Abstract
We have recently identified lens epithelium-derived growth factor (LEDGF/p75, also known as PSIP1) as a component of the homologous recombination DNA repair machinery. Through its Pro-Trp-Trp-Pro (PWWP) domain, LEDGF/p75 binds to histone marks associated with active transcription and promotes DNA end resection by recruiting DNA endonuclease retinoblastoma-binding protein 8 (RBBP8/CtIP) to broken DNA ends. Here we show that the structurally related PWWP domain-containing protein, hepatoma-derived growth factor-related protein 2 (HDGFRP2), serves a similar function in homologous recombination repair. Its depletion compromises the survival of human U2OS osteosarcoma and HeLa cervix carcinoma cells and impairs the DNA damage-induced phosphorylation of replication protein A2 (RPA2) and the recruitment of DNA endonuclease RBBP8/CtIP to DNA double strand breaks. In contrast to LEDGF/p75, HDGFRP2 binds preferentially to histone marks characteristic for transcriptionally silent chromatin. Accordingly, HDGFRP2 is found in complex with the heterochromatin-binding chromobox homologue 1 (CBX1) and Pogo transposable element with ZNF domain (POGZ). Supporting the functionality of this complex, POGZ-depleted cells show a similar defect in DNA damage-induced RPA2 phosphorylation as HDGFRP2-depleted cells. These data suggest that HDGFRP2, possibly in complex with POGZ, recruits homologous recombination repair machinery to damaged silent genes or to active genes silenced upon DNA damage.
INTRODUCTION
Potentially lethal DNA double strand breaks (DSBs) are mostly repaired by non-homologous end joining or homologous recombination (1,2). Non-homologous end joining ligates DSB ends with no or limited processing, while homologous recombination involves extensive and energy-consuming DNA end resection and uses sister chromatids as templates to ensure error-free repair. Non-homologous end joining can occur throughout the cell cycle, whereas homologous recombination is restricted to S and G2 phases of the cell cycle when sister chromatids are available. The choice of this repair process during S- and G2-phases of the cell cycle depends on the chromatin context in which the DNA DSB occurs, with histone marks of active transcription units being crucial for the recruitment of the homologous recombination repair machinery preferentially to these sites (3,4). We have recently shown that this process is facilitated by PC4 and SFRS1 interacting protein 1 (PSIP1; also known as LEDGF/p75), which interacts with H3K36me3 and other methyl-lysine histone marks associated with active transcription via its N-terminal Pro-Trp-Trp-Pro (PWWP) domain, and enhances the recruitment of the resection-promoting DNA endonuclease retinoblastoma-binding protein 8 (RBBP8/CtIP) to these sites (4).
LEDGF/p75 and its alternatively spliced isoform LEDGF/p52 belong to the hepatoma-derived growth factor (HDGF) related protein (HDGFR) family (5). All family members share a highly conserved N-terminal region termed ‘homologous to the amino terminus of HDGF’ (HATH), which contains a PWWP domain of 70 amino acids (6). The PWWP domain belongs to the Tudor domain ‘Royal family’ and has been described as a potential histone methylation reader (7). It is implicated in various nuclear processes such as epigenetic regulation (8,9), chromatin structure modulation (10,11) and DNA repair (12,13). HDGFRP2 (also known as HRP-2) is the only other HDGFRP family member sharing both N-terminal PWWP domain and C-terminal integrase-binding domain (IBD) with LEDGF/p75. Both LEDGF/p75 and HDGFRP2 have been extensively studied for their ability to tether the HIV integrase (IN) to active transcription units (14–16). LEDGF/p75 binds to IN via its IBD and promotes HIV integration in the body of genes, most likely mediated by binding of its PWWP domain to methylated histones associated with actively transcribed genes (17). HDGFRP2, however, has been shown to have a substantially lower binding affinity towards HIV integrase (8) but still compensates for depletion of LEDGF/p75 and helps to redirect integration to active transcription units (18).
Whereas the cellular function of HDGFRP2 has so far not been addressed, we could recently demonstrate an essential role of LEDGF/p75 in DNA DSB repair by homologous recombination (4). The determining step in pathway choice after DNA DSB formation is DNA end resection, which commits the cell to homologous recombination. A complex of Mre11, Rad50 and Nbs1 (MRN) together with DNA endonuclease RBBP8/CtIP performs the initial resection (19,20), followed by long-range resection of up to 10 kb from the initial DNA DSB mediated by EXO1 in concert with DNA2 and BLM helicase (21,22). The resulting single stranded DNA is rapidly covered by replication protein A 32 kDa subunit (RPA2, also known as RP-A p32), which in turn is phosphorylated by the ataxia-telangiectasia and Rad3-related (ATR). A BRCA1-dependent exchange of phosphorylated RPA2 with Rad51 then initiates strand invasion and thereby subsequent DNA resolvase- and ligase-mediated completion of homologous recombination. We showed earlier that LEDGF/p75 via its PWWP domain associates with transcriptionally active units of the genome and upon DNA damage, binds to RBBP8/CtIP and promotes its recruitment to DNA DSBs (4). Since LEDGF/p75 and HDGFRP2 share both the PWWP domain and the IBD, we hypothesized a similar involvement of HDGFRP2 in DNA DSB repair and investigated its role in this process.
MATERIALS AND METHODS
Cell culture and treatments
All cells were grown in Dulbecco's Modified Eagle Medium supplemented with 6% fetal calf serum, 25 U.I./ml penicillin and 25 μg/ml streptomycin. U2OS DR-GFP and U2OS-GFP-RBBP8/CtIP cells were selected with 5 μg/ml puromycin.
Cells were treated with 1 μM camptothecin (Sigma) for 1 h, washed and incubated to recover for the indicated times. Ionizing radiation was performed in an X-ray generator (Yxlon, Y.Smart 160E/1.5, 160 kV).
Transfections
Forward or reverse transfections of siRNAs were done with 20 or 30 nM siRNA using RNAiMax (Invitrogen). The following siRNA sequences (sense, 5′-3′) were used in this study:
Control siRNA (C): sequence not available (Qiagen, AllStars Negative Control siRNA)
PSIP1 (L): GCAAUGAGGAUGUGACUAA
HDGFRP2 #3 (H3): GAGAUCAAGUUUGCCCUAA
HDGFRP2 #4 (H4): GACUCAGACUCAGACAAGA
HDGFRP2 #6 (H6): CCGUGAAGGUGGAGCGGAC
RBBP8: GCUAAAACAGGAACGAAUC
POGZ #1 (P1): GGUGUUACAGGCGAAAAUAGC
POGZ #2 (P2): GACAGUUGGAGUUCCACAA
POGZ #3 (P3): CAGGUACCCAGUUUGUUAA
POGZ #4 (P4): GUCAUGCCGGCCACUCUUA
pFlag-HDGFRP2 plasmid (23) was kindly provided by Eric Poeschla (Mayo Clinic College of Medicine, Rochester, MN, USA). H4 siRNA-resistant Flag-HDGFRP2 construct Flag-HDGFRP2* was established by introducing three point mutations (C528T_A531G_C534T; NCBI reference sequence NM_001001520.1) to pFlag-HDGFRP2 using QuikChange Lightening Site-directed Mutagenesis kit (Agilent Technologies, # 210518) according to the manufacturer's instruction. The mutagenic primers used were 5′-GGATGGAGAGCGACTCAGATTCGGATAAGAGTAGCGACAACAGTG-3′ (forward) and 5′-CACTGTTGTCGCTACTCTTATCCGAATCTGAGTCGCTCTCCATCC-3′ (reverse). The resulting M13 mutant plasmid was verified by DNA sequencing using primer sequence 5′-AGGACCTGTTCCCCTACGAC-3′.
Immunoblotting
Cells were lysed in 1× Laemmli sample buffer (for CoIPs and peptide pulldown: 2× Laemmli sample buffer). Proteins were separated using sodium dodecyl sulphate-polyacrylamide gel electrophoresis, transferred on a nitrocellulose membrane, incubated with designated primary and appropriate secondary antibodies (Dako, Agilent) and incubated with ECL detection reagents (ECL Plus: Amersham, GE Healthcare Life Sciences) before analysis using the imaging system ImageQuant LAS-4000 (GE Healthcare Life Sciences). The following primary antibodies were used for immunoblotting: β-actin (Sigma, A2228), CBX1 (Merck Millipore, mab3448), CHK2 (Santa Cruz, sc-17747), CHK1 (Santa Cruz, sc-8408), CHK1 (P-S317) (Cell Signalling, 2344P), CHK2 (P-T68) (Cell Signaling 2661), Cyclin A2 (Santa Cruz, sc-751), γH2AX (Abcam, ab2893), GAPDH (Serotec AbD, MCA4740), HDGFRP2 (Santa Cruz sc-55223), LEDGF (Bethyl, A300–848), POGZ (Santa Cruz, sc-102062), RPA2 (Abcam, ab2175), RPA2 (P-S4/S8) (Bethyl A300–245A) and RBBP8/CtIP (Bethyl, A300–488A).
Immunocytochemistry
For confocal microscopy, an LSM-510 META (Carl Zeiss Microimaging Inc.) coupled with an upright Zeiss-Axioimager and equipped with an oil immersion objective (Plan-Apochromat 40×/1.3 and 63×/1.4) was used. Dual colour confocal images were acquired with lines 488 and 543 nm for excitation of Alexa Fluor 488 and Alexa Fluor 594 dyes and band-pass filters 505–530 nm and 560–615 nm, respectively. Image acquisition and analysis were carried out with LSM-ZEN software. Briefly, cells were optionally pre-extracted by incubation for 5 min at 4 C in CSK buffer (25 mM Hepes pH7.4, 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2 300 mM Sucrose and 0.5% Triton-X-100), followed by fixation in 3.7% formaldehyde for 12 min and staining with appropriate primary and according secondary antibodies. The following primary antibodies were used for immunocytochemistry: Cyclin A2 (Abcam, ab16 726), γH2AX (Abcam, ab2893), HDGFRP2 (Santa Cruz sc-55 223 and ProteinTech 15 134–1-AP), LEDGF (Bethyl, A300–848), RPA2 (Abcam, ab16 850) and POGZ (Santa Cruz Biotechnology, sc-102 062). Secondary antibodies AlexaFluor488 donkey anti-rabbit, AlexaFluor488 donkey anti-mouse, AlexaFluor594 donkey anti-rabbit and AlexaFluor594 donkey anti-mouse were from Invitrogen, Molecular Probes.
Immunoprecipitation
Cells were grown as subconfluent monolayer in 10 cm cell culture dishes, lysed and nuclear extracts were prepared essentially following the instructions for the Nuclear Complex CoIP kit (Active Motif), but diluted in RIPA lysis buffer (50 mM TRIS HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS), 2 mM MgCl2). Extracts were precleared and afterwards incubated with the designated primary antibodies: IgG (rabbit, Agilent Technologies/Dako), HDGFRP2 (rabbit, Proteintech) on a rotator for 4 h at 4°C. Anti-rabbit magnetic beads (Life Technologies, Dynabeads M-280 anti-rabbit IgG) were added and incubated for 2 h at 4°C. Bead-bound protein complexes were washed three times in RIPA wash buffer (50mM TRIS HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.25% sodium deoxycholate), boiled in 2× Laemmli sample buffer and analysed by immunoblotting.
Laser micro-irradiation
UV laser micro-irradiation was performed essentially as described (24,25). U2OS-GFP-RBBP8/CtIP were transfected with indicated siRNAs for 72 h. For the last 24 h, the culture medium was supplemented with 10 μM BrdU to sensitize the cells to DSB generation by UVA laser (λ = 337 nm). Micro-irradiated cells were followed by real-time confocal microscopy for 15 minutes, after which stripe-forming cells were counted.
Histone peptide pulldown
Biotin-conjugated histone 3 (H3) peptides (Eurogentech) were immobilized on streptavidin magnetic beads (Life Technologies, Dynabeads M-280 Streptavidin) and incubated with nuclear extracts prepared as in the immunoprecipitation procedure but resolved in binding buffer (150 mM Hepes-KOH (pH7.6), 200 mM KCl, 1 mM MgCl2, 1 mM EDTA, 10% Glycerol, 0.5% NP-40, 1 mM DTT + Protease Inhibitors) for 2 h at 4°C. Bound proteins were washed five times in binding buffer, boiled in 2× SDS sample buffer and analysed by immunoblotting.
Homologous recombination assay
The homologous recombination assay was performed essentially as described previously (26). U2OS cells expressing the integrated homologous recombination reporter DR-GFP were transfected with indicated siRNAs for 48 h, an I-SceI expression vector (pCBA-I-SceI) and a vector expressing monomeric red fluorescent protein (pCS2-mRFP) were transfected in a 2:1 ratio for 24 h. Cells were harvested, fixed and subjected to flow cytrometric analysis to quantify GFP positive and transfected (RFP positive) cells (BD Biosciences, FACSVerse). The threshold for GFP positivity was set to 1% background level in control cells not transfected with I-SceI. The percentage of double positive cells of all RFP positive cells was used as the measure of homologous recombination, and that value in control siRNA-transfected cells was set to 100% in each experiment.
Cell viability assays
Cells were transfected with indicated siRNA for 48 h, washed and incubated with PrestoBlue reagent according to the manufacturer's instructions (Thermo Fisher Scientific). The fluorescence was measured in a microplate reader at 571 nm and at the reference wavelength of 610 nm. Values were corrected for background fluorescence and normalized to the transfection control. Total cell count was determined by staining the cells with Hoechst-33342 and using Celigo cytometer (Brooks Life Science Systems, measuring at 461 nm) and its software to analyse the data. Values of total cell count were normalized to the transfection control performed in triplicate parallel to each experiment.
Proximity ligation assay (PLA)
After indicated treatments, U2OS cells (14 000 cells/well in Falcon tissue culture slides) were fixed and permeabilized using 4% paraformaldehyde and 0.5% Triton X-100, blocked with 3% bovine serum albumin and incubated with primary antibodies including rabbit anti-HDGFRP2 (Proteintech; 15 134–1-AP), goat anti-HDGFRP2 (Santa Cruz; sc-55 223), murine anti-POGZ (Sigma-Aldrich; SAB1407181), rabbit anti-H3K9me3 (Abcam, ab8898) and murine anti-CtIP (GeneTex, GTX70264). Protein–protein interactions were analysed using Duolink-based in situ proximity ligation assay (PLA) (Sigma-Aldrich) according to the manufacturer's instructions. Signals were quantified using a confocal LSM-780 microscope and the Duolink ImageTool. The quantification of the PLA signals is shown as median and presented as floating-bar plots.
Cell cycle analysis
U2-OS cells were reverse transfected with 20 μM siRNAs using Lipofectamine® RNAiMAX, treated with 0.04 μg/ml nocodozale or DMSO 24 h later, and washed in phosphate buffered saline, fixed with methanol on ice for 15 min 48 h after the transfection. The fixed cells were resuspended in FxCycle PI/RNase Staining Solution (Thermo Fischer Scientific) and analysed by a BD FACSVerse and FlowJo software.
Statistical analysis
P-values were calculated with the two tailed student's t-test except for the PLAs where the Mann–Whitney U test was used.
RESULTS
HDGFRP2 depletion induces cell death
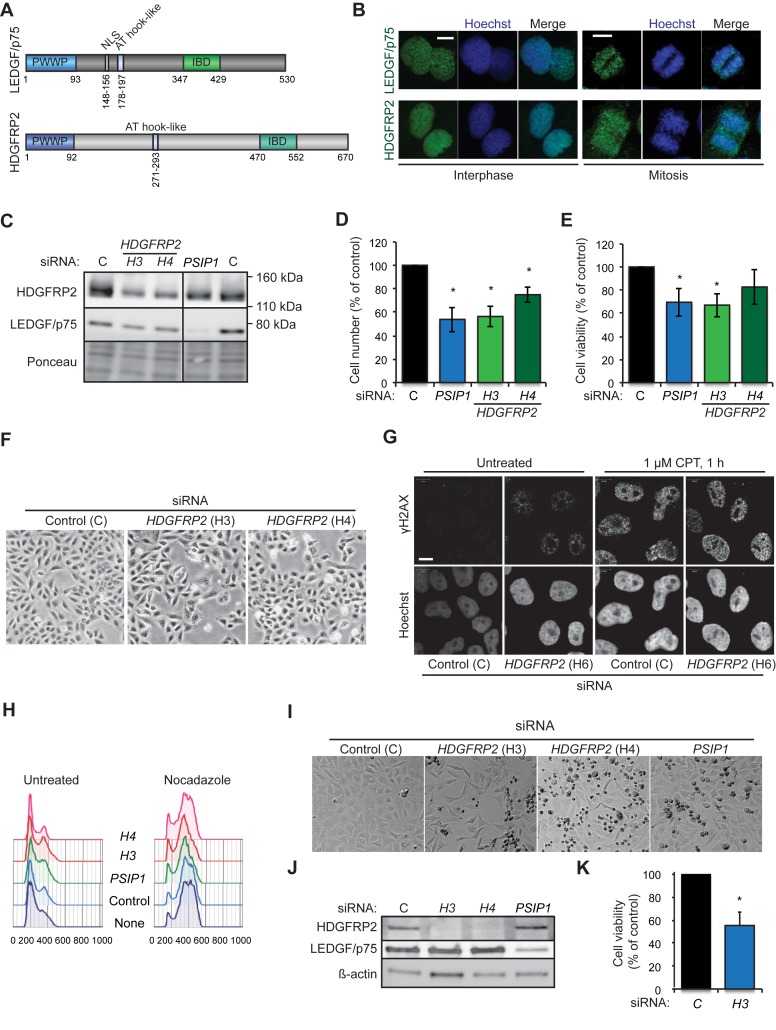
Prompted by the recently identified role of chromatin-binding LEDGF/p75 in the DNA DSB repair and cell survival (4), we hypothesized a similar nuclear role for the structurally related HDGFRP2. In line with their analogous chromatin- and DNA-binding domains (Figure 1A), both LEDGF/p75 and HDGFRP2 displayed punctate and strictly nuclear localization in interphasic U2OS (Figure 1B) and HeLa (data not shown) cells. Contrary to LEDGF/p75, which remained tightly bound to the chromatin during mitosis, HDGFRP2 dissociated partly from condensed chromosomes in mitotic cells (Figure 1B).
Figure 1.
HDGFRP2 is essential for the survival of U2OS and HeLa cells. (A) Schematic representation of the protein structures of LEDGF/p75 and HDGFRP2. (B) Representative confocal images of interphasic (left) and mitotic (right) U2OS cells fixed and stained for LEDGF/p75 and HDGFRP2. Scale bars, 10 μm. (C) Representative immunoblots of indicated proteins from lysates of U2OS cells transfected with non-targeting control (C), H3 and H4 (HDGFRP2) and PSIP1 (LEDGF) siRNAs for 72 h. Ponceau staining is shown as a control for equal loading. The vertical line indicates where the blot was cut to remove non-essential bands after staining and exposure. (D and E) The number (D) and viability (E) of U2OS cells transfected as in (C) were analysed by Celigo cytometer and PrestoBlue assay, respectively. (F) Representative brightfield images of U2OS cells transfected as in (C). (G) Representative confocal images of U2OS cells transfected with non-targeting control (C) or H6 (HDGFR2) siRNAs for 72 h and left untreated or treated with 1 μM camptothecin (CPT) for 1 h. DNA damage foci were detected by staining for γH2AX and nuclei were stained with Hoechst. Scale bars, 10 μm. See Supplementary Figure S1 for an immunoblot demonstrating the efficacy of H6 siRNA. (H) Cell cycle profiles of U2OS cells transfected as in (C) and left untreated (left) or treated with 4 μg/ml nocodazole for the last 24 h (right) before fixation and staining with propidium iodide (PI). PI staining was quantified using flow cytometry and analysed by FlowJo software. (I and J) Representative brightfield images of HeLa cells transfected with indicated siRNAs for 72 h (I) and immunoblots of proteins extracted from the same cells (J). Note an increase in condensed dead cells (small and dark) in samples treated with HDGFRP2 and PSIP1 siRNAs. (K) The viability of HeLa cells transfected with indicated siRNAs for 72 h was analysed by PrestoBlue assay. Error bars, SEM of two (D and E) or three (K) independent triplicate experiments. *P < 0.05 when compared with cells transfected with control siRNA in parallel.
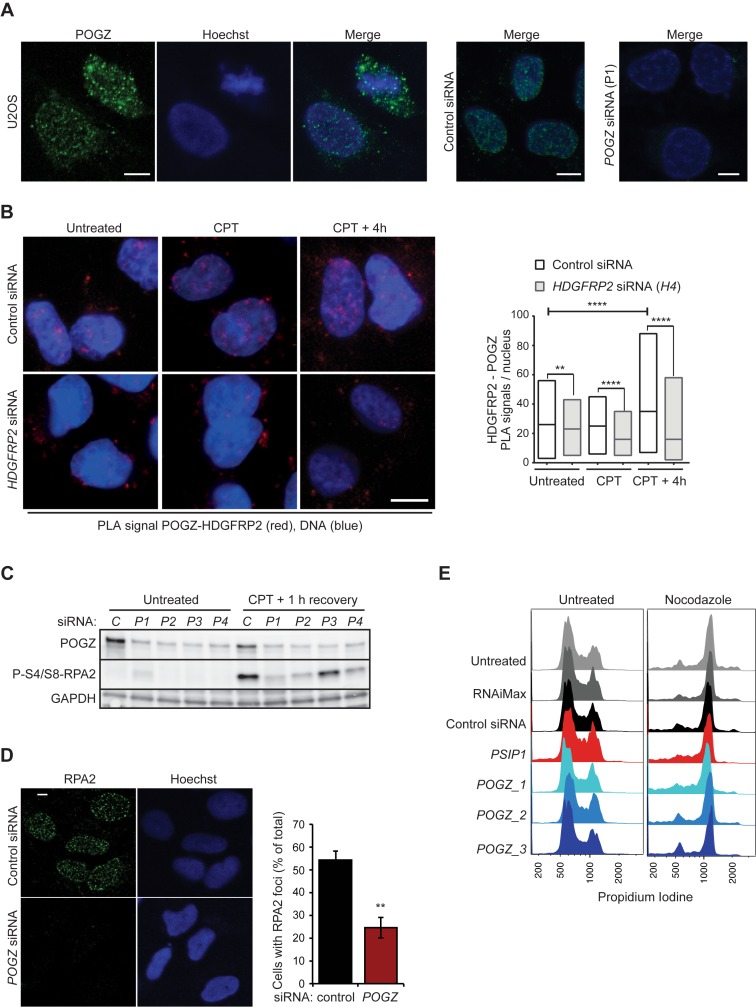
To investigate the consequences of HDGFRP2 depletion on cell survival, we treated U2OS cells with siRNAs targeting the HDGFRP2 encoding HDGFRP2 gene (Figure 1C). Akin to an siRNA targeting the LEDGF encoding PSIP1 gene, treatment of cells with HDGFRP2 siRNAs for 72 h led to an increase in cell death as demonstrated by cell counting (Figure 1D), PrestoBlue cell viability assay (Figure 1E) and light microscopy (Figure 1F). HDGFRP2 depletion was associated with spontaneous DNA damage as indicated by appearance of γH2AX foci in the absence of DNA damaging agents (Figure 1G), but altered neither the proportion of replicating cells in normal growth conditions nor progression through G1 and S phases of cell cycle when the progression through mitosis was inhibited by nocodazole treatment (Figure 1H). HDGFRP2 depletion induced a similar cell death phenotype in HeLa cells (Figure 1I–K). Taken together, these data suggest that the reduced cell number upon HDGFRP2 depletion is due to the induction of cell death rather than disturbances in cell cycle progression.
HDGFRP2 depletion impairs DNA repair by homologous recombination
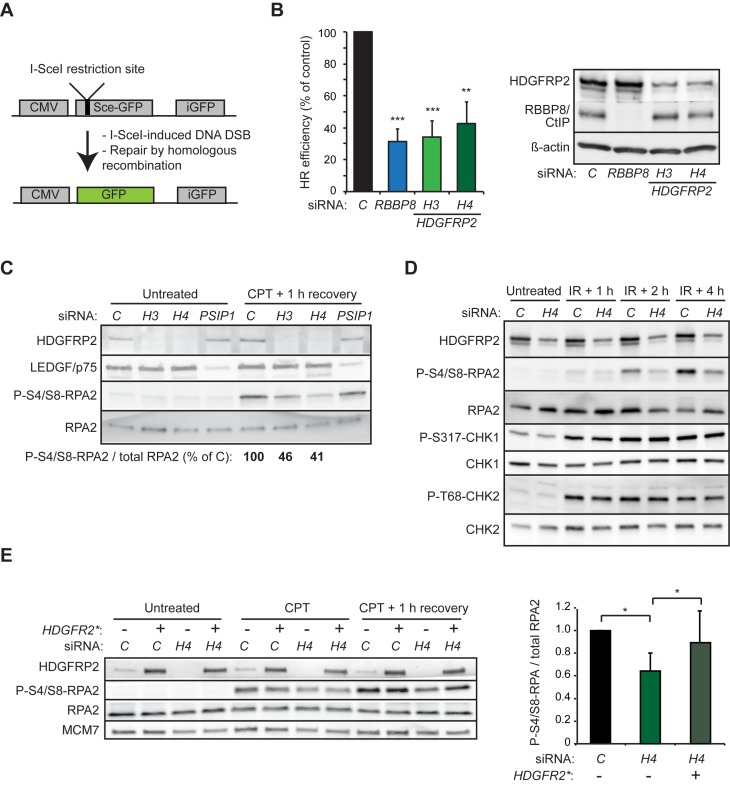
To study whether the pro-survival effect of HDGFRP2 reflected its role in DNA DSB repair, we made use of an assay directly measuring homologous recombination efficiency in U2OS cells carrying an integrated GFP-based homologous recombination reporter construct (DR-GFP) (26). This construct codes for an inactive GFP harbouring the restriction site for the I-SceI endonuclease, and a truncated wild-type GFP that can serve as a template during recombination (Figure 2A). Following I-SceI-mediated cleavage of the restriction site, recombination events will result in restored and active GFP, which can be used as a direct measure of recombinational DNA repair efficiency. In this model system, knockdown of HDGFRP2 by six independent siRNAs reduced homologous recombination efficiency, four of them reducing it to a level comparable to that observed in cells depleted for RBBP8/CtIP or LEDGF/p75 (Figure 2B and Supplementary Figure S1).
Figure 2.
Knockdown in HDGFRP2 inhibits DNA DSB repair by homologous recombination. (A) The homologous repair reporter contains an I-SceI restriction site, which upon I-SceI expression is cleaved to generate a DNA DSB. DSB repair by homologous recombination using truncated wild-type GFP (iGFP) as a template results in an intact GFP gene. (B) U2OS DR-GFP cells were transfected with control (C), RBBP8 or HDGFRP2 (H3 and H4) siRNAs for 48 h, followed by transfection with I-Sce-I and RFP (internal control) for 24 h. The efficiency of homologous recombination was determined by the GFP/RFP ratio analysed by flow cytometry. Representative immunoblots of indicated proteins from lysates of the same cells are shown on right. Error bars, SEM of four independent experiments. when compared with cells transfected with control siRNA in parallel. (C) Representative immunoblots of indicated proteins from lysates of U2OS cells transfected with control (C), HDGFRP2 (H3 and H4) or PSIP1 siRNAs for 72 h and left untreated or subjected to treatment with 1 μM camptothecin (CPT) for 1 h followed by 1 h recovery. The quantification of the indicated bands is presented below. The experiment was repeated at least four times with similar results. (D) Representative immunoblots of indicated proteins from lysates of U2OS cells transfected with control (C) or HDGFRP2 (H4) siRNAs and left untreated or subjected to 5 Gy ionizing radiation (IR) followed by 1, 2 or 4 h recovery. The experiment was repeated at least four times with similar results. Two experiments with H3 and H6 siRNAs gave similar results. (E) Representative immunoblots of indicated proteins from lysates of U2OS cells transfected with H4 siRNA-resistant mutant of HDGFRP2 (HDGFRP2*, +) or empty vector (−) together with control (C) or HDGFRP2 (H4) siRNAs for 72 h and left untreated or subjected to treatment with 1 μM camptothecin (CPT) for 1 h followed by 1 h recovery (left). The quantification of P-S4/S8-RPA2 relative to total RPA2 is shown on the right. Error bars, SD of three independent experiments. *P < 0.05, **P < 0.01 when compared with cells transfected with control siRNA in parallel (B) or as indicated (E).
Together with other DNA repair factors, including BRCA1 and MRN complex, RBBP8/CtIP catalyses an initial 5′-3′ DNA resection of the broken DNA ends generating 3′ single-stranded DNA (ssDNA) overhangs, which are rapidly coated by the heteromeric ssDNA-binding RPA complex (27–29). The RPA-coated ssDNA then triggers assembly of the homologous recombination repair signalling machinery governed by Serine/threonine-protein kinase ATR, which leads to the phosphorylation of multiple target proteins, including the RPA2 subunit of RPA (30). In order to assess the role of HDGFRP2 in the initial steps of homologous recombination, we first monitored the phosphorylation of RPA2 at S4/S8, a site whose phosphorylation depends on ssDNA-induced activation of ATR (31). U2OS cells depleted for HDGFRP2 had a clearly reduced capacity to phosphorylate this site in response to DNA damage induced by irradiation or inhibition of topoisomerase I by camptothecin (Figure 2C and D). On the contrary, the irradiation-induced ATR-mediated phosphorylation of CHK1 at S317, which does not depend on ssDNA (32), as well as the serine-protein kinase ATM-mediated phosphorylation of CHK2 at T68, were not affected by the lack of HDGFRP2. Importantly, the reduced camptothecin-induced RPA phosphorylation in HRP2 depleted cells was effectively reverted by the expression of a siRNA-resistant HDGFRP2 mutant (Figure 2E). Thus, HDGFRP2 appears to regulate DNA DSB repair by homologous recombination without affecting the activities of ATR and ATM kinases.
HDGFRP2 depletion inhibits the recruitment of RBBP8/CtIP to DNA DSB sites
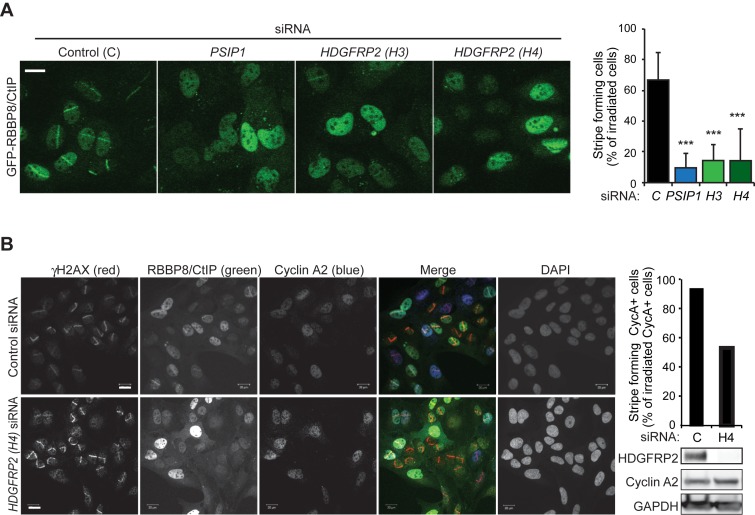
We next tested whether HDGFRP2 regulates homologous recombination in a manner similar to LEDGF/p75, which binds RBBP8/CtIP and promotes its recruitment to the sites of DNA DSBs (4). For this purpose, we depleted U2OS cells that express GFP-RBBP8/CtIP for HDGFRP2 and followed GFP-RBBP8/CtIP recruitment to laser micro-irradiated DNA DSB tracks. Depletion of HDGFRP2 by two independent siRNAs led to a significant reduction in the GFP-RBBP8/CtIP stripe formation that was comparable to that observed in cells depleted for LEDGF/p75 (Figure 3A). Because DNA repair by homologous recombination takes place preferentially in S and G2 phase cells, altered cell cycle progression could affect this assay. To ensure that this was not the case, we repeated the assay and calculated the proportion of micro-irradiated GFP-RBBP8/CtIP stripes in cells expressing cyclin A2, whose expression increases in S phase and peaks in G2 phase. As expected, GFP-RBBP8/CtIP stripes formed almost exclusively in cyclin A2 positive cells and depletion of HDGFRP2 clearly reduced their formation and intensity in micro-irradiated, cyclin A2 positive cells (Figure 3B). It should also be noted that HDGFRP2 depletion did not affect the cell cycle distribution or the expression of cyclin A2, LEDGF/p75, RBBP8/CtIP or RAD51 in U2OS cells (Figure 3B; Supplementary Figure S1). These data suggest that HDGFRP2 promotes the recruitment of RBBP8/CtIP to the sites of DNA DSBs.
Figure 3.
HDGFRP2 is essential for the recruitment of RBBP8/CtIP DNA DSBs. (A) Representative confocal images of U2OS GFP-RBBP8/CtIP cells transfected with indicated siRNAs for 72 h and subjected to micro-laser irradiation followed by 15 min incubation (left), and quantification of the percentage of GFP-RBBP8/CtIP-stripe forming cells of all micro-irradiated cells (right). Error bars, SEM of three independent experiments. *** P < 0.005 when compared with cells transfected with control siRNA in parallel. Scale bar, 10 μm. (B) Representative confocal images of U2OS GFP-RBBP8/CtIP cells transfected with indicated siRNAs for 72 h, irradiated as in (A) and stained with indicated antibodies and DAPI (nuclei) (left), quantification of the percentage of GFP-RBBP8/CtIP-stripe forming Cyclin A2-positive cells of all Cyclin A2-positive micro-irradiated cells (right, upper panel) and immunoblots of indicated proteins from the cell lysates (right, lower panel). Scale bar, 20 μm.
HDGFRP2 associates with heterochromatin-binding proteins and binds heterochromatin-associated histone marks
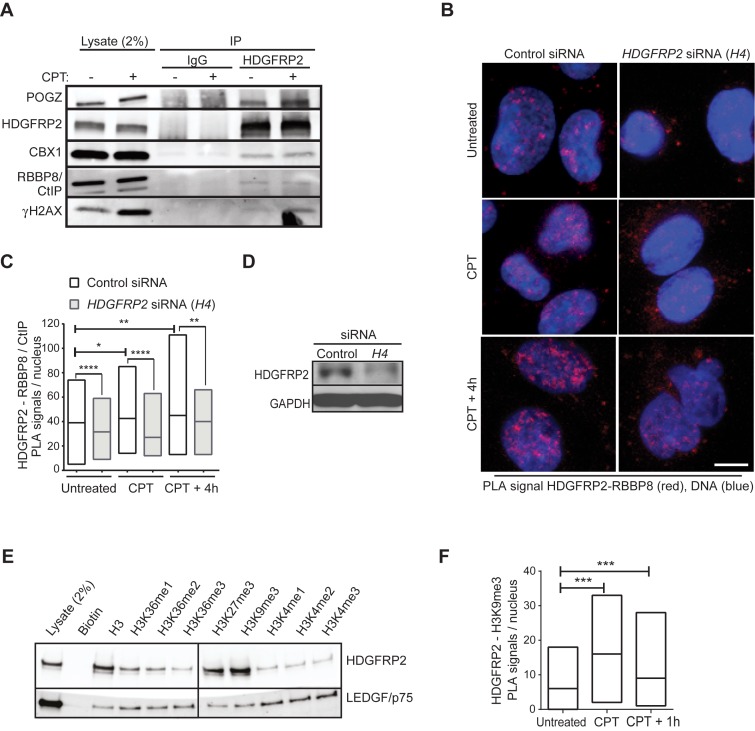
Prompted by the necessity of HDGFRP2 for the effective recruitment of RBBP8/CtIP to DNA damage sites, we next investigated whether HDGFRP2 associated with RBBP8/CtIP. Indeed, RBBP8/CtIP antibodies repeatedly detected small amounts of RBBP8/CtIP in complex with HDGFRP2 as demonstrated by immunoblotting of proteins that co-immunoprecipitated with endogenous HDGFRP2 (Figure 4A). Further analysis of co-immunoprecipitated proteins confirmed the previously reported interaction between HDGFRP2 and heterochromatin-associated pogo transposable element derived protein with zinc finger domain (POGZ) (33,34). The heterochromatin-binding protein, chromobox homologue 1 (CBX1; or heterochromatin protein 1 homologue ß) was also identified as an HDGFRP2-associated protein (Figure 4A). Furthermore, HDGFRP2 associated with γHA2X, which marks DNA damage sites (Figure 4A). Notably, the association of HDGFRP2 with both POGZ and γHA2X was enhanced by 1 h camptothecin treatment, whereas that of RBBP8/CtIP appeared unchanged (Figure 4A). PLA employing antibodies against HDGFRP2 and RBBP8/CtIP verified the close proximity of HDGFRP2 and RBBP8/CtIP in nuclei of untreated cells, which was significantly enhanced during 1 h camptothecin treatment and up to 4 h of recovery (Figure 4B-D). Since it is unclear whether HDGFRP2 recruits RBBP8/CtIP to the site of DNA damage or whether the two proteins are recruited together (e.g. in response to changes in histone marks) we next studied the association of HDGFRP2 with modified histones.
Figure 4.
HDGFRP2 associates with RBBP8/CtIP, heterochromatin-binding proteins and histone marks of transcriptionally silent chromatin. (A) Lysates of U2OS cells left untreated or treated with 1 μM camptothecin (CPT) for 1 h were immunoprecipitated with HDGFRP2 antibody (HDGFRP2) or IgG control and analysed by immunoblotting of indicated proteins. Similar results were obtained in a minimum of three independent experiments. (B–D) Co-localization of HDGFRP2 and RBBP8/CtiP was analysed using rabbit anti-HDGFRP2 and murine anti-RBBP8/CtiP antibodies and Duolink-based in situ proximity ligation assay (PLA) with appropriate secondary antibodies. U2OS cells were first transfected with control (C) or HDGFRP2 (H4) siRNA for 72 h and then left untreated or subjected to treatment with 1 μM camptothecin (CPT) for 1 h with or without a 4 h recovery. Representative confocal images of PLA-stained cells, in which co-localization of the antibodies used results in red (624 nm) fluorescence are shown in (B). Median values of nuclear PLA signals from a minimum of 75 (control siRNA) cells per condition presented as floating-bar plots are shown in (C) and immunoblot demonstrating the efficacy of H4 siRNA in (D). The specificity of the PLA signal was confirmed by the significant reduction of the signal by H4 siRNA. Note, that the remaining HDGFR2 protein in H4 siRNA-treated cells can also result in the PLA signal. DNA was stained with DAPI to visualize the nuclei. (E) Biotin-conjugated histone peptides were incubated with U2OS lysates, followed by a pulldown with streptavidin-coated magnetic beads. Representative immunoblot of indicated proteins. The vertical line indicates where the blot was cut to remove non-essential bands after staining and exposure. (F) Co-localization of HDGFRP2 and H3K9me3 was analysed using goat anti-HDGFRP2 and rabbit anti-H3K9me3 antibodies and Duolink-based in situ PLA with appropriate secondary antibodies in U2OS cells left untreated or treated with 1 μM camptothecin (CPT) for 1 h with or without 1 h recovery. Median values of nuclear PLA signals from a minimum of 75 cells per condition (right) are shown as floating-bar plots. DNA was stained with DAPI to visualize the nuclei. Representative confocal images are shown in Supplementary Figure S2. Scale bars, 10 μm. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Both POGZ and CBX1 have been reported to bind strongly to histone marks characteristic of transcriptionally repressed genes, e.g. H3K9me3 and H3K27me3 (34–36). Similarly, HDGFRP2 had high affinity to biotinylated histone H3K9me3 and H3K27me3 peptides in vitro (Figure 4E). Supporting the specificity of HDGFRP2 binding to transcriptionally silent chromatin, binding to histone peptides characteristic for transcriptionally active units (e.g. H3K4me3 and H3K36me3) was much weaker. In contrast, LEDGF/p75 showed higher binding affinity to the histone marks of active chromatin than those of silent chromatin (Figure 4E). In order to investigate the dynamics of the association of HDGFRP2 with silent chromatin, we performed PLA in untreated and camptothecin-treated U2OS cells employing specific antibodies to endogenous HDGFRP2 and H3K9me3. The nuclear proximity ligation signal verified the close proximity of HDGFRP2 and H3K9me3 in untreated cells, and a significant increase in the signal upon camptothecin treatment suggested that co-localization of these proteins was regulated by DNA damage (Figure 4F and Supplementary Figure S2). These data suggest that LEDGF/p75 and HDGFRP2 function in two separate complexes.
POGZ binds to HDGFRP2 and is involved in DNA DSB repair by homologous recombination
POGZ has been described as a modulator of the dissociation of chromobox protein homologue 5 (CBX5, also known as heterochromatin protein 1 homologue α) from chromosome arms during mitosis (37), and has recently been found to be in complex with three CBX isoforms (CBX1, CBX3 and CBX5) as well as with HDGFRP2 in human and mouse tissues (13,34). Because the two POGZ binding partners, HDGFRP2 (Figures 2 and 3) and CBX1 (38,39), have been shown to regulate DNA DSB repair by homologous recombination, we next investigated the role of POGZ in this process. POGZ protein localized mainly to nucleus of interphase U2OS cells but was distributed more diffusely in mitotic cells, with only a small proportion remaining localized close to the mitotic chromosomes (Figure 5A). PLA employing specific antibodies against POGZ and HDGFRP2 confirmed their co-localization in the nuclei of untreated cells and revealed a significant increase in this co-localization during the recovery phase after camptothecin treatment (Figure 5B). In order to test the putative involvement of POGZ in the DNA damage repair by homologous recombination, we investigated the effect of POGZ depletion on RPA2 phosphorylation after DNA DSB induction by camptothecin treatment in U2OS cells. Four independent POGZ siRNAs reduced the level of camptothecin-induced RPA2 phosphorylation considerably as compared to control siRNA-treated cells (Figure 5C). Paralleling these observations, POGZ depletion greatly impaired RPA2 foci formation after camptothecin treatment (Figure 5D). Notably, the efficient siRNA-mediated depletion of POGZ in U2OS cells did not alter the cell cycle profiles (Figure 5E), even though POGZ has been reported to be essential for correct mitotic progression (37). These observations strongly suggest a novel role for POGZ in DNA DSB repair by homologous recombination.
Figure 5.
POGZ depletion impairs DNA DSB repair by homologous recombination. (A) Representative confocal images of U2OS cells, fixed and stained for POGZ and DNA (Hoechst). Cells transfected with control and POGZ (P1) siRNAs for 72 h and stained similarly are shown to verify the specificity of the POGZ staining. (B) Co-localization of HDGFRP2 and POGZ association was analysed using rabbit anti-HDGFRP2 and murine anti-POGZ antibodies and Duolink-based in situ PLA in U2OS cells first transfected with indicated siRNAs for 72 h and then left untreated or subjected to treatment with 1 μM camptothecin (CPT) for 1 h with or without 4 h recovery. Representative confocal images (left) and median values of nuclear PLA signals in a minimum of 75 (control siRNA) or 60 (HDGFRP2 (H4) siRNA) cells per condition (right) are shown as floating-bar plots. Note, that the remaining signal in H4 siRNA transfected cells can be due to the remaining HDGFR2 protein (see Figure 4D) or unspecific background staining. (C) Representative immunoblots of indicated proteins from lysates of U2OS cells transfected with non-targeting control siRNA (C) or four independent POGZ siRNAs (P1–4) for 72 h and left untreated or treated with 1 μM camptothecin (CPT) for 1 h followed by an 1 h recovery. (D) Representative confocal images of U2OS transfected with control and POGZ siRNA (P1) for 72 h, treated with 1 μM camptothecin (CPT) followed by a 30 min recovery and stained for RPA2 (left) and quantification of cells with RPA2 foci (right). Error bars, SEM of two independent experiments. (E) Cell cycle profiles of U2OS cells left untreated, treated with transfection medium (RNAiMax) alone or with indicated siRNAs for 72 h and left untreated (left) or treated with 4 μg/ml nocodazole for the last 24 h (right) before fixation and staining with propidium iodide (PI). PI staining was quantified using flow cytometry and FlowJo analysis. Scale bars, 10 μm. **P < 0.01; ***P < 0.001; ****P < 0.0001.
DISCUSSION
We have earlier identified LEDGF/p75 as a key player in the DNA end resection during DNA DSB repair by homologous recombination, where it enhances the phosphorylation of RPA2 and the tethering of RBBP8/CtIP to DNA DSBs (4). Here we show that a structurally related member of the HDGF family, HDGFRP2, performs a similar function during homologous recombination repair. This conclusion is based on ample data showing that the siRNA-mediated depletion of HDGFRP2 results in (i) significantly reduced homologous recombination repair activity in cells carrying an integrated GFP-based homologous recombination reporter construct, (ii) reduced RPA2 phosphorylation after DNA damage induction by camptothecin treatment or ionizing radiation, and iii) reduced recruitment of GFP-RBBP8/CtIP to laser-generated DSB tracks. The defective homologous recombination phenotype is likely to be a consequence of the specific depletion of HDGFRP2 because several independent siRNAs targeting HDFGFRP2 induce a similar phenotype and a siRNA-resistant HDGFRP2 cDNA rescues the RPA2 phosphorylation defect in HDGFRP2 depleted cells. Hence, LEDGF/p75 and HDGFRP2 appear to promote homologous recombination repair by a similar mechanism.
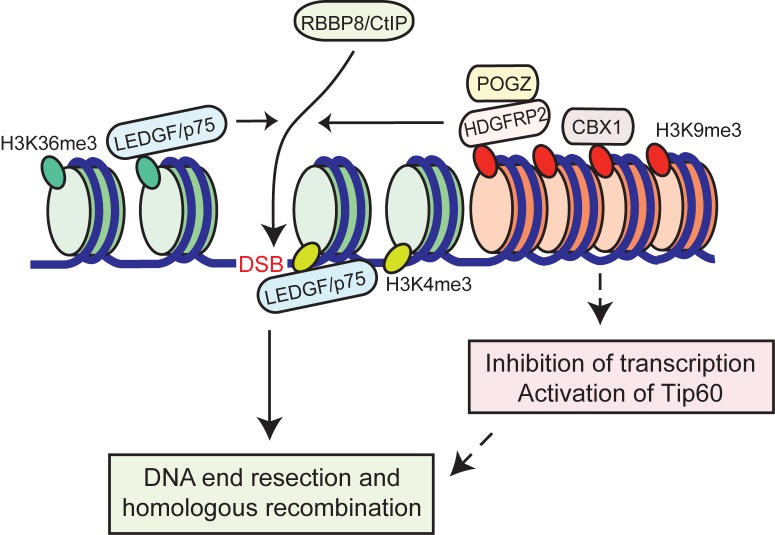
If the functions of LEDGF/p75 and HDGFRP2 in homologous recombination were identical, one would expect the depletion of a single protein to result in only a moderate phenotype. However, depletion of either protein alone reduces the RBB8/CtIP recruitment by over 80% and the homologous recombination activity by over 60%, suggesting that the two proteins perform similar but not identical functions. In line with this, there is a clear difference in chromatin association between HDGFRP2 and LEDGF/p75. Whereas LEDGF/p75 preferentially binds to histone peptides characteristic of active chromatin (e.g. H3K36me3) in vitro (4), HDGFRP2 shows preferential binding to histone peptides characteristic of transcriptionally inactive chromatin (e.g. H3K9me3 and H3K27me3). The physiological relevance of the in vitro binding assay is supported by the close cellular co-localization of HDGFRP2 and H3K9me3 observed using the PLA as well as by the co-immunoprecipitation of heterochromatin-associated proteins, CBX1 and POXZ1 (34–36), with HDGFRP2. Thus, LEDFG/p75 and HDGFRP2 might be functionally redundant in DNA DSB repair but act in separate regions of chromatin, i.e. euchromatin and heterochromatin, respectively. It could be advantageous for the cell to utilize two different proteins for active transcription units and heterochromatin as these areas have different repair kinetics. Whereas actively transcribed chromatin quickly responds to DNA damage with signalling events like histone ubiquitination and subsequent initiation of recombinational repair, DSBs in heterochromatin are thought to first move outside of a CBX3 domain to complete repair by homologous recombination (40). The global effect of HDGFRP2 depletion on DNA damage-induced RPA phosphorylation as well as the requirement of HDGFRP2 for the repair of a CMV promoter-driven actively transcribed GFP gene argue, however, against the heterochromatin-restricted role for HDGFRP2. Instead, the binding of HDGFRP2 complex to histone marks of silent chromatin may recruit HDGFRP2 to genes silenced around the sites of DNA damage, a phenomenon that has recently been found to be essential for efficient DNA repair of damaged active genes (41–44). In such a scenario, LEDGF/p75 and HDGFRP2 could serve spatially and kinetically distinct complementary functions even around the same DNA DSBs (Figure 6). Supporting this hypothesis, camptothecin treatment significantly increased the co-localization of HDGFRP2 with H3K9me3.
Figure 6.
Model of the function of LEDGF/p75 and HDGFRP2 in homologous recombination DNA repair. Repair of a DNA double strand break (DSB) by HR in active chromatin requires the histone H3K4me3- and H3K36me3-binding protein LEDGF/p75 to recruit RBBP8/CtIP and ensure efficient DNA end resection. The structurally highly similar protein HDGFRP2 binds preferentially to histone marks of silent genes (e.g. histone H3K9me3) and contributes, together with its interaction partner POGZ, to efficient DSB repair by HR. HDGFRP2 facilitates RBBP8/CtIP recruitment to the damage site and may play a role in the separate heterochromatic chromatin compartment or in chromatin silenced around the DNA DSB break. Its binding to silent chromatin may reflect its role e.g. in the inhibition of transcription and activation of Tip60 around the damage site.
In addition to identifying HDGFRP2 as a protein regulating homologous recombination repair, our data also identify its binding partner, POGZ, as a protein involved in this process. This conclusion is based on the inhibition of camptothecin-induced RPA2 phosphorylation and foci formation by POGZ RNAi. It remains, however, to be studied whether HDGFRP2 and POGZ function as a complex in the repair process. Supporting this hypothesis, our data from PLA demonstrates a significant increase in the co-localization of these two proteins following camptothecin treatment. Until now, the only function assigned to POGZ is related to the dissociation of CBX5 from chromosome arms (37). Various CBX isoforms, on the other hand, have been implicated in both negative and positive regulation of DNA DSB repair (39). It is, however, unclear whether they act directly in the repair process or indirectly e.g. by dissociation-induced changes in the chromosomal structure that alter the accessibility of other repair factors (45,46). The current model envisions that a transient dissociation of CBX family proteins from chromatin opens up the chromatin structure, increases the pool of available CBX protein and exposes the histone marks to other H3K9me3 binding proteins, e.g. histone acetyltransferase Tip60 that activates the ATM-dependent repair (47). These events could be paralleled by an association of CBX isoforms to DSB sites to promote local chromatin changes that help stabilizing loose ends or restrict transcription. The relatively strong associations between HDGFRP2, POGZ and CBX1 and the enhanced association of the first two upon DNA damage suggest that these proteins may act in concert to regulate homologous recombination either directly or by inducing changes in the chromatin conformation.
In summary, our data introduce two heterochromatin-binding proteins, HDGFRP2 and POGZ, as proteins that regulate the DNA DSB repair by homologous recombination upstream of RPA2 phosphorylation. Further investigations are needed to define their exact roles in this process.
Supplementary Material
Acknowledgments
We thank Karina Grøn Henriksen for excellent technical assistance, Claus Storgaard Sørensen, Claudia Lukas and Jiri Lukas for valuable advice, and Eric Poeschla for the Flag-HDGFRP2 plasmid.
Footnotes
Present addresses:
Annika Baude, Epigenomics and Cancer Risk Factors, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany.
Tania Løve Aaes, Unit of Molecular Signalling and Cell Death, VIB Inflammation Center, Ghent University, Technologiepark 927, 9052 Ghent, Belgium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish National Research Foundation; Danish Medical Research Council; Danish Cancer Society; Vancouver Prostate Centre. Funding for open access charge: Danish Cancer Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chapman J.R., Taylor M.R., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 2.Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 3.Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P., et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daugaard M., Baude A., Fugger K., Povlsen L.K., Beck H., Sorensen C.S., Petersen N.H., Sorensen P.H., Lukas C., Bartek J., et al. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 2012;19:803–810. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
- 5.Dietz F., Franken S., Yoshida K., Nakamura H., Kappler J., Gieselmann V. The family of hepatoma-derived growth factor proteins: characterization of a new member HRP-4 and classification of its subfamilies. Biochem. J. 2002;366:491–500. doi: 10.1042/BJ20011811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izumoto Y., Kuroda T., Harada H., Kishimoto T., Nakamura H. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem. Biophys. Res. Commun. 1997;238:26–32. doi: 10.1006/bbrc.1997.7233. [DOI] [PubMed] [Google Scholar]
- 7.Maurer-Stroh S., Dickens N.J., Hughes-Davies L., Kouzarides T., Eisenhaber F., Ponting C.P. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanov P., Devroe E., Silver P.A., Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 9.Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R.Z., Ragozin S., Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huen M.S., Huang J., Leung J.W., Sy S.M., Leung K.M., Ching Y.P., Tsao S.W., Chen J. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol. Cell. 2010;37:854–864. doi: 10.1016/j.molcel.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maltby V.E., Martin B.J., Schulze J.M., Johnson I., Hentrich T., Sharma A., Kobor M.S., Howe L. Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Mol. Cell. Biol. 2012;32:3479–3485. doi: 10.1128/MCB.00389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Reddy B., Thompson J., Wang H., Noma K., Yates J.R. 3rd, Jia S. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol. Cell. 2009;33:428–437. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberl H.C., Spruijt C.G., Kelstrup C.D., Vermeulen M., Mann M. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell. 2013;49:368–378. doi: 10.1016/j.molcel.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 14.Llano M., Saenz D.T., Meehan A., Wongthida P., Peretz M., Walker W.H., Teo W., Poeschla E.M. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 15.Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 16.Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 17.Gijsbers R., Vets S., De Rijck J., Ocwieja K.E., Ronen K., Malani N., Bushman F.D., Debyser Z. Role of the PWWP domain of lens epithelium-derived growth factor (LEDGF)/p75 cofactor in lentiviral integration targeting. J. Biol. Chem. 2011;286:41812–41825. doi: 10.1074/jbc.M111.255711. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Schrijvers R., Vets S., De Rijck J., Malani N., Bushman F.D., Debyser Z., Gijsbers R. HRP-2 determines HIV-1 integration site selection in LEDGF/p75 depleted cells. Retrovirology. 2012;9:84. doi: 10.1186/1742-4690-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limbo O., Chahwan C., Yamada Y., de Bruin R.A., Wittenberg C., Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nimonkar A.V., Ozsoy A.Z., Genschel J., Modrich P., Kowalczykowski S.C. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanegas M., Llano M., Delgado S., Thompson D., Peretz M., Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 24.Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukas C., Falck J., Bartkova J., Bartek J., Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 26.Pierce A.J., Hu P., Han M., Ellis N., Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukas J., Lukas C., Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 28.Polo S.E., Jackson S.P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernstein K.A., Rothstein R. At loose ends: resecting a double-strand break. Cell. 2009;137:807–810. doi: 10.1016/j.cell.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 32.Kousholt A.N., Fugger K., Hoffmann S., Larsen B.D., Menzel T., Sartori A.A., Sorensen C.S. CtIP-dependent DNA resection is required for DNA damage checkpoint maintenance but not initiation. J. Cell Biol. 2012;197:869–876. doi: 10.1083/jcb.201111065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartholomeeusen K., Christ F., Hendrix J., Rain J.C., Emiliani S., Benarous R., Debyser Z., Gijsbers R., De Rijck J. Lens epithelium-derived growth factor/p75 interacts with the transposase-derived DDE domain of PogZ. J. Biol. Chem. 2009;284:11467–11477. doi: 10.1074/jbc.M807781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vermeulen M., Eberl H.C., Matarese F., Marks H., Denissov S., Butter F., Lee K.K., Olsen J.V., Hyman A.A., Stunnenberg H.G., et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen A.L., Oulad-Abdelghani M., Ortiz J.A., Remboutsika E., Chambon P., Losson R. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 36.Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 37.Nozawa R.S., Nagao K., Masuda H.T., Iwasaki O., Hirota T., Nozaki N., Kimura H., Obuse C. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 2010;12:719–727. doi: 10.1038/ncb2075. [DOI] [PubMed] [Google Scholar]
- 38.Liu H., Galka M., Mori E., Liu X., Lin Y.F., Wei R., Pittock P., Voss C., Dhami G., Li X., et al. A method for systematic mapping of protein lysine methylation identifies functions for HP1beta in DNA damage response. Mol. Cell. 2013;50:723–735. doi: 10.1016/j.molcel.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soria G., Almouzni G. Differential contribution of HP1 proteins to DNA end resection and homology-directed repair. Cell Cycle. 2013;12:422–429. doi: 10.4161/cc.23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiolo I., Minoda A., Colmenares S.U., Polyzos A., Costes S.V., Karpen G.H. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruhlak M., Crouch E.E., Orlov M., Montano C., Gorski S.A., Nussenzweig A., Misteli T., Phair R.D., Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- 42.Kakarougkas A., Ismail A., Chambers A.L., Riballo E., Herbert A.D., Kunzel J., Lobrich M., Jeggo P.A., Downs J.A. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell. 2014;55:723–732. doi: 10.1016/j.molcel.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shanbhag N.M., Rafalska-Metcalf I.U., Balane-Bolivar C., Janicki S.M., Greenberg R.A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ui A., Nagaura Y., Yasui A. Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Mol. Cell. 2015;58:468–482. doi: 10.1016/j.molcel.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 45.Ayoub N., Jeyasekharan A.D., Bernal J.A., Venkitaraman A.R. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 46.Soria G., Polo S.E., Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol. Cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Kaidi A., Jackson S.P. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature. 2013;498:70–74. doi: 10.1038/nature12201. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.