Abstract
Genomic DNA replication is tightly controlled to ensure that DNA replication occurs once per cell cycle; loss of this control leads to genomic instability. Geminin, a DNA replication inhibitor, plays an important role in regulation of DNA replication. To investigate the role of human geminin in the maintenance of genomic stability, we eliminated geminin by RNA interference in human cancer cells. Depletion of geminin led to overreplication and the formation of giant nuclei in cells that had wild-type or mutant p53. We found that overreplication caused by depletion of geminin activated both Chk1 and Chk2, which then phosphorylated Cdc25C on Ser216, resulting in its sequestration outside the nucleus, thus inhibiting cyclin B-Cdc2 activity. This activated G2/M checkpoint prevented cells with overreplicated DNA from entering mitosis. Addition of caffeine, UCN-01, or inhibitors of checkpoint pathways or silencing of Chk1 suppressed the accumulation of overreplicated cells and promoted apoptosis. From these results, we conclude that geminin is required for suppressing overreplication in human cells and that a G2/M checkpoint restricts the proliferation of cells with overreplicated DNA.
The cell cycle is precisely regulated to ensure the proper duplication and segregation of chromosomes into daughter cells (15, 28). DNA replication has to be tightly controlled to ensure that it occurs once and only once per cell cycle (1, 10). Alteration of this regulation leads to genomic instability (26). Replication begins at discrete sites in DNA and requires the assembly of multisubunit complexes at these replication origins. Replication origins initially associate with a six-subunit origin recognition complex (2). The origin recognition complex facilitates the subsequent recruitment of additional components of the replication initiation complex, beginning with Cdc6 and Cdt1 in the late mitosis or early G1 phase (11, 18). Both Cdc6 and Cdt1 facilitate the subsequent recruitment of the six-subunit MCM2-7 complex to replication origins for DNA replication initiation (20, 25).
Geminin, originally identified as a substrate of the anaphase-promoting complex, inhibits DNA replication by binding Cdt1 and preventing the loading of MCM proteins onto chromatin (22, 34, 40). Geminin is expressed from S phase to late mitosis. Thus, the appearance of geminin in S phase could contribute to the prevention of rereplication by inhibiting Cdt1 activity. Depletion of geminin by RNA interference in Xenopus laevis leads to a G2 phase arrest but does not yield overreplicated DNA (21). However, overreplication is observed in Drosophila melanogaster cells in which geminin is eliminated by RNA interference (24). Chk1 is activated in both systems, suggesting an important role of geminin in maintaining genomic stability. Additionally, in Xenopus embryos, geminin was shown to induce uncommitted embryonic cells to differentiate as neurons, suggesting that geminin's role may not be limited to regulation of DNA replication (17). Since cancer is marked by genomic instability, and given geminin's properties, it is necessary to investigate the role of human geminin in the maintenance of genomic stability in human cancer cells.
In our first attempt to disrupt the geminin-Cdt1 balance, we overexpressed Cdt1 with its cofactor Cdc6 in human cancer cells (37). Only p53− cells show conspicuous rereplication. The rereplication is accompanied by the activation of ATM/ATR and Chk2 protein kinases. In p53+ cells this results in the activation of p53, leading to cell cycle inhibition and apoptosis. The activation of checkpoint pathways following rereplication induced by overexpressed Cdt1 and Cdc6 suggests that mammalian cells use these surveillance systems to limit the damage from rereplication (37).
Various checkpoints play a significant role in maintenance of genomic stability (33). The Chk2- and p53-dependent checkpoint pathway induced by overexpression of Cdt1 and Cdc6 is different from the G2/mitosis checkpoint pathway that prevents onset of mitosis before S phase is completed. In response to genotoxic stress, Chk1 and Chk2 are activated by the ATM/ATR-mediated pathway. Active Chk1 and Chk2 then negatively regulate Cdc25C by phosphorylating it on Ser216 (27, 30), resulting in its inhibition or sequestration in the cytoplasm (14, 26). Cdc25C is a phosphatase that removes the inhibitory phosphates from Cdc2 and activates cyclinB-Cdc2, a crucial step in regulating the entry of cells into mitosis (26, 33). Failure to activate the G2/M checkpoint results in genomic instability and cell death (26). Thus, the checkpoint pathway induced by overexpression of Cdt1 and Cdc6 is different from the Chk1 activation induced by geminin depletion in Xenopus or Drosophila cells. To resolve the difference between these results, we decided to investigate the effects of geminin depletion in human cells in culture.
In this study, we therefore depleted endogenous geminin from human cancer cells by RNA interference. Depletion of geminin led to overreplication and formation of giant nuclei containing more than 4N DNA content regardless of the p53 status of the cells. Overreplication in geminin-depleted cells activated both Chk1- and Chk2-mediated G2/M checkpoint which arrests cells with giant nuclei before mitosis. Abolition of the G2/M checkpoint suppressed the accumulation of overreplicated cells and caused cell death through apoptosis. Moreover, silencing of Cdt1 partially suppressed the overreplication in geminin-depleted cells, suggesting that this overreplication is Cdt1 dependent. Collectively, these results suggest that geminin plays an important role in maintaining genomic stability by preventing rereplication. Even if geminin is bypassed, the resulting rereplication triggers checkpoint pathways that prevent the duplication of cells with an abnormal DNA complement.
MATERIALS AND METHODS
Cell lines and drugs.
Human colorectal cancer cell lines HCT116 (p53+/+) and HCT116 (p53−/−) were grown in 10% fetal bovine serum and 1% penicillin-streptomycin in McCoy's 5A modified medium (Cellgro). Human lung cancer cell line H1299 was grown in Dulbecco's modified Eagle's medium (Cellgro) containing 10% fetal bovine serum and 1% penicillin-streptomycin. The concentration of caffeine (Sigma) used was 5 mM; 7-hydroxystaurosporine was used at 300 nM.
siRNAi and PCR primers.
Short interfering (siRNA) oligonucleotides (Dharmacon) were made to the following target sequences (sense): geminin (GEM), UGCCAACUCUGGAAUCAAA; Cdt1, GUACCCCCGAGGCCCCAGA; Chk1 (CHK1), UCGUGAGCGUUUGUUGAAC; Chk2 (CHK2), GAACCUGAGGACCAAGAAC; p53 (P53), AAGACUCCAGUGGUAAUCUAC; and control oligonucleotide (GL2), AACGUACGCGGAAUACUUCGA. Transfections were performed with 200 nM siRNA oligonucleotide duplexes with Oligofectamine (Invitrogen) according to the instructions of the manufacturer. PCR primers were made to following sequences: lamin B2 (forward), AGAATCCGATCATGCACCTGT; lamin B2 (reverse), ACGGCGATCTGCACTTTCA; centromere (forward), AGCAGCTCCTTTGGAGACATA; centromere (reverse), GGCTTCCTTTGCCAAACTTT; telomere (forward), TATGCTGCCACCTGTACATGC; and telomere (reverse), ACATCCTCCCCCTCCCTTT.
Antibodies, immunoblotting, and immunofluorescence.
Rabbit anti-geminin and rabbit anti-Cdt1 were raised as described earlier (40). Rabbit anti-Chk1, rabbit anti-phosphohistone H3 (Ser10), mouse anti-phospho-H2AX (serine 139) (Upstate), mouse anti-Chk2, mouse anti-β-actin (Sigma), rabbit anti-p53, rabbit anti-phospho-Chk1 (Ser317), rabbit anti-phospho-Chk2 (Thr68), rabbit anti-Cdc2, rabbit anti-phospho-Cdc2, rabbit anti-Cdc25C, rabbit anti-phospho-Cdc25C (Ser216) (Cell Signaling Technology), rabbit anti-cyclin A (H432), and mouse anti-cyclin B1 (H433) (Santa Cruz) were used for immunoblotting and immunofluorescence. For Western blotting, cell extracts were prepared by lysis in 0.2% NP-40-50 mM Tris-HCl (pH 7.4)-150 mM NaCl-1 mM EDTA-1 mM Na3VO4 and protease inhibitor cocktail (Sigma). Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and probed. Immunofluorescence was performed as follows. Cells were fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min and permeabilized with 0.2% Triton X-100 for 10 min at room temperature. Coverslips were blocked with 3% bovine serum albumin in PBST (PBS with 0.02% Tween 20) and incubated with primary antibody for 1 h at room temperature. Cells were then washed and incubated with tetramethylrhodamine isothiocyante- or fluorescein isothiocyanate-conjugated secondary antibody (Dako Corporation). Cells were mounted with solution containing 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Laboratories) before examination under the microscope.
FACS analysis.
Cells were collected by trypsinization and fixed with 70% ethanol overnight at 4°C. After fixation, cells were centrifuged and stained in 1 ml of propidium iodide solution (0.05% NP-40, 50 ng of propidium iodide per ml, and 10 μg of RNase A per ml). The labeled cells were analyzed on a Becton Dickinson flow cytometer with Cellquest software. For fluorescence-activated cell sorting (FACS) analysis with both propidium iodide and bromodeoxyuridine, cells were labeled with 15 μM bromodeoxyuridine (Sigma) for 30 min before harvest as described above. Fixed cells were permeabilized in 1 ml of 2N HCl-0.5% Triton X-100 for 1 h at room temperature. After washing in 1 ml of PBS with 1% Tween 20 and 0.2% bovine serum albumin, cells were incubated with fluorescein isothiocyanate-conjugated anti-bromodeoxyuridine antibody (BD Pharmingen) for 30 min at room temperature. Propidium iodide solution was added to stain cells after washing by 1 ml of PBS. The procedure for immunofluorescent detection of phosphorylated histone H3 has been described (41)
Bromodeoxyuridine immunostaining.
Cells transfected with siRNA were treated with 15 μM bromodeoxyuridine (Sigma) for 30 min before harvest. After fixation with 4% paraformaldehyde in PBS for 20 min at room temperature, cells were permeabilized with 0.2% Triton X-100 for 15 min at 4°C, followed by treatment with 1.5 N HCl for 30 min at room temperature. After washing three times with 1× PBS-1% goat serum (Jackson Immunoresearch), cells were incubated with Alexa Fluor 594-conjugated antibromodeoxyuridine (Molecular Probes) for 1 h. Finally, cells were fixed in mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Laboratories) for microscopic observation.
Measuring cell growth and DNA synthesis.
The number of viable cells was estimated with an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] cell growth assay kit (Promega) according to the manufacturer's instructions.
DNA synthesis was estimated by measuring [3H]thymidine incorporation. Briefly, cells were plated in 12-well plates and cultured. The medium was then changed to warm fresh Dulbecco's modified Eagle's medium (Cellgro) with 10% dialyzed fetal bovine serum. The cells were pulse labeled with 5 μCi of [3H]thymidine per ml for 30 min. The medium was removed, and the cells were washed two times with PBS, once with ice-cold 10% trichloroacetic acid containing 0.2 M sodium pyrophosphate, and twice with 95% ethanol, and then lysed with 200 μl of solution containing 1% SDS and 10 mM NaOH; 200-μl aliquots were analyzed in a liquid scintillation counter. The resulting 3H counts per minute normalized to viable cell numbers obtained from the MTT assay represented DNA synthesis.
RESULTS
Overreplication is seen regardless of p53 status in geminin-depleted cells.
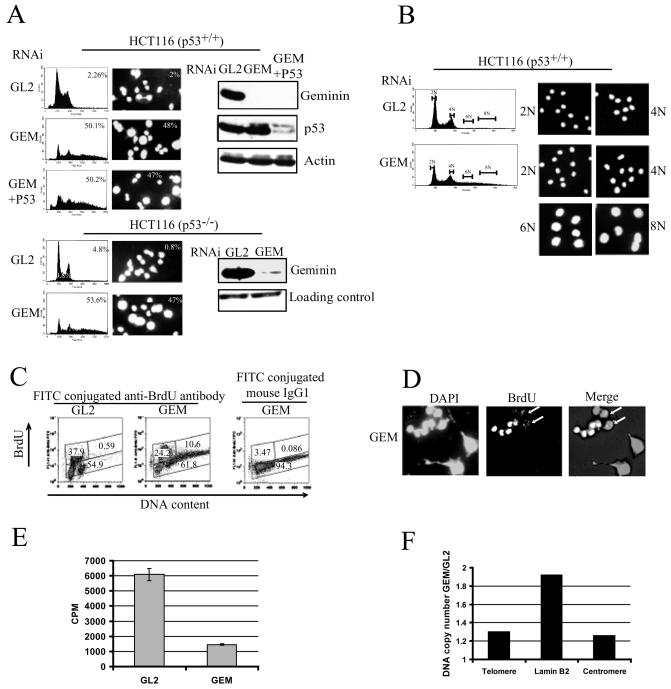
To better understand the role of geminin in regulation of DNA replication, RNA interference was performed to silence the expression of the protein in human colon cancer cell lines HCT116 (p53+/+) and HCT116 (p53−/−). The latter is derived from the former by targeted disruption of both alleles of p53 by homologous recombination (5). As seen in Fig. 1A, geminin levels were significantly reduced in cells transfected with the GEM siRNA oligonucleotide but not in cells transfected with the control siRNA oligonucleotide (GL2). Flow cytometry analysis revealed that nearly 50% of GEM-transfected cells contained more than 4N DNA content with a continuous peak extending from 4N to 8N, suggesting that overreplication occurs in geminin-depleted cells. We also observed DNA overreplication in other cell lines such as HCC1937 (human breast cancer) and H1299 (human lung cancer) (data not shown).
FIG. 1.
Depletion of geminin leads to overreplication in a p53-independent manner in human cancer cells. (A) Overreplication occurs in geminin-depleted cells. HCT116 cells were transfected with control siRNA oligonucleotide GL2 or geminin siRNA oligonucleotide GEM or cotransfected with GEM and P53 oligonucleotide and harvested 72 h after transfection. The left panels show a histogram of cells transfected with siRNA oligonucleotide GL2, GEM, or GEM and P53 (72 h) and then stained with propidium iodide for DNA content before flow cytometry analysis; 10,000 cells were counted for FACS. y axis, cell count; x axis, propidium iodide (PI) fluorescence. The middle panels show nuclei of cells treated as described above and stained with DAPI. The percentage of cells with more than G2/M DNA content or the percentage of cells with giant nuclei (at least twofold bigger than control nuclei) is indicated; 500 cells were counted for each panel. The right panels show expression of geminin, p53, and actin determined by immunoblotting. The loading control was a protein which cross-reacts with anti-geminin antibody. (B) Cells with overreplicated DNA have giant nuclei. HCT116 (p53+/+) cells treated as in panel A were fixed and then sorted into classes with 2N, 4N, 6N, and 8N DNA content by FACS (1,000 cells were sorted for each class). Sorted cells were stained with DAPI and examined under a fluorescence microscope. (C) Cells with overreplicated DNA fail to incorporate bromodeoxyuridine. HCT116 (p53+/+) cells were treated as in panel A. Bromodeoxyuridine (BrdU) was added to label cells for 30 min before harvest. Fixed cells were stained with fluorescein isothiocyanate-conjugated mouse antibromodeoxyuridine antibody or fluorescein isothiocyanate-conjugated mouse immunoglobulin G1 as a negative control and propidium iodide (PI). The percentage of cells in each gate is indicated in the panel; 10,000 cells were counted for FACS analysis. (D) HCT116 (p53+/+) cells treated as in panel A were treated with 15 μM bromodeoxyuridine for the last 30 min and stained with DAPI and antibromodeoxyuridine antibody. (E) HCT116 (p53+/+) cells treated as in panel A were treated with [3H]thymidine (5 μCi/ml) for last 30 min. y axis, 3H counts per minute normalized against the number of viable cells obtained from the MTT assay. (F) HCT116 (p53+/+) cells treated as in panel A were harvested, and cell concentrations were counted. Equal numbers of cells (1.4 × 106) were used to isolate genomic DNA and for PCR. The y axis represents the ratio of the intensity of PCR products in GEM-transfected cells to that in GL2-transfected cells. The x axis indicates the primer pairs used for quantitative PCR.
To determine if overreplication changes the nuclear morphology, we examined the DAPI-stained nuclei in geminin-depleted cells by fluorescence microscopy. Nearly 50% of GEM-transfected cells had a single giant nucleus (Fig. 1A). The percentage of cells with giant nuclei was very close to that with more than 4N DNA content detected by FACS analysis, suggesting that cells with overreplicated DNA may have giant nuclei. To confirm this, we sorted cells by 2N, 4N, 6N, and 8N DNA content and stained them with DAPI. As shown in Fig. 1B, cells with 6N or 8N DNA content had giant nuclei.
To examine whether DNA replication was continuing in cells with giant nuclei, we labeled cells with bromodeoxyuridine and monitored the incorporation by flow cytometry analysis. The results showed that cells with giant nuclei (containing more than 4N DNA) incorporated bromodeoxyuridine at a much lower level than cells with smaller nuclei (containing less than 4N DNA) (Fig. 1C). Moreover, even among the population with greater than 4N DNA, cells with more DNA content (towards the right end of the x axis) had less bromodeoxyuridine incorporation (Fig. 1C). These results suggest that cells may activate checkpoint pathways to suppress overreplication due to depletion of geminin. The reduced bromodeoxyuridine incorporation in cells with giant nuclei was further supported by a bromodeoxyuridine immunostaining assay. None of the giant nuclei were positive for bromodeoxyuridine, compared to 37% (n = 200) of the small nuclei and 34% (n = 224) of GL2-transfected cells (Fig. 1D). Among the middle-sized nuclei (smaller than giant nuclei but bigger than small nuclei), we still saw partial bromodeoxyuridine incorporation (indicated by arrows), suggesting that there is DNA synthesis during early steps of rereplication (Fig. 1D). We also measured DNA synthesis by [3H]thymidine incorporation. As shown in Fig. 1E, DNA synthesis in geminin-transfected cells was only about 25% of that in GL2-transfected cells.
Since the DNA content in geminin-depleted cells was distributed over a broad region between 4N and 8N, we assumed that the DNA overreplication shown in Fig. 1A is partial overreplication and not a complete duplication of the genome. We next tried to estimate the extent of the overreplication by quantitative PCR measurement of the copy number of three regions on chromosome 19, including lamin B2, a known DNA replication origin (4), centromere, and telomere. The PCR products were quantified, and the ratio of products from geminin-depleted cells to control cells was calculated for three primers. Figure 1F displays the results.
This analysis revealed that the lamin B2 locus had nearly twofold more DNA than in control cells. Telomere or centromere loci were overreplicated less, suggesting that the overreplication is not evenly distributed on the chromosome. Since only 50% of the cells have greater than 4N DNA (Fig. 1A), the twofold increase in DNA at the lamin B2 locus suggests that there may be a fourfold enrichment of the locus in the rereplicated cells. Thus, there were about two to three cycles of rereplication at an early replicating origin in geminin-depleted cells. The absence of runaway rereplication (with many cycles of replication) and the decreased bromodeoxyuridine incorporation in the giant nuclei suggest that continued DNA synthesis may be inhibited by the activated checkpoint pathways that we describe later.
Overreplication was observed in both p53+ and p53− cells (Fig. 1A), indicating that overreplication in geminin-depleted cells is not adversely affected by p53, contrary to what we observed upon overexpression of Cdt1 and Cdc6 (37). To further address this, we cosilenced p53 with geminin by siRNA transfection. Although p53 was significantly reduced in geminin-depleted cells, no further rereplication was observed (Fig. 1A). Additionally, the phosphorylation of p53 on Ser15 and Ser 20 was not observed in geminin-depleted cells (data not shown).
H2AX, Chk1, and Chk2 are activated in geminin-depleted cells.
The overreplication and giant nuclei shown in Fig. 1 suggest that depletion of geminin causes genomic instability. We next examined whether the DNA damage checkpoint was activated. One of the earliest substrates of phosphorylation after DNA damage is a variant form of the histone H2AX (6, 39). We examined H2AX phosphorylation using phosphorylation-specific anti-phospho-H2AX (Ser139) antibody by both Western blotting and immunofluorescence assays following geminin RNA interference. Phosphorylated H2AX foci accumulated in most of the cells with giant nuclei, 86% in HCT116 (p53+/+) cells and 80% in HCT116 (p53−/−) cells, but not in cells with small nuclei or GL2-transfected cells (Fig. 2A, top panel). This result was confirmed by Western blotting, which showed that H2AX was phosphorylated in geminin-depleted HCT116 (p53+/+) cells (Fig. 2A, lower panel).
FIG. 2.
DNA damage checkpoints are activated in geminin-depleted cells. (A) H2AX was activated in geminin-depleted cells. HCT116 cells treated as in Fig. 1A were either immunostained or immunoblotted with phospho-H2AX antibody (P-H2AX, Ser 139) antibody; 95 cells with giant nuclei and 120 cells with small nuclei were counted to determine the percentage of cells with phosphorylated H2AX foci in HCT116 cells. (B) Both Chk1 and Chk2 are phosphorylated in geminin-depleted cells. HCT116 cells treated as in Fig. 1A were immunostained or immunoblotted for the indicated proteins after separation on a 10% polyacrylamide gel. P-Chk1, phospho-Chk1 (Ser317); P-Chk2, phospho-Chk2 (Thr68) antibody. We counted 105 cells with giant nuclei and 100 cells with small nuclei to determine the percentage of cells with phosphorylated Chk1 and Chk2 foci in HCT116 cells.
Histone H2AX has been shown to play a critical role in recruitment of repair factors to nuclear foci after DNA damage (29). Phosphorylation of H2AX in geminin-depleted cells indicates a possible activation of the DNA damage checkpoint. In response to DNA damage, both Chk1 and Chk2 are activated in ATM (ataxia telangiectasia mutated) and ATR (ATM-Rad3 related) kinase-mediated pathways (26). We next examined the phosphorylation of both Chk1 and Chk2 in geminin-depleted cells. In HCT116 (p53+/+) cells, phosphorylated Chk1 foci were observed in 88% of the cells with giant nuclei and phosphorylated Chk2 foci were observed in 80% of cells with giant nuclei (Fig. 2B, top panel). Phosphorylation of both Chk1 and Chk2 was confirmed by Western blotting (Fig. 2B, lower panel). Similar results were also obtained in HCT116 (p53−/−) cells (Fig. 2B) and in H1299 cell lines that are naturally mutated for p53 (data not shown).
Collectively, our data imply that overreplication in geminin-depleted cells activates both Chk1- and Chk2-mediated damage checkpoint pathways. We next tested whether the intra-S-phase DNA damage checkpoint was activated in geminin-depleted cells. It has been shown that NBS1, a component of the double-strand break repair protein complex MRE11/RAD50/NBS1, is phosphorylated by ATM in response to double-strand breaks, and this phosphorylation is required for activation of the intra-S-phase checkpoint (16, 33, 35). In geminin-depleted cells, NBS1 was not phosphorylated, as determined by both Western blot and immunofluorescence assays (data not shown), suggesting that the NBS1-mediated intra-S-phase DNA damage checkpoint normally activated by double-strand breaks was not activated in geminin-depleted cells.
G2/mitosis checkpoint is activated in geminin-depleted cells.
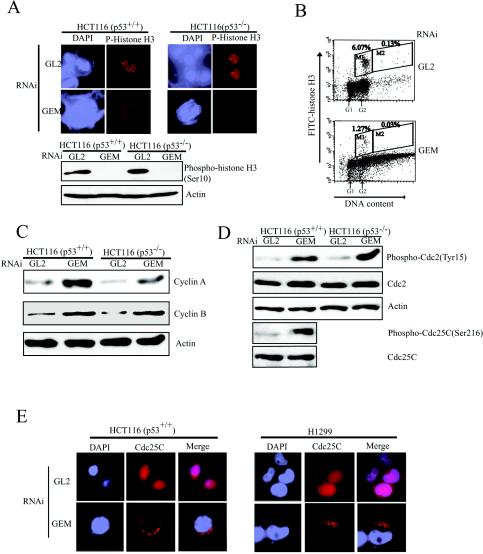
To test whether the G2/M or the spindle checkpoint is activated in geminin-depleted cells, we examined the cell cycle stage of the cells with overreplicated DNA and giant nuclei, G2 or mitosis. Cells transfected with either the GEM or GL2 siRNA oligonucleotide were immunostained with anti-phosphohistone H3 antibody to identify mitotic cells (41, 42) (Fig. 3A). Quantitative analyses revealed that none of the cells with giant nuclei had phosphohistone H3 foci, but 3% of cells with small nuclei had phosphohistone H3 foci in GEM-transfected cells (Table 1). In control siRNA GL2-transfected cells, about 15% of cells had phosphohistone H3 foci. This result was confirmed by Western blotting, which showed a high level of phosphorylated histone H3 in GL2-transfected cells but no histone H3 phosphorylation in geminin-depleted cells (Fig. 3A, lower panel).
FIG. 3.
G2/M checkpoint is activated in geminin-depleted cells. (A) Cells with giant nuclei arrest before mitosis. HCT116 cells treated as in Fig. 1A were either immunostained or immunoblotted with phosphohistone H3 antibody. (B) HCT116 (p53+/+) cells treated as in Fig. 1A were costained for DNA content and phosphorylated histone H3 and analyzed by flow cytometry. Cells with 4N DNA content and phosphorylated histone H3 were gated in M1, and cells with more than 4N DNA and phosphorylated histone H3 were gated in M2. The cell percentage of each gate is indicated. (C) HCT116 cells treated as in Fig. 1A were immunoblotted for the indicated proteins. (D) HCT116 cells treated as in Fig. 1A were immunoblotted for the indicated proteins. (E) Cdc25C is sequestered in the cytoplasm. HCT116 (p53+/+) and H1299 (p53−) cells treated as in Fig. 1A were immunostained for Cdc25C; 120 cells with giant nuclei were counted.
TABLE 1.
Mitotic index in cells transfected with GL2 or GEM siRNAa
| Cells | % Mitotic cells (no. of cells counted)
|
||
|---|---|---|---|
| GL2- transfected cells with small nuclei | GEM- transfected cells with small nuclei | GEM- transfected cells with giant nuclei | |
| HCT116 (p53+/+) | 15 (300) | 3 (100) | 0 (120) |
| HCT116 (p53−/−) | 10 (350) | 2 (100) | 0 (120) |
Cells with phosphohistone H3 foci were regarded as mitotic cells.
To further examine whether cells with overreplicated DNA are in mitosis, we identified mitotic cells by costaining cells with propidium iodide for DNA and with anti-phosphohistone H3 antibody for mitotic cells (41). In geminin-depleted cells, 0.03% of cells with more than 4N DNA content were in mitosis, compared to 1.27% of cells with 4N DNA content (Fig. 3B). Additionally, both cyclin A and cyclin B were elevated in geminin-depleted cells (Fig. 3C). These results indicate that cells with giant nuclei are arrested before mitosis, most likely in G2. No differences were noted in the p53−/− HCT116 cells.
Next we tested whether the G2/M checkpoint was activated in geminin-depleted cells. In response to incomplete DNA replication, Chk1 and/or Chk2 prevents Cdc2 dephosphorylation at Tyr15 by negatively regulating Cdc25C. As shown in Fig. 3C, inhibitory phosphorylation of Cdc25C at Ser216 was increased in geminin-depleted cells but Cdc25C protein levels were stable. Consistent with this result, inhibitory phosphorylation of Cdc2 at Tyr15 was elevated in geminin-depleted cells (Fig. 3D). It is likely that increased phosphorylation of Cdc2 at Tyr15 is due to the inhibitory phosphorylation of Cdc25C at Ser216. Since sequestering Cdc25C in the cytoplasm also prevents activation of Cdc2, we examined the cellular localization of Cdc25C in geminin-depleted cells. Cdc25C foci appeared in the cytoplasm of 92% of geminin-depleted H1299 (p53−) cells (Fig. 3E, right panel). Cdc25C was perinuclear in 86% of geminin-depleted HCT116 (p53+/+) cells (Fig. 3E, left panel). Collectively, the data shown in Fig. 3 suggest that overreplication in geminin-depleted cells activates a Chk1/Chk2-mediated checkpoint, which then inhibits the activity of Cdc2 by phosphorylating Cdc25C at Ser216, resulting in its sequestration outside the nucleus. Regardless of p53 status, activation of this G2/M checkpoint pathway in geminin-depleted cells prevents cells with overreplicated DNA from entering mitosis.
Abolition of the G2/M checkpoint suppresses the accumulation of overreplicated cells and promotes apoptosis.
If overreplication arrested cells in G2 phase by the G2/M checkpoint, we hypothesized that abolition of the G2/M checkpoint will have a deleterious effect on these cells. We first examined the time course of overreplication after GEM transfection. The results showed that the percentage of overreplicated cells between 48 and 60 h after GEM transfection was about 46%, similar to that at 72 h (data not shown). Since both caffeine and UCN-01 have been shown to abolish the G2/M checkpoint (31, 38), we therefore added either caffeine or UCN-01 to cells at 57 h after transfection and harvested at 72 h. As shown in Fig. 4A, both caffeine and UCN-01 decreased the percentage of cells with overreplicated DNA. Additionally, caffeine reduced the phosphorylation of Cdc25C at Ser216 and phosphorylation of Cdc2 at Tyr15 (Fig. 4C), supporting the conclusion that the G2/M checkpoint was activated in geminin-depleted cells.
FIG. 4.
Abolition of G2/M checkpoint suppresses overreplication. HCT116 (p53+/+) cells were transfected with control siRNA oligonucleotide GL2 or geminin siRNA oligonucleotide (GEM) or cotransfected with the siRNA oligonucleotide for CHK1, CHK2, or both CHK1 and CHK2 and harvested 72 h after transfection. HCT116 (p53+/+) cells treated as in Fig. 1A for 57 h were treated with either caffeine or UCN-01 for another 15 h before harvest. The percentage of cells with overreplicated DNA was analyzed by FACS. The x axis indicates the siRNA oligonucleotides or chemical used together with either GL2 or GEM. (B) HCT116 (p53+/+) cells treated as in panel A were immunoblotted for the indicated proteins. (C) HCT116 (p53+/+) cells transfected with either GL2 or GEM were treated with either caffeine or UCN-01 for the last 15 h before harvest 72 h after transfection. The cells were then immunoblotted for the indicated proteins. (D) HCT116 (p53+/+) cells transfected with either GL2 or GEM were treated with caffeine for the last 15 h before harvest 72 h after transfection. Cells were then immunoblotted for indicated proteins or stained with propidium iodide for DNA content before flow cytometry analysis.
Abolition of the G2/M checkpoint in cells with damaged DNA can induce apoptotic cell death (3, 23). To detect apoptosis, we used immunoblotting to look for the presence of poly(ADP-ribose) polymerase 1 (PARP-1) cleavage products in geminin-depleted cells with or without caffeine. PARP-1 is a zinc finger nuclear protein that is the target of caspase-3 and caspase-7, which cleave the protein to 24- and 85-kDa fragments during apoptosis (12, 36). As shown in Fig. 4D, PARPp24 and PARPp85 were detected in geminin-depleted cells after caffeine treatment, suggesting that abolition of the G2/M checkpoint resulted in apoptosis in geminin-depleted cells. This result was confirmed by FACS analysis, which showed that the cell population with sub-G1 DNA content increased to 41% in geminin-depleted cells after caffeine treatment (Fig. 4D, lower panel). Silencing of geminin was reported to cause apoptotic cell death in Drosophila cells (24). However, we did not detect PARPp24 and PARPp85 in geminin-depleted human cells (Fig. 4D), indicating that depleting geminin does not activate apoptotic pathways in human cells, most likely due to activation of the G2/M checkpoint.
To validate the experiments performed with chemical inhibitors, we downregulated Chk1, Chk2, or both Chk1 and Chk2 by RNA interference in geminin-depleted cells. Silencing of Chk1, Chk2, or both Chk1 and Chk2 with geminin partially suppressed the percentage of cells with overreplicated DNA (Fig. 4A). The Western blot showed a good depletion of Chk1, Chk2, and geminin by RNA interference (Fig. 4B). Additionally, downregulation of Chk1 or both Chk1 and Chk2 reduced the phosphorylation of Cdc2 at Tyr15, but this was not seen upon downregulation of Chk2 alone, suggesting that Chk1 is the major player in the G2/M checkpoint in geminin-depleted cells (Fig. 4B).
Downregulation of Cdt1 suppresses overreplication in geminin-depleted cells.
Geminin has been shown to inhibit DNA replication by inhibiting Cdt1 (34, 40). We therefore examined the Cdt1 levels in geminin-depleted cells to determine whether there was an excess of Cdt1. Our data indicated that Cdt1 protein levels decreased in the final population of geminin-depleted cells regardless of p53 status (Fig. 5A). Cdt1 protein levels are cell cycle regulated, with a peak in G1 and degradation upon entry into S phase (25). Since geminin-depleted cells arrest at G2 phase (Fig. 3), it is possible that downregulation of Cdt1 in geminin-depleted cells is due to the G2 arrest, although we cannot exclude the possibility that geminin directly regulates Cdt1 protein levels (24). The decrease in Cdt1 is most likely an end-stage situation because replication and rereplication are unlikely to occur without this essential replication activator.
FIG. 5.
Cdt1 is required for overreplication in geminin-depleted cells. (A) Cdt1 protein level is downregulated in geminin-depleted cells. HCT116 cells treated as in Fig. 1A were immunoblotted for the indicated proteins. (B) HCT116 (p53+/+) and HCT116 (p53−/−) cells were cotransfected with siRNA oligonucleotide GL2 (control) or GEM (geminin) and GL2 or cotransfected with GL2 or GEM and CDT1 for 72 h. The percentage of cells with overreplicated DNA was analyzed by FACS. The x axis indicates the siRNA oligonucleotide used together with either GL2 or GEM.
To test whether Cdt1 is required for overreplication in geminin-depleted cells, we cosilenced Cdt1 with geminin in both HCT116 (p53+/+) and HCT116 (p53−/−) cells. As shown in Fig. 5B, cosilencing of Cdt1 partially suppressed the overreplicating population, suggesting that Cdt1 is required for overreplication in geminin-depleted cells.
DISCUSSION
Geminin was initially identified as a DNA replication inhibitor (22) and was later shown to play a role in maintaining genomic stability in both Xenopus and Drosophila cells (21, 24). Our data confirmed a similar role of geminin in mammalian cells by showing that geminin depletion leads to overreplicated DNA and G2/M arrest by activation of a G2/M checkpoint, although there are some differences that could be mammalian cell specific (Fig. 6). Moreover, our data revealed interesting differences in checkpoint pathways induced by geminin depletion versus Cdt1 and Cdc6 overexpression.
FIG. 6.
Rereplication by depletion of geminin activates the G2/M checkpoint. Rereplication caused by depletion of geminin activates Chk1, and active Chk1 phosphorylates Cdc25C on Ser216, resulting its sequestration outside nucleus, thus inhibiting cyclin B-Cdc2 activity. This active G2/M checkpoint prevents cells with overreplicated DNA from entering mitosis.
Geminin inhibits DNA replication by inhibiting Cdt1 activity (22, 34, 40). For this reason, overreplication in geminin-depleted cells should activate the same pathway as that observed in cells with rereplication by overexpressing Cdt1. We have shown that p53 inhibits the accumulation of rereplicated cells by inducing p21 and PIG3 in cells overexpressing Cdt1 and Cdc6 (37). Therefore, it is surprising to see overreplication in p53+ cells upon depletion of geminin. One difference could be that Cdc6 levels were not increased when geminin was depleted. We have recently discovered that excess Cdc6 produces double-strand DNA breaks and activates p53 without discernible rereplication (N. Wagle and A. Dutta, unpublished data). Therefore, a “pure” decrease in the geminin-Cdt1 ratio might not have the same double-strand breaks and p53 activation. It is also likely that geminin depletion and Cdt1 excess might activate different checkpoints. Consistent with this, addition of caffeine to cells overexpressing Cdt1 increased rereplication (data not shown) instead of suppressing rereplication, as shown here. Further work is required to clarify these issues.
We did not observe phosphorylation of p53 at either Ser15 or Ser20 after geminin depletion, sites which are shown to be phosphorylated and to contribute to p53 activation after DNA damage (7-9, 32). Phosphorylation of p53 at these sites appears to function in the G1/S checkpoint in response to DNA damage (9), although a role of phosphorylation of p53 at these sites in the G2 checkpoint cannot be excluded. Given the evidence that overreplication in geminin-depleted cells appeared to activate the G2/M checkpoint (Fig. 3 and 4), we are not surprised by the lack of phosphorylation of p53 at these sites.
Several lines of evidence indicate that the G2 arrest response comprises an early activation stage as well as a subsequent maintenance phase, with p53 signaling implicated in the latter (5, 19). After γ irradiation, HCT116 (p53+/+) cells arrested in G2 phase through activation of the Chk1-Cdc25C-Cdc2 pathways, while HCT116 (p53−/−) cells progressed into mitosis and exhibited a G1 DNA content (5), indicating that p53 is required for sustaining G2 arrest after DNA damage. However, our data showed that geminin-depleted cells remained arrested in G2 phase in the absence of p53 (Fig. 3), suggesting that a distinct p53-independent pathway exists in geminin-depleted cells to sustain the G2 arrest. It is not clear how the G2/M checkpoint was sustained in geminin-depleted cells.
In geminin-depleted cells, the end-stage giant nuclei showed a suppression of DNA synthesis. Moreover, both Chk1 and Chk2 were activated in geminin-depleted cells, the G2/M checkpoint was activated, and the cells were prevented from entering mitosis. How is further DNA synthesis suppressed in geminin-depleted cells? Chk1 is required for inhibiting DNA replication at late origins in response to stalled DNA replication forks in human cells (13). Figure 1E shows that lamin B2, a known early replication origin in human cells, has more DNA content than the telomere or centromere in geminin-depleted cells, suggesting that a similar suppression of late origins might be true in cells with depleted geminin. It is likely that active Chk1 suppresses rereplication at early origins by the same mechanisms used to suppress firing of late origins, resulting in inhibition of total DNA synthesis in the giant nuclei. The very fact that there is significant rereplication before this shutdown occurs suggests that the signal for the checkpoint accumulates as rereplication proceeds and has to exceed a certain threshold before further DNA synthesis is inhibited. Our best guess for the signal is an accumulation of stalled replication forks, which are expected to increase with the progressive increase of onion skin structures at the rereplicating loci.
In geminin-depleted cells, both Chk1 and Chk2 are activated. However, cosilencing of Chk2 produced less suppression of overreplication than cosilencing of Chk1 in geminin-depleted cells (Fig. 4), suggesting that Chk1 plays a major role in G2/M checkpoint activation. Our results are consistent with previous observation in Drosophila and Xenopus cells (21, 24). In Drosophila cells, Chk2 knockout does not show a significant effect on geminin deficiency-induced overreplication (24). Activation of Chk2 is often observed in cells with DNA double-stand breaks and activation of Chk1 in cells with single-stranded DNA (26). A minor role of Chk2 in geminin-depleted cells may suggest that there are no double-strand breaks in geminin-depleted cells. This is supported by comet assay results, in which a comet tail representing broken DNA was not observed in geminin-depleted cells (data not shown). It is interesting that similar checkpoint activation was observed in hydroxyurea-treated cells, in which both Chk1 and Chk2 are activated but only Chk1 is necessary for this intra-S-phase checkpoint activation (13). Our data suggest that overreplication without an excess of Cdc6 may activate the same checkpoint pathway as incomplete replication.
In D. melanogaster, extensive cell death is not observed in Chk1 knockout cells depleted of geminin. Addition of caffeine to geminin-depleted human cells, however, led to apoptosis (Fig. 4D), suggesting that human cells with overreplicated DNA underwent aberrant mitosis which caused cell death. Additionally, silencing geminin caused apoptotic cell death in Drosophila cells but not in human cells (Fig. 4D) (24). These differences may indicate that human cells have a more vigilant checkpoint pathway that responds to overreplication by a G2/M arrest. It is only when this checkpoint-induced arrest is bypassed that the cells commit suicide. Thus, there are at least two protective responses in human cells that limit the deleterious consequences of overreplication following geminin depletion.
Acknowledgments
We thank members of the Dutta laboratory for technical support and helpful discussions. We thank T. M. Friedrich for providing us with UCN-01.
This work was supported by grant CA60499 from NIH to A.D.
REFERENCES
- 1.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard, E. J., R. J. Muschel, V. J. Bakanauskas, and W. G. McKenna. 1996. Reducing the radiation-induced G2 delay causes HeLa cells to undergo apoptosis instead of mitotic death. Int. J. Radiat. Biol. 69:575-584. [DOI] [PubMed] [Google Scholar]
- 4.Biamonti, G., M. Giacca, G. Perini, G. Contreas, L. Zentilin, F. Weighardt, M. Guerra, G. Della Valle, S. Saccone, and S. Riva. 1992. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol. Cell. Biol. 12:3499-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 6.Burma, S., B. P. Chen, M. Murphy, A. Kurimasa, and D. J. Chen. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462-42467. [DOI] [PubMed] [Google Scholar]
- 7.Canman, C. E., and D. S. Lim. 1998. The role of ATM in DNA damage responses and cancer. Oncogene 17:3301-3308. [DOI] [PubMed] [Google Scholar]
- 8.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 9.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 14:278-288. [PMC free article] [PubMed] [Google Scholar]
- 10.Diffley, J. F. 2001. DNA replication: building the perfect switch. Curr. Biol. 11:R367-R370. [DOI] [PubMed] [Google Scholar]
- 11.Dimitrova, D. S., T. A. Prokhorova, J. J. Blow, I. T. Todorov, and D. M. Gilbert. 2002. Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell Sci. 115:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duriez, P. J., and G. M. Shah. 1997. Cleavage of poly(ADP-ribose) polymerase: a sensitive parameter to study cell death. Biochem. Cell Biol. 75:337-349. [PubMed] [Google Scholar]
- 13.Feijoo, C., C. Hall-Jackson, R. Wu, D. Jenkins, J. Leitch, D. M. Gilbert, and C. Smythe. 2001. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J. Cell Biol. 154:913-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves, P. R., C. M. Lovly, G. L. Uy, and H. Piwnica-Worms. 2001. Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 protein binding. Oncogene 20:1839-1851. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, J., H. Tauchi, S. Sakamoto, A. Nakamura, K. Morishima, S. Matsuura, T. Kobayashi, K. Tamai, K. Tanimoto, and K. Komatsu. 2002. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 12:1846-1851. [DOI] [PubMed] [Google Scholar]
- 17.Kroll, K. L., A. N. Salic, L. M. Evans, and M. W. Kirschner. 1998. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development 125:3247-3258. [DOI] [PubMed] [Google Scholar]
- 18.Lei, M., and B. K. Tye. 2001. Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci. 114:1447-1454. [DOI] [PubMed] [Google Scholar]
- 19.Levedakou, E. N., W. K. Kaufmann, D. A. Alcorta, D. A. Galloway, and R. S. Paules. 1995. p21CIP1 is not required for the early G2 checkpoint response to ionizing radiation. Cancer Res. 55:2500-2502. [PubMed] [Google Scholar]
- 20.Maiorano, D., J. Moreau, and M. Mechali. 2000. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 404:622-625. [DOI] [PubMed] [Google Scholar]
- 21.McGarry, T. J. 2002. Geminin deficiency causes a Chk1-dependent G2 arrest in Xenopus. Mol. Biol. Cell 13:3662-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGarry, T. J., and M. W. Kirschner. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93:1043-1053. [DOI] [PubMed] [Google Scholar]
- 23.McKenna, W. G., E. J. Bernhard, D. A. Markiewicz, M. S. Rudoltz, A. Maity, and R. J. Muschel. 1996. Regulation of radiation-induced apoptosis in oncogene-transfected fibroblasts: influence of H-ras on the G2 delay. Oncogene 12:237-245. [PubMed] [Google Scholar]
- 24.Mihaylov, I. S., T. Kondo, L. Jones, S. Ryzhikov, J. Tanaka, J. Zheng, L. A. Higa, N. Minamino, L. Cooley, and H. Zhang. 2002. Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol. Cell. Biol. 22:1868-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishitani, H., Z. Lygerou, T. Nishimoto, and P. Nurse. 2000. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404:625-628. [DOI] [PubMed] [Google Scholar]
- 26.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 27.Peng, C. Y., P. R. Graves, R. S. Thoma, Z. Wu, A. S. Shaw, and H. Piwnica-Worms. 1997. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277:1501-1505. [DOI] [PubMed] [Google Scholar]
- 28.Pines, J., and T. Hunter. 1994. The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J. 13:3772-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez, Y., C. Wong, R. S. Thoma, R. Richman, Z. Wu, H. Piwnica-Worms, and S. J. Elledge. 1997. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277:1497-1501. [DOI] [PubMed] [Google Scholar]
- 31.Schlegel, R., and A. B. Pardee. 1986. Caffeine-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science 232:1264-1266. [DOI] [PubMed] [Google Scholar]
- 32.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289-300. (Erratum, Genes Dev. 15::750.) [PMC free article] [PubMed]
- 33.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 34.Tada, S., A. Li, D. Maiorano, M. Mechali, and J. J. Blow. 2001. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tauchi, H., S. Matsuura, J. Kobayashi, S. Sakamoto, and K. Komatsu. 2002. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene 21:8967-8980. [DOI] [PubMed] [Google Scholar]
- 36.Tewari, M., L. T. Quan, K. O'Rourke, S. Desnoyers, Z. Zeng, D. R. Beidler, G. G. Poirier, G. S. Salvesen, and V. M. Dixit. 1995. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell 81:801-809. [DOI] [PubMed] [Google Scholar]
- 37.Vaziri, C., S. Saxena, Y. Jeon, C. Lee, K. Murata, Y. Machida, N. Wagle, D. S. Hwang, and A. Dutta. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11:997-1008. (Erratum, Mol. Cell 11:1415.) [DOI] [PubMed]
- 38.Wang, Q., S. Fan, A. Eastman, P. J. Worland, E. A. Sausville, and P. M. O'Connor. 1996. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J. Natl. Cancer Inst. 88:956-965. [DOI] [PubMed] [Google Scholar]
- 39.Ward, S. M. 2001. A recessive allele inhibiting saponin synthesis in two lines of Bolivian quinoa (Chenopodium quinoa Willd.). J. Hered. 92:83-86. [DOI] [PubMed] [Google Scholar]
- 40.Wohlschlegel, J. A., B. T. Dwyer, S. K. Dhar, C. Cvetic, J. C. Walter, and A. Dutta. 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290:2309-2312. [DOI] [PubMed] [Google Scholar]
- 41.Xu, B., S. Kim, and M. B. Kastan. 2001. Involvement of Brca1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol. Cell Biol. 21:3445-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, B., S. T. Kim, D. S. Lim, and M. B. Kastan. 2002. Two molecularly distinct G2/M checkpoints are induced by ionizing irradiation. Mol. Cell. Biol. 22:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]