Abstract
High affinity antisense oligonucleotides (ASOs) containing bicylic modifications (BNA) such as locked nucleic acid (LNA) designed to induce target RNA cleavage have been shown to have enhanced potency along with a higher propensity to cause hepatotoxicity. In order to understand the mechanism of this hepatotoxicity, transcriptional profiles were collected from the livers of mice treated with a panel of highly efficacious hepatotoxic or non-hepatotoxic LNA ASOs. We observed highly selective transcript knockdown in mice treated with non-hepatotoxic LNA ASOs, while the levels of many unintended transcripts were reduced in mice treated with hepatotoxic LNA ASOs. This transcriptional signature was concurrent with on-target RNA reduction and preceded transaminitis. Remarkably, the mRNA transcripts commonly reduced by toxic LNA ASOs were generally not strongly associated with any particular biological process, cellular component or functional group. However, they tended to have much longer pre-mRNA transcripts. We also demonstrate that the off-target RNA knockdown and hepatotoxicity is attenuated by RNase H1 knockdown, and that this effect can be generalized to high affinity modifications beyond LNA. This suggests that for a certain set of ASOs containing high affinity modifications such as LNA, hepatotoxicity can occur as a result of unintended off-target RNase H1 dependent RNA degradation.
INTRODUCTION
Antisense oligonucleotides (ASO) are short synthetic oligonucleotides designed to hybridize with a target RNA and modulate gene expression in a sequence-dependent manner (1,2). There are multiple mechanisms by which an ASO can modulate gene expression including modifying RNA intermediary metabolism, inhibition of translation, disruption of RNA structures, blocking RNA–protein interaction and promotion of RNA degradation. Two common mechanisms for promoting RNA degradation include RNase H and siRNAs. RNase H is an endogenous nuclease that cleaves the RNA strand of a DNA–RNA heteroduplex. ASOs that support the RNase H mechanism of action should contain at least 5 consecutive DNA oligonucleotides, with 7–10 being optimal (3). To take advantage of high affinity and stable nucleic acid analogs, a gap design is often used in which the DNA oligonucleotides are flanked by various sugar modified nucleotides (4,5). These are often referred to as ‘gapmer’ oligonucleotides.
Gapmer ASOs with 2′-O-methoxyethyl (MOE) residues possess excellent pharmacokinetic properties (6–8) and exhibit robust pharmacological activity in several animal models of human disease when administered systemically with repeated weekly subcutaneous (SC) injection (9,10). At the higher doses examined in rodents, minimal to moderate increases in serum transaminases (ALT, AST) are observed and have been attributed to mild proinflammatory effects occasionally observed in subchronic or chronic rodent studies (11). These observations have generally not translated to primate studies or human clinical trials, and pharmacologic activity as well as tolerability of several MOE ASOs have been demonstrated in the clinic (12–14).
More recently, bicyclic nucleic acid (BNA) modifications such as locked nucleic acid (LNA) or constrained ethyl (cEt) (15–17) have been used in place of MOE in gapmer ASOs. These modifications offer greater binding affinity to RNA, translating to increased potency and the ability to use shorter sequences (12–16 nucleotides). Both of these properties have the potential to improve the overall therapeutic index of antisense drug by realizing a 5–10-fold greater potency compared to MOE containing ASOs and decreased class effects due to the shorter length of the ASO (17,18).
These cEt and LNA ASOs have also started to enter early clinical development (19–21). While addition of cEt or LNA modifications to gapmer ASOs can markedly improve ASO potency compared to related MOE gapmer ASOs (18), several authors have suggested that the gain in potency provided by the addition of high affinity modifications such as LNA might occur, in some cases, at the expense of tolerability as evidenced by reports of severe hepatotoxicity caused by some LNA gapmer ASOs in mice (18,22–27). Although the incidence of hepatotoxicity in mice for LNA ASOs is higher than other chemistries, it is possible to identify LNA ASOs that are very effective at reducing expression of targeted transcripts without producing hepatotoxicity (16,28–30). However, some evidence of severe dose limiting hepatic and nephro-toxicity have also been reported in human clinical trials of some LNA gapmer ASOs (19,20,31).
While hepatotoxic ASOs are easily detected in rodent studies by monitoring serum transaminases levels and, therefore, would be unlikely to enter clinical development, a clear understanding of the mechanism leading to these effects is warranted to improve the development of antisense oligonucleotide therapeutics that are safe and well tolerated in patients. Swayze et al. (18) have previously demonstrated that LNA ASO mediated hepatotoxicity can arise independently of the modulation of the target mRNA. Additionally, transcription profiling studies comparing multiple hepatotoxic LNA ASOs have shown little overlap between transcriptional or biological pathway changes preceding the onset of toxicity suggesting the lack of a common pathological mechanism underlying LNA ASO toxicity (22). Yet, liver toxicity is strongly associated with the sequence content of LNA gapmer ASOs and several groups have recently reported that hepatotoxicity of many LNA ASOs could be predicted with a fairly high level of accuracy solely on the basis of sequence composition using machine learning approaches (23,25,26). These disparate observations highlight that the mechanism leading to LNA ASO mediated hepatotoxicity is still not well understood (22,23,25,26).
Since the hepatotoxic properties of LNA ASOs have been the most widely reported to date (22,23,25,26), most of the focus of these manuscripts is aimed at describing the properties of hepatotoxic LNA ASOs with the principal objective to determine whether hepatotoxic LNA ASOs have a shared common mechanism of hepatotoxicity. Additionally, we show evidence that these observations likely generalize to some extent to other high affinity ASO chemistries, such as cEt ASOs, by substituting LNA modifications with cEt modifications in several hepatotoxic ASO sequences. We examined the hepatic transcriptional profiles from mice treated with a panel of either well tolerated or severely hepatotoxic LNA ASOs. Our focus was on describing the transcriptional changes preceding any serum transaminase elevations or evidences of liver injury. In this study, we demonstrated that hepatotoxic BNA ASOs shared a unique property of down-regulating transcripts with very long pre-mRNA sequences (>125 kb). The magnitude of the down-regulation of those very long pre-mRNA transcripts was strongly correlated with the severity of liver as measured by transaminases elevation. Furthermore, we also demonstrated that both the down-regulation of the very long transcripts and the liver injuries was mediated in large part through an RNase H1 dependent mechanism.
MATERIALS AND METHODS
Oligonucleotide design and synthesis
LNA, cEt and MOE phosphoramidites were either purchased from commercial suppliers or manufactured by Isis Pharmaceuticals. Oligonucleotides were prepared similar to that described previously (18) on either an Amersham AKTA 10 or AKTA 100 oligonucleotide synthesizer. All the ASO tested were 16 bases long. The first three bases and last three bases of chimeric ASO have LNA, cEt or 2′ MOE modification and the ASO also have a phosphorothioate backbone. The sequences are detailed in Table 1. This chimeric design has been shown to provide both increased nuclease resistance and mRNA affinity, while maintaining the robust RNase H terminating mechanism utilized by these types of ASO (32). Among the ASOs selected for this report, there was little evidence of sequence bias between well tolerated and hepatotoxic LNA ASOs at any site for any nucleotide in the gap-aligned sequences (Supplementary Figure S1).
Table 1. Gapmer ASO sequences and their on-target mRNA transcripts.
| Isis No | Alternative ID | Sequence | Modification | Gene | Genbank number | On-target Species | First Toxic Dose (mg/kg) >1000 IU/ml ALT (96 h) |
|---|---|---|---|---|---|---|---|
| 569713 | ASO ctrl | 5′-GACGCGCCTGAAGGTT-3′ | LNA | N/A, control | N/A | Mouse | >300 |
| 571035 | FVII-1 | 5′-CAGATATAGGACTGGA-3′ | LNA | F7 | NM_010172.3 | Human | >300 |
| 571033 | FXI-3 | 5′-ATCCAGAGATGCCTCC-3′ | LNA | F11 | NM_028066.1 | Mouse | >300 |
| 569714 | FXI-4 | 5′-GGCCACCACGCTGTCA-3′ | LNA | F11 | NM_028066.1 | Mouse | >300 |
| 571034 | FXI-5 | 5′-TGCCACCGTAGACACG-3′ | LNA | F11 | NM_028066.1 | Mouse | >300 |
| 569715 | SOD1–1 | 5′-GGACACATTGGCCACA-3′ | LNA | Sod1 | NM_011434.1 | Mouse | >300 |
| 569716 | FVII-2 | 5′-CCCTGGTGTACACCCC-3′ | LNA | F7 | NM_010172.3 | Mouse | 300 |
| 569717 | PTEN | 5′-ATCATGGCTGCAGCTT-3′ | LNA | Pten | NM_008960.2 | Mouse | 33 |
| 569718 | FVII-3 | 5′-TGGTCCCTGCAGTACT-3′ | LNA | F7 | NM_010172.3 | Mouse | 100 |
| 569719 | FXI-1 | 5′-GTCTGTGCATCTCTCC-3′ | LNA | F11 | NM_028066.1 | Mouse | 11 |
| 443919 | FXI-1-M | 5′-GTCTGTGCATCTCTCC-3′ | 2’ MOE | F11 | NM_028066.1 | Mouse | 300 |
| 464917 | FXI-1-C | 5′-GTCTGTGCATCTCTCC-3′ | cEt | F11 | NM_028066.1 | Mouse | 11 |
| 569720 | FXI-2 | 5′-GTCAGTATCCCAGTGT-3′ | LNA | F11 | NM_028066.1 | Mouse | 100 |
| 444011 | FXI-2-M | 5′-GTCAGTATCCCAGTGT-3′ | 2’ MOE | F11 | NM_028066.1 | Mouse | 300 |
| 465178 | FXI-2-C | 5′-GTCAGTATCCCAGTGT-3′ | cEt | F11 | NM_028066.1 | Mouse | 100 |
| 569721 | SOD1–2 | 5′-TGAGGTCCTGCACTGG-3′ | LNA | Sod1 | NM_011434.1 | Mouse | 33 |
| 529933 | SOD1–2-M | 5′-TGAGGTCCTGCACTGG-3′ | 2’ MOE | Sod1 | NM_011434.1 | Mouse | 300 |
| 508031 | SOD1–2-C | 5′-TGAGGTCCTGCACTGG-3′ | cEt | Sod1 | NM_011434.1 | Mouse | 33 |
| 554219 | BIRC5 | 5′- CTCAATCCATGGCAGC-3′ | LNA | Birc5 | NM_001168.2 | Human | 300 |
The 16-mer antisense oligonucleotides were synthesized with the indicated modifications (underlined sequences). The center (gap) of the antisense oligonucleotides (not underlined) consist of deoxynucleotide bases and phosphorothioate backbone linkages.
Animal treatment
Animal experiments were approved by the Animal Welfare Committee and conducted according the guidelines of the American Association for the Accreditation of Laboratory Animal Care. Male Balb/c, mice were obtained from Charles River Laboratories. All animals were housed in temperature-controlled conditions under a light/dark photocycle with food and water supplied ad libitum. Animal weights were monitored prior to dosing throughout the live phase of the study. Compounds were dissolved in phosphate buffered saline (PBS), filter sterilized and administered by s.c. injection in a volume corresponding to 10 μl/g animal weight. Immediately prior to sacrifice, mice were anesthetized with isoflurane and terminal bleed was performed by cardiac puncture. Plasma was isolated from whole blood and analyzed for clinical chemistries. Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were determined using a Beckman Coulter AU480 bioanalyzer (Brea, CA, USA). Liver samples were collected for RNA and protein for analysis.
Anti-RNase H1 ASO pretreatment
Male Balb/c mice were treated with 50 mg/kg of antisense 5–10–5 MOE Gapmer ASO against RNase H1 (ISIS 385005) or non-coding control 5–10–5 MOE Gapmer ASO (ISIS 104838; Table 2). The MOE ASOs were administered s.c. on day 0, 2, 4, 7, 9, 11, 13, 15 and 17. Seventy two hours after the last dose of MOE ASO, a single dose of LNA ASOs (ISIS 569717, ISIS 569719 and ISIS 569721) was administered s.c. at 25 and 75 mg/kg.
Table 2. MOE Gapmer ASO sequences and their on-target mRNA transcripts.
| Isis No | Sequence | mRNA target name | Target species | Gene ID |
|---|---|---|---|---|
| 104838 | 5′-GCTGATTAGAGAGAGGTCCC-3′ | TNF-α | Human | 7124 |
| 284754 | 5′-GTCCTTTATTCACCCTAACT-3′ | Ptprk | Mouse | 19272 |
| 292684 | 5′-CCCTCTGTTATTATTCTCAA-3′ | Ppp3Ca | Mouse | 19055 |
| 385005 | 5′-AGGCAAAGAAATCTATTAGT-3′ | RNaseh1 | Mouse | 19819 |
| 481067 | 5′-TCTTAAAACCAACATGCCAA-3′ | Fto | Mouse | 26383 |
| 518094 | 5′-TTCCGGCGCAGAGATGTGCA-3′ | Rptor | Mouse | 74370 |
| 566054 | 5′-CAAGCTTTTAATTCCCTTGT-3′ | Iqgap2 | Mouse | 544963 |
The 20-mer antisense oligonucleotides were synthesized with the indicated MOE modifications (underlined sequences). The center (gap) of the antisense oligonucleotides (not underlined) consist of deoxynucleotide bases and phosphorothioate backbone linkages.
ASO mixture Study
Male Balb/c mice were treated with a mixture containing of up to 5 different 5–10–5 MOE Gapmer ASO against Rptor, Fto, Ppp3ca, Ptprk and Iqgap2, or control ASO. A control MOE ASO, ISIS 1048438 was used to keep a consistent total ASO dose at 250 mg/kg across all treatment groups. The MOE ASOs were administered at 50 mg/kg s.c. on day 0, 2, 4, 7, 9, 11 and 13 (Table 2).
RNA extraction and qRT–PCR analysis
RNA was extracted from the liver samples using RNeasy columns (Qiagen, Valencia, Ca) according to manufacturer's protocol. RNA samples were analyzed by fluorescence-based quantitative real-time polymerase chain reaction (qRT–PCR) using an Applied Biosystems 7700 sequence detector. Target RNA levels were normalized to the total RNA concentration determined using ribogreen. Primers and probes for analysis of the expression of different genes were designed using Primer Express Software (Applied Biosciences, Carlsbad, CA, USA) (Supplementary Table S1). For the analysis, 100 ng of total RNA was used.
RNA preparation and microarray experiments
The Mouse WG-6 v2 Expression BeadChip (Illumina, San Diego, CA, USA) was processed in accordance with the manufacturer's instructions. Two hundred nanograms of total RNA was used for cRNA in vitro transcription and labeling with the TotalPrep™ RNA Labeling Kit using Biotinylated-UTP (Ambion, Austin, TX, USA). Hybridization is carried out in accordance with the Illumina Hybridization System Manual. The raw and background subtracted matrices extracted from Illumina Beadstudio are subject to a modified quantile normalization procedure where in the quantile distribution is estimated from the project samples, and negative values in the quantile distribution are scaled to fall within the range of 1 and 2. The resulting quantile distributions are then log transformed and then used to normalize each individual array following the typical quantile normalization procedure.
Microarray analysis
Background subtracted and quantile normalized microarray intensity data (see above) were imported into Genespring GX 12.6 (Agilent, Redwood City, CA, USA) software for analysis. The gene expression level measured for each gene on the array was compared with the corresponding gene's median expression level in the untreated control samples. Differentially expressed genes were identified as those genes with a statistically significant 2-fold or higher change in expression based on a corrected FDR ≤ 0.01.
Arrays data from the liver of mice treated with several ASO for 24 h (n = 4) were compared with vehicle treated samples at the same time point. The Log2 transformed and normalized ratio data were subjected to pair-wise comparison at the various time points assayed by Student t-test (P ≤0.01 and multiple testing correction Benjamini–Hochberg false discovery rate (FDR ≤ 0.01 (33)), transcripts were further filtered by selecting transcripts modulated by a given ASO at 2-fold in relation to vehicle treated sample.
Identification of putative perfect and mismatched binding sites
Using the transcript sequences defined by the refseq gene annotations associated with MGSv37 (mm9) build of the mouse genome we interrogated the number and quality of putative binding sites for each oligo tested across all genes for which we had gene expression measurements. Since ASOs can act on both pre-mRNA and mature mRNA molecules the longest annotated full length pre-mRNA sequences was searched. A simple searching algorithm was implemented that tallied the number of perfect Watson–Crick hybridization (e.g. A:T, G:C) pairs for each ASO-length sized site along the transcript. For each site we retained the number hybridization pairs observed (e.g. matches) as well as the length of the longest substring within the site for which every nucleotide formed a perfect Watson–Crick pairing.
Western blot analysis
Liver samples were homogenized in PBS containing Protease Inhibitor Cocktail (Calbiochem, San Diego, CA, USA). Protein samples were separated on a Tris-glycine 4–20% gel (Invitrogen, Carlsbad, CA, USA) and transferred to a PVDF membrane (Invitrogen). Membranes were incubated at room temperature in blocking buffer consisting of 5% non-fat dry milk in TBST for 1 h, then incubated with rabbit primary antibodies (1:1000 and 1:2500, respectively) against Raptor (Bethyl Laboratories, Montgomery, TX) or Ppp3ca (Lifespan Biosciences, Seattle, WA, USA). After thorough washing with TBST, horseradish peroxidase-conjugated donkey anti-rabbit secondary antibodies (1:10 000 dilution in TBST; Jackson ImmunoResearch, West Grove, PA, SUA) were applied, and the blots were developed by using ECL-plus reagent (GE Healthcare Lifesciences, Piscataway, NJ, USA).
Statistical analysis
The data of in vivo studies are expressed as the mean ± standard deviation of the mean (SDM). Comparisons of in vivo data between two groups were analyzed with an unpaired Student's t-test. Multiple comparisons were evaluated with one-way analysis of variance (ANOVA), followed by a post hoc Dunnett's test with JMP version 12 (SAS Institute, Cary, NC). A P ≤ 0.05 was considered significant.
RESULTS
BNA ASOs can induce acute hepatotoxicity after a single dose administration
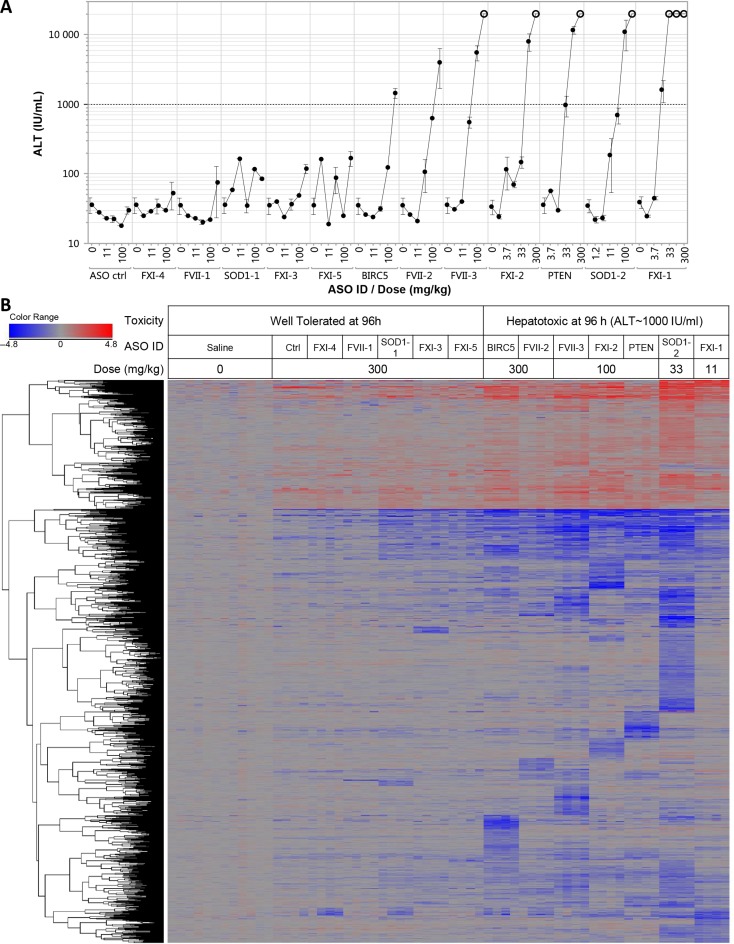
We characterized a panel of 3–10–3 gapmer LNA ASOs with little similarity in sequence composition targeting a range of mRNA transcripts for their ability to induce hepatotoxicity following single administration in Balb/c mice (Tables 1 and 3, Supplementary Figure S1). Serum transaminase levels (ALT and AST) were measured daily up to 96 h after subcutaneous administration. Single administration of increasing doses of ASOs resulted in 7 out of 13 LNA ASOs producing severe serum transaminases increase (ALT > 1000 IU/ml) or death in some animals at doses ranging from 11 mg/kg to 300 mg/kg as early as 48 h post treatment (Figure 1A, only 96 h data are shown). The remaining 6 LNA ASOs were well tolerated at doses up to 300 mg/kg, as all the treated animals showed ALT or AST levels within the normal range compared to vehicle treated animal throughout the observation period (96 h). In contrast to well tolerated LNA ASOs, hepatotoxic LNA ASOs consistently displayed extensive hepatocellular necrosis consistent with an acute transaminase release, while well-tolerated LNA ASOs did not show any evidence of hepatic injury by hematoxylin and eosin stain (Supplementary Figure S2). We substituted the LNA modifications in three of the most hepatotoxic ASOs described herein with either cEt or 2′ MOE modifications seeking to assess whether the presence of cEt or 2′ MOE in place of LNA modification would affect the severity of the increase in ALT (Supplementary Figure S3A). Substituting LNA with 2′ MOE resulted in a complete suppression of ALT increase within the dose range and the time frame assayed. However, the reduced hepatotoxicity also paralleled a reduction in on-target potency (Supplementary Figure S3A). In contrast, substituting LNA with cEt had essentially no impact on potency for either on or off-target transcripts, and only modestly reduced the magnitude of the ALT increase (Supplementary Figure S3A–B).
Table 3. Mean ALT levels measured daily up to 96 h. Number of mRNA transcripts modulated by each LNA ASOs as a function of dose 24 h after administration.
| ALT (IU/ml) | Number of transcripts modulated at 24 h | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ASO ID | Alternative ID | Dose (mg/kg) | Toxicity (96 h) | 24 h | 48 h | 72 h | 96 h | Down (−2x, P-value ≤ 0.01) | Up (2x, P-value ≤ 0.01) |
| 569713 | ASO ctrl | 33 | Safe | 49 | 21 | 20 | 23 | 0 | 1 |
| 300 | Safe | 32 | 25 | 25 | 27 | 19 | 14 | ||
| 569714 | FXI-4 | 33 | Safe | 23 | 18 | 25 | 32 | 0 | 1 |
| 300 | Safe | 41 | 26 | 32 | 30 | 27 | 8 | ||
| 571035 | FVII-1 | 33 | Safe | 24 | 22 | 24 | 19 | 1 | 0 |
| 300 | Safe | 76 | 28 | 63 | 93 | 11 | 4 | ||
| 569715 | SOD1—1 | 33 | Safe | 32 | 26 | 30 | 39 | 1 | 0 |
| 300 | Safe | 34 | 20 | 30 | 87 | 121 | 20 | ||
| 571033 | FXI-3 | 33 | Safe | 28 | 20 | 24 | 33 | 1 | 0 |
| 300 | Safe | 32 | 22 | 65 | 121 | 111 | 10 | ||
| 571034 | FXI-5 | 33 | Safe | 34 | 21 | 25 | 56 | 1 | 0 |
| 300 | Safe | 237 | 29 | 86 | 193 | 96 | 10 | ||
| 554219 | BIRC5 | 33 | Safe | 187 | 22 | 37 | 33 | 20 | 9 |
| 300 | Severe | 29 | 69 | 1239 | 1475 | 418 | 114 | ||
| 569716 | FVII-2 | 33 | Safe | 31 | 20 | 28 | 128 | 5 | 1 |
| 300 | Severe | 30 | 39 | 4038 | 4690 | 282 | 43 | ||
| 569718 | FVII-3 | 33 | Moderate | 25 | 22 | 100 | 461 | 140 | 8 |
| 100 | Severe | 29 | 123 | 3880 | 5034 | 568 | 173 | ||
| 569720 | FXI-2 | 33 | Safe | 22 | 23 | 35 | 125 | 87 | 34 |
| 100 | Severe | 26 | 129 | 4960 | 10 179 | 392 | 167 | ||
| 569717 | PTEN | 33 | Severe | 44 | 36 | 187 | 1143 | 86 | 4 |
| 100 | Severe | 27 | 130 | 2067 | 10 262 | 397 | 101 | ||
| 569721 | SOD1–2 | 33 | Severe | 30 | 337 | 878 | 2288 | 867 | 394 |
| 569719 | FXI-1 | 11 | Severe | 50 | 59 | 183 | 1486 | 318 | 159 |
| 33 | Severe | 47 | 3983 | 10 771 | 20 000 | 1301 | 998 | ||
Figure 1.
(A) Plasma ALT levels from of Balb/c mice treated with increasing doses of LNA ASO were measured at 96 h (single subcutaneous injection). Animals found dead prior to 96 h were arbitrarily assigned an ALT value of 20 000 IU/ml if they had shown increase ALT at 48 or 72 h (open circles). (B) Oneway hierarchical clustering of transcripts significantly modulated by LNA ASOs. A single dose for each LNAASO is shown. The highest tested dose (300 mg/kg) for safe ASOs (ALT < 200 IU/mL at 96 h) is represented, whereas the lowest dose producing an increase in ALT level of approximately 1000 IU/ml is represented for toxic ASO (Union of unpaired Student's t-test assuming equal variance, 2-fold change, FDR ≤ 0.001, 1788 Transcripts). Transcription profiles from 12 saline treated livers and 4 treated livers per ASO/dose group are represented. Down-regulated transcripts are represented in shades of blue proportional to the intensity of change, while up-regulated transcripts are represented in shades of red proportional to the intensity of change. Unchanged transcripts are colored in gray.
Characterization of liver transcriptome perturbation by LNA ASOs prior the onset of transaminitis
To investigate the underlying mechanism leading to LNA ASOs mediated acute liver injury, the hepatic transcript levels were assessed in both well tolerated and toxic LNA ASOs using whole transcriptome expression profiling at a time previously shown to result in target reduction, but prior to the manifestation of any phenotypically observable toxicities. Balb/c mice were treated with 13 different LNA ASOs at two different dose levels based on the dose range finding study (Figure 1A and Table 2). All the LNA ASOs were dosed at 33 mg/kg as well as at another variable dose that was determined by relative tolerability in previous experiment (up to 300 mg/kg). The higher dose selected was either 300 mg/kg if transaminases levels remained within the normal range compared to vehicle treated animals in the dose response or the lowest dose producing approximately 1000 IU/ml increase in ALT by 96 h. Animals from the various treatment groups (n = 4) were sacrificed at 24 h following LNA ASO dosing for transcriptome analysis or at 96 h post-dosing for characterization of ALT and liver pathology. The lack of increase in serum transaminases in any of the treatment groups were confirmed 24 h post dosing (Table 1). RNA expression analysis was performed in livers collected 24 h after dosing.
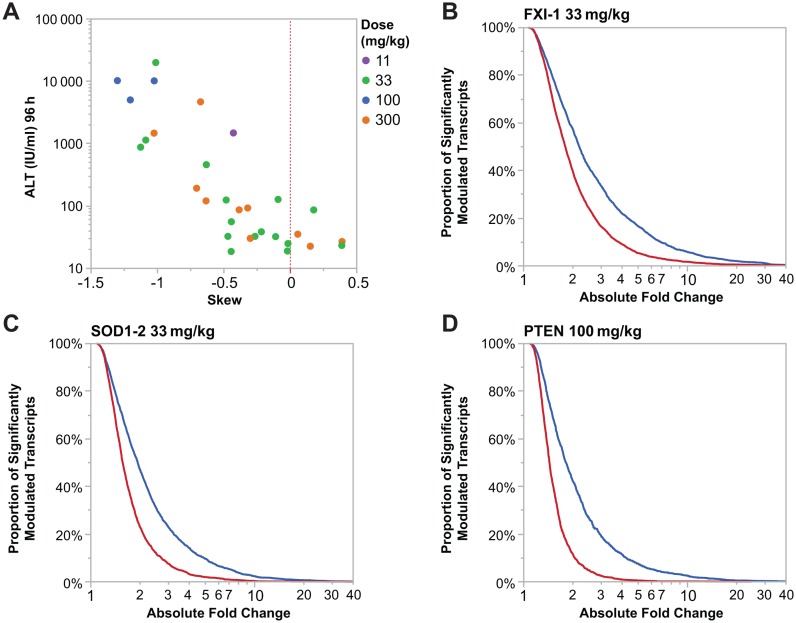
Even in absence of overt toxicity at 24 h, the number of transcripts significantly modulated (P-value ≤ 0.01, ≥2x absolute fold change compared to vehicle treated animals) was strongly correlated with the magnitude of the ALT increase observed at 96 h (r2 = 0.69, Table 3). Furthermore, more transcripts are down-regulated than are up-regulated by hepatotoxic LNA ASOs (Table 3). Although many fewer transcripts were modulated by well tolerated LNA ASOs than by hepatotoxic ASOs, the bias toward down-regulation also held true following treatment with those well tolerated LNA ASOs. Figure 2A quantifies the degree of biased expression change. If an equal number of transcript levels increased and decreased after treatment with LNA ASOs the distribution of log2 expression ratios is expected to have a normal distribution. We can calculate the degree to which the observed distribution is biased to either up-regulated or down-regulated genes by calculating the skew in the distribution. Increasingly negative skew values would suggest an increasing bias toward down-regulation while positive values would suggest a bias toward up-regulation. We observe a clear correlation of increased negative skew values and hepatotoxicity as measured by ALT for all hepatotoxic compounds. Additionally the magnitude of skewness observed is dose dependent. Additionally, we show graphically in Figure 2B–D that for three hepatotoxic compounds there are many more genes down-regulated than up-regulated regardless of the fold-change threshold chosen.
Figure 2.
(A) Increased ALT levels in mice is correlated with an increased number of genes with lowered expression. Skew was calculated using standard methods as implemented in the python scipy package. Each dot represents an LNA ASO at a specific dose represented by the color of the dot. (B–D) Cumulative distribution function plot demonstrating that hepatotoxic LNA ASOs (FXI-1, SOD1–2 and PTEN LNA, respectively) are down-regulating a greater proportion of significantly modulated transcripts (P-value ≤ 0.05) and to a greater intensity (absolute fold change) than significantly up-regulated transcripts (P-value ≤ 0.05).
Unsupervised one-way hierarchical clustering of the union of all transcripts significantly perturbed in liver 24 h after any LNA ASO treatment (1788 transcripts, P-value ≤ 0.01, ≥2x absolute fold change compared to vehicle treated animals) revealed several interesting modulation patterns (Figure 1B). First, the pattern of transcripts down-regulation seems to be mostly sequence specific with little overlap between transcript clusters across the various hepatotoxic LNA ASOs. Only a small subset of transcripts was commonly down-regulated across all toxic LNA ASOs. Second, in contrast to down-regulated transcripts whose modulations were largely sequence specific, most up-regulated transcripts tend to be modulated more consistently across LNA ASOs with little evidence of clusters of transcript discriminating various toxic LNA ASOs (Figure 1B). Up-regulations of those transcripts seems to be more of a reflection of the severity and response to the hepatic injury. Third, the vast majority of clusters or collection of transcripts whose expression were altered appeared to be unique to one or few treatment groups particularly among down-regulated transcripts. Interestingly, transcripts or clusters significantly up-regulated by one ASO were rarely significantly down-regulated by another ASO and vice versa. Finally, even at a high dose of 300 mg/kg, well tolerated LNA ASOs modulated a fairly small number of transcripts. Moreover, a smaller, more therapeutically relevant, dose of 33 mg/kg for well tolerated LNA ASOs did not have a significant impact on transcript modulation in contrast to hepatotoxic LNA ASOs assayed at the same dose (Table 3).
Few biological pathways or functions are perturbed systematically across toxic LNA ASOs
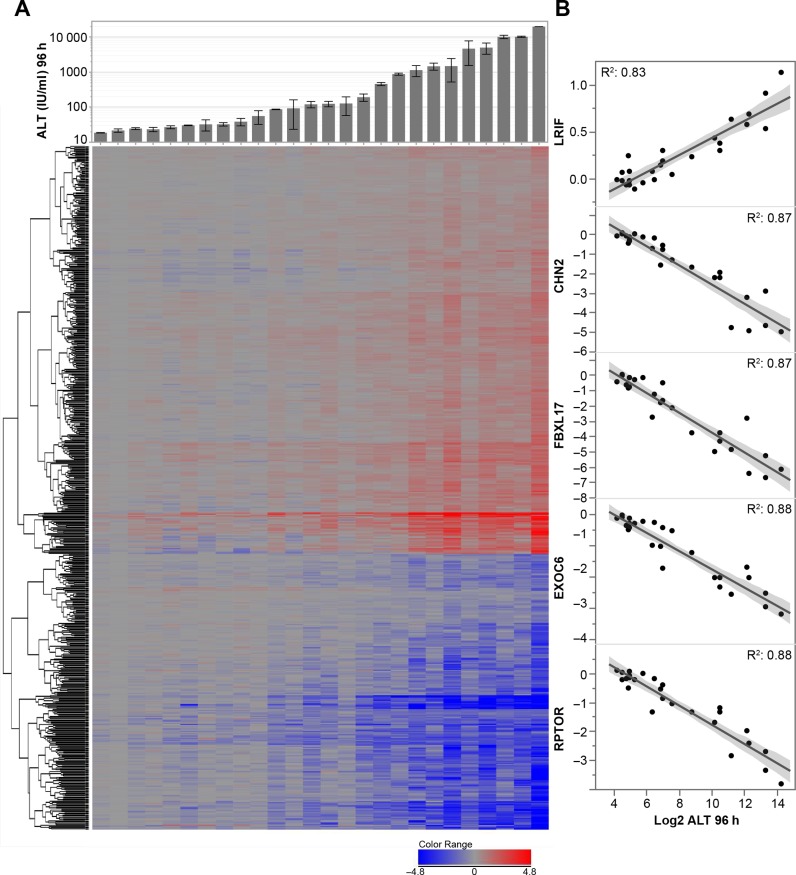
We correlated the average relative changes in gene expression obtained 24 h post treatment with average changes in serum transaminases observed 96 h post treatment across all treatment conditions (Figure 3AB). Transcripts positively (371) or negatively (251) correlated (Prob > 0.000001; r2 > 0.54) to the increase in ALT observed at 96 h were analyzed for their association with known canonical biological pathway (IPA) (Supplementary Figure S4A). Few canonical pathways that were either down-regulated or up-regulated were strongly enriched with transcripts associated with increased ALT and only a small number of transcripts seemed to contribute to most of those pathways. Even if pathway enrichment is assessed independently for each treatment condition, only a very small number of pathways were modulated significantly in most toxic LNA ASOs without being also modulated by well tolerated LNA ASOs (Supplementary Figure S4B). Overall, none of the modulated pathways in common across multiple hepatotoxic ASOs suggest shared mechanism of hepatotoxicity.
Figure 3.
(A) Upper panel: Mean of plasma ALT (IU/ml) measured 96 h post treatment. Bottom panel: Oneway hierarchical clustering of transcripts significantly modulated in the liver at 24 h post treatment by LNA ASOs in correlation with ALT increase at 96 h. (Prob > 0.000001, r2 > 0.54). Thirteen LNA ASOs were tested at 1 or 2 doses (min 11 mg/kg, max 300 mg/kg as detailed in Table 3). Each column represents the average expression for a given ASO/dose. Up-regulated transcripts are represented in shades of red (n = 353) and down-regulated transcripts in shades of blue (n = 240). (B) Top 5 mRNA transcripts whose modulation 24 h post treatment is the most correlated to ALT increase 96 h post treatmentselected from panel A. Normalized liver mRNA transcripts level 24 h post ASO treatment (y-axis) versus Log2 of serum ALT level 96 h post treatment (x-axis) for the R2 is shown.
Large scale down-regulation of very long pre-mRNA strongly predict serum transaminase increase
Since we were unable to identify a common molecular pathway, process or function that could adequately explain the gene expression changes observed, we assessed whether some unifying ‘genomic’ properties of those perturbed transcripts could shed some light on the general mechanism of LNA ASO mediated hepatotoxicity. Jackson et al. (34–38) have previously demonstrated that some siRNA oligonucleotides could exert non-specific miRNA-like effects. The pattern of transcriptional down-regulation caused by toxic LNA ASOs was reminiscent of those caused by siRNAs, suggesting the large-scale pattern of transcript down-regulation might be the result of promiscuous antisense mediated cleavage of off-target transcripts.
We looked for evidence of ASO sequence similarity (similar to miRNA seed matches) among down-regulated transcripts in 3′ untranslated regions of transcripts by conducting a Sylamer analysis (39,40). This approach is often used to assess target engagement of miRNA or off-target engagement by siRNA. We did not observe any evidence of full or partial ASO sequence match enrichment for any of the toxic LNA ASO among down-regulated transcripts in 3′ untranslated regions of transcripts as would be seen with siRNA and miRNA. The absence of enrichment held true even when transcripts region analyzed included entire mRNA or even pre-mRNA suggesting that Ago2 mediated off-target transcript down-regulation following treatment with siRNA is unlikely to play a role in off-target transcript down-regulation caused by toxic LNA ASOs.
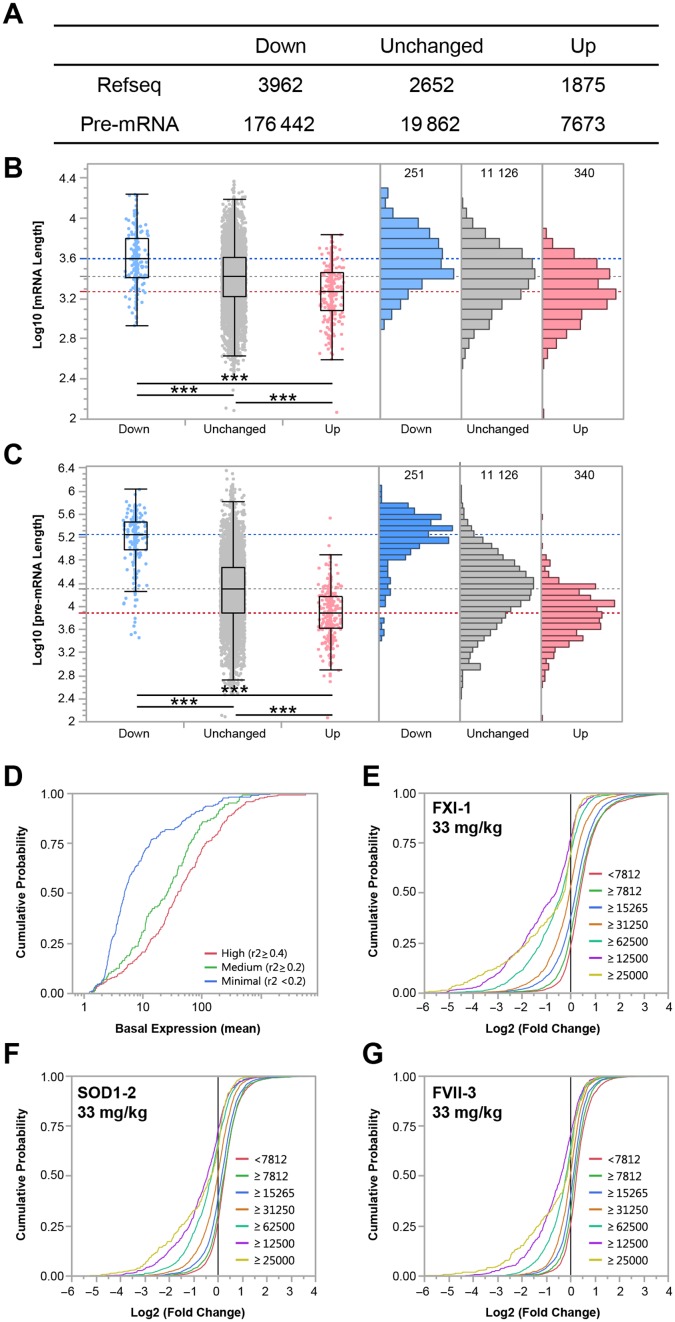
Next, we compared the length of both ‘mature’ mRNA and ‘immature’ pre-mRNA transcripts modulated after LNA ASO treatments in correlation with ALT increase (Figure 4A–C). Unexpectedly, the median length of mRNA transcripts whose down-regulation was strongly correlated the ALT increase (Prob > 0.000001; r2 > 0.54) at 96 h was significantly longer than unaffected transcripts (1.5-fold, P-value < 0.0001, Figure 4B). In contrast, the median length of up-regulated transcripts (Prob > 0.000001; r2 > 0.54) was significantly shorter than unchanged transcripts (P-value < 0.0001, Figure 4AB). This phenomenon was even more pronounced if the length of pre-mRNA transcripts is taken into consideration (Figure 4C). The median length of pre-mRNA transcripts down-regulated in correlation with ALT increase was 8.9 times longer than unchanged pre-mRNA transcripts (P-value < 0.0001). More striking, up-regulated pre-mRNA transcripts were 23 times shorter than transcripts down-regulated in correlation with ALT increase (P-value < 0.0001). In addition, we noted that among the transcripts with very long pre-mRNA sequences (>125 000 b), down-regulated transcripts strongly negatively correlated to ALT increase (r2 > 0.4) were expressed at a basal level in untreated animals higher than lesser correlated transcripts (Figure 4D). Interestingly, if all transcripts down-regulated by a given LNA ASO at a given dose are taken into consideration, regardless whether those transcripts are correlated with an increase in ALT level, then the magnitude of down-regulation of a given transcript tend to be correlated to the length of the pre-mRNA (Figure 4E–G).
Figure 4.
(A) Median length of mRNA and pre-mRNA transcripts expressed on the microarray as a function of the modulation: ‘Down’ represents the number of transcripts down-regulated 24 h post treatment (240) negatively correlated to ALT increase at 96 h, ‘Up’ represents the number of transcripts up-regulated 24 h post treatment (353) positively correlated to ALT increase at 96 h (Prob > 0.000001, r2 > 0.54). Distribution of mRNA (B) and pre-mRNA (C) transcripts expressed on array. The blue horizontal line represents the median length of down-regulated transcripts, in gray unchanged transcripts and in red, up-regulated transcripts. (D) Cumulative probability plot of the basal expression levels of long (pre-mRNA length > 125 000 b) down-regulated transcripts with high (red), medium (green) or minimal correlation to ALT increase, shows transcripts with low level of basal expression tend to exhibit a low level of correlation with ALT increase following treatment with hepatotoxic ASOs. (E–G) Cumulative probability plot of fold change of modulated transcripts binned by pre-mRNA length for three hepatotoxic LNA ASOs (FXI-1, SOD1–2 and FVII-3 LNA at 33 mg/kg, respectively) treatment group are represented. The proportion of transcripts as well as the intensity of transcript down-regulation increases as a function of the length of pre-mRNA transcripts.
Down-regulation of long pre-mRNA Transcripts by hepatotoxic ASOs is similar across comparable high affinity modifications
We selected several long pre-mRNA transcripts whose down-regulation were strongly correlated with ALT increases. We compared the down-regulation profiles of those transcripts by RT-PCR following single dose administration of three different pairs of hepatotoxic ASO sequences containing either LNA or cEt modifications, which have comparable binding affinity enhancement profiles in in vitro studies (17). As with on-target knockdown, long off-target transcripts were similarly down-regulated by a given hepatotoxic sequence in a dose responsive manner regardless of the nature of the high affinity modification (LNA or cEt) present on the ASO (Supplementary Figure S3B).
Hepatotoxicity of LNA ASOs is RNase H1 dependent
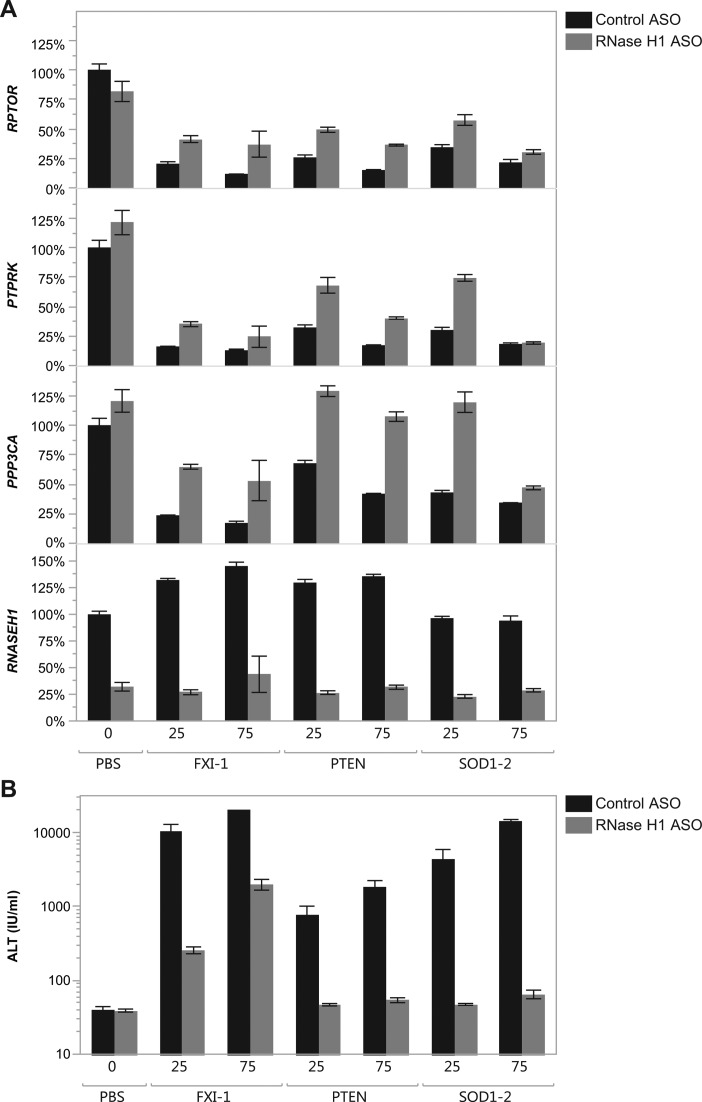
The broad down-regulation of transcripts by toxic LNA ASOs in a sequence-specific manner as well as the preferentially down-regulation of long pre-mRNAs suggested the possibility of a promiscuous antisense mechanism which would require the formation of mismatched LNA ASO: RNA duplex followed by RNase H1 cleavage. To directly test if the non-specific knockdown observed is RNase H1 dependent, we pretreated Balb/c mice with 3 doses per week for 3 weeks with a MOE gapmer ASO targeting RNase H1 (ISIS 385005) or an ASO control (ISIS 104838) followed 72 h after the last dose by treatment with hepatotoxic LNA ASOs (41). Anti-RNase H1 MOE ASO pretreatment resulted in about 75% reduction in RNase H1 mRNA level compared to control treated MOE ASO (Figure 5A). Pretreatment with either RNase H1 MOE ASO or a control oligonucleotide alone did not produce any evidence of liver injury (Figure 5B). While pretreatment with the control oligonucleotide did not affect the severity of the toxicity caused by any of the LNA ASOs tested, pretreatment with the RNase H1 MOE ASO lead to a strong reduction in transaminase increase of toxic LNA ASOs (25 and 75 mg/kg) (Figure 5B). The levels of three transcripts with long pre-mRNAs (Rptor, Ptprk and Ppp3ca) previously strongly correlated to ASO mediated ALT increase were measured in the liver by RT-PCR 24 h after dosing with the LNA ASOs (Figure 5A). In the case of all three hepatotoxic LNA ASOs, the down-regulation of the three off-target transcripts (Rptor, Ptprk and Ppp3ca) was mitigated by pretreatment with an anti-RNase H1 ASO (Figure 5A).
Figure 5.
Effect of anti-RNase H1 MOE ASO pretreatment on LNA ASO mediated hepatotoxicity as measure by plasma ALT increase. Mice were pretreated with either ISIS 104838 (black, control MOE ASO) or ISIS 385005 (gray, anti-RNase H1 MOE ASO) prior to treatment with hepatotoxic LNA ASOs at 25 and 75 mg/kg. (A) mRNA transcript levels were measured by RT–PCR in the liver of mice treated for 24 h with the hepatotoxic LNA ASOs at 25 or 75 mg/kg. (B) Plasma ALT levels are measured 72 h after LNA ASO administration.
RNase H1 can exert its activity by cleaving the ASO:RNA duplex within the unmodified region of the ASO (the ‘gap’). We hypothesized that modifications to the gap region of the ASO by the addition of BNA (cEt) modification already present in the wing region of the ASO would adversely affect RNase H1 ability to cleave the duplex within the gap. To test this hypothesis, we inserted in the gap of 2 hepatotoxic cEt gapmer ASOs 3 cEt modification in position 3, 5 and 8 (relative to the 3′ wing gap junction, Supplementary Figure S5A). The resulting gap impaired ASOs (483526 and 483520) showed reduction on-target mRNA cleavage activity as well as abrogated hepatotoxicity compared to parent ASOs (457852 and 482051, respectively, Supplementary Figure S5BC).
Off-target knock-down is not correspondent with quality or prevalence of recognizable perfect or mismatched ASO binding sites
Given that on- and off-target transcript knockdown was observed to be RNase H1 dependent, we next asked if observed knockdown can be ascribed to ASO site match statistics. Given that thermodynamic parameters for estimating RNA:ASO duplex stability are currently unknown and the rules governing ASO transcript scanning and hybridization are also poorly defined we exhaustively searched every possible ASO-sized site across all genes and simply counted the number of expected perfect matched Watson–Crick base pairs (see Methods and Materials). This simple heuristic, although may underestimate the number of true hybridization events as structural bulges in either the ASO or transcript are not identified, it does provide a lower-bound on the number of putative ASO binding sites. For the toxic ASOs where we see pervasive off-target activity, we observe a striking correlation between target knockdown and either the presence of a single higher-quality putative site (Supplementary Figure S5A) or the number of putative sites (Supplementary Figure S5C). However, since long transcripts inherently will have both more putative sites and higher-quality ‘best sites’ (Supplementary Figure S6D–G) these observations are not independent from our initial observation that long genes appear to be more dramatically effected by toxic ASOs than shorter genes. Interestingly for safe ASOs, although there appears to be relatively similar numbers of putative off-target sites, genes harboring these sites are not affected (Supplementary Figure S6H–T).
Targeted down-regulation of several long pre-mRNA transcripts by mixtures of MOE ASOs mimics LNA ASOs hepatotoxicity
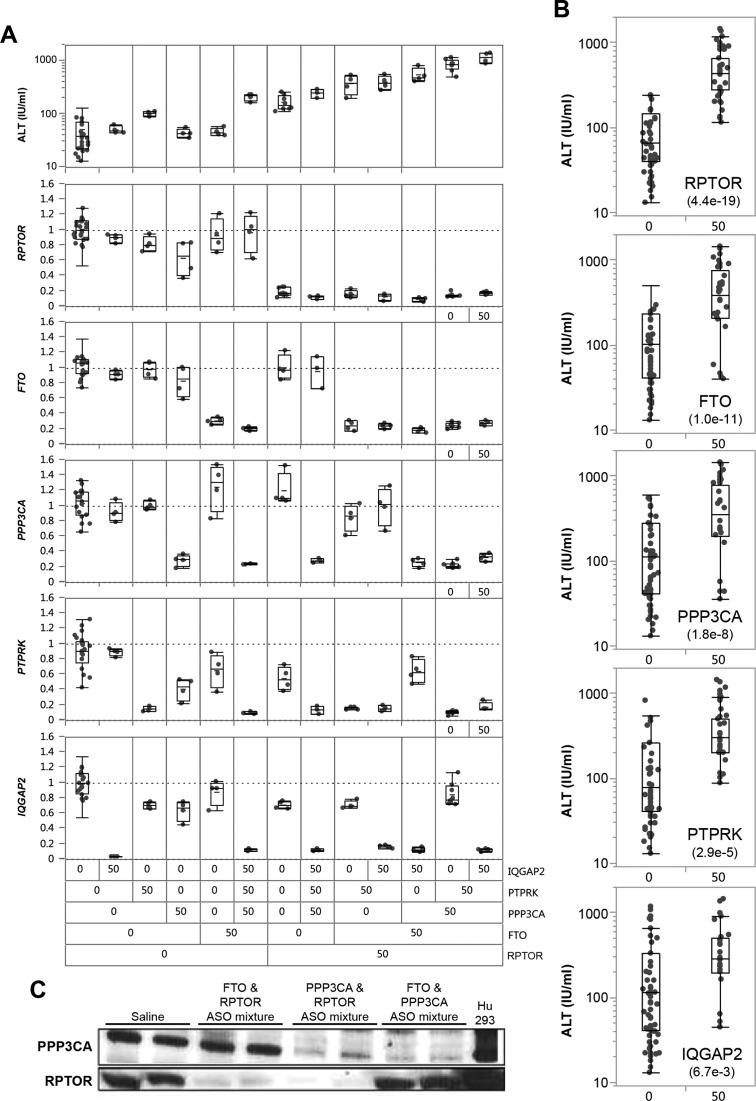
Those findings strongly suggested that increases in transaminases and the severe liver necrosis observed following treatment with toxic LNA ASOs (18,22,25,26) are in large part dependent on the presence of functional RNase H1 in hepatocytes. Yet, it remains unclear how the down-regulation of a broad set of transcripts that is variable from one toxic LNA ASO to another would result in acute liver injury. We hypothesized that the combined reduction of a set of proteins encoded by long pre-mRNA transcripts, many of which may be associated with important cellular functions, would be sufficient to cause toxicity. Taken separately, a specific reduction in any single protein might not be sufficient to cause an adverse effect, however, the combined decrease in multiple proteins resulting through RNase H1 mediated down-regulation of their associated long transcripts would compromise the survival of the affected cells. Depending on the number and nature of the proteins reduced by each toxic LNA ASO, the phenotype, the severity and the kinetics of the toxicity would vary. In order to determine whether toxicity could be triggered by a core set of reduced proteins, we have identified five protein coding transcripts with long pre-mRNAs (RPTOR, FTO, PTPRK, IQGAP2 and PPP3CA) reduced in response to toxic ASO treatment. These transcripts were singled out from a broader panel of highly correlated transcripts on the basis of that knockout mice for the genes encoding the transcripts were embryonic or post-natally lethal, or for their reported involvement in key metabolic function (42–47). MOE gapmer ASOs specifically targeting each of these transcripts were identified and validated in mice. Targeting each transcript individually with a dose of the MOE ASO that produced 80% or greater reduction of the targeted transcript for 15 days failed to significantly modulate the other targets or produce evidence of hepatotoxicity (Figure 6A). However, mice treated with combinations of MOE ASOs targeting several of these selected transcripts (up to five MOE ASOs administered as mixtures) showed clear signs of synergistic induction of hepatotoxicity. For these experiments the total amount of ASO delivered to the mice was held constant to 250 mg/kg by adding the control ASO to the mixture. In order to assess the individual contribution of each down-regulated transcript toward the induction of hepatotoxicity, we perform a series of Student's t-tests comparing the effect of the presence or absence of a given MOE ASO on ALT increase regardless of the presence of any other MOE ASO in the mixture (Figure 6B). The presence of RPTOR, FTO or PPP3CA ASOs to any of the mixtures of MOE ASOs most significantly contributed to the onset of hepatotoxicity (P-values for Student t-test assuming equal variance were 2 × 10e−12, 5.4 × 10e−12, 7.6 × 10e−10, respectively). In contrast, the presence of IQGAP2 seemed to offer little to no contribution to the hepatotoxicity of MOE ASOs mixtures (P-value = 4.4 × 10 – 3). The reduction of Rptor and Ppp3ca was also confirmed at the protein level (Figure 6C).
Figure 6.
Different combination of MOE ASO mixtures to modulate levels of various combination of mRNA target reduction results in synergistic induction of hepatotoxicity in conjunction with targeted mRNA and liver protein reduction. (A) Effect of MOE ASO mixtures on plasma ALT levels (top panel) and five different mRNA target down-regulation (bottom five panels). MOE ASO targets are ordered according to t-test significance (derived from panel B) from bottom to top. ‘0’ and ‘50’ indicates the absence or presence of a given MOE ASO in the mixture. (B) Effect of specific mRNA down-regulation by MOE ASOs on the circulating serum ALT levels regardless of the presence of other MOE ASOs present in the mixture. The level of significance is indicated lower left corner below the target name. (C) Specific protein reduction (Rptor and Ppp3ca) in the liver of mice treated with ASO mixtures measured by western blot.
Well Tolerated ASOs that contain high affinity modifications can be identified
Next, we also tested three additional 3–10–3 cEt ASOs (ISIS 549139, ISIS 549144 and ISIS 549148) in balb/c mice at 50 mg/kg/week for 12 weeks. Those ASOs were designed to have no less than three mismatches in any transcripts in the transcriptome. All three ASOs were very well tolerated causing minimal increase in ALT and minimal reduction in four of the very long transcripts down-regulated by hepatotoxic BNA ASOs (Supplementary Figure S7AB). Furthermore, ASOs that contain high affinity modifications such as cEt that are suitable for clinical development have been identified through careful sequence selection and traditional preclinical toxicity screening in rodent and primate species (16,48). This further demonstrates that safe and effective ASOs containing high affinity modifications can be identified by employing appropriate and rigorous screening paradigms.
DISCUSSION
Despite the increased likelihood of hepatotoxicity associated with ASOs containing high affinity modifications in mice, it is important to emphasize that these effects are highly dependent on the sequence context, and thorough selection and screening can produce well tolerated and efficacious ASOs that take advantage of the increased affinity provided by the addition of BNA modifications as LNA and cEt. Recently two cEt ASOs (AZD5321, https://clinicaltrials.gov/ct2/show/NCT02144051 and AZD9150, https://clinicaltrials.gov/ct2/show/NCT01839604) have entered phase I clinical trial in patients with androgen receptor tumors and hepatocellular carcinoma, respectively. To date, some patients have been treated with up to 1000 mg/kg for at least 3 cycles of treatment with cEt ASOs without evidence of dose limiting hepatotoxicity (unpublished data) demonstrating that stringent preclinical ASO selection can result in well tolerated and potent BNA ASOs (16). Those findings suggest that the mechanism of toxicity described herein should not preclude the development of therapeutic BNA ASOs as a class as hepatotoxicity is predictable in preclinical animal models as well as in silico (23), therefore, avoidable and monitorable through routine liver function assessment for ASO entering early clinical development. Recognizing and understanding this mechanism of hepatotoxicity will dramatically reduce the number of ASOs incorporating high affinity modifications that need to be screened to identify well tolerated and potent therapies in man.
We sought to better understand the mechanism of toxicity for ASOs incorporating high affinity modifications using LNA as the test system. We characterized 13 different LNA ASOs causing varying degree of hepatotoxicity in acute single dose mouse studies. Three of the most hepatotoxic LNA ASO sequences described herein were also modified by substituting LNA with cEt. Specific down-regulation of on-target transcripts such as F7, F11 or SOD1 is unlikely to have contributed directly to the liver injuries as a consequence of exaggerated pharmacology, as other ASOs directed at those targets have been previously shown to be well tolerated even when the level of on-target knockdown was greater than what has been observed in the present study (49,50).
In order to gain some insight into the mechanism of toxicity of those LNA ASOs, we assessed the liver mRNA transcription profiles 24 h after treatment and prior the appearances of any signs of liver injuries. Similarly to reports by Kakiuchi–Kiyota et al. (22), we did not observed biological pathways commonly modulated by hepatotoxic LNA ASOs. However, we noted that the median length of the down-regulated pre-mRNA transcripts correlated to the ALT increase was much longer than transcripts unaffected by the ASO treatments. Furthermore, across all liver expressed genes the degree of transcript knock-down across several hepatotoxic ASOs was also proportional to their pre-mRNA length. We can also recapitulate these observations using data from an independent study (GSE53230) by Kakiuchi-Kiyota et al. (22), Supplementary Figure S8A–B).
To understand if these observations were unique to LNAs or general to all high-affinity modifications, we prepared and tested cEt versions of the most toxic LNA ASOs described in this study by substituting LNA modifications with cEt modifications. While we found only a modest reduction in hepatotoxicity, the ASOs displayed similar on-target mRNA reduction, as well as off-target reduction across a panel of very long pre-mRNA transcripts (Supplementary Figure S3). These observations suggested that the mechanism of toxicity leading to increased transaminases following treatment with some LNA ASOs is not unique to this type modification but rather might be a common feature of ASOs incorporating high affinity modifications, including cEt. In this context, toxicity is driven by the combination of the presence of high affinity modifications with the sequence context.
The role played by RNase H1 in the mechanism of hepatotoxicity of some ASO was further demonstrated by selectively reducing RNase H1 levels by 75% by pretreating mice with MOE ASOs targeted against it. The severity of the liver injuries as well as the knockdown of long off-target transcript caused by some hepatotoxic LNA ASOs was markedly ameliorated following pre-treatment with RNase H1 MOE ASOs.
Next, we demonstrated the liver injuries might be caused by reduction of specific proteins encoded by long protein coding pre-mRNA transcripts, whose perturbations can lead to embryonic or post-natal lethality or are involved in key metabolic function (42–47). While individual reduction in gene expression was generally well tolerated, combined MOE ASO mediated reduction of at least three different targets (e.g. RPTOR, FTO and PPP3CA) caused a synergistic increase in liver injury. Although the severity of the injuries was milder and the onset longer than observed with the hepatotoxic BNA ASOs, we speculate that progressively knocking down of more of the protein coding transcripts would likely result in further synergistic increase in severity of the liver injuries. Together, those observations reinforce the hypothesis that the toxicity of BNA ASOs originates from the reduction of a set of key long RNA transcripts.
Since the hepatotoxicity of some BNA ASOs was largely driven through an RNase H1 dependent mechanism, we hypothesized that promiscuous off-target transcript knockdown are likely driven through ASO-directed RNase H1 catalyzed cleavage of those transcripts at one or more sites. Transcripts down-regulation appears to be affected largely as a function of their pre-mRNA length likely as a result of unintended hybridization and RNase H1 mediated cleavage. The very long length of some pre-mRNA transcripts is hypothesized to increase the likelihood of find one or more near-matches to a given ASO that are sufficiently amenable to RNase H1 activity (Supplementary Figure S6). While partially matched ASO binding sites would be unlikely to be the same across multiple BNA ASOs, the length of those transcripts strongly increases the chance of finding even moderately active binding sites for a broad range of ASO sequences. While the quality of most of those sites (Supplementary Figure S6A) would not be necessarily be better than those found in shorter transcripts and most of the putative sites on their own might exhibit only minimal antisense activity, the sum antisense activity of those sites could results in a pronounced knockdown. In contrast, short transcripts are much less likely to contain even partial sequence matches for a broad range of ASOs that would also be amenable to RNase H1 activity, which translate in more off-target short transcripts down-regulation specificity.
Given that many of the knocked down off-target transcripts do not contain a perfect complementary sequence for the ASOs in our study (Supplementary Figure S6) our observations are consistent with a model in which an ASO:partial-match hybrid is catalyzing transcript cleavage and subsequent degradation in vivo. Furthermore, since BNA ASOs have increased duplex stability it is possible for a variety of reasons to design shorter ASO which retain high potency (51–53). For example, typical LNA or cEt ASOs currently in clinical development now are between 15–17 nucleotides long as opposed 19–21 nucleotides long for ASOs incorporating MOE modifications. The number of perfect or near-match complementary sites in the transcriptome is therefore much increased for the shorter LNA or cEt ASOs. Yet the number of near-match complementary sites for a given transcript itself is not enough to explain why well tolerated BNA ASOs are well tolerated while other ASOs are severely hepatotoxic even though the number of near matches is similar between hepatotoxic and well-tolerated BNA ASOs. This suggests that there is an inherent ASO-sequence dependent property of some ASOs that facilitates RNase H1 mediated cleavage at mismatched sites in vivo. This is also supported by work presented by Hagedorn et al. (23) where they showed that potential for hepatotoxicity could be predicted in silico with a good degree of confidence solely using sequence content of ASOs; perhaps the presences of ‘hepatotoxic’ or ‘protective’ motifs in BNA ASOs increase or decrease the tolerance of RNase H1 for cleaving at mismatched RNA:DNA duplexes.
A hypothesis that attempts to provide a mechanistic explanation of how ASO-inherent sequence motifs could lead to shifts in ASO tolerability can be formulated based on work of Lima et al. where they have previously shown that modifications affecting the structure of matched heteroduplexes influenced the location of the cleavage sites and the kinetic of cleavage by RNase H1 (54,55). Therefore, the ability for RNase H1 to retain some activity against the mismatched heteroduplex might be affected also by the location of the mismatch(es) within the heteroduplex as well as by its impact on the structure of the mismatched heteroduplex (56). A slight gain in RNase H1 tolerance for mismatched duplexes provided by the appropriate sequence content (i.e. presence of ‘hepatotoxic’ motifs or absence of ‘protective’ motifs) would enable variable but often marginal gain in RNase H1 off-target cleavage activity. Conversely, a slight gain in RNase H1 intolerance to mismatched duplexes provided by the presence of ‘protective’ motifs (or at least absence of ‘hepatotoxic motifs’) would increase the specificity of the ASO toward its on-target transcript and minimize off-target knockdown. In this model, long pre-mRNA or very long intergenic non-coding RNA (vlincRNA) transcripts would often be most sensitive to that phenomenon and most often seen across seemingly unrelated ASO sequences (57). In essence, we postulate BNA ASO mediated hepatotoxicity is the result of cellular ‘death by a thousand cuts’.
A recent position paper from the Oligonucleotide Safety Working Group (OSWG) (58) recommends using in silico approaches to identify putative off-target for a given ASO sequences. While in silico methods can identify putative off-target mismatches, the principal difficulty resides in setting an appropriate threshold with a low false positive rate of off-target mismatches identification and then validating those in a relevant setting. A stringent threshold at 1 or 2 mismatch per 16 nucleotide long ASO sequence can identify up to several hundred putative off-target transcripts (Supplementary Table S2). In the case of well-tolerated BNA ASOs described herein, the majority of transcripts containing such a putative site were unaffected, while many of the transcripts with a putative site were down-regulated by the most hepatotoxic LNA ASOs. While not all of the long pre-mRNAs reduced by toxic sequences are identified using the in silico prediction methods envisioned by the recommendation from OSWG (58), alternative in silico methods can identify ASOs capable of eliciting toxicity ((23), Hart et al., unpublished data). These observations illustrate the challenge of predicting which partially matched sites might be susceptible to RNase H mediated cleavage without some empirical validation or without updated in silico screening strategies (23).
As a consequence of those observations, the off-target activity profiles of ASOs and the potential toxicity that might result from some unintended activity should be carefully vetted when developing high affinity ASO therapeutics (58). When this taken into consideration and applied to a stringent preclinical screening process, well tolerated and efficacious BNA ASOs can be identified and provide safe and effective therapies to patients (16).
Supplementary Material
Acknowledgments
The authors thank Dr Brett Monia, Dr Frank Rigo, Dr Aaron Chang, Dr Peter Lindsey and Dr Aimee Jackson for their helpful comments and insights. The authors also thank Heather Murray and Dr Walt Lima for assistance with the RNase H1 experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Isis Pharmaceuticals.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yacyshyn B., Bowen-Yacyshyn M.B., Shanahan W. The clinical experience of antisense therapy to ICAM-1 in Crohn's disease. Curr. Opin. Mol. Ther. 1999;1:332–335. [PubMed] [Google Scholar]
- 2.Bennett F.C. In: Antisense Drug Technologies, Second Edition. Crooke S.T., editor. Vol. 2. LLC, Boca Raton: Taylor & Francis Group; 2007. pp. 273–303. Vol. [Google Scholar]
- 3.Monia B.P., Lesnik E.A., Gonzalez C., Lima W.F., McGee D., Guinosso C.J., Kawasaki A.M., Cook P.D., Freier S.M. Evaluation of 2′ modified oligonucleotides containing deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 4.Crooke S.T., Vickers T., Lima W., Wu H. In: Antisense drug technology, principles, strategies and applications. Crooke S.T., editor. Vol. 2. Boca Raton, FL: Taylor & Francis Group; 2007. pp. 5–46. Vol. [Google Scholar]
- 5.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 6.Geary R.S. Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin. Drug Metab. Toxicol. 2009;5:381–391. doi: 10.1517/17425250902877680. [DOI] [PubMed] [Google Scholar]
- 7.Geary R.S., Yu R.Z., Watanabe T., Henry S.P., Hardee G.E., Chappell A., Matson J., Sasmor H., Cummins L., Levin A.A. Pharmacokinetics of a tumor necrosis factor-alpha phosphorothioate 2′-O-(2-methoxyethyl) modified antisense oligonucleotide: comparison across species. Drug Metab. Dispos. 2003;31:1419–1428. doi: 10.1124/dmd.31.11.1419. [DOI] [PubMed] [Google Scholar]
- 8.Yu R.Z., Geary R.S., Monteith D.K., Matson J., Truong L., Fitchett J., Levin A.A. Tissue disposition of a 2′-O-(2-methoxy) ethyl modified antisense oligonucleotides in monkeys. J. Pharm. Sci. 2004;93:48–59. doi: 10.1002/jps.10473. [DOI] [PubMed] [Google Scholar]
- 9.Mullick A.E., Fu W., Graham M.J., Lee R.G., Witchell D., Bell T.A., Whipple C.P., Crooke R.M. Antisense oligonucleotide reduction of apoB-ameliorated atherosclerosis in LDL receptor-deficient mice. J. Lipid Res. 2011;52:885–896. doi: 10.1194/jlr.M011791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merki E., Graham M.J., Mullick A.E., Miller E.R., Crooke R.M., Pitas R.E., Witztum J.L., Tsimikas S. Antisense oligonucleotide directed to human apolipoprotein B-100 reduces lipoprotein(a) levels and oxidized phospholipids on human apolipoprotein B-100 particles in lipoprotein(a) transgenic mice. Circulation. 2008;118:743–753. doi: 10.1161/CIRCULATIONAHA.108.786822. [DOI] [PubMed] [Google Scholar]
- 11.Henry S.P., Taylor J., Midgley L., Levin A.A., Kornbrust D.J. Evaluation of the toxicity of ISIS 2302, a phosphorothioate oligonucleotide in a 4-week study in CD-1 mice. Antisense Nucleic Acid Drug Dev. 1997;7:473–481. doi: 10.1089/oli.1.1997.7.473. [DOI] [PubMed] [Google Scholar]
- 12.Kastelein J.J., Wedel M.K., Baker B.F., Su J., Bradley J.D., Yu R.Z., Chuang E., Graham M.J., Crooke R.M. Potent reduction of apolipoprotein B and low-density lipoprotein cholesterol by short-term administration of an antisense inhibitor of apolipoprotein B. Circulation. 2006;114:1729–1735. doi: 10.1161/CIRCULATIONAHA.105.606442. [DOI] [PubMed] [Google Scholar]
- 13.Gaudet D., Brisson D., Tremblay K., Alexander V.J., Singleton W., Hughes S.G., Geary R.S., Baker B.F., Graham M.J., Crooke R.M., et al. Targeting APOC3 in the familial chylomicronemia syndrome. N. Engl. J. Med. 2014;371:2200–2206. doi: 10.1056/NEJMoa1400284. [DOI] [PubMed] [Google Scholar]
- 14.Buller H.R., Bethune C., Bhanot S., Gailani D., Monia B.P., Raskob G.E., Segers A., Verhamme P., Weitz J.I., the, F.X.I.A.S.O.T.K.A.I Factor XI Antisense Oligonucleotide for Prevention of Venous Thrombosis. N. Engl. J. Med. 2014;372:232–240. doi: 10.1056/NEJMoa1405760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wengel J. Synthesis of 3‘-C- and 4‘-C-Branched Oligodeoxynucleotides and the Development of Locked Nucleic Acid (LNA) Acc. Chem. Res. 1999;32:301–310. [Google Scholar]
- 16.Burel S.A., Han S.R., Lee H.S., Norris D.A., Lee B.S., Machemer T., Park S.Y., Zhou T., He G., Kim Y., et al. Preclinical evaluation of the toxicological effects of a novel constrained ethyl modified antisense compound targeting signal transducer and activator of transcription 3 in mice and cynomolgus monkeys. Nucleic Acid Ther. 2013;23:213–227. doi: 10.1089/nat.2013.0422. [DOI] [PubMed] [Google Scholar]
- 17.Seth P.P., Siwkowski A., Allerson C.R., Vasquez G., Lee S., Prakash T.P., Wancewicz E.V., Witchell D., Swayze E.E. Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 2009;52:10–13. doi: 10.1021/jm801294h. [DOI] [PubMed] [Google Scholar]
- 18.Swayze E.E., Siwkowski A.M., Wancewicz E.V., Migawa M.T., Wyrzykiewicz T.K., Hung G., Monia B.P., Bennett C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bianchini D., Omlin A., Pezaro C., Lorente D., Ferraldeschi R., Mukherji D., Crespo M., Figueiredo I., Miranda S., Riisnaes R., et al. First-in-human Phase I study of EZN-4176, a locked nucleic acid antisense oligonucleotide to exon 4 of the androgen receptor mRNA in patients with castration-resistant prostate cancer. Br. J. Cancer. 2013;109:2579–2586. doi: 10.1038/bjc.2013.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Poelgeest E.P., Swart R.M., Betjes M.G., Moerland M., Weening J.J., Tessier Y., Hodges M.R., Levin A.A., Burggraaf J. Acute kidney injury during therapy with an antisense oligonucleotide directed against PCSK9. Am. J. Kidney Dis. 2013;62:796–800. doi: 10.1053/j.ajkd.2013.02.359. [DOI] [PubMed] [Google Scholar]
- 21.Raetz E.A., Morrison D., Romanos-Sirakis E., Gaynon P., Sposto R., Bhojwani D., Bostrom B.C., Brown P., Eckroth E., Cassar J., et al. A phase I study of EZN-3042, a novel survivin messenger ribonucleic acid (mRNA) antagonist, administered in combination with chemotherapy in children with relapsed acute lymphoblastic leukemia (ALL): a report from the therapeutic advances in childhood leukemia and lymphoma (TACL) consortium. J. Pediatr. Hematol. Oncol. 2014;36:458–463. doi: 10.1097/MPH.0b013e3182a8f58f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kakiuchi-Kiyota S., Koza-Taylor P.H., Mantena S.R., Nelms L.F., Enayetallah A.E., Hollingshead B.D., Burdick A.D., Reed L.A., Warneke J.A., Whiteley L.O., et al. Comparison of hepatic transcription profiles of locked ribonucleic acid antisense oligonucleotides: evidence of distinct pathways contributing to non-target mediated toxicity in mice. Toxicol. Sci. 2014;138:234–248. doi: 10.1093/toxsci/kft278. [DOI] [PubMed] [Google Scholar]
- 23.Hagedorn P.H., Yakimov V., Ottosen S., Kammler S., Nielsen N.F., Hog A.M., Hedtjarn M., Meldgaard M., Moller M.R., Orum H., et al. Hepatotoxic potential of therapeutic oligonucleotides can be predicted from their sequence and modification pattern. Nucleic Acid Ther. 2013;23:302–310. doi: 10.1089/nat.2013.0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seth P.P., Jazayeri A., Yu J., Allerson C.R., Bhat B., Swayze E.E. Structure activity relationships of alpha-L-LNA modified phosphorothioate gapmer antisense oligonucleotides in animals. Mol. Ther. Nucleic Acids. 2012;1:e47. doi: 10.1038/mtna.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanton R., Sciabola S., Salatto C., Weng Y., Moshinsky D., Little J., Walters E., Kreeger J., DiMattia D., Chen T., et al. Chemical modification study of antisense gapmers. Nucleic Acid Ther. 2012;22:344–359. doi: 10.1089/nat.2012.0366. [DOI] [PubMed] [Google Scholar]
- 26.Burdick A.D., Sciabola S., Mantena S.R., Hollingshead B.D., Stanton R., Warneke J.A., Zeng M., Martsen E., Medvedev A., Makarov S.S., et al. Sequence motifs associated with hepatotoxicity of locked nucleic acid–modified antisense oligonucleotides. Nucleic Acids Res. 2014;42:4882–4891. doi: 10.1093/nar/gku142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dullea R., Salatto C., Sciabola S., Chen T., Dimattia D., Gandhok H., Kreeger J., Weng Y., Clark T., Vage C., et al. Study of CRTC2 pharmacology using antisense oligonuceotides. Nucleic Acid Ther. 2014;24:127–138. doi: 10.1089/nat.2013.0456. [DOI] [PubMed] [Google Scholar]
- 28.Durig J., Duhrsen U., Klein-Hitpass L., Worm J., Hansen J.B., Orum H., Wissenbach M. The novel antisense Bcl-2 inhibitor SPC2996 causes rapid leukemic cell clearance and immune activation in chronic lymphocytic leukemia. Leukemia. 2011;25:638–647. doi: 10.1038/leu.2010.322. [DOI] [PubMed] [Google Scholar]
- 29.Koch T.R.C., Hansen H.F., Hansen B., Straarup E.M., Kauppinen S. In: Therapeutic Oligonucleotides. Kurreck J., editor. RSC Biomolecular Sciences; 2008. pp. 103–141. [Google Scholar]
- 30.Westergaard M.H.H.F., Thrue C.A., Hansen J.B., Kjaerulff L.S., Straarup E.M., Olsen O., Dalby L.W., Hildebrandt-Eriksen L., Kearney P. SPC2968—A novel Hif-1{alpha} antagonist for treatment of clear cell renal cell carcinoma. J. Clin. Oncol. 2006;24:14581. [Google Scholar]
- 31.Tolcher A.W., Patnaik A., Papadopoulos K.P., Agnew J., Lokiec F.M., Rezai K., Kalambakas S., Buchbinder A. Proceedings of the 102nd Annual Meeting of AACR, LB-409 (abstract) Orlando, Florida: 2011. [Google Scholar]
- 32.McKay R.A., Miraglia L.J., Cummins L.L., Owens S.R., Sasmor H., Dean N.M. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-α expression. J. Biol. Chem. 1999;274:1715–1722. doi: 10.1074/jbc.274.3.1715. [DOI] [PubMed] [Google Scholar]
- 33.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statistical Soc. 1995;57:289–300. [Google Scholar]
- 34.Jackson A.L., Linsley P.S. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 36.Burchard J., Jackson A.L., Malkov V., Needham R.H., Tan Y., Bartz S.R., Dai H., Sachs A.B., Linsley P.S. MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA. 2009;15:308–315. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 38.Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dongen S., Abreu-Goodger C., Enright A.J. Detecting microRNA binding and siRNA off-target effects from expression data. Nat. Methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 41.Burel S.A., Machemer T., Ragone F.L., Kato H., Cauntay P., Greenlee S., Salim A., Gaarde W.A., Hung G., Peralta R. Unique MOE-DNA chimeric oligonucleotide induces an atypical MDA5 dependent induction of type I interferon response. J. Pharmacol. Exp. Ther. 2012;342:150–162. doi: 10.1124/jpet.112.193789. [DOI] [PubMed] [Google Scholar]
- 42.Guertin D.A., Stevens D.M., Thoreen C.C., Burds A.A., Kalaany N.Y., Moffat J., Brown M., Fitzgerald K.J., Sabatini D.M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Asano A., Tsubomatsu K., Jung C.G., Sasaki N., Agui T. A deletion mutation of the protein tyrosine phosphatase kappa (Ptprk) gene is responsible for T-helper immunodeficiency (thid) in the LEC rat. Mamm. Genome. 2007;18:779–786. doi: 10.1007/s00335-007-9062-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhang B.W., Zimmer G., Chen J., Ladd D., Li E., Alt F.W., Wiederrecht G., Cryan J., O'Neill E.A., Seidman C.E., et al. T cell responses in calcineurin A alpha-deficient mice. J. Exp. Med. 1996;183:413–420. doi: 10.1084/jem.183.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt V.A., Chiariello C.S., Capilla E., Miller F., Bahou W.F. Development of hepatocellular carcinoma in Iqgap2-deficient mice is IQGAP1 dependent. Mol. Cell. Biol. 2008;28:1489–1502. doi: 10.1128/MCB.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischer J., Koch L., Emmerling C., Vierkotten J., Peters T., Bruning J.C., Ruther U. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 47.Park D., Park J., Park S.G., Park T., Choi S.S. Analysis of human disease genes in the context of gene essentiality. Genomics. 2008;92:414–418. doi: 10.1016/j.ygeno.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Pandey S.K., Wheeler T.M., Justice S.L., Kim A., Younis H., Gattis D., Jauvin D., Puymirat J., Swayze E.E., Freier S.M. Identification and characterization of modified antisense oligonucleotides targeting dmpk in mice and nonhuman primates for the treatment of Myotonic Dystrophy type 1. J. Pharmacol. Exp. Ther. 2015;355:310–321. doi: 10.1124/jpet.115.226969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhattacharjee G., Revenko A.S., Crosby J.R., May C., Gao D., Zhao C., Monia B.P., MacLeod A.R. Inhibition of vascular permeability by antisense-mediated inhibition of plasma kallikrein and coagulation factor 12. Nucleic Acid Ther. 2013;23:175–187. doi: 10.1089/nat.2013.0417. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H., Lowenberg E.C., Crosby J.R., MacLeod A.R., Zhao C., Gao D., Black C., Revenko A.S., Meijers J.C., Stroes E.S., et al. Inhibition of the intrinsic coagulation pathway factor XI by antisense oligonucleotides: a novel antithrombotic strategy with lowered bleeding risk. Blood. 2010;116:4684–4692. doi: 10.1182/blood-2010-04-277798. [DOI] [PubMed] [Google Scholar]
- 51.Kierzek E., Ciesielska A., Pasternak K., Mathews D.H., Turner D.H., Kierzek R. The influence of locked nucleic acid residues on the thermodynamic properties of 2′-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 2005;33:5082–5093. doi: 10.1093/nar/gki789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You Y., Moreira B.G., Behlke M.A., Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Straarup E.M., Fisker N., Hedtjarn M., Lindholm M.W., Rosenbohm C., Aarup V., Hansen H.F., Orum H., Hansen J.B., Koch T. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38:7100–7111. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lima W.F., Rose J.B., Nichols J.G., Wu H., Migawa M.T., Wyrzykiewicz T.K., Vasquez G., Swayze E.E., Crooke S.T. The positional influence of the helical geometry of the heteroduplex substrate on human RNase H1 catalysis. Mol. Pharmacol. 2007;71:73–82. doi: 10.1124/mol.106.025429. [DOI] [PubMed] [Google Scholar]
- 55.Lima W.F., Rose J.B., Nichols J.G., Wu H., Migawa M.T., Wyrzykiewicz T.K., Siwkowski A.M., Crooke S.T. Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol. Pharmacol. 2007;71:83–91. doi: 10.1124/mol.106.025015. [DOI] [PubMed] [Google Scholar]
- 56.Ostergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Bennett C.F., et al. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41:9634–9650. doi: 10.1093/nar/gkt725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.St Laurent G., Shtokalo D., Dong B., Tackett M.R., Fan X., Lazorthes S., Nicolas E., Sang N., Triche T.J., McCaffrey T.A., et al. VlincRNAs controlled by retroviral elements are a hallmark of pluripotency and cancer. Genome Biol. 2013;14:R73. doi: 10.1186/gb-2013-14-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindow M., Vornlocher H.P., Riley D., Kornbrust D.J., Burchard J., Whiteley L.O., Kamens J., Thompson J.D., Nochur S., Younis H., et al. Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat. Biotechnol. 2012;30:920–923. doi: 10.1038/nbt.2376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.