo et al. (15), who observed that silencing of a telomeric reporter gene was more sensitive to nicotinamide inhibition than silencing of a gene at the NTS1 region of rDNA. However, their comparison was made across two different strain backgrounds, whereas our assays were all conducted in isogenic strains. We have observed differences in nicotinamide sensitivity between different strain backgrounds (J. Tanny, unpublished observations), and therefore we believe that our results more accurately reflect the effects of nicotinamide at the various loci.
In examining the sensitivities of various silent loci to nicotinamide, we found that the NTS1 and NTS2 regions of rDNA show different responses. Silencing at NTS1, like telomeric silencing, was completely abolished by growth on a medium containing high concentrations of nicotinamide, but an NTS2 reporter gene still showed residual repression (Fig. 8C). We showed, by using an activity-defective Sir2 mutant (sir2-H364Y), that this difference was partly due to a component of silencing at NTS2 that is independent of Sir2 activity (Fig. 9). These results indicate that some silencing at NTS2 cannot be accounted for by the activity of the RENT complex. The nature of the Sir2 activity-independent silencing at NTS2 is currently unclear, but it seems likely to involve a structural role for Sir2 in the formation of silent chromatin at this locus.
The locus-specific regulation of Sir2 activity by different protein-protein interactions could be a useful way to modulate silencing at the various loci in response to the environment. The elevated nicotinamide sensitivity of RENT relative to that of Sir2/Sir4 could render it more sensitive to changes in cellular NAD metabolism, which are known to respond to environmental stimuli. For example, PNC1 is highly induced in response to a variety of stress conditions, which could lead to strengthening of silencing at the NTS1 region of the rDNA repeats relative to other silenced loci (1). Silencing at NTS1 has been proposed to counteract rDNA recombination, a phenomenon that is implicated in yeast aging (13, 58). It may be necessary for changes in the metabolic state of the cell to be rapidly integrated with life-span control mechanisms such as the regulation of rDNA recombination without drastically affecting silencing at other genomic locations.
The results presented here provide the first detailed biochemical characterization of native NAD-dependent deacetylases and have furthered our understanding of how the activity of this class of enzymes might be controlled in vivo. The purified Sir2 complexes described here will be useful in mechanistic experiments that address how Sir2 activity is coupled to the assembly of silent chromatin.
Acknowledgments
This work was supported by grants from the NIH (GM61641) and the Ellison Medical Foundation. D.M. is a scholar of the Leukemia and Lymphoma Society.
We thank Alison Farrell and Julie Huang for providing purified proteins; Adam Rudner and Julie Huang for providing yeast strains; Roy Frye, Ju-Fang Chang, and Tom Ellenberger for providing plasmids; and Geeta Narlikar, Andy Saurin, Nicole Francis, and Robert Kingston for providing reagents and advice regarding nucleosome reconstitution. We also thank André Verdel, Brian Hall, and Julie Huang for critical reading of the manuscript and members of the D. Moazed lab for helpful discussions.
REFERENCES
- 1.Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, and D. A. Sinclair. 2003. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423:181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, C. M., M. Kaeberlein, S. I. Imai, and L. Guarente. 2002. Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity. Mol. Biol. Cell 13:1427-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avalos, J. L., I. Celic, S. Muhammad, M. S. Cosgrove, J. D. Boeke, and C. Wolberger. 2002. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell 10:523-535. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148-151. [DOI] [PubMed] [Google Scholar]
- 5.Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves, and D. A. Sinclair. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277:45099-45107. [DOI] [PubMed] [Google Scholar]
- 6.Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592-604. [DOI] [PubMed] [Google Scholar]
- 7.Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel, and M. J. Curcio. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11:255-269. [DOI] [PubMed] [Google Scholar]
- 8.Carmen, A. A., L. Milne, and M. Grunstein. 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277:4778-4781. [DOI] [PubMed] [Google Scholar]
- 9.Chang, J. F., B. E. Hall, J. C. Tanny, D. Moazed, D. Filman, and T. Ellenberger. 2003. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure (Cambridge) 11:637-649. [DOI] [PubMed] [Google Scholar]
- 10.Chang, J. H., H. C. Kim, K. Y. Hwang, J. W. Lee, S. P. Jackson, S. D. Bell, and Y. Cho. 2002. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J. Biol. Chem. 277:34489-34498. [DOI] [PubMed] [Google Scholar]
- 11.Cockell, M. M., S. Perrod, and S. M. Gasser. 2000. Analysis of sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics 155:2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuperus, G., R. Shafaatian, and D. Shore. 2000. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 19:2641-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Defossez, P. A., R. Prusty, M. Kaeberlein, S. J. Lin, P. Ferrigno, P. A. Silver, R. L. Keil, and L. Guarente. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3:447-455. [DOI] [PubMed] [Google Scholar]
- 14.Frye, R. A. 1999. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260:273-279. [DOI] [PubMed] [Google Scholar]
- 15.Gallo, C. M., D. L. Smith, Jr., and J. S. Smith. 2004. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 24:1301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghidelli, S., D. Donze, N. Dhillon, and R. T. Kamakaka. 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 20:4522-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein, A. L., X. Pan, and J. H. McCusker. 1999. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast 15:507-511. [DOI] [PubMed] [Google Scholar]
- 18.Gottlieb, S., and R. E. Esposito. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771-776. [DOI] [PubMed] [Google Scholar]
- 19.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751-762. [DOI] [PubMed] [Google Scholar]
- 20.Grewal, S. I., and S. C. Elgin. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12:178-187. [DOI] [PubMed] [Google Scholar]
- 21.Grewal, S. I., and D. Moazed. 2003. Heterochromatin and epigenetic control of gene expression. Science 301:798-802. [DOI] [PubMed] [Google Scholar]
- 22.Guarente, L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021-1026. [PubMed] [Google Scholar]
- 23.Herskowitz, I., J. Rine, and J. Strathern. 1992. Mating-type determination and mating-type interconversion in Saccharomyces cerevisiae, p. 583-656. In E. W. Jones, J. R. Pringle, and J. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 24.Holmes, S. G., A. B. Rose, K. Steuerle, E. Saez, S. Sayegh, Y. M. Lee, and J. R. Broach. 1997. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics 145:605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie, S. P. Gygi, and D. Moazed. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, J., and D. Moazed. 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 17:2162-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 28.Imbalzano, A. N., H. Kwon, M. R. Green, and R. E. Kingston. 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370:481-485. [DOI] [PubMed] [Google Scholar]
- 29.Ivy, J. M., A. J. Klar, and J. B. Hicks. 1986. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol. Cell. Biol. 6:688-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson, M. D., and J. M. Denu. 2002. Structural identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta-NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 277:18535-18544. [DOI] [PubMed] [Google Scholar]
- 31.Jackson, M. D., M. T. Schmidt, N. J. Oppenheimer, and J. M. Denu. 2003. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J. Biol. Chem. 278:50985-50998. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, C. A., D. A. White, J. S. Lavender, L. P. O'Neill, and B. M. Turner. 2002. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J. Biol. Chem. 277:9590-9597. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, L. M., G. Fisher-Adams, and M. Grunstein. 1992. Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J. 11:2201-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landry, J., J. T. Slama, and R. Sternglanz. 2000. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 278:685-690. [DOI] [PubMed] [Google Scholar]
- 35.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laroche, T., S. G. Martin, M. Tsai-Pflugfelder, and S. M. Gasser. 2000. The dynamics of yeast telomeres and silencing proteins through the cell cycle. J. Struct. Biol. 129:159-174. [DOI] [PubMed] [Google Scholar]
- 37.Lin, S. J., E. Ford, M. Haigis, G. Liszt, and L. Guarente. 2004. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 18:12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 39.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 40.Moazed, D., A. Kistler, A. Axelrod, J. Rine, and A. D. Johnson. 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 94:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.North, B. J., B. L. Marshall, M. T. Borra, J. M. Denu, and E. Verdin. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11:437-444. [DOI] [PubMed] [Google Scholar]
- 42.Onyango, P., I. Celic, J. M. McCaffery, J. D. Boeke, and A. P. Feinberg. 2002. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. USA 99:13653-13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palladino, F., T. Laroche, E. Gilson, A. Axelrod, L. Pillus, and S. M. Gasser. 1993. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75:543-555. [DOI] [PubMed] [Google Scholar]
- 44.Park, H., and R. Sternglanz. 1999. Identification and characterization of the genes for two topoisomerase I-interacting proteins from Saccharomyces cerevisiae. Yeast 15:35-41. [DOI] [PubMed] [Google Scholar]
- 45.Parsons, X. H., S. N. Garcia, L. Pillus, and J. T. Kadonaga. 2003. Histone deacetylation by Sir2 generates a transcriptionally repressed nucleoprotein complex. Proc. Natl. Acad. Sci. USA 100:1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 47.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 48.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 49.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Sauve, A. A., I. Celic, J. Avalos, H. Deng, J. D. Boeke, and V. L. Schramm. 2001. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry 40:15456-15463. [DOI] [PubMed] [Google Scholar]
- 52.Sauve, A. A., and V. L. Schramm. 2003. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry 42:9249-9256. [DOI] [PubMed] [Google Scholar]
- 53.Schwer, B., B. J. North, R. A. Frye, M. Ott, and E. Verdin. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 55.Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed, Z. W. Chen, J. Jang, H. Charbonneau, and R. J. Deshaies. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233-244. [DOI] [PubMed] [Google Scholar]
- 56.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sinclair, D., K. Mills, and L. Guarente. 1998. Aging in Saccharomyces cerevisiae. Annu. Rev. Microbiol. 52:533-560. [DOI] [PubMed] [Google Scholar]
- 58.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91:1033-1042. [DOI] [PubMed] [Google Scholar]
- 59.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 60.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, J. S., C. B. Brachmann, L. Pillus, and J. D. Boeke. 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149:1205-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Starai, V. J., I. Celic, R. N. Cole, J. D. Boeke, and J. C. Escalante-Semerena. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390-2392. [DOI] [PubMed] [Google Scholar]
- 63.Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies, A. D. Johnson, and D. Moazed. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245-256. [DOI] [PubMed] [Google Scholar]
- 64.Stryer, L. 1995. Biochemistry, 4th ed. W. H. Freeman and Co., New York, N.Y.
- 65.Suka, N., Y. Suka, A. A. Carmen, J. Wu, and M. Grunstein. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8:473-479. [DOI] [PubMed] [Google Scholar]
- 66.Tanner, K. G., J. Landry, R. Sternglanz, and J. M. Denu. 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97:14178-14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanny, J. C., G. J. Dowd, J. Huang, H. Hilz, and D. Moazed. 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99:735-745. [DOI] [PubMed] [Google Scholar]
- 68.Tanny, J. C., and D. Moazed. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson, J. S., X. Ling, and M. Grunstein. 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369:245-247. [DOI] [PubMed] [Google Scholar]
- 70.Tong, J. K., C. A. Hassig, G. R. Schnitzler, R. E. Kingston, and S. L. Schreiber. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395:917-921. [DOI] [PubMed] [Google Scholar]
- 71.Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. Gygi, S. I. Grewal, and D. Moazed. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Visintin, R., E. S. Hwang, and A. Amon. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398:818-823. [DOI] [PubMed] [Google Scholar]
- 73.Workman, J. L., and R. E. Kingston. 1991. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science 258:1780. [DOI] [PubMed] [Google Scholar]
- 74.Workman, J. L., I. C. A. Taylor, R. E. Kingston, and R. G. Roeder. 1991. Control of class II gene transcription during in vitro nucleosome assembly. Methods Cell Biol. 35:419-447. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]
- 76.Zhao, K., X. Chai, A. Clements, and R. Marmorstein. 2003. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat. Struct. Biol. 10:864-871. [DOI] [PubMed] [Google Scholar]
- 77.Zhao, K., X. Chai, and R. Marmorstein. 2003. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2′-O-acetyl ADP ribose and histone peptide. Structure (Cambridge) 11:1403-1411. [DOI] [PubMed] [Google Scholar]