Abstract
In eukaryotes the primary cleavage of the precursor rRNA (pre-rRNA) occurs in the 5′ external transcribed spacer (5′ETS). In Saccharomyces cerevisiae and animals this cleavage depends on a conserved U3 small nucleolar ribonucleoprotein particle (snoRNP), including fibrillarin, and on other transiently associated proteins such as nucleolin. This large complex can be visualized by electron microscopy bound to the nascent pre-rRNA soon after initiation of transcription. Our group previously described a radish rRNA gene binding activity, NF D, that specifically binds to a cluster of conserved motifs preceding the primary cleavage site in the 5′ETS of crucifer plants including radish, cauliflower, and Arabidopsis thaliana (D. Caparros-Ruiz, S. Lahmy, S. Piersanti, and M. Echeverria, Eur. J. Biochem. 247:981-989, 1997). Here we report the purification and functional characterization of NF D from cauliflower inflorescences. Remarkably NF D also binds to 5′ETS RNA and accurately cleaves it at the primary cleavage site mapped in vivo. NF D is a multiprotein factor of 600 kDa that dissociates into smaller complexes. Two polypeptides of NF D identified by microsequencing are homologues of nucleolin and fibrillarin. The conserved U3 and U14 snoRNAs associated with fibrillarin and required for early pre-rRNA cleavages are also found in NF D. Based on this it is proposed that NF D is a processing complex that assembles on the rDNA prior to its interaction with the nascent pre-rRNA.
The rRNAs 18S, 5.8S, and 25S are encoded by tandemly repeated single transcriptional units. Transcription of these units by RNA polymerase I (RNA Pol I) produces a primary transcript (pre-rRNA) containing the rRNA flanked by external spacer sequences (ETS) and internal spacer sequences. The pre-rRNA is then subjected to a complex maturation process that involves the accurate removal of the spacers and the modification of numerous rRNA residues (49). In vivo, all these events are accomplished by large small nucleolar ribonucleoprotein particle (snoRNP) complexes that transiently interact with the pre-rRNA in the nucleolus (47).
One of the earliest processing events on the pre-rRNA is an endonucleolytic cut in the 5′ETS upstream from the 18S rRNA. This primary pre-RNA cleavage is conserved in all eukaryotes, but its position within the 5′ETS is distinct in each species. In Saccharomyces cerevisiae it occurs at site A0, located 90 nucleotides upstream from the 5′ end of the 18S rRNA (49). Genetic studies have shown that A0 cleavage depends on the U3 snoRNP, a large complex containing the C/D snoRNA U3 and associated nucleolar proteins (16). This implicates a base-pair interaction of U3 with the 5′ETS that may stabilize or promote a pre-RNA structure required for processing and subsequent production of 18S rRNA (5, 6). The U3 snoRNP proteins, including Nop1p (fibrillarin in vertebrates), and three other core proteins associated with all C/D snoRNAs are essential for this cleavage. In addition, there are proteins which are specific to U3 snoRNP and are not found associated with the methyl guide C/D snoRNAs (data summarized in reference 49)
In vertebrates and other systems the primary pre-rRNA cleavage occurs at site A′, found hundreds of nucleotides upstream from the 18S rRNA (29, 38). S. cerevisiae does not have an A′-like site, but in some species, like Xenopus laevis (9), and also in trypanosomes (27), in addition to A′ sites a second cleavage in the 5′ETS occurs at a site similar to yeast A0. The reproduction of cleavage at A′ by acellular extracts confirmed the essential role of U3 in mammals (29) and Xenopus (38). This also revealed other snoRNAs that stimulate this cleavage in vitro, like U14, a conserved C/D snoRNA, and two H/ACA snoRNAs, U17 and E3 (18). These studies also demonstrated an essential role for nucleolin, an abundant nucleolar protein that is not a component of snoRNPs. Nucleolin is implicated in ribosome biogenesis but also has other functions (23). The vertebrate nucleolin is an RNA binding protein characterized by an N-terminal acidic stretch, a central domain with four RNA binding domains (rRNA recognition motif [RRM] type), and a glycine-arginine-rich (RGG) C-terminal domain (23). Nucleolin specifically binds to the 5′ETS on the pre-RNA (20) and is required for primary pre-rRNA cleavage in vitro (21). Nucleolin is not conserved, but genes encoding proteins with similar structural organization are found in other eukaryotes. In S. cerevisiae the Nsr1 gene encodes a nucleolin-like protein, characterized by only two RRMs, which is required for normal pre-rRNA processing and 18S rRNA synthesis (33).
In plants little is known about the pre-rRNA processing pathway, and only the primary cleavage in the 5′ETS has been mapped in some species (13, 41, 42). Some conserved factors controlling this event in Arabidopsis thaliana and other plants have been identified, like U3 (36), U14 (32), and fibrillarin (4). In addition to fibrillarin, genes encoding all other common proteins associated with all C/D snoRNAs and H/ACA snoRNAs (49) have been found (10) or predicted in the Arabidopsis genome (M. Echeverria, unpublished data). Notably in Arabidopsis, all these factors are encoded by multigene families. In many cases, although not always, gene copies encode very similar snoRNAs or nucleolar proteins that fulfill redundant functions. For instance, in Arabidopsis two duplicated genes encode fibrillarin functional homologues (4). On the other hand, additional factors controlling pre-rRNA processing in other systems exhibit significant divergence in plants. This is the case of the plant nucleolin-like proteins, described for alfalfa (8) and pea (48). These have structural organizations similar to that of nucleolin but have only two RRMs and are therefore more related to yeast Nsr1p. The role of the plant nucleolin-like proteins in pre-rRNA synthesis has not been investigated. Even greater divergence is found for some of the yeast U3 snoRNP-specific proteins, like Mpp10p (17), for which no homologue can be predicted by alignment in the Arabidopsis genome (Echeverria, unpublished).
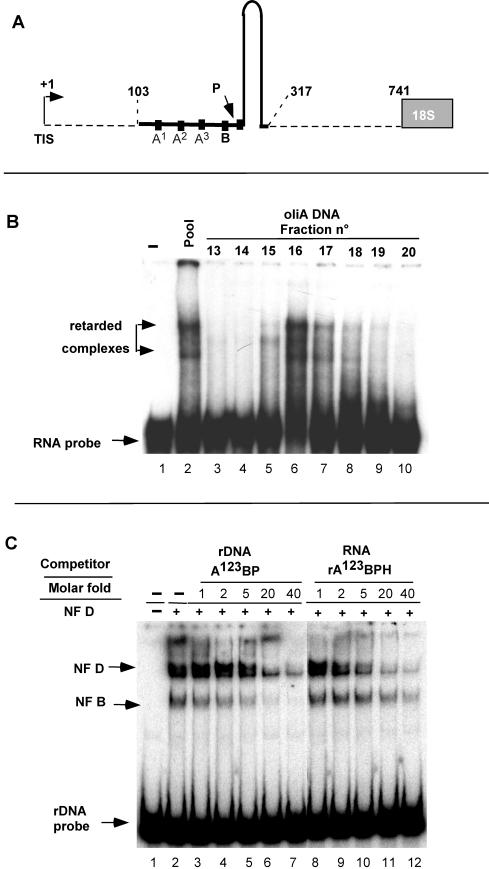
We are interested in pre-RNA processing and the factors that control primary pre-RNA cleavage in crucifer plants. This family includes Arabidopsis, a model plant for genetic studies, and other species like cauliflower, which provide a source of proteins for biochemical studies. By use of electrophoretic mobility shift assays (EMSA), two sequence-specific rRNA gene (rDNA) binding activities, NF D and NF B, had been found in radish extracts that interact with a cluster of motifs, A123B (see Fig. 1A), found in the radish 5′ETS region. DNase I footprinting analysis with the separated radish NF D and NF B factors showed that each binds to the rDNA A123B cluster: the NF D binding site encompasses motifs A123 and overlaps the NF B binding site, which encompasses motif B (11). Based on their rDNA binding specificities it was suggested that these factors could be involved in pre-rRNA synthesis or processing (11).
FIG. 1.
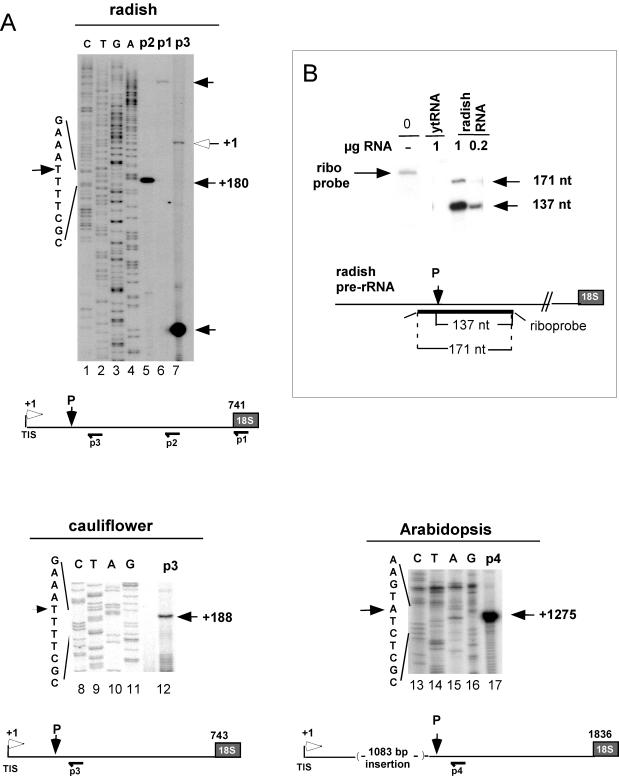
Primary pre-rRNA cleavage site in the 5′ETS of crucifer plants. (A) Mapping by primer extension of pre-rRNA cleavages in the 5′ETSs of radish (Raphanus sativus), cauliflower (Brassica oleracea), and Arabidopsis (A. thaliana). Reverse transcriptions were performed on total seedling RNAs from primers p1 to p4 as indicated. P3 is complementary to both the radish and the cauliflower 5′ETS sequences. Open and closed arrows indicate the TIS and the primary processing site (P), respectively. CTGA are 5′ETS DNA sequences made from corresponding primers (p2 for radish). The scheme of the 5′ETS with the position of TIS, the cleavage signal P, and the primers used for mapping is shown to scale. Nucleotides are numbered from the TISs that were previously mapped (7, 13, 25). (B) Mapping of the primary processing site in the radish 5′ETS by RNase protection analysis. RNaseA/T1 protection was done with a radiolabeled RNA probe complementary to the 5′ETS radish sequence, including the P site detected by primer extension. The riboprobe sequence complementary to the 5′ETS is indicated. Eight additional nucleotides from vector sequences are indicated by thin lines at the ends of the riboprobe. The expected sizes of protected fragments are shown. The assay was carried out with the indicated amount of total RNA from radish seedlings or yeast tRNA as indicated. A control lane loaded with the untreated riboprobe is shown (lane 0). Following the RNase reaction the protected fragments were analyzed in a 6% sequencing gel. The sizes of protected bands evaluated from a marker ladder run in parallel (not shown) are indicated on the right of the gel.
To identify these factors, we purified them from cauliflower inflorescences, a tissue highly enriched in meristematic cells. Notably, the purification of these factors revealed that they are structurally related: NF D is an unstable complex which dissociates into smaller subcomplexes, including NF B. We describe this work here and focus on the functional characterization of NF D. We show that NF D is a high-molecular-weight ribonucleoprotein complex that includes nucleolar factors involved in pre-rRNA processing: the nucleolin-like protein, fibrillarin, and C/D snoRNAs U3 and U14. Finally, we provide evidence that the highly purified NF D fraction reproduces the primary pre-RNA cleavage in vitro. It is proposed that NF D could represent a pre-rRNA processing complex related to U3 snoRNP that assembles on the rDNA prior to its binding to the nascent transcript.
MATERIALS AND METHODS
Preparation of cauliflower whole-cell extracts.
Whole-cell extracts were prepared from chilled (4°C) cauliflower inflorescences purchased at a local supermarket, with all steps performed at 4°C. After the stalks and large stems were removed, 150 g of inflorescence was added to 200 ml of ice-cold buffer I (50 mM Tris-HCl [pH 8.5], 10 mM MgCl2, 10% sucrose, 20 glycerol, 1 mM EDTA) supplemented just prior to use with a cocktail of protease inhibitors and reducing agents (2.5 μg of antipain/ml, 0.35 μg of bestatin/ml, 0.5 μg of leupeptin/ml, 4.0 μg of pepstatin A/ml, 10 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride). The tissue was homogenized at maximum speed in a Waring blender using five pulses of 5 s each. The homogenate was filtered through two layers of Miracloth (Calbiochem), and 1/10 sample volume of 3.8 M (NH4)2SO4 was added. The sample was mixed on a stirring motor for 30 min and centrifuged at 95,800 × g for 1 h. The supernatant was precipitated with solid (NH4)2SO4 added to 0.33 g/ml over a 30-min period, followed by incubation at 4°C for 1 h. Precipitated proteins were recovered by centrifugation at 15,300 × g for 30 min and resuspended in 20 ml of buffer II-100 (50 mM Tris-HCl [pH 8.0], 6 mM MgCl2, 15% glycerol, 1 mM EDTA, 2% NP-40, 100 mM KCl) supplemented with the inhibitors and reducing agents added to buffer I. The resuspended fraction was dialyzed against 500 ml of 3× buffer II-100.
Purification of NF D.
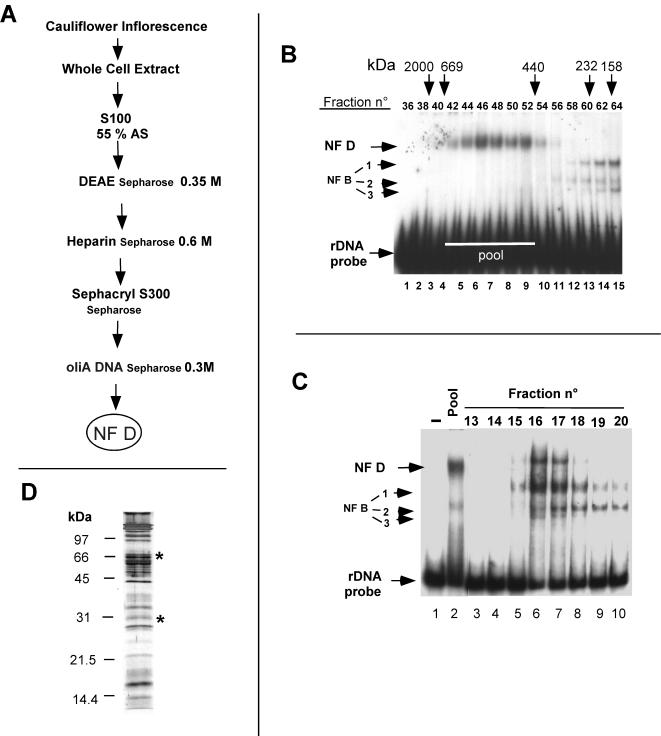
The dialyzed extract was then fractionated through four chromatographic steps (see Fig. 4A). NF D activity was detected by EMSA with an rDNA A123BP probe (see Fig. 3A). The dialyzed fraction (∼20 ml) was loaded onto a 25-ml DEAE-Sepharose CL-6B (Pharmacia) column equilibrated in buffer II-100. The flowthrough was collected, and the column was washed with buffer II-100. Bound proteins were step eluted with buffer II-350 (buffer II-350 mM KCl). NF D activity eluted in a single peak. The peak fractions were pooled, diluted with 1 volume of buffer II, and loaded onto a 25-ml heparin-Sepharose (Pharmacia) column equilibrated in buffer II-175 (buffer II-175 mM KCl). After a washing with buffer II-175, the NF D activity was recovered in a single peak by step elution with buffer II-600 (buffer II-600 mM KCl). Peak fractions were pooled and loaded directly onto a 120-ml Hi-Prep 16/60 Sephacryl S300 HR (Pharmacia) column equilibrated and run in buffer II-100. The fractions containing NF D activity (see Fig. 4B) were pooled and subjected to chromatography on a 1-ml oliA DNA-Sepharose chromatography column (see below) equilibrated in buffer II-100. The column was washed with buffer II-100, and the NF D fraction was eluted with buffer II-300 (see Fig. 4C). The fractions corresponding to the peak of integral NF D activity were pooled and represent the NF D pure fraction used for characterization.
FIG. 4.
Purification of NF D. (A) Scheme of purification of NF D. Details are given in Materials and Methods. (B) Sephacryl S300 elution profile of NF D activity detected by EMSA with the rDNA A123BP probe. Pool indicates fractions loaded on the oliA DNA-Sepharose column. (C) oliA DNA-Sepharose elution profile of NF D activity detected with the rDNA A123BP probe. (D) Protein profile of pure NF D separated by SDS-PAGE and revealed by silver staining. The asterisks indicate bands identified by Western blotting as AtNuc-L1 and AtFib.
FIG. 3.
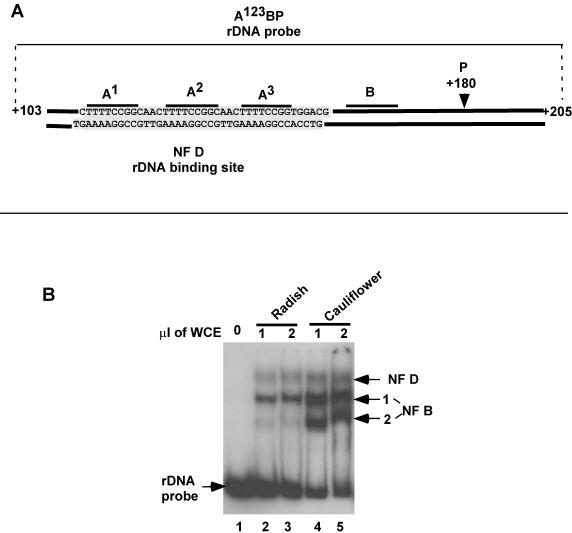
NF D and NF B activities in crucifer extracts. (A) Diagram of the rDNA probe A123BP used in the EMSA to detect NF D and NF B. The probe encompasses genomic 5′ETS rDNA nucleotides from +103 to +205. It has no additional sequences except for flanking restriction sites (11). The nucleotide sequence shown is the rDNA binding site of NF D mapped by DNase I footprinting (data from reference 11). (B) EMSA with the A123BP probe and the indicated amounts of whole-cell extracts (WCE) from radish seedlings or cauliflower inflorescences. Arrows on the right show NF D and NF B1-2 complexes.
A purified NF B fraction was prepared by pooling S300 fractions eluting below 250 kDa containing NF B free of NF D activity (Fig. 4B). NF B was subsequently purified through an oliB-Sepharose DNA chromatography column as described previously (11).
Preparation of oliA DNA-Sepharose.
The oliA DNA-Sepharose chromatography column was prepared by coupling oligomerized double-stranded DNA oligonucleotides AfD2 and AfD2r to CNBr-activated Sepharose (Sigma) as described previously (28).
Protein sequencing.
For protein sequencing, a preparative gel was run loaded with an NF D/NF B purified fraction extracted from 1 kg of cauliflower. The purification protocol used in this case was modified by excluding Sephacryl S300 chromatography. The pool corresponding to the fractions of the peak of NF D/NF B activity eluting from heparin chromatography was directly loaded into an oliA DNA chromatography column. To reduce nonspecific interactions, calf thymus DNA was added to the heparin pool prior to loading onto the affinity column, as previously described (11). The NF D/NF B activities eluted as a single peak from the oliA DNA-Sepharose column were pooled, precipitated with acetone, and separated on a preparative 10% polyacrylamide gel by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (30). Protein bands visualized by Coomassie blue staining were excised and prepared for in situ proteolysis by trypsin according to the method of Rosenfeld et al. (45). The tryptic peptides were isolated by reverse-phase high-performance liquid chromatography on a C8 column (2 by 100 mm) eluted with an acetonitrile gradient in 0.1% trifluoroacetic acid. After a second purification on a C18 column, peptides were sequenced on a Procise sequencer (Perkin-Elmer, Foster City, Calif.) using the manufacturer's pulsed liquid program.
SDS-PAGE and Western blotting.
For SDS-PAGE and Western blot analysis, 500 μl of purified fractions of NF D or NF B was concentrated by acetone precipitation. Protein pellets were resuspended in Laemmli loading buffer and subjected to SDS-12.5% PAGE (30). After electrophoresis proteins were visualized by silver staining.
For Western blotting, proteins separated by SDS-PAGE were transferred to nitrocellulose membranes (Bio-Rad) according to the manufacturer's instructions, using a Bio-Rad Protein II apparatus. The membrane was then blotted with a 1:2,500 dilution of primary antibody. Immunoreactive proteins were detected by using the Amersham Pharmacia Biotech ECL kit (RPN 2195) for enhanced chemiluminescence.
Expression of recombinant proteins and antibody production.
A cDNA fragment encoding amino acids 279 to 457 of AtNuc-L1 (see Fig. 6A) was amplified by PCR from an expressed sequence tag (EST) clone (GenBank accession number N65625). This fragment was cloned into the BamHI site of plasmid pET16b (Novagen, Madison, Wis.) to produce the His-tagged ΔAtNuc-L1 recombinant fusion protein. By using a similar approach, a cDNA fragment encoding amino acids 63 to 309 of AtFib1 (see Fig. 6A) was amplified from an EST (accession number AF233443) and was cloned into the BamHI site of plasmid pET16b to create a His-ΔAtFib1 fusion protein. The recombinant fusion proteins were produced and purified by using an Ni2+ column following Novagen's instructions. Rabbit polyclonal antibodies against His-ΔAtNuc-L1 and His-ΔAtFib1 fusion proteins were customer made by Eurogentec (Seraing, Belgium). The immunoglobulin G fractions from the antisera were purified through a HiTrap rProtein A affinity column from Amersham Pharmacia Biotech (Wikstroms, Sweden).
FIG. 6.
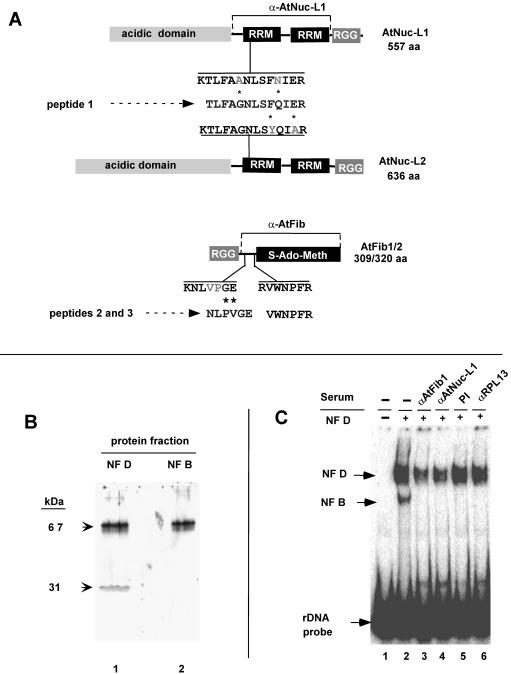
AtNuc-L1 and AtFib are components of NF D. (A) Alignment of sequences from peptides 1 to 3 derived from the 67- and 31-kDa polypeptides to predicted AtNuc-L1 (GenBank accession no. AC084414), AtNuc-L2 (GenBank accession no. AP001303), and AtFib1 and AtFib2 (4). Protein structures and domains are schematized to scale. The positions of similar peptide sequences of Arabidopsis proteins in predicted AtNuc-L1, AtNuc-L2, and AtFib1/AtFib 2 are shown. Asterisks indicate changes between the cauliflower and Arabidopsis peptide sequences. RRM, RNA recognition motif (1); RGG, arginine-glycine-rich domain. The horizontal lines indicated by α-AtNuc-L1 and α-AtFib show the protein regions that were overexpressed and used as antigens to raise specific antibodies. (B) Western blot of NF D and NF B fractions with antibodies against AtNuc-L1 or AtFib. Total proteins from purified NF D or NF B separated fractions as indicated were used for Western blotting (see Materials and Methods) with antibodies directed against AtNuc-L1 or AtFib. The purified NF B fraction corresponds to a pool of fractions eluting below 250 kDa in a Sephacryl S300 column and subsequently purified by oliB DNA chromatography as previously reported (11). (C) Effects of anti-AtNuc-L1 (α-AtNuc-L1) and α-AtFib antibodies in DNA binding activity of NF D. The purified NF D was subjected to immunodepletion (see Materials and Methods) with antibody α-AtNuc-L1, α -AtFib, or α -RPL13 or PI (preimmune serum). After treatment, immunodepleted extracts were assayed by EMSA with the rDNA A123BP probe.
Immunodepletion of NF D extracts.
The immunoglobulin G fraction (10 to 20 μg) against His-ΔAtNuc-Ll or His-ΔAtFib1 was coupled to 100 μl of Dynabeads protein A magnetic beads according to the manufacturer's instructions (Dynal Biotech). After being coupled, the magnetic beads were pretreated with bovine serum albumin (BSA) (1 mg/ml) at 4°C for 1 h to reduce nonspecific binding before use. For immunodepletion, 15 μl of NF D purified fraction was incubated with 15 μl of the corresponding magnetic beads for 2 h at 4°C. The beads were removed with a magnet, and the supernatant fraction was tested for NF D rDNA binding activity.
Plasmid constructs.
pA123BP and pA123BPH, respectively, contain radish 5′ETS rDNA sequences from +103 to +205 and from +103 to +317 (relative to the transcription start site). Both 5′ETS sequences were cloned as an EcoRI/HindIII restriction fragment into plasmid pGem-3Zf(+) (Promega).
DNA and RNA binding assays.
For DNA binding assays the EcoRI/HindIII DNA fragment of pA123BP was labeled by using Klenow fragment and [α-32P]dATP and [α-32P]dCTP. Then 12 fmol of gel-purified fragment was mixed with 6 to 12 μl of NF D purified fraction in a 15-μl reaction mixture containing 50 mM Tris HCl (pH 8), 6 mM MgCl2, 15% glycerol, 1 mM EDTA, 2% NP-40, and 100 mM KCl. Reaction mixtures were supplemented with 100 ng of dI/dC or 1 μg of BSA for S300- or oliA-Sepharose-purified fractions, respectively. Binding reaction mixtures were incubated on ice for 20 min, and the products were analyzed by EMSA as described previously (11).
For RNA binding assays [32P]CTP-labeled rA123BPH RNA (see Fig. 5A) was produced by transcription of a HindIII-linearized pA123BPH template with the Riboprobe in vitro transcription systems (Promega). After being labeled RNA probes were purified by electrophoresis on a polyacrylamide gel. The purified RNA probe (30,000 cpm) was mixed with 6 μl of protein fraction in a 15-μl reaction mixture containing 50 mM Tris HCl (pH 8), 6 mM MgCl2, 15% glycerol, 1 mM EDTA, 2% NP-40, 100 mM KCl, 5 μg of yeast tRNA/ml, 5 μg of BSA/ml, and 40 U of RNasin (Promega). After 20 min of incubation on ice reaction products were analyzed by EMSA (11).
FIG. 5.
NF D binds to the 5′ETS RNA. (A) The RNA probe rA123BPH, encompassing nucleotides +103 to +317 from the radish 5′ETS, is shown by the continuous line. The A123B motifs, the P site, and the predicted hairpin are indicated. (B) The rRNA binding activity in the oliA DNA-Sepharose-eluting fractions detected by EMSA with the rRNA A123BPH probe. Fractions are from the same column analyzed for Fig. 4C. (C) Competition for binding of NF D to rDNA probe A123BP with increasing amounts of unlabeled rDNA A123BP or rA123BPH as indicated.
Primer extension and RNase A/T1 protection assays.
Total RNAs from cauliflower inflorescences, A. thaliana, and radish seedlings were extracted with Trizol reagent (Invitrogen) according to the manufacturer's instructions. All samples were then treated with RQ-DNase (Promega) to eliminate contaminant DNA. Primer extension analysis was done as previously described (4) using 5 to 10 μg of RNAs and specific 5′end-labeled primers. Products of the reaction were analyzed on an 8% polyacrylamide-7 M urea sequencing gel.
For RNase A/T1 mapping the riboprobe was produced by in vitro transcription of the linearized antisense rDNA sequence (see Fig. 1B) cloned in the EcoRI/HindIII site of the pGem3Z vector (Promega), incorporating [α-32P]CTP. The radiolabeled RNA probe was purified on a denaturing 8% polyacrylamide gel, and an RNase protection assay was done as described previously (24).
Mapping the in vitro A′ cleavage site.
Unlabeled rA123BPH RNA was prepared using as a template 1 μg of linearized pA123BPH transcribed with the Riboprobe in vitro transcription systems (Promega). Then, 2 μl of A123BPH RNA (30 ng/μl) was mixed with 6 μl of NF D purified fraction in a 15-μl reaction buffer as described above. After 5 min of incubation on ice, the reaction mixtures were moved to room temperature and incubated for an additional 25 min. Processing reactions were terminated by the addition of 285 μl of stop solution (50 mM Tris HCl [pH 8], 0.15 M NaCl, 250 mM sodium acetate, 6 mM EDTA, 0.5% SDS, 0.1 mg of yeast tRNA/ml). The RNA products were extracted with phenol-chloroform and used as the substrate for primer extension with 5′end-labeled primer p4.
Detection of small RNAs by RT-PCR.
RNAs were extracted from 150 μl of purified NF D fraction with phenol-chloroform-ethanol-precipitated RNA and resuspended in 20 μl of water. Then, 2 μl was used as the template for reverse transcription PCR (RT-PCR). RNA samples and the corresponding pairs of primers, 5′AtU3 and 3′AtU3 for U3 snoRNA, 5′AtU14 and 3′AtU14 for U14, 5′AtR82 and 3′AtR82 for snoR82, and 5′AtU6 and 3′AtU6 for U6. RT-PCR was performed by using SuperScript One-Step RT-PCR with the Platinum Taq system (Invitrogen).
Primers and DNA oligonucleotides used in this work.
The following primers and oligonucleotides were used in this work: AfD2, CAACTTTTCCGGCAACTTTTCCGGTGGACG; AfD2r, GTTGCGTCCACCGGAAAAGTTGCCGGAAAA; p1, CTACTGGCAGGATCAACCAGGTAG; p2, GTTGGTCTGTAGTTGGCTGCCCGAGC; p3, CGTTCAATTGCCCCACTCACATCA; p4, CATCAATCGTTCCAACTAATCTAC; 5′AtU3, CGACCTTACTTGAACAGGATCTGTTG; 3′AtU3, CTGTCAGACCGCCGTGCGT; 5′AtU6, GGACCATTTCTCGATTTATGCG; 3′AtU6, CAGGGAAGCCCCTGTAGGC; 5′AtR82, GCTTCTTTGATTGGGTC; 3′AtR82: GTGCCGGTAGATTAAGG; 5′AtU14, GCCGCCTAAGAGCTTTCGCC; 3′AtU14, TCAGACATCCAAGGAAGGATT.
RESULTS
The primary cleavage site in the 5′ETS of crucifers maps within a cluster of repeated motifs.
The primary cleavage in the 5′ETS of various crucifer pre-rRNAs was mapped by primer extension. In radish, using three different primers, a single pre-RNA cleavage signal was detected in the 5′ETS, mapping at site P (Fig. 1A, lanes 5 to 7). This site is located 180 nucleotides downstream from the transcription initiation site (TIS) and 561 nucleotides upstream from the 18S rRNA. This was confirmed by an RNase protection assay that mapped the primary processing site to a similar position (Fig. 1B). The same processing site on the radish pre-rRNA had been mapped previously by S1 nuclease analysis (13). Thus, site P corresponds to site A′ in vertebrates (9, 29) and trypanosomes (27). It is nevertheless not possible to exclude that other cleavage sites occur in the crucifer 5′ETS in addition to P but that these cannot be detected due to rapid processing of the other intermediates.
Notably, site P mapped in a UUUUCGCGC element found just downstream from a cluster of four UUUUCG-rich motifs named A123B (Fig. 2A). A search for RNA secondary structures with Mfold (50) predicts an ∼120-nucleotide hairpin located just downstream from the cleavage site (Fig. 2B). A similar organization was found for the primary pre-rRNA cleavage site P in cauliflower pre-rRNA. This was mapped 188 nucleotides downstream from the TIS (Fig. 1A, lane 12), just downstream from the conserved A123B cluster (Fig. 2A) and associated with a predicted hairpin structure (results not shown).
FIG. 2.
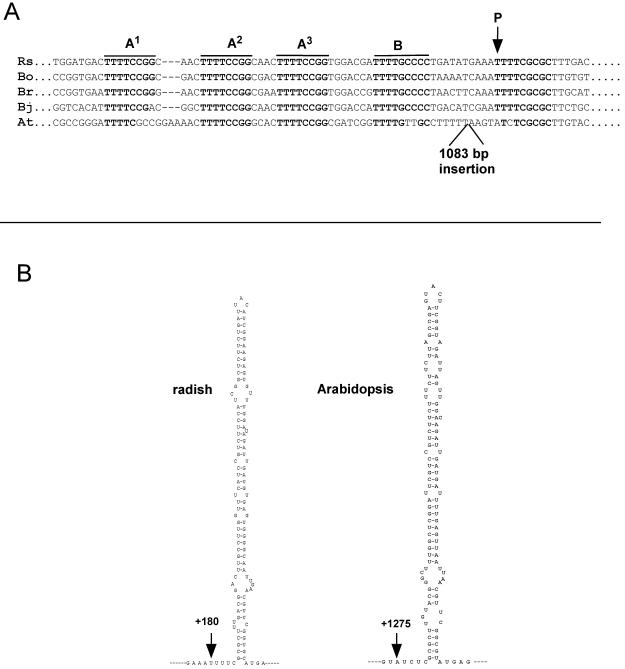
The A123B cluster and predicted structures flanking the primary processing site in the 5′ETSs of crucifers. (A) Alignment of the sequence encompassing the primary pre-rRNA cleavage site and the A123B cluster in crucifers. The A1, A2, A3, B, and P motifs are overlined. Rs, R. sativus (13); Bo, B. oleracea (7); Br, Brassica rapa (12); Bj, Brassica juncea (26); At, A. thaliana (25). EMBL accession numbers: Rs, Z11677; Bo, X60324; Br, S78172; Bj, X73032; At, X52631. (B) The hairpin structures predicted downstream from the P site are shown for radish and Arabidopsis. Secondary structures were predicted by using Mfold (50).
Comparison of 5′ETS sequences reveals that the position of site P and the structural organization of this site are conserved in all crucifers (Fig. 2A). This is also the case for A. thaliana, although in this species P was mapped 1,275 nucleotides downstream from the TIS (Fig. 1A, lane 17). The processing at this site was confirmed by RNase mapping analysis (result not shown). Analysis of the 5′ETS shows that this is due to an “insertion” of 1,083 nucleotides made up of repeats between the A123B cluster and site P (25). In spite of this insertion the A123B cluster (Fig. 2A) and the predicted hairpin (Fig. 2B) associated with P are conserved. In Arabidopsis most rDNA units that produce the bulk of 18S, 5.8S, and 25S rRNAs contain this 1,083-bp insertion, as estimated by PCR analysis (F. Barneche, unpublished data). Only a very minor fraction display some length heterogeneity in the 5′ETS insertion (34).
The conservation of an A123B cluster preceding site P in crucifers, including Arabidopsis, suggests that it may have a role either in pre-rRNA processing or rDNA transcription in crucifers.
Identification of a complex that binds to the A123 cluster on rDNA.
Based on their rDNA binding affinity for the A123B cluster, two distinct factors, NF D and NF B, had been characterized in radish seedling extracts (11). These activities can be detected by an EMSA with an rDNA A123BP probe (Fig. 3A) in whole-cell extracts from crucifers like radish (Fig. 3B, lanes 2 and 3) and cauliflower (lanes 4 and 5). NF D forms a single diffuse complex of lower electrophoretic mobility, while NF B forms various complexes of higher mobility, NF B1, NF B2, or even more. As shown below, the NF B complexes are produced by dissociation of NF D. This dissociation process is difficult to control. It increases throughout purification as well as upon storage of purified NF D. Therefore, the relative ratio of NF D to NF B forms is variable depending on the preparation.
The DNase I footprinting analysis with the radish NF D purified fraction revealed that this factor interacts with both rDNA strands on the A123 cluster (Fig. 3A) (11). The A123 cluster is close to site P in all crucifer 5′ETSs, except in Arabidopsis. In this species it is separated from site P by the large 5′ETS insertion (Fig. 2B). It was confirmed by EMSA that the radish NF D fraction has similar affinity for the radish and Arabidopsis rDNA A123 clusters (Echeverria, unpublished). This shows that the interaction of NF D with the A123 cluster on the rDNA has been conserved in Arabidopsis in spite of the large 5′ETS insertion.
Purification of cauliflower NF D.
NF D had been partially purified from radish seedlings (11). Further purification of the radish factor resulted in NF D dissociation and loss of activity due to the dilution of the fractions. To circumvent this, NF D was extracted from cauliflower inflorescences, a meristematic tissue enriched in dividing cells with a high protein content.
Basically, the purification of cauliflower NF D followed the procedure used for the radish factor. NF D activity was monitored by EMSA with the rDNA A123BP probe. Whole-cell extracts from cauliflower inflorescences were fractionated through various chromatographic steps, including size-exclusion (Sephacryl S300) and DNA-affinity chromatographies (Fig. 4A). The Sephacryl S300 separates NF D, eluting in the range from 650 to 450 kDa (Fig. 4B, lanes 4 to 9), from NF B forms (lanes 13 to 15), which are less than 250 kDa. The high-molecular-mass fractions with NF D activity devoid of NF B were pooled and purified by oliA DNA chromatography (Fig. 4A). This column was made of double-stranded DNA corresponding to oligomerized A motifs mimicking the A123 cluster coupled to Sepharose (see Materials and Methods). The oliA DNA column equilibrated with 100 mM KCl retained all input NF D activity (Fig. 4C, lane 2). After a washing, this activity was eluted by a 300 mM KCl step in a single peak (lanes 5 to 8). Nevertheless, much of the NF D complex loaded on the column was dissociated with a concomitant increase in NF B1-3 complexes (compare lane 2 and lanes 5 to 10). This shows that NF D is an unstable complex of ∼600 kDa that dissociates, generating smaller NF B complexes.
The fractions with the highest “integral” NF D activity eluting from the oliA DNA column (like fractions 16 and 17 in Fig. 4C) represent purified NF D used for its characterization. This fraction is highly purified but has low protein content and is very unstable. To visualize the polypeptides found in purified NF D total proteins from this fraction, fractions were concentrated, separated by SDS-PAGE, and visualized by silver staining (Fig. 4D). This revealed overall 30 polypeptides from ∼20 to ∼200 kDa. The stoichiometry of these is difficult to estimate because reactivity of proteins to silver staining is variable and does not always reflect their abundance.
NF D binds to 5′ETS RNA.
It was previously reported that NF D does not bind to an RNA probe encompassing the A123BP sequence (11). Nevertheless, this probe excluded the ∼120 nucleotides predicted to fold into a hairpin structure (Fig. 2B). We therefore tested whether NF D can interact with the RNA probe rA123BPH, including these sequences (Fig. 5A). The fractions eluting from the oliA DNA chromatography (Fig. 4C) were tested by EMSA with the RNA probe. As shown, a single peak of RNA binding activity with a maximum in fractions 16 and 17 (Fig. 5B, lanes 5 to 8) coeluted perfectly with the rDNA binding activity detected in this column (Fig. 4C, lanes 5 to 8). The RNA retardation profile shows two major and some minor retarded bands, reminiscent of multiple complexes observed with the rDNA probe.
To confirm that the 5′ETS RNA binding activities reside in the NF D rDNA binding complex, a competition analysis was carried out. Increasing amounts of unlabeled rA123BPH displaced the binding of NF D to the rDNA probe (Fig. 5C, lanes 8 to 12), with efficiency similar to that of the unlabeled rDNA A123BP competitor (lanes 3 to 7). It was noted that the binding of NF B1 to the rDNA probe was less affected by the RNA than by the DNA competitor, showing that the subcomplex has less affinity for the RNA than NF D. These results show that NF D can also bind to the 5′ETS RNA from crucifers.
Nucleolin-like protein and fibrillarin are two components of NF D.
The next objective was to identify major polypeptides forming NF D. To achieve this, the purification procedure was adapted to scale up the amount of protein recovered. Considering that NF B derives from NF D and that Sephacryl S300 chromatography resulted in important loss of NF D activity, this step was eliminated. Thus, a pool of NF D and NF B activity eluting in a single peak from heparin chromatography (Fig. 4A) was directly loaded on the oliA DNA affinity column. In these conditions a purified NF D/NF B fraction was obtained in sufficient quantity to visualize polypeptides separated by SDS-PAGE by Coomassie blue staining. Major bands, including 67- and 31-kDa polypeptides, were excised and in gel digested by trypsin. This produced internal peptides that were sequenced by Edman degradation. The peptide sequences were then used in a search using TBLASTN (3) for the corresponding proteins in the Arabidopsis ESTs and genomic sequences in data banks. The identification of the 67- and the 31-kDa polypeptides is reported here.
From the 67-kDa protein, only one peptide sequence of significant length could be obtained that matched two Arabidopsis gene products, here called AtNuc-L1 (GenBank accession no. AG29744) and AtNuc-L2 (GenBank accession no. BAB02219.1) (Fig. 6A). The genes are duplicated genes that encode proteins similar to nucleolin and which have been found in alfalfa (8) and pea (48). AtNuc-L1 and AtNuc-L2, predicted to have 557 and 636 amino acids, respectively, differ mainly in the number of acidic stretches in the N-terminal domain (Fig. 6A). In both AtNuc-L1 and AtNuc-L2 the peptide sequence is preceded by a lysine, a residue usually recognized by trypsin for cleavage. The cauliflower peptide has two amino acid changes compared with the AtNuc-L1 and AtNuc-L2 peptides. These changes are conservative for AtNuc-L1, while only one is conservative for AtNuc-L2 (Fig. 6A). In addition, many cognate ESTs or cDNAs from the AtNuc1 gene are found in data banks, while none are found for AtNuc-L2. Indeed, we could not detect expression of AtNuc-L2 by RT-PCR in Arabidopsis seedlings, suggesting that under normal conditions this gene is not expressed (J. Sáez-Vasquez, unpublished result). Therefore, we consider that the 67-kDa protein component of cauliflower NF D could be the homologue of the predicted AtNuc-L1. The mass of the AtNuc-L1 protein is ∼59 kDa, which differs from the 67-kDa polypeptide isolated by SDS-PAGE. This oversize migration is typical of nucleolin-like proteins and is probably due to the high content of charged residues in this protein (23).
From the 31-kDa polypeptide, two peptide sequences were obtained. Both matched AtFib1 and AtFib2 (Fig. 6A), the Arabidopsis fibrillarins encoded by duplicated genes (4). One of the cauliflower peptides had an inversion of two residues (PV versus VP) relative to the AtFib sequence. AtFib1 and AtFib 2 proteins are structurally and functionally very similar (4), and the peptides probably derive from both isoforms of cauliflower-predicted homologues.
To confirm the presence of AtNuc-L1 and AtFib1/2 in NF D, antibodies to AtFib and AtNuc-L1 were raised against the corresponding proteins (see Materials and Methods). To maximize the specificity of recognition, the recombinant proteins used as antigens did not include the RGG domain found in many nucleolar proteins and the acidic stretch in AtNuc-L1 (Fig. 6A). The Western blot of the purified NF D fraction shows that each antibody recognizes a single band corresponding to the 31- and 67-kDa polypeptides (Fig. 6B, lane 1). A control with preimmune serum for each of the antibodies gave no signal (result not shown). A similar Western blot with a purified fraction of NF B1-2 devoid of NF D (see Materials and Methods) detected only nucleolin, showing that this complex lacks fibrillarin (Fig. 6B, lane 2).
To confirm that nucleolin and fibrillarin are components of the NF D rDNA binding activity, the purified fraction was submitted to one round of immunodepletion with antibody anti-AtFib or anti-AtNuc-L1. Removal of AtNuc-L1 and AtFib was estimated by Western blot analysis to be 30 to 40% (result not shown). Greater depletion of these proteins by multiple rounds of immunodepletion was not possible due to the extreme lability of NF D activity. The partial depletion of AtFib and AtNuc-L1 correlates with a significant reduction in the rDNA binding activity of NF D; this was specifically detected in the extracts immunodepleted with anti-AtFib (Fig. 6C, lane 3) and anti-AtNuc-L1 (lane 4). No significant reduction was detected in NF D fractions treated either with a preimmune serum (lane 5) or with an antiserum directed against ribosomal protein RPL13 from Brassica napus (lane 6) (46). These data further support the conclusion that AtNuc-L1 and AtFib1/2 are components of the rDNA binding NF D complex.
Identification of U3 and U14 C/D snoRNAs in purified NF D.
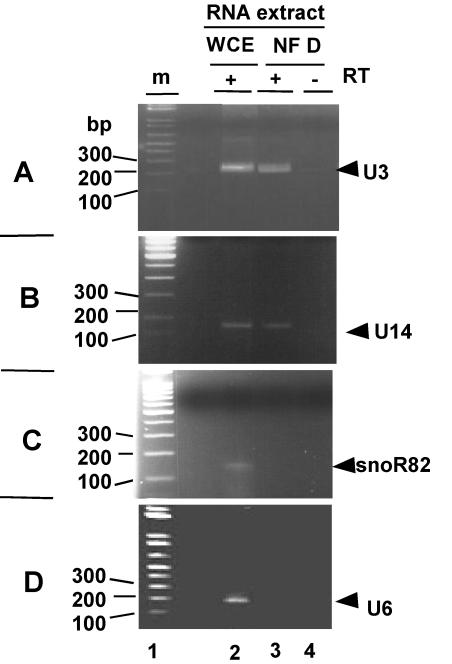
The identification of AtFib1/2 in NF D suggested that U3 snoRNA could be in this fraction. This was tested by an RT-PCR assay. In Arabidopsis there are three genes expressing nearly identical U3 isoforms (36). A pair of primers designed to amplify all three isoforms produced an RT-PCR band of the expected size for U3 when tested on total RNA from cauliflower (Fig. 7A, lane 2). A similar product was amplified by RT-PCR from the RNA extracted from purified NF D (lane 3) but was not detected when reverse transcriptase was omitted from the assay (lane 4). This shows that U3 is found in the purified NF D fraction. U14 is another conserved C/D snoRNA associated with fibrillarin and implicated in early pre-RNA cleavages in S. cerevisiae (49) and animals (18). In Arabidopsis, conserved U14 isoforms have been found (32). By use of a similar RT-PCR-based approach, U14 was also detected in the NF D fraction (Fig. 7B).
FIG. 7.
Identification of C/D snoRNAs in NF D by RT-PCR. Specific primers were designed to detect U3, U14, R82, and U6 based on Arabidopsis gene sequences (see Materials and Methods). RT-PCR was carried out with the corresponding primer pairs and either total RNA from cauliflower inflorescence or RNA extracted from purified NF D fraction as indicated. In control reaction mixtures reverse transcriptase was omitted from the RT-PCR assay, as indicated.
The presence of Arabidopsis snoR82 (35), an H/ACA snoRNA which does not associate with fibrillarin, was also tested. The cauliflower snoR82 homologue was detected in total RNA (Fig. 7C, lane 2) but not in the RNA fraction extracted from NF D (lane 3).
Finally, U6, an abundant spliceosomal RNA unrelated to snoRNPs, detected in total RNA from cauliflower (Fig. 7D, lane 2), was not found in the NF D fraction (lane 3).
Altogether these data reveal that U3 and U14 are components of NF D, probably through their interaction with AtFib 1/2. To assess the role of U3 and U14 within NF D, purified fractions were treated with microccocal nuclease to inactivate the RNA component. Nevertheless, the subsequent addition of EGTA to inactivate the Ca2+-dependent nuclease resulted in complete loss of NF D activity. Therefore, it was not possible to evaluate the contribution of the RNA component to the integrity of NF D or its rDNA binding activity.
NF D reproduces primary pre-rRNA cleavage in vitro.
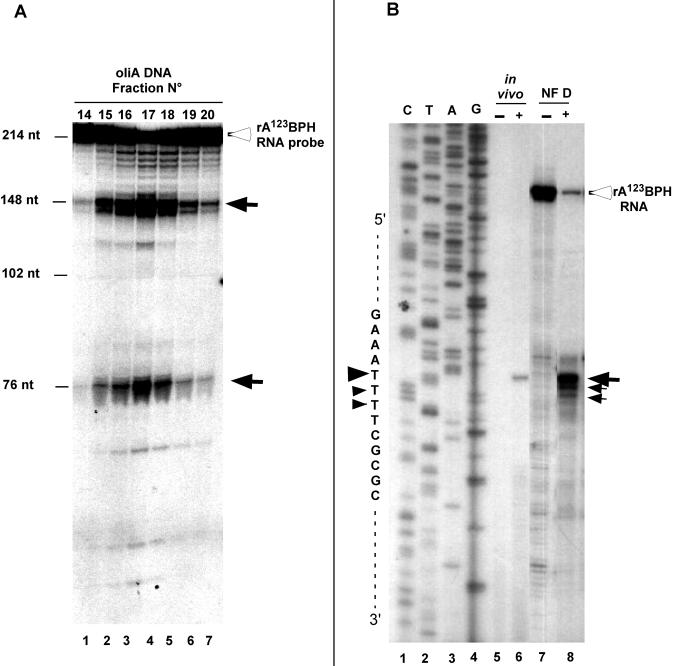
NF D binds to the 5′ETS RNA and contains essential components implicated in primary pre-rRNA cleavage. Therefore, whether it can reproduce this event in vitro was tested. Fractions displaying NF D activity eluting from the oliA affinity chromatography column (Fig. 4C) were incubated with the rA123BPH probe. The products of the reaction were then analyzed on a sequencing gel. Clearly, an endonucleolytic activity coelutes with rDNA and 5′ETS RNA binding activity of NF D that cleaves the probe in two products of ∼148 and ∼76 nucleotides, as expected for cleavage at site P (Fig. 8A).
FIG. 8.
Specific cleavage of rA123BPH RNA by NF D fraction. (A) Fractions of the peak of NF D activity eluting from oliA DNA chromatography (shown in Fig. 4C) were incubated in a cleavage reaction buffer with RNA probe rA123BPH (see Materials and Methods). The open arrowhead indicates full-length RNA probe. Closed arrows indicate cleavage products. Size markers are indicated on the left. (B) Cleavage reaction was carried out with unlabeled rA123BPH RNA substrate in the presence (lane 8) or absence (lane 7) of purified NF D (see Materials and Methods). After incubation, products of the reaction were analyzed by primer extension using primer p3 (shown in Fig. 1). The cleavage site on the pre-rRNA produced in vivo was mapped with the same primer on total RNA extracted from radish seedlings (lane 6). A control was done without RNA added to the reaction mixture (lane 5). CTAG indicates the DNA sequence of the radish 5′ETS made with the same primer. Closed arrows indicate cleavage signals according to their intensities; closed arrowheads indicate the positions of the cleavage signals on the sequence.
To precisely map the cleavage site on rA123BPH, the purified NF D fraction was incubated with unlabeled rA123BPH and the RNA products of the reaction were analyzed by primer extension (Fig. 8B). The NF D fraction efficiently cleaved the rA123BPH substrate (Fig. 8, lane 8), yielding a major cleavage signal that maps to the P site detected in vivo (lane 6).
These results strongly suggest that NF D is associated with an endonuclease activity that specifically cleaves the 5′ETS RNA at the P site in vitro.
DISCUSSION
NF D is a pre-rRNA processing complex.
In crucifers the pre-RNA primary cleavage was located in a P site at the 5′-terminal end of the 5′ETS, similar to site A′ in the vertebrate 5′ETS. Structural analysis shows that P is always associated with a cluster of motifs and a predicted hairpin structure in crucifers. A large complex, NF D, was purified from cauliflower and specifically interacts with the rDNA cluster just upstream from the P site. Evidence was provided showing that NF D also binds to the pre-rRNA and can cleave it at the P site in vitro. These data strongly suggest that NF D could be implicated in 5′ETS primary cleavage in crucifers.
One component identified in NF D corresponds to cauliflower homologues of AtFib1 and AtFib2 proteins. AtFib1 and AtFib2 are very similar, and both complement a yeast NOP1 null mutant (4). It seems unlikely that NF D is constituted by any of these isoforms exclusively. Nop1p is the nucleolar rRNA methylase in eukaryotes that associates with all C/D snoRNAs (49). In S. cerevisiae, Nop1p is also essential for cleavages at sites A0, A1, and A2 in the pre-rRNA (49). Nop1p has no endonuclease activity, and its role in the cleavage reaction is probably mediated by the U3 snoRNP. As expected, after identification of AtFib1/AtFib2 homologues, the U3 snoRNA was found in purified NF D. This fraction accurately cleaved the A123BPH RNA at site P. Due to the inhibitory effect of EGTA on NF D, we could not test the importance of an RNA moiety for NF D integrity or in its cleavage activity. The A123BPH RNA had the predicted hairpin conserved in all crucifer 5′ETSs (Fig. 2B) but did not include any sequence with significant complementarity to the U3 hinge region. Cleavage at A′ in vertebrates absolutely requires the U3 snoRNP (5, 6, 21, 29, 38). However, it is not known whether a direct U3-5′ETS interaction is required for this cleavage in vitro or in vivo. Interestingly, in trypanosomes, which have both A′- and A0-like sites in the 5′ETS, a direct interaction is required for cleavage at A0, as in S. cerevisiae (5, 6), but is dispensable for A′ cleavage in vivo (27). A U3-5′ETS interaction could be dispensable for A′ cleavage when another element, for instance the predicted hairpin downstream from A′ in crucifers, is recognized by a factor recruiting the U3 snoRNP. It will be important to assess the contributions of the predicted hairpin and the 5′ETS-U3 interaction for A′ pre-rRNA cleavage in Arabidopsis and higher eukaryotes in vivo.
NF D is a large complex, copurifying with 30 polypeptides overall (Fig. 4D), that could be related to the yeast U3 snoRNP (16). In addition to AtFib1 and AtFib2 one would expect to find all C/D snoRNA core proteins which are conserved associated with fibrillarin, as well as U3-specific associated proteins (49). However, NF D is different from the yeast U3 snoRNP as it also contains AtNuc-L1, a nucleolin-like protein, and U14, a C/D snoRNA implicated in early pre-rRNA cleavages, including A′ in vertebrates (18).
The presence of AtNuc-L1 in NF D is highly relevant. This protein could act as an assembly factor for this processing complex and direct its binding to pre-RNA. This role has already been shown for nucleolin, which is essential for A′ cleavage (21). The vertebrate protein binds to pre-RNA (20), interacts with the U3 snoRNP (21), and directs assembly of a processing complex on the pre-rRNA (22). A survey of the complete Arabidopsis genome showed two genes most related to nucleolin, encoding AtNuc-L1 and AtNc-L2, which are similar to nucleolin-like proteins described in other plants (8, 48). Only the AtNuc-L1 gene is expressed at a level which can account for nucleolin abundance in the nucleolus. We could not detect expression of AtNuc-L2 by RT-PCR (Sáez-Vasquez, unpublished), while a gene previously described as encoding a nucleolin homologue (15) is indeed much more divergent. AtNuc-L1 differs from nucleolin because it has only two RRMs, like the yeast Nsr1p (33). Notably, the specific binding of nucleolin to the 5′ETS RNA implicates only two contiguous RRMs, out of the four RRMs found in this protein (2), while the interaction with U3 snoRNP and the processing factors implicate the N-terminal acidic domain (21, 22). In addition, genetic analysis of the yeast Nsr1p showed that, like nucleolin, it is also required for pre-rRNA processing and 18S rRNA synthesis (33). Thus, AtNuc-L1 could fulfill functions similar to those of nucleolin and Nsr1 in pre-rRNA cleavage.
Finally, NF D copurifies with an endonuclease activity that cleaves the pre-rRNA at site P (Fig. 8). To confirm that the RNase activity is stably associated with NF D, we tried to assess it in extracts depleted for AtFib and AtNuc-L1. Nevertheless, the cleavage activity is extremely labile and it was totally inactivated upon prolonged incubation necessary for the immunodepletion treatment (Sáez-Vasquez, unpublished). In any case NF D would direct or control the endonuclease cleavage at site P in the 5′ETS pre-rRNA.
NF D and rDNA transcription.
DNase I footprinting analysis previously showed that NF D binds to double-stranded rDNA, interacting with both strands of the A123 cluster (11). The data presented here support fibrillarin, nucleolin, and the pre-rRNA cleavage activity as components of this rDNA binding factor: the rDNA binding activity of NF D was efficiently displaced by the A123BPH RNA substrate (Fig. 5C), the binding of NF D to rDNA was reduced by treatment of the purified extracts with antibodies against AtNuc-L1 and AtFib1 (Fig. 6C), and the 5′ETS rRNA cleavage activity was associated with the purest NF D fractions (Fig. 8A and B).
Altogether this suggests that NF D is a pre-rRNA processing complex that could assemble on the rDNA just upstream from the P site in crucifers. A fraction of this complex, including AtNuc-L1, would then dissociate from the rDNA to bind to the nascent transcript. In this model AtNuc-L1 is a good candidate to direct the consecutive binding of NF D first to rDNA and then to pre-rRNA. Nucleolin has strong rDNA binding activity, although without any sequence specificity (39). In addition, nucleolin is found in different complexes that exhibit sequence-specific DNA binding activity (23). One example is LR1, a sequence-specific DNA binding transcriptional regulator in animals. LR1 is a heterodimer formed by nucleolin and hnRNP D, another RNA binding protein (14). Thus, it is conceivable that in the case of NF D its specific binding to rDNA could be determined by the interaction of AtNuc-L1 with a partner that remains unidentified.
The binding of NFD to rDNA could establish a link between rDNA transcription and early pre-rRNA processing. Such a link is indeed observed in the nucleolus. The nascent pre-rRNAs synthesized at the fibrillar center interphase are extended into the dense fibrillar component where pre-rRNA maturation occurs (31). In all species the early pre-rRNA processing complexes can be visualized by electron microscopy on the 5′ end of the nascent transcript very soon after transcription initiation (16, 37).
At the molecular level, accumulating data for vertebrates also implicate nucleolin in rDNA transcription. Overexpression of nucleolin in Xenopus oocytes repressed RNA Pol I transcription and affected the packaging of nascent pre-rRNA and the formation of 40S subunits (44). Interestingly, the effect of nucleolin was dependent on RNA Pol I transcription (43). In Xenopus, U8 and U22 snoRNAs, implicated in pre-rRNA cleavages, are assembled during the transcriptional process, although processing begins only when the full-length transcript has been transcribed (40). In S. cerevisiae an RNA Pol I holoenzyme fraction has been found that contains pre-rRNA processing activities, including Nop1p and snoRNAs U3 and U14, as well as other components (19). Thus, NF D and AtNuc-L1 could link both processes through assembly of a pre-rRNA processing complex on the rDNA. It would thus be of great interest to search for an RNA Pol I-NF D interaction.
The most elusive question is the nature of the endonuclease activity that catalyzes the primary pre-RNA cleavage in eukaryotes. NF D represents the first highly purified high-molecular-weight complex that reproduces this event in vitro. The purification of NF D provides a biochemical system to define the role of nucleolin and fibrillarin and should identify other factors regulating this event through identification of the other NF D-associated polypeptides by mass spectrometry. This should demonstrate whether NF D is also implicated in subsequent steps of pre-rRNA maturation, the regulation of this complex, and the link to RNA Pol I transcription.
Acknowledgments
D. Caparros-Ruiz was supported by EEC program RTD Actions (training grant ERB4001GT972623) and a grant from Communauté de Travail des Pyrénées. J. Sáez-Vasquez was initially supported by a Poste de Chercheur-Associé au CNRS and ECOS-Sud (grant no. C00B03).
We are grateful to Richard Cooke for correction of the English.
REFERENCES
- 1.Alba, M. M., and M. Pages. 1998. Plant proteins containing the RNA-recognition motif. Trends Plant Sci. 3:15-20. [Google Scholar]
- 2.Allain, F. H., P. Bouvet, T. Dieckmann, and J. Feigon. 2000. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 19:6870-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Barneche, F., F. Steinmetz, and M. Echeverria. 2000. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275:27212-27220. [DOI] [PubMed] [Google Scholar]
- 5.Beltrame, M., Y. Henry, and D. Tollervey. 1994. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 22:5139-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrame, M., and D. Tollervey. 1995. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 14:4350-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett, R. I., and A. G. Smith. 1991. The complete nucleotide sequence of the intergenic spacer region of an rDNA operon from Brassica oleracea and its comparison with other crucifers. Plant Mol. Biol. 16:1095-1098. [DOI] [PubMed] [Google Scholar]
- 8.Bogre, L., C. Jonak, M. Mink, I. Meskiene, J. Traas, D. T. Ha, I. Swoboda, C. Plank, E. Wagner, E. Heberle-Bors, and H. Hirt. 1996. Developmental and cell cycle regulation of alfalfa nucMs1, a plant homolog of the yeast Nsr1 and mammalian nucleolin. Plant Cell 8:417-428. [PMC free article] [PubMed] [Google Scholar]
- 9.Borovjagin, A. V., and S. A. Gerbi. 2001. Xenopus U3 snoRNA GAC-box A′ and box A sequences play distinct functional roles in rRNA processing. Mol. Cell. Biol. 21:6210-6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calikowski, T. T., T. Meulia, and I. Meier. 2003. A proteomic study of the arabidopsis nuclear matrix. J. Cell. Biochem. 90:361-378. [DOI] [PubMed] [Google Scholar]
- 11.Caparros-Ruiz, D., S. Lahmy, S. Piersanti, and M. Echeverria. 1997. Two ribosomal DNA-binding factors interact with a cluster of motifs on the 5′ external transcribed spacer, upstream from the primary pre-rRNA processing site in a higher plant. Eur. J. Biochem. 247:981-989. [DOI] [PubMed] [Google Scholar]
- 12.Da Rocha, P. S., and H. Bertrand. 1995. Structure and comparative analysis of the rDNA intergenic spacer of Brassica rapa. Implications for the function and evolution of the Cruciferae spacer. Eur. J. Biochem. 229:550-557. [DOI] [PubMed] [Google Scholar]
- 13.Delcasso-Tremousaygue, D., F. Grellet, F. Panabieres, E. D. Ananiev, and M. Delseny. 1988. Structural and transcriptional characterization of the external spacer of a ribosomal RNA nuclear gene from a higher plant. Eur. J. Biochem. 172:767-776. [DOI] [PubMed] [Google Scholar]
- 14.Dempsey, L. A., L. A. Hanakahi, and N. Maizels. 1998. A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J. Biol. Chem. 273:29224-29229. [DOI] [PubMed] [Google Scholar]
- 15.Didier, D. K., and H. J. Klee. 1992. Identification of an Arabidopsis DNA-binding protein with homology to nucleolin. Plant Mol. Biol. 18:977-979. [DOI] [PubMed] [Google Scholar]
- 16.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunbar, D. A., S. Wormsley, T. M. Agentis, and S. J. Baserga. 1997. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol. 17:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enright, C. A., E. S. Maxwell, G. L. Eliceiri, and B. Sollner-Webb. 1996. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA 2:1094-1099. [PMC free article] [PubMed] [Google Scholar]
- 19.Fath, S., P. Milkereit, A. V. Podtelejnikov, N. Bischler, P. Schultz, M. Bier, M. Mann, and H. Tschochner. 2000. Association of yeast RNA polymerase I with a nucleolar substructure active in rRNA synthesis and processing. J. Cell Biol. 149:575-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghisolfi-Nieto, L., G. Joseph, F. Puvion-Dutilleul, F. Amalric, and P. Bouvet. 1996. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J. Mol. Biol. 260:34-53. [DOI] [PubMed] [Google Scholar]
- 21.Ginisty, H., F. Amalric, and P. Bouvet. 1998. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 17:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ginisty, H., G. Serin, L. Ghisolfi-Nieto, B. Roger, V. Libante, F. Amalric, and P. Bouvet. 2000. Interaction of nucleolin with an evolutionarily conserved pre-ribosomal RNA sequence is required for the assembly of the primary processing complex. J. Biol. Chem. 275:18845-18850. [DOI] [PubMed] [Google Scholar]
- 23.Ginisty, H., H. Sicard, B. Roger, and P. Bouvet. 1999. Structure and functions of nucleolin. J. Cell Sci. 112:761-772. [DOI] [PubMed] [Google Scholar]
- 24.Goodall, G. J., K. Wiebauer, and W. Filipowicz. 1990. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 181:148-161. [DOI] [PubMed] [Google Scholar]
- 25.Gruendler, P., I. Unfried, K. Pascher, and D. Schweizer. 1991. rDNA intergenic region from Arabidopsis thaliana. Structural analysis, intraspecific variation and functional implications. J. Mol. Biol. 221:1209-1222. [DOI] [PubMed] [Google Scholar]
- 26.Gupta, V., G. Lakshmisita, M. S. Shaila, V. Jagannathan, and M. Lakshmikumaran. 1992. Characterization of species specific repeated DNA sequences from Brassica nigra. Theor. Appl. Genet. 84:397-402. [DOI] [PubMed] [Google Scholar]
- 27.Hartshorne, T., W. Toyofuku, and J. Hollenbaugh. 2001. Trypanosoma brucei 5′ETS A′-cleavage is directed by 3′-adjacent sequences, but not two U3 snoRNA-binding elements, which are all required for subsequent pre-small subunit rRNA processing events. J. Mol. Biol. 313:733-749. [DOI] [PubMed] [Google Scholar]
- 28.Kadonaga, J. T., and R. Tjian. 1986. Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. USA 83:5889-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kass, S., K. Tyc, J. A. Steitz, and B. Sollner-Webb. 1990. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 60:897-908. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Lazdins, I. B., M. Delannoy, and B. Sollner-Webb. 1997. Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma 105:481-495. [DOI] [PubMed] [Google Scholar]
- 32.Leader, D. J., G. P. Clark, J. Watters, A. F. Beven, P. J. Shaw, and J. W. Brown. 1997. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 16:5742-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, W. C., D. Zabetakis, and T. Melese. 1992. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol. Cell. Biol. 12:3865-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luschnig, C., A. Bachmair, and D. Schweizer. 1993. Intraspecific length heterogeneity of the rDNA-IGR in Arabidopsis thaliana due to homologous recombination. Plant Mol. Biol. 22:543-545. [DOI] [PubMed] [Google Scholar]
- 35.Marker, C., A. Zemann, T. Terhorst, M. Kiefmann, J. P. Kastenmayer, P. Green, J. P. Bachellerie, J. Brosius, and A. Huttenhofer. 2002. Experimental RNomics: identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol. 12:2002-2013. [DOI] [PubMed] [Google Scholar]
- 36.Marshallsay, C., T. Kiss, and W. Filipowicz. 1990. Amplification of plant U3 and U6 snRNA gene sequences using primers specific for an upstream promoter element and conserved intragenic regions. Nucleic Acids Res. 18:3459-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mougey, E. B., M. O'Reilly, Y. Osheim, O. L. Miller, Jr., A. Beyer, and B. Sollner-Webb. 1993. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 7:1609-1619. [DOI] [PubMed] [Google Scholar]
- 38.Mougey, E. B., L. K. Pape, and B. Sollner-Webb. 1993. A U3 small nuclear ribonucleoprotein-requiring processing event in the 5′ external transcribed spacer of Xenopus precursor rRNA. Mol. Cell. Biol. 13:5990-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olson, M. O., Z. M. Rivers, B. A. Thompson, W. Y. Kao, and S. T. Case. 1983. Interaction of nucleolar phosphoprotein C23 with cloned segments of rat ribosomal deoxyribonucleic acid. Biochemistry 22:3345-3351. [DOI] [PubMed] [Google Scholar]
- 40.Peculis, B. A. 2001. snoRNA nuclear import and potential for cotranscriptional function in pre-rRNA processing. RNA 7:207-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry, K. L., and P. Palukaitis. 1990. Transcription of tomato ribosomal DNA and the organization of the intergenic spacer. Mol. Gen. Genet. 221:103-112. [DOI] [PubMed] [Google Scholar]
- 42.Piller, K. J., S. R. Baerson, N. O. Polans, and L. S. Kaufman. 1990. Structural analysis of the short length ribosomal DNA variant from Pisum sativum L. cv. Alaska. Nucleic Acids Res. 18:3135-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roger, B., A. Moisand, F. Amalric, and P. Bouvet. 2003. Nucleolin provides a link between RNA polymerase I transcription and pre-ribosome assembly. Chromosoma 111:399-407. (First published 11 February 2003 [Online].) [DOI] [PubMed]
- 44.Roger, B., A. Moisand, F. Amalric, and P. Bouvet. 2002. Repression of RNA polymerase I transcription by nucleolin is independent of the RNA sequence that is transcribed. J. Biol. Chem. 277:10209-10219. (First published 31 December 2001 [Online].) [DOI] [PubMed]
- 45.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 46.Saez-Vasquez, J., P. Gallois, and M. Delseny. 2000. Accumulation and nuclear targeting of BnC24, a Brassica napus ribosomal protein corresponding to a mRNA accumulating in response to cold treatment. Plant Sci. 156:35-46. [DOI] [PubMed] [Google Scholar]
- 47.Shaw, P. J., and E. G. Jordan. 1995. The nucleolus. Annu. Rev. Cell Dev. Biol. 11:93-121. [DOI] [PubMed] [Google Scholar]
- 48.Tong, C. G., S. Reichler, S. Blumenthal, J. Balk, H. L. Hsieh, and S. J. Roux. 1997. Light regulation of the abundance of mRNA encoding a nucleolin-like protein localized in the nucleoli of pea nuclei. Plant Physiol. 114:643-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 50.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]