Abstract
Objective
Matrix-assisted autologous chondrocyte implantation is frequently applied to replace damaged cartilage in order to support tissue regeneration or repair and to prevent progressive cartilage degradation and osteoarthritis. Its application, however, is limited to primary defects and contraindicated in the case of osteoarthritis that is partially ascribed to dedifferentiation and phenotype alterations of chondrocytes obtainable from patients’ biopsies. The differentiation state of chondrocytes is reflected at the level of structural gene (COL2A1, ACAN, COL1A1) and transcription factor (SOX9, 5, 6) expression.
Methods/Design
We determined the mRNA abundances of COL2A1, ACAN, and COL1A1as well as SOX9, -5, and -6 of freshly isolated and passaged collagen I implant–derived and osteoarthritic chondrocytes via reverse transcription–polymerase chain reaction. Moreover, we analyzed the correlation of structural and transcription factor gene expression. Thus, we were able to evaluate the impact of the mRNA levels of transcription factors on the expression of cartilage-specific structural genes.
Results
Significant differences were obtained (1) for freshly isolated osteoarthritic versus collagen I implant–derived chondrocytes, (2) due to passaging of the respective cell sources, (3) for osteoarthritic versus nonosteoarthritic chondrocytes, and (4) for COL2A1 versus ACAN expression with respect to the coherence with SOX9, -5, and -6 transcript levels.
Conclusion
Our results might contribute to a better understanding of the transcriptional regulation of structural gene expression of chondrocytes with implications for their use in matrix-assisted autologous chondrocyte implantation.
Keywords: chondrocytes, osteoarthritis, transcription factor, collagen, correlation
Introduction
Early treatment of articular cartilage defects is the prerequisite to prevent the onset of mechanically mediated progressive cartilage degradation and inflammation-driven osteoarthritis. A promising approach is the tissue engineering–based matrix-assisted autologous chondrocyte implantation (MACI) applying biocompatible and degradable scaffolds seeded with autologous chondrocytes to primarily replace damaged tissue and to subsequently support regeneration or functionally equivalent tissue repair. A variety of scaffolds of distinct chemical composition, microstructure, and fabrication design have already been applied in orthopedic surgery.1 Among these, collagen-based sponges and hydrogels have become prominent in the past 2 decades.2-4 Although cartilage-specific type II collagen is supposed to be superior by preserving the chondrocyte-specific phenotype, type I collagen is predominantly utilized for practical reasons at the present time (Freyria et al.3 and citations therein). Apart from the biomechanical aspect, the differentiation state and biosynthetic performance of chondrocytes is a crucial determinant for cartilage tissue formation. Indeed, MACI is contraindicated in case of osteoarthritis which is partially ascribed to dedifferentiation and phenotype alterations of chondrocytes accompanying and mediating disease progression.5-7 Furthermore, a common problem is that sufficient numbers of chondrocytes are rarely obtainable due to the tissue-characteristically low cellularity and the small size of patients’ biopsies which inevitably necessitates cell expansion in vitro. However, cell culture conditions (e.g., monolayer cultivation) adapted to foster proliferation unavoidably implicate the gradual dedifferentiation and loss of chondrogenic potential of chondrocytes.8-10 Alternatively, encapsulating chondrocytes in a 3-dimensional environment (e.g., micromass, alginate, collagen) has been proven to redifferentiate phenotype, indicating the significance of cell-cell and cell-matrix interactions in this context.9
The transcription factor SRY (sex determining region-Y)-box, of COL2A1 (collagen type II, alpha 1) and COL1A1 (collagen type I, alpha 1) abbreviated as SOX9, SOX5, and SOX6 are master regulators of cartilage formation in the course of skeletogenesis.11 SOX9 controls mesenchymal condensation as well as chondrogenic lineage commitment and differentiation by, for example, inducing SOX5 and SOX6 expression.12,13 Cooperatively, they are pivotal for the transcription of chondrocyte-specific genes like COL2A1 and aggrecan (ACAN).12,14-16 Moreover, SOX9 prevents hypertrophic differentiation, endochondral ossification, and vascularization and is thereby crucial for the maintenance of noncalcified articular cartilage as functional unit of diarthrodial joints.17,18 However, SOX9 transcription seems to be necessary but not sufficient for COL2A1 and ACAN expression reflected by the inverse correlation in adult/normal in contrast to osteoarthritic and fetal tissue-derived chondrocytes indicating its differential role in developing, adult/normal, and diseased tissues.19 Indeed, the activity of SOX9 is regulated by, for example, phosphorylation, sumoylation, and nuclear translocation on biochemical or mechanical stimuli (reviewed in Chajra et al.4 and Kanazawa et al.20).
However, the impact of SOX9 in conjunction with SOX5 and SOX6 transcription on articular cartilage-specific gene expression by collagen I matrix–embedded chondrocytes assigned for and osteoarthritic cartilage-derived chondrocytes excluded from MACI has not been covered yet. Therefore, we determined SOX9, SOX5, and SOX6 as well as COL2A1 and ACAN transcript levels of nonosteoarthritic collagen implant–derived and osteoarthritic chondrocytes (CIC and OAC, respectively) after enzymatic isolation and during cultivation and passaging. Furthermore, we correlated transcription factor and structural gene expression in order to correlate transcription factor and COL2A1 and ACAN mRNA abundances.
Materials and Methods
Tissue Harvest and Cell Culture
Human osteoarthritic articular cartilage was obtained from patients (n = 4; age 61-80 years) undergoing total knee arthroplasty. Collagen I matrices seeded with nonosteoarthritic chondrocytes assigned for MACI (n = 3; CaRes implants; age 21, 31, 59 years) were provided by Arthro Kinetics (Krems, Austria). Fabrication in brief: Chondrocytes are isolated enzymatically from patients’ biopsies, seeded into a collagen I matrix (rat tail) without prior expansion in 2 dimensions, cultivated in medium with autologous serum for 10 to 14 days, and delivered subsequently to the clinician as ready-to-use implants. A second implant destined for reserve is routinely fabricated for each patient. Unutilized implants were used for investigations. The acquisition of tissues and implants occurred with patients’ informed consent and was approved by the Regional Committee for Ethics in Medical Research.
For chondrocyte isolation, articular cartilage was minced into 2 mm3 pieces prior to enzymatic digestion with Liberase Blendzyme 3 (0.2 WU/mL, Roche Diagnostics GmbH, Mannheim, Germany) in medium (GIBCO DMEM/F12 GlutaMAX-I, Invitrogen, LifeTech Austria, Vienna, Austria) with antibiotics (penicillin 200 U/mL; streptomycin 0.2 mg/mL, and amphotericin B 2.5 µg/mL (Sigma-Aldrich Chemie GmbH, Steinheim, Germany)) under permanent agitation for 18 to 22 hours at 37°C. Implants were minced and digested with collagenase II (100 U/mL, GIBCO, Invitrogen, LifeTech Austria, Vienna, Austria) for 3 hours at 37°C. Subsequently, cell suspensions were passed through a 40 µm filter (BD, Franklin Lakes, NJ) to remove debris, washed with phosphate-buffered saline (PBS), centrifuged (10 minutes, 500 × g, room temperature) and resuspended in PBS. Cell number and viability was determined via trypan blue staining (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) using a hemocytometer and 2 × 105 cells were subjected to RNA isolation and reverse transcription–polymerase chain reaction (RT-PCR) giving rise to the samples of freshly isolated osteoarthritic chondrocytes (OAC-FI) and collagen implant–derived chondrocytes (CIC-FI).
The remaining cells were seeded into 75 cm2 culture flasks (Nunc, Rochester, NY) at a density of 104 cells/cm2 and further cultivated in medium supplemented with antibiotics (see above), 10% fetal calf serum (PAA Laboratories GmbH, Linz, Austria), and l-ascorbic acid (50 µg/mL; Sigma- Aldrich Chemie GmbH, Steinheim, Germany) at 37°C in a humid environment with 5% CO2. Medium was changed every 3 days. For passaging, cells grown to 80% confluency were harvested by use of accutase (1.5 mL/flask; PAA Laboratories GmbH, Linz, Austria), counted, and seeded again or subjected to RNA isolation and RT-PCR. By that, samples of 4 passages of osteoarthritic (OAC-P0-3) as well as collagen implant–derived chondrocytes (CIC-P0-3) were gathered.
Total RNA Isolation, Reverse Transcription, and Real-Time Polymerase Chain Reaction
All kits used were from Roche (Roche Diagnostics GmbH, Mannheim, Germany). Total RNA isolation was performed by use of the High Pure RNA Isolation Kit from 2x105 cells according to the manufacturer’s instructions including DNAse I-digestion. RNA was stored at −80°C until it was used for reverse transcription.
Complementary DNA was synthesized using the First Strand cDNA Synthesis Kit for RT-PCR (AMV) and Random Primer p(dN)6 according to the manufacturer’s instructions. Transcript levels of COL1A1, COL2A1, ACAN, SOX9, SOX6, SOX5 as well as GAPDH as reference gene for normalization of expression levels were determined.
Real-time PCR was performed with FastStartTaqMan Probe Master and with gene-specific primers (Eurofins MWG Synthesis GmbH, Ebersberg, Germany) in triplicate on the iCycler iQ (Bio-Rad Laboratories, Hercules, CA). Probes and primers (Table 1) were selected by use of Universally Probe Library System and by applying in silico PCR (Roche). The primer-dependent optimal annealing temperature was determined experimentally. Real-time PCR was carried out as follows: Initial denaturation step at 95°C for 10 minutes, further denaturation at 95°C for 30 seconds, an annealing step at 55°C to 62°C optimized for the respective primers for 30 seconds, a polymerization step at 72°C for 15 seconds.
Table 1.
Sequences of Primers for Real-Time Polymerase Chain Reaction.
| Gene | Forward (fw) and Reverse (rv) Primer | Roche Probe Number | Identification |
|---|---|---|---|
| GAPDH | fw: ctctgctcctcctgttcgac | #60 | ENSG00000111640.3 |
| rv: acgaccaaatccgttgactc | |||
| ACAN | fw: cctccccttcacgtgtaaaa | #76 | ENSG00000157766.7 |
| rv: gctccgcttctgtagtctgc | |||
| COL2A1 | fw: gtgtcagggccaggatgt | #75 | NM_001844.4 |
| rv: tcccagtgtcacagacacagat | |||
| COL1A1 | fw: gggattccctggacctaaag | #67 | NM_000088.2 |
| rv: ggaacacctcgctctccag | |||
| SOX9 | fw: tacccgcacttgcacaac | #61 | ENSG00000125398.2 |
| rv: tctcgctctcgttcagaagtc | |||
| SOX5 | fw: tttacctcaggagtttgaaagga | #60 | NM_006940.4 |
| rv: gcttgtcaccatggctacct | |||
| SOX6 | fw: tcaaaggcaatttaccagtgatt | #45 | NM_033326.3 |
| rv: ggcttgcttggaagacattc |
The data resulting from the fluorescence measurement were relatively quantified without efficiency correction according to R = 2 – ∆Ct [MEAN target-MEAN reference] method.21
Statistics
Statistics was done via SPSS (IBM SPSS, Armonk, NY, USA). Data were normally distributed (Komolgorov-Smirnov test) and statistically analyzed using the t test for independent variables. Correlation data are represented as Pearson coefficients. Statistically significant differences are indicated by asterisks (**P < 0.01; *P < 0.05). Pearson coefficients of >0.8 were considered as strong, 0.5 to 0.8 as moderate correlation.
Results
Transcription Factor and Structural Gene Expression of Freshly Isolated and Passaged CI-Derived and OA Chondrocytes
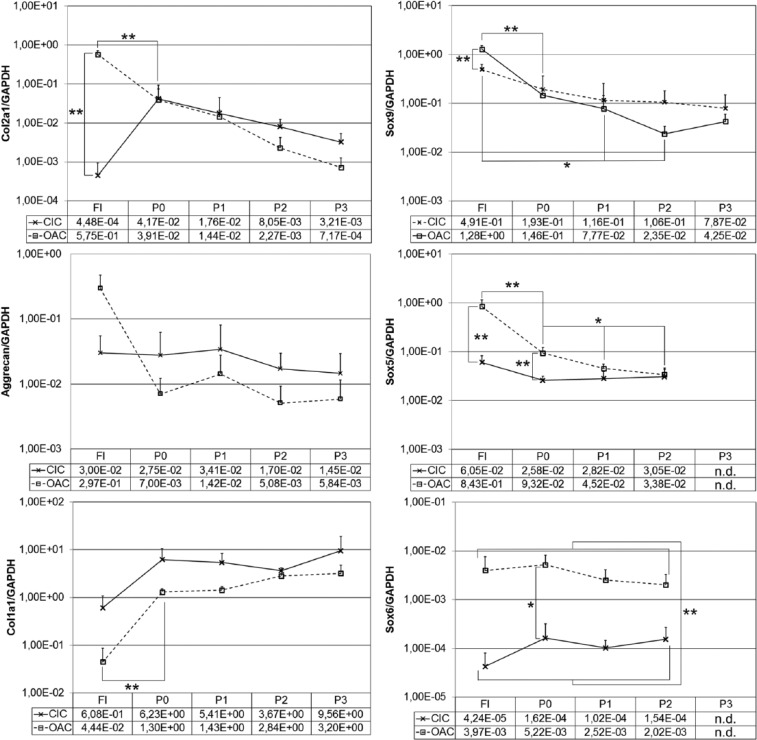
Results are depicted in Figure 1. Freshly isolated collagen implant-derived chondrocytes (CIC-FI) expressed pronouncedly less COL2A1 (1.283-fold), moderately less ACAN (10-fold) and moderately more COL1A1 (14-fold) than osteoarthritic chondrocytes (OAC-FI) resulting in COL2A1/COL1A1 ratios of 7.4 × 10−4 and 1.3 × 101 in CIC-FI and OAC-FI, respectively. Concomitantly, SOX9, SOX5, and SOX6 transcript levels were decreased 2.6-fold, 14-fold, and 94-fold, respectively, in CIC-FI compared with OAC-FI.
Figure 1.
Gene expression of freshly isolated (FI) and passaged (P0-P3(P2)) collagen implant–derived chondrocytes (CIC) and osteoarthritic chondrocytes (OAC) (**P < 0.01; *P < 0.05; n.d.: not determined).
After the first cultivation period (P0) of collagen implant–derived chondrocytes (CIC-P0), COL2A1 and SOX9 as well as SOX5 transcript levels were obviously recoupled indicated by a pronouncedly increased COL2A1 mRNA level (93-fold) even though those of SOX9 and SOX5 were slightly and to a similar extent reduced compared to CIC-FI (2.5-fold and 2.3-fold, respectively). Notably, SOX6 was increased 4-fold in CIC-P0 compared with CIC-FI indicating that the pronouncedly restricted COL2A1 transcription observed in collagen-embedded chondrocytes might be at least partially a consequence of the low SOX6 mRNA abundance. In contrast, osteoarthritic chondrocytes passaged once (OAC-P0) showed a substantially decreased COL2A1 transcript level (15-fold) concomitant with 5-fold lower SOX9 as well as SOX5 transcript levels compared with OAC-FI. By that, collagen implant–derived (CIC) and osteoarthritic chondrocytes (OAC) exhibited almost equal expression levels of COL2A1 and SOX9 after the first cultivation period. However, SOX5 and SOX6 levels remained significantly lower in CIC.
Intriguingly, ACAN transcript levels of CIC proved to be stable almost irrespective of alterations of transcription factor abundances occurring during cultivation and passaging indicating the involvement of other transcription factors or nontranscriptional regulatory mechanisms. In general, transcription factor mRNA levels remained stable (SOX6) or were slightly but progressively reduced (SOX9 and SOX5) during passaging of OAC as well as CIC. Interestingly, passaged collagen implant–derived chondrocytes (CIC-P) consistently retained a lower SOX6 level than osteoarthritic chondrocytes (OAC-P) throughout cultivation and passaging (P0-P2) manifesting a stably established and fundamental difference of those cell sources.
Detailed Correlation Analysis of CI-Derived versus OA and Freshly Isolated versus Passaged Chondrocytes
In order to extend the global view, we determined the correlation of structural gene and transcription factor expression within the subgroups CIC-FI, CIC-P, OAC-FI, and OAC-P to factor in intrasubgroup variability or coherence (Table 2). COL2A1 and ACAN expression positively correlated in CIC but not in OAC. SOX9 expression correlated with COL2A1 and ACAN in CIC-FI, which was maintained or even enforced during passaging (CIC-P). Notably, there was no correlation of SOX9 and COL2A1 as well as a negative correlation with ACAN in OAC-FI which was established or lost, respectively, during passaging suggesting a differential contribution of SOX9 to structural gene expression of the different cell sources. SOX9 expression negatively correlated with SOX5 and SOX6 in CIC-FI but positively in OAC-FI.
Table 2.
Correlation of Gene Expression Represented as Pearson Coefficient From 12 Data Points.a
| COL2A1 |
ACAN |
SOX9 |
SOX5 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| CIC | OAC | CIC | OAC | CIC | OAC | CIC | OAC | ||
| ACAN | FI | 0.87 | −0.45 | ||||||
| P | 0.79** | 0.34 | |||||||
| SOX9 | FI | 0.96 | −0.3 | 0.7 | −0.68 | ||||
| P | 0,91** | 0.73** | 0.89** | 0.2 | |||||
| SOX5 | FI | −0.97 | −0.74 | −0.71 | −0.19 | –1.00* | 0.85 | ||
| P | 0.1 | 0.74** | 0.14 | 0.08 | 0.24 | 0.75** | |||
| SOX6 | FI | −0.82 | 0.234 | −0.43 | –0.97* | −0.95 | 0.81 | 0.94 | 0.39 |
| P | −0.51 | −0.11 | −0.4 | −0.54 | −0.29 | 0.26 | 0.31 | 0.49 | |
CIC = collagen implant–derived chondrocytes; OAC = osteoarthritic chondrocytes; ACAN = aggrecan; FI = freshly isolated; P, passaged.
Strong correlation is depicted in bold and moderate correlation in italic. Significant correlation is marked by asterisks (**P < 0.01; *P < 0.05).
Discussion
Several restraints of MACI application arise from cell quality issues like dedifferentiation of osteoarthritic chondrocytes or due to expansion in monolayer which is frequently inevitable to obtain sufficient cell numbers. Therefore, efforts have been made to characterize gene expression of articular chondrocytes and dedifferentiated or osteoarthritic chondrocytes to evaluate their chondrogenic potential.9,10,22 Notably, a better understanding of regulatory mechanisms underlying structural gene expression of already utilized or potential cell sources for MACI would help improve the quality of cells or extend their applicability (e.g., in osteoarthritis). Although the view of the regulatory network has become more and more sophisticated in recent years,8,23-25 the transcription factors SOX9, -5, and -6 occupy center stage in cartilage development, chondrocyte differentiation, and maintenance.11-13 However, their impact on structural gene expression seems to depend on the cell source. Aigner et al.19 reported that SOX9 and type II collagen expression did not correlate in normal articular cartilage and that decreased SOX9 transcription occurred concomitantly with significantly increased collagen II expression in osteoarthritic chondrocytes indicating the involvement of, for example, posttranscriptional regulatory mechanisms26,27 of COL2A1 expression.
In this study, we investigated the impact of mRNA abundances of SOX9, -5, and -6 on chondrocyte-specific COL2A1 and ACAN expression of freshly isolated and passaged nonosteoarthritic collagen I implant–derived (CIC-FI and CIC-P) and osteoarthritic chondrocytes (OAC-FI and OAC-P) via RT-PCR. We supposed that COL2A1 and ACAN expression of the different cell sources under investigation might differentially rely on transcription factor mRNA abundances and hence might be differentially regulated.
Overall, our results concerning COL2A1 and ACAN expression by freshly isolated osteoarthritic chondrocytes and their alterations with passaging were in accordance with published data. COL2A1 and COL1A1 were concomitantly expressed by freshly isolated osteoarthritic chondrocytes. Whereas COL1A1 transcript abundance reached a steady-state level in passage 2, COL2A1 decreased almost progressively with the number of passages. Indeed, high expression of type I and type II collagen has been reported to be associated with osteoarthritic cartilage5,28,29 and the shift of collagen type expression is a well-known attribute of cultivation-induced dedifferentiation of chondrocytes.10 Intriguingly, ACAN and COL2A1 mRNA levels were decreased pronouncedly during the first passage. However, no tendency for further progressive down-regulation but rather a fluctuating expression of ACAN was observed. Several authors reported that ACAN expression was downregulated during dedifferentiation although to a substantially lesser extent than that of COL2A1 and some other cartilage-specific genes.10,30-32 Notably, the studies were performed with cells from healthy cartilage indicating that our results might reflect a characteristic of osteoarthritic chondrocytes. In contrast, nonosteoarthritic collagen I implant-derived chondrocytes showed a robust ACAN expression.
Significant differences of COL2A1, ACAN, and COL1A1 transcript abundances were observed between freshly isolated osteoarthritic and collagen I implant–derived chondrocytes. The latter showed pronouncedly lower COL2A1, moderately decreased ACAN as well as moderately higher COL1A1 mRNA levels, which co-occurred with slightly moderately, and substantially decreased levels of SOX9, SOX5, and SOX6, respectively, compared with freshly isolated osteoarthritic chondrocytes. Taken together, this phenotype might be assigned to the specific microenvironment of chondrocytes embedded within collagen I. In general, transcription factor mRNA abundances differed significantly irrespective of the cell source (SOX9 > SOX5 » SOX6), which is an interesting detail assuming the impact of stoichiometry at the protein level for binding to cartilage-specific enhancers, the supposed redundancy of SOX5 and -6, and studies using transfection of SOX9, -5, and -6 to investigate their role in chondrogenesis.33,34 Remarkably, COL2A1 mRNA abundance of collagen I implant–derived chondrocytes was found to be tremendously increased at the end of the first cultivation period which co-occurred with a slight decrease of SOX9 and 5 but a moderate increase of SOX6 transcript abundances compared with cells freshly isolated from implants. Moreover, SOX6 levels remained constantly beyond that of osteoarthritic chondrocytes throughout cultivation. Taking into consideration, that cell source-associated differences of gene expression of, for example, COL2A1 and COL1A1 are rapidly equilibrated after cell isolation from the respective matrices (cartilage, collagen I matrix) and that cultivation-induced dedifferentiation proceeds similarly afterward, we assume that the observed difference of SOX6 levels indeed specifically distinguish nonosteoarthritic from osteoarthritic chondrocytes. SOX9 has been described as pivotal for SOX5 and SOX6 expression.12,13 However, detailed analysis of transcription factor mRNA abundances revealed that SOX9 expression of freshly isolated implant-derived chondrocytes correlated negatively with SOX5 and SOX6 expression in contrast to osteoarthritic chondrocytes indicating differential regulation. Although, our current study is only limited to the assessment of SOX trio (SOX5, 6, 9) gene expression level. A direct correlation of gene expression to the intracellular protein levels of type II collagen and aggrecan would further validate our findings toward the transcriptional regulation of structural gene expression in chondrocytes. In our current study, the enzymatic digestion for obtaining chondrocytes from collagen I implant–derived chondrocytes is performed for 3 hours with collagenase II. Whereas, freshly isolated chondrocytes from osteoarthritic cartilage are digested with Liberase Blendzyme for up to 22 hours as performed in a study led by Jakob et al.35 In this study, enzymatic digestion for 22 hours with collagenase resulted in a 5-fold increase of cell yield than digestion for up to 6 hours. But it has to be taken into account that digestion for 6 hours had less impact on the change in gene expression compared with the native cartilage than digestion for up to 22 hours as reported in another study.36 However, a recent study demonstrated the optimal incubation period for an efficient cell yield, viability and no significant differences in changes to gene expression was dependent on the collagenase concentration of 0.2% (w/v) for a time period of 10 hours.37 Similarly, it has to be taken into account that, in our current study, we compared freshly isolated OA chondrocytes from elderly patients with healthy chondrocytes from younger patients. However, a study reporting the differences in age of chondrocyte donors has identified that the glycosaminoglycan, proliferation rate and the collagen type II was not correlated with age and only with a slight decrease but not significantly.38 Although, our study design could have included a control condition with OA chondrocytes being embedded on collagen gels for a direct comparison with chondrocytes from younger patients in collagen gels. Our study results, however, demonstrate the possibility of utilizing chondrocytes from elderly patients as a reparative cell source for implications in MACI constructs based on the higher expression of chondrogenic redifferentiation markers in OA chondrocytes.
In summary, we report pronounced differences (1) concerning freshly isolated osteoarthritic versus collagen I implant–derived chondrocytes, (2) due to passaging of the respective cell sources, (3) concerning osteoarthritic versus nonosteoarthritic chondrocytes, and (4) concerning COL2A1 versus ACAN expression with respect to the coherence with SOX9, -5, and -6 transcript levels. Our results might contribute to a better understanding of the transcriptional regulation of structural gene expression of chondrocytes with interesting implications for their use in MACI.
Footnotes
Informed Consent: Written informed consent was obtained from all subjects before the study.
Acknowledgments and Funding: This work was supported by the government of Lower Austria (WST3-T-96/004-2008) and cofunded by the European Regional Development Fund.
Declaration of Conflicting Interests: The Author(s) declare(s) that there are no conflicts of interest.
Ethical Approval: Ethical approval for this study was obtained from the Regional Committee for Ethics in Medical Research (approval nos. GS4-EK-4/062-2009 and GS4-EK-4/130-2011).
References
- 1. Raghunath J, Rollo J, Sales KM, Butler PE, Seifalian AM. Biomaterials and scaffold design: key to tissue-engineering cartilage. Biotechnol Appl Biochem. 2007;46:73-84. [DOI] [PubMed] [Google Scholar]
- 2. Hunziker EB. Articular cartilage repair: basic science and clinical progress. A review of the current status and prospects. Osteoarthritis Cartilage. 2002;10:432-63. [DOI] [PubMed] [Google Scholar]
- 3. Freyria AM, Ronzière MC, Cortial D, Galois L, Hartmann D, Herbage D, et al. Comparative phenotypic analysis of articular chondrocytes cultured within type I or type II collagen scaffolds. Tissue Eng. 2009;15:1233-45. [DOI] [PubMed] [Google Scholar]
- 4. Chajra H, Rousseau CF, Cortial D, Ronzière MC, Herbage D, Mallein-Gerin F, et al. Collagen-based biomaterials and cartilage engineering. Application to osteochondral defects. Biomed Mater Eng. 2008;18(1 Suppl):S33-45. [PubMed] [Google Scholar]
- 5. Aigner T, Zhu Y, Chansky HH, Matsen FA, 3rd, Maloney WJ, Sandell LJ. Reexpression of type IIA procollagen by adult articular chondrocytes in osteoarthritic cartilage. Arthritis Rheum. 1999;42:1443-50. [DOI] [PubMed] [Google Scholar]
- 6. Nerlich AG, Wiest I, von der Mark K. Immunohistochemical analysis of interstitial collagens in cartilage of different stages of osteoarthrosis. Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;63:249-55. [DOI] [PubMed] [Google Scholar]
- 7. von der Mark K, Kirsch T, Nerlich A, Kuss A, Weseloh G, et al. Glückert K. Type X collagen synthesis in human osteoarthritic cartilage. Indication of chondrocyte hypertrophy. Arthritis Rheum 1992;35:806-11. [DOI] [PubMed] [Google Scholar]
- 8. Lin L, Shen Q, Zhang C, Chen L, Yu C. Assessment of the profiling microRNA expression of differentiated and dedifferentiated human adult articular chondrocytes. J Orthop Res. 2011;29:1578-84. [DOI] [PubMed] [Google Scholar]
- 9. Barlic A, Drobnic M, Malicev E, Kregar-Velikonja N. Quantitative analysis of gene expression in human articular chondrocytes assigned for autologous implantation. J Orthop Res. 2008;26:847-53. [DOI] [PubMed] [Google Scholar]
- 10. Cheng T, Maddox NC, Wong AW, Rahnama R, Kuo AC. Comparison of gene expression patterns in articular cartilage and dedifferentiated articular chondrocytes. J Orthop Res. 2012;30:234-45. [DOI] [PubMed] [Google Scholar]
- 11. Gadjanski I, Spiller K, Vunjak-Novakovic G. Time-dependent processes in stem cell-based tissue engineering of articular cartilage. Stem Cell Rev. 2012;8:863-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 2002;16:2813-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akiyama H, Stadler HS, Martin JF, Ishii TM, Beachy PA, Nakamura T, et al. Misexpression of Sox9 in mouse limb bud mesenchyme induces polydactyly and rescues hypodactyly mice. Matrix Biol. 2007;26:224-33. [DOI] [PubMed] [Google Scholar]
- 14. Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, et al. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183:108-21. [DOI] [PubMed] [Google Scholar]
- 15. Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, et al. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J Biol Chem. 2000;275:10738-44. [DOI] [PubMed] [Google Scholar]
- 16. Bridgewater LC, Lefebvre V, de Crombrugghe B. Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J Biol Chem. 1998;273:14998-5006. [DOI] [PubMed] [Google Scholar]
- 17. Zhao Q, Eberspaecher H, Lefebvre V, De Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377-86. [DOI] [PubMed] [Google Scholar]
- 18. Hattori T, Müller C, Gebhard S, Bauer E, Pausch F, Schlund B, et al. SOX9 is a major negative regulator of cartilage vascularization, bone marrow formation and endochondral ossification. Development. 2010;137:901-11. [DOI] [PubMed] [Google Scholar]
- 19. Aigner T, Gebhard PM, Schmid E, Bau B, Harley V, Poschl E. SOX9 expression does not correlate with type II collagen expression in adult articular chondrocytes. Matrix Biol 2003;22:363-72. [DOI] [PubMed] [Google Scholar]
- 20. Kanazawa T, Furumatsu T, Hachioji M, Oohashi T, Ninomiya Y, Ozaki T. Mechanical stretch enhances COL2A1 expression on chromatin by inducing SOX9 nuclear translocalization in inner meniscus cells. J Orthop Res. 2012;30:468-74. [DOI] [PubMed] [Google Scholar]
- 21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402-8. [DOI] [PubMed] [Google Scholar]
- 22. Gebhard PM, Gehrsitz A, Bau B, Soder S, Eger W, Aigner T. Quantification of expression levels of cellular differentiation markers does not support a general shift in the cellular phenotype of osteoarthritic chondrocytes. J Orthop Res. 2003;21:96-101. [DOI] [PubMed] [Google Scholar]
- 23. Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285:24381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones SW, Watkins G, Le Good N, Roberts S, Murphy CL, Brockbank SM, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-α and MMP13. Osteoarthritis Cartilage. 2009;17:464-72. [DOI] [PubMed] [Google Scholar]
- 25. Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008;3:e3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sim H, Argentaro A, Harley VR. Boys, girls and shuttling of SRY and SOX9. Trends Endocrinol Metab. 2008;19:213-22. [DOI] [PubMed] [Google Scholar]
- 27. Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master-regulator SOX9 by miRNA-145. J Biol Chem. 2012;287:916-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martin I, Jakob M, Schafer D, Dick W, Spagnoli G, Heberer M. Quantitative analysis of gene expression in human articular cartilage from normal and osteoarthritic joints. Osteoarthritis Cartilage. 2001;9:112-8. [DOI] [PubMed] [Google Scholar]
- 29. Nerlich AG, Brenner RE, Wiest I, Lehmann H, Yang C, Müller PK, et al. Immunohistochemical localization of interstitial collagens in bone tissue from patients with various forms of osteogenesis imperfecta. Am J Med Genet. 1993;45:258-9. [DOI] [PubMed] [Google Scholar]
- 30. Diaz-Romero J, Nesic D, Grogan SP, Heini P, Mainil-Varlet P. Immunophenotypic changes of human articular chondrocytes during monolayer culture reflect bona fide dedifferentiation rather than amplification of progenitor cells. J Cell Physiol. 2008;214:75-83. [DOI] [PubMed] [Google Scholar]
- 31. Zaucke F, Dinser R, Maurer P, Paulsson M. Cartilage oligomeric matrix protein (COMP) and collagen IX are sensitive markers for the differentiation state of articular primary chondrocytes. Biochem J. 2001;358:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Darling EM, Athanasiou KA. Rapid phenotypic changes in passaged articular chondrocyte subpopulations. J Orthop Res. 2005;23:425-32. [DOI] [PubMed] [Google Scholar]
- 33. Han Y, Lefebvre V. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol Cell Biol. 2008;28:4999-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang HN, Park JS, Woo DG, Jeon SY, Do HJ, Lim HY, et al. Chondrogenesis of mesenchymal stem cells and dedifferentiated chondrocytes by transfection with SOX Trio genes. Biomaterials. 2011;32:7695-704. [DOI] [PubMed] [Google Scholar]
- 35. Jakob M, Demarteau O, Schafer D, Stumm M, Heberer M, Martin I. Enzymatic digestion of adult human articular cartilage yields a small fraction of the total available cells. Connect Tissue Res. 2003;44:173-80. [DOI] [PubMed] [Google Scholar]
- 36. Hayman DM, Blumberg TJ, Scott CC, Athanasiou KA. The effects of isolation on chondrocyte gene expression. Tissue Eng. 2006;12:2573-81. [DOI] [PubMed] [Google Scholar]
- 37. Oseni AO, Butler PE, Seifalian AM. Optimization of chondrocyte isolation and characterization for large-scale cartilage tissue engineering. J Surg Res. 2013;181:41-8. [DOI] [PubMed] [Google Scholar]
- 38. Barbero A, Grogan S, Schafer D, Heberer M, Mainil-Varlet P, Martin I. Age related changes in human articular chondrocyte yield, proliferation and post-expansion chondrogenic capacity. Osteoarthritis Cartilage. 2004;12:476-84. [DOI] [PubMed] [Google Scholar]