Abstract
Proline-rich antimicrobial peptides (PrAMPs) produced as part of the innate immune response of animals, insects and plants represent a vast, untapped resource for the treatment of multidrug-resistant bacterial infections. PrAMPs such as oncocin or bactenecin-7 (Bac7) interact with the bacterial ribosome to inhibit translation, but their supposed specificity as inhibitors of bacterial rather than mammalian protein synthesis remains unclear, despite being key to developing drugs with low toxicity. Here, we present crystal structures of the Thermus thermophilus 70S ribosome in complex with the first 16 residues of mammalian Bac7, as well as the insect-derived PrAMPs metalnikowin I and pyrrhocoricin. The structures reveal that the mammalian Bac7 interacts with a similar region of the ribosome as insect-derived PrAMPs. Consistently, Bac7 and the oncocin derivative Onc112 compete effectively with antibiotics, such as erythromycin, which target the ribosomal exit tunnel. Moreover, we demonstrate that Bac7 allows initiation complex formation but prevents entry into the elongation phase of translation, and show that it inhibits translation on both mammalian and bacterial ribosomes, explaining why this peptide needs to be stored as an inactive pro-peptide. These findings highlight the need to consider the specificity of PrAMP derivatives for the bacterial ribosome in future drug development efforts.
INTRODUCTION
Antimicrobial peptides (AMPs) represent a large and diverse group of molecules that form part of the innate immune response of a variety of invertebrate, plant and animal species (1). While many AMPs kill bacteria by disrupting the bacterial cell membrane, there is growing evidence that some AMPs have intracellular targets (1). Members of one such class of non-membranolytic peptides are referred to as proline-rich AMPs (PrAMPs) and are present in the hemolymph of several species of insects and crustaceans, as well as in the neutrophils of many mammals (2). PrAMPs exhibit potent antimicrobial activity against a broad range of bacteria, especially Gram-negative, and are therefore considered as potential lead candidates for the development of therapeutic antimicrobial agents (3). Well-characterized insect PrAMPs include the apidaecins produced by bees (Apis melifera) and wasps (Apis Vespidae), pyrrhocoricin from firebugs (Pyrrhocoris apterus), drosocins from fruit flies (Drosophila), metalnikowins from the green shield bug (Palomena prasina) and the milkweed bug (Oncopeltus fasciatus) oncocins (2,4,5). PrAMPs are synthesized as inactive precursors, which undergo proteolysis to release the active peptide. In contrast to the active insect peptides, which are generally <21 amino acids in length, the active mammalian mature forms tend to be much longer; for example, the porcine PR-39 is 39 residues long, whereas the bovine bactenecin-7 (Bac7), which is also found in sheep and goats, is 60 residues long (2). Nevertheless, C-terminally truncated versions of the mammalian PrAMPs retain antimicrobial activity (6–9) and exhibit high sequence similarity with the insect PrAMPs. Indeed, the Bac7(1–16) and Bac7(1–35) derivatives corresponding to the first 16 and 35 residues of Bac7, respectively, display similar, if not improved, antimicrobial activities compared to the full-length processed Bac7 peptide (6,10,11). For instance, Bac7(1–35) reduces mortality from Salmonella typhimurium in a mouse model of infection (12) as well as in a rat model for septic shock (13).
The insect-derived PrAMPs apidaecin and oncocin, as well as the mammalian Bac7, penetrate the bacterial cell membrane mainly via the SbmA transporter present in many Gram-negative bacteria (10,14). Early studies identified interactions between both insect and mammalian PrAMPs and DnaK, suggesting that this molecular chaperone was the common intracellular target (2,15). However, subsequent studies questioned the relevance of this interaction by demonstrating that these PrAMPs also display an equally potent antimicrobial activity against bacterial strains lacking the dnaK gene (16–18). Instead, apidaecin, oncocin and Bac7 were shown to bind to the ribosome and inhibit translation (17,19). Subsequent crystal structures of the oncocin derivative Onc112 in complex with the bacterial 70S ribosome revealed that this peptide binds with a reverse orientation in the ribosomal tunnel and blocks binding of the aminoacyl-tRNA to the A-site (20,21). However, there are no crystal structures to date of a mammalian PrAMP in complex with the ribosome.
Here we present 2.8–2.9 Å resolution X-ray structures of the Thermus thermophilus 70S (Tth70S) ribosome in complex with either the mammalian Bac7 derivative Bac7(1–16) or the insect-derived PrAMPs metalnikowin I or pyrrhocoricin. The structures reveal that Bac7(1–16), metalnikowin I and pyrrhocoricin bind within the ribosomal tunnel with a reverse orientation compared to a nascent polypeptide chain, as observed previously for oncocin (20,21). In contrast to the insect PrAMPs oncocin, metalnikowin I and pyrrhocoricin, the mammalian Bac7(1–16) utilizes multiple arginine side chains to establish stacking interactions with exposed nucleotide bases of the rRNA, and we show that its unique N-terminal RIRR motif is critical for inhibiting translation. Like oncocin, metalnikowin I and pyrrhocoricin, the binding site of Bac7 overlaps with that of the A-tRNA, consistent with our biochemical studies indicating that Bac7(1–16) allows 70S initiation complex formation, but prevents subsequent rounds of translation elongation. Furthermore, we demonstrate that Bac7(1–35) displays activity in a mammalian in vitro translation system, providing a possible explanation for why Bac7 is produced as a pre-pro-peptide that is targeted to large granules and phagosomes, thus avoiding direct contact between the active peptide and the mammalian ribosome.
MATERIALS AND METHODS
Peptide synthesis and purification
The Bac7 N-terminal fragments Bac7(1–16; RRIRPRPPRLPRPRPR), Bac7(1–35; RRIRPRPPRLPRPRPRPLPFPRPGPRPIPRPLPFP) and Bac7(5–35; PRPPRLPRPRPRPLPFPRPGPRPIPRPLPFP) were synthesized on solid phase and purified by reversed-phase HPLC as described previously (22). Their concentrations were determined as reported previously (4). All peptides, with a purity of at least 95%, were stored in milliQ water at −80°C until use. The Onc112 peptide was obtained from an earlier study (21). Metalnikowin I (VDKPDYRPRPRPPNM) and pyrrhocoricin (VDKGSYLPRPTPPRPIYNRN) were synthesized to 97.5 and 98.1% purity by NovoPro Bioscience (China).
Purification of T. thermophilus 70S ribosomes
Tth70S ribosomes were purified as described earlier (23) and resuspended in buffer containing 5 mM HEPES-KOH, pH 7.5, 50 mM KCl, 10 mM NH4Cl and 10 mM Mg(CH3COO)2 to yield a final concentration of ∼30 mg/ml. Tth70S ribosomes were flash frozen in liquid nitrogen and kept at −80°C for storage.
Preparation of mRNA, tRNAiMet and YfiA
Synthetic mRNA containing a Shine-Dalgarno sequence and an AUG start codon followed by a phenylalanine codon (5′-GGC AAG GAG GUA AAA AUG UUC UAA -3′) was purchased from Eurogentec. Escherichia coli tRNAiMet was overexpressed in E. coli HB101 cells and purified as described previously (24). YfiA was overexpressed in BL21 Star cells and purified as described previously (25).
Complex formation
A quaternary complex containing Tth70S ribosomes, mRNA, deacylated tRNAiMet and Bac7(1–16) peptide was prepared by mixing of 5 μM Tth70S ribosomes with 10 μM mRNA and 50 μM Bac7(1–16), and incubating at 55°C for 10 min. After addition of 20 μM tRNAiMet, the mixture was incubated at 37°C for 10 min. The sample was then incubated at room temperature for at least 15 min and centrifuged briefly prior to use. Ternary complexes containing 50 μM metalnikowin I or pyrrhocoricin, 5 μM Tth70S ribosomes and 50 μM YfiA were formed by incubation for 30 min at room temperature. The final buffer conditions were 5 mM HEPES-KOH, pH 7.6, 50 mM KCl, 10 mM NH4Cl and 10 mM Mg(CH3COO)2.
Crystallization
Published conditions were used as a starting point for screening crystallization conditions by vapour diffusion in sitting-drop trays at 20°C (23,26). Crystallization drops consisted of 3 μl of quaternary or ternary complexes and 3–4 μl of reservoir solution containing 100 mM Tris–HCl, pH 7.6, 2.9% (v/v) PEG 20,000, 7–10% (v/v) 2-methyl-2,4-petanediol (MPD) and 175 mM arginine. Crystals appeared within 2–3 days and grew to ∼1000 × 100 × 100 μm within 7–8 days. For cryoprotection, the concentration of MPD was increased in a stepwise manner to yield a final concentration of 40% (v/v). The ionic composition during cryoprotection was 100 mM Tris–HCl, pH 7.6, 2.9% (v/v) PEG 20,000, 50 mM KCl, 10 mM NH4Cl and 10 mM Mg(CH3COO)2. Crystals were flash frozen in a nitrogen cryostream at 80 K for subsequent data collection.
Data collection and processing
Diffraction data for Bac7(1–16) were collected at PROXIMA-2A, a beamline at the SOLEIL synchrotron (Saclay, France) equipped with an ADSC Q315 detector. A complete dataset was obtained by merging 0.25° oscillation data collected at 100 K with a wavelength of 0.98011 Å from multiple regions of the same crystal. Diffraction data for metalnikowin I and pyrrhocoricin were collected at PROXIMA-1, a beamline at the SOLEIL synchrotron equipped with a DECTRIS PILATUS 6M detector. Complete datasets were obtained by merging 0.1° oscillation data collected at 100 K with a wavelength of 0.97857 Å from multiple regions of the crystal. Initial data processing, including integration and scaling, was performed with X-ray Detector Software (XDS) (27). The data could be indexed in the P212121 space group, with unit-cell dimensions approximating 210 × 450 × 625 Å and an asymmetric unit containing two copies of the Tth70S ribosome.
Model building and refinement
Initial phases were obtained by molecular replacement performed with Phaser (28). The search model was obtained from a high-resolution structure of the Tth70S ribosome (PDB ID: 4Y4O) (29) where the RNA backbone had been further improved with the ERRASER-Phenix pipeline (30), using the deposited structure factors. Restrained crystallographic refinement was carried out with Phenix (31) and consisted of a single cycle of rigid-body refinement followed by multiple cycles of positional and individual B-factor refinement. Rigid bodies comprised four domains from the small 30S subunit (head, body, spur and helix h44) and three domains from the large 50S subunit (body, L1 stalk and the C terminus of ribosomal protein L9). Non-crystallographic symmetry restraints between the two copies of the Tth70S ribosome in the asymmetric unit were also applied during refinement. After confirming that a single tRNA was bound to the P site or that YfiA was present at the decoding center, and that additional density corresponding to the PrAMPs was visible within the exit tunnel in a minimally biased FO–FC map, models of the corresponding PrAMPs were built in Coot (32). The models for the tRNA and mRNA were obtained from a high-resolution structure of the Tth70S ribosome pre-attack complex (PDB ID: 1VY4). The model for YfiA was obtained from a high resolution Tth70S ribosome structure (PDB ID: 4Y4O). Further refinement and model validation was carried out in Phenix (31) and on the MolProbity server (33), respectively. In the final models, 0.56–0.95% of protein residues were classified as Ramachandran outliers, and 92.4–94.3% had favourable backbone conformations (Supplementary Table S1). Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5F8K (Bac7(1–16)), 5FDU (Metalnikowin I) and 5FDV (Pyrrhocoricin).
In vitro translation assays
Escherichia coli lysate-based transcription-translation coupled assay (RTS100, 5Prime) were performed as described previously for other translational inhibitors (34). Briefly, 6 μl reactions, with or without PrAMP were mixed according to the manufacturer's description and incubated for 1 h at 30°C with shaking (750 rpm). A total of 0.5 μl of each reaction were stopped with 7.5 μl kanamycin (50 μg/μl). The effect of Bac7(1–35) on eukaryotic translation was determined using Rabbit Reticulocyte Lysate System (Promega). A total of 6 μl reactions, with or without Bac7(1–35) were mixed according to the manufacturer′s description and incubated for 1 h at 30°C with shaking (300 rpm). A total of 5 μl of each reaction were stopped in 5 μl kanamycin (50 μg/μl). All samples were diluted with 40 μl of Luciferase assays substrate (Promega) into a white 96-well chimney flat bottom microtiter plate (Greiner). The luminescence was then measured using a Tecan Infinite M1000 plate reader. Relative values were determined by defining the luminescence value of the sample without inhibitor as 100%.
Toe-printing assay
The position of the ribosome on the mRNA was monitored with a toe-printing assay (35) based on an in vitro–coupled transcription-translation system with the PURExpress in vitro protein synthesis kit (NEB), as described previously (21,36). Briefly, each translation reaction consisted of 1 μl solution A, 0.5 μl Δisoleucine amino acid mixture, 0.5 μl tRNA mixture, 1.5 μl solution B, 0.5 μl (0.5 pmol) hns37aa template: (5′-ATTAATACGACTCACTATAGGGATATAAGGAGGAAAACATatgAGCGAAGCACTTAAAattCTGAACAACCTGC GTACTCTTCGTGCGCAGGCAAGACCGCCGCCGC TTGAAACGCTGGAAGAAATGCTGGAAAAATTA GAAGTTGTTGTTtaaGTGATAGAATTCTATCGTTAATAAGCAAAATTCATTATAAC-3′, with start codon ATG, catch isoleucine codon ATT and stop codon TAA in bold, the hns37aa ORF underlined and toe-print primer binding site in italics) and 0.5 μl additional agents (nuclease-free water, water dissolved Bac7(1–35) Bac7(1–16), Bac7(5–35) (1, 10 or 100 μM final concentration) or antibiotics (100 μM thiostrepton, 50 μM edeine, 50 μM clindamycin final concentration)). Translation was performed in the absence of isoleucine at 37°C for 15 min at 500 rpm in 1.5 ml reaction tubes. After translation, 2 pmol Alexa647-labelled NV-1 toe-print primer (5′-GGTTATAATGAATTTTGCTTATTAAC-3′) was added to each reaction. Reverse transcription was performed with 0.5 μl of AMV RT (NEB), 0.1 μl dNTP mix (10 mM) and 0.4 μl Pure System Buffer and incubated at 37°C for 20 min. Reverse transcription was quenched and RNA degraded by addition of 1 μl 10 M NaOH and incubation for at least 15 min at 37°C and then was neutralized with 0.82 μl of 12 M HCl. 20 μl toe-print resuspension buffer and 200 μl PN1 buffer were added to each reaction before treatment with a QIAquick Nucleotide Removal Kit (Qiagen). The Alexa647-labelled DNA was then eluted from the QIAquick columns with 80 μl of nuclease-free water. A vacuum concentrator was used to vaporize the solvent and the Alexa647-labelled DNA was then dissolved into 3.5 μl of formamide dye. The samples were heated to 95°C for 5 min before being applied onto a 6% polyacrylamide (19:1) sequencing gel containing 7 M urea. Gel electrophoresis was performed at 40 W and 2000 V for 2 h. The GE Typhoon FLA9500 imaging system was subsequently used to scan the polyacrylamide gel.
Filter binding assay
Filter binding assays were performed as described previously (34,37). Briefly, 3 pmol of 70S ribosomes purified from BL21 E. coli strain were exposed to 30 pmol of radiolabelled [14C]-Erythromycin (Perkin Elmer; 110 dpm/pmol) in presence of 1x filter binding buffer (10 mM HEPES/KOH [pH 7.4], 30 mM MgCl2, 150 mM NH4Cl and 6 mM β-mercaptoethanol) for 15 min at 37°C. Our controls indicated that approximately 65% of the 70S ribosomes (2 pmol) contained [14C]-Erythromycin previous to the addition of the different PrAMPs. The PrAMPs were diluted in nuclease-free water to a concentration of 1 mM, 100 μM and 10 μM. 2 μl of each PrAMP stock dilution (Onc112, Bac7(1–35), Bac7(1–16) and Bac7(5–35)) were transferred to the respective tube resulting in final concentrations of 100, 10 and 1 μM. Reactions were incubated for an additional 25 min at 37°C. Afterwards the 20 μl samples were passed through a HA-type nitrocellulose filter from Millipore (0.45 μm pore size) and the filter subsequently washed three times with 1 ml 1× filter binding buffer. Scintillation counting was performed in the presence of Rotiszint® eco plus Scintillant. All reactions were performed in duplicate and results were analysed using GraphPad Prism 5. Error bars represent the standard deviation from the mean.
Disome formation assay
The disome formation assay was performed as described previously (38,39). Briefly, in vitro translation of the 2xermBL construct was performed using the Rapid Translation System RTS 100 E. coli HY Kit (Roche). Translations were carried-out for 1 h at 30°C and then analysed on 10–55% sucrose density gradients (in a buffer containing 50 mM HEPES-KOH, pH 7.4, 100 mM KOAc, 25 mM Mg(OAc)2, 6 mM β-mercaptoethanol) by centrifugation at 154 693 × g (SW-40 Ti, Beckman Coulter) for 2.5 h at 4°C.
RESULTS
The N-terminus of Bac7 adopts a compact conformation
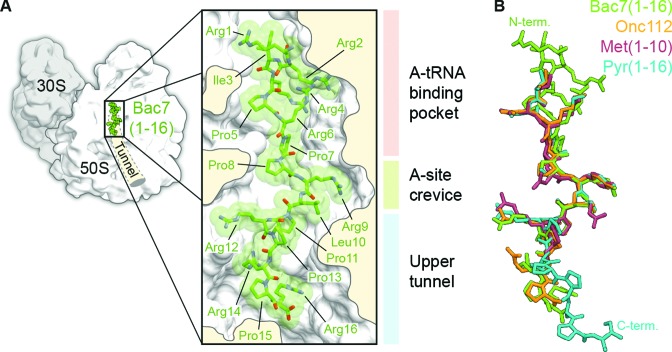
We obtained a structure referred to here as Tth70S-Bac7 from co-crystals of Tth70S ribosomes in complex with deacylated tRNAiMet, a short mRNA and Bac7(1–16) (Supplementary Table S1). In addition, we obtained two additional structures, Tth70S-MetI and Tth70S-Pyr, from co-crystals of Tth70S ribosomes in complex with YfiA and either metalnikowin I or pyrrhocoricin, respectively (Supplementary Table S1). The quality of the electron density in the minimally biased FO–FC difference maps calculated after refinement of a model comprising Tth70S ribosomes and tRNAiMet/mRNA or YfiA, made it possible to build a model for the entire Bac7(1–16; RRIRPRPPRLPRPRPR), the first 10 (of 15; VDKPDYRPRPRPPNM) residues of metalnikowin I (MetI) and the first 16 (of 20; VDKGSYLPRPTPPRPIYNRN) residues of pyrrhocoricin (Pyr), as well as to position several neighbouring solvent molecules (Supplementary Figure S1). Like the insect-derived Onc112 peptide (20,21), MetI, Pyr and Bac7(1–16) all bind to the ribosomal exit tunnel in a reverse orientation relative to the nascent polypeptide chain and make extensive interactions with three distinct regions of the large 50S ribosomal subunit: the A-tRNA binding pocket, the A-site crevice and the upper section of the nascent polypeptide exit tunnel (Figure 1A, B and Supplementary Figure S1). A nearly identical, extended backbone conformation is seen for residues 7–13 of Bac7(1–16) and residues 4–10 of Onc112, Met1 and Pyr, with Arg9 of Bac7(1–16) substituting for Tyr6 of Onc112, Met1 and Pyr (Figure 1B). The structural similarity however does not extend to the N-terminus of Bac7(1–16), where the first six residues adopt a structure that deviates substantially from that of the shorter N-terminus of the insert-derived PrAMPs. Indeed, arginine residues within this region are arranged such that the side chain of Arg6 is sandwiched between the side chains of Arg2 and Arg4 to form a compact, positively charged structure (Figure 1A and B). The binding site of Bac7(1–16) suggests that the additional C-terminal residues of Bac7(1–35) and of the full-length Bac7 (60 residues) would occupy the entire length of the ribosomal tunnel. Consistently, a photocrosslinkable derivative of Bac7(1–35) has been cross-linked to two ribosomal proteins of ∼16 and 25 kDa (19), which we suggest to be L22 and L4, respectively, based on their size and close proximity to the Bac7(1–16) binding site (Supplementary Figure S2). Compared to Onc112 and Met1, additional density for the C-terminal PRPR motif (residues 13–16) of Pyr is observed extending deeper into the tunnel (Figure 1 and Supplementary Figure S1). With the exception of Arg14 for which no density is observed, the PRPR motif is quite well ordered despite not forming any obvious direct interactions with the ribosome.
Figure 1.
Binding site of Bac7(1–16) on the ribosome and comparison with Onc112. (A) Overview and closeup view of a cross-section of the Tth70S ribosomal exit tunnel showing the Bac7(1–16) peptide (RRIRPRPPRLPRPRPR) in green and highlighting the three regions of interaction with the ribosome: the A-tRNA binding pocket (light pink), the A-site crevice (light green) and the upper section of the exit tunnel (light blue). (B) Structural comparison of Bac7(1–16) (green) with Onc112 (orange)(20,21), Met1(1–10) (burgundy) and Pyr(1–16) (cyan), highlighting the distinct structure of the Bac7 N-terminus (N-term) and the Pyr C-terminus (C-term).
Bac7 makes extensive interactions with the 50S ribosomal subunit
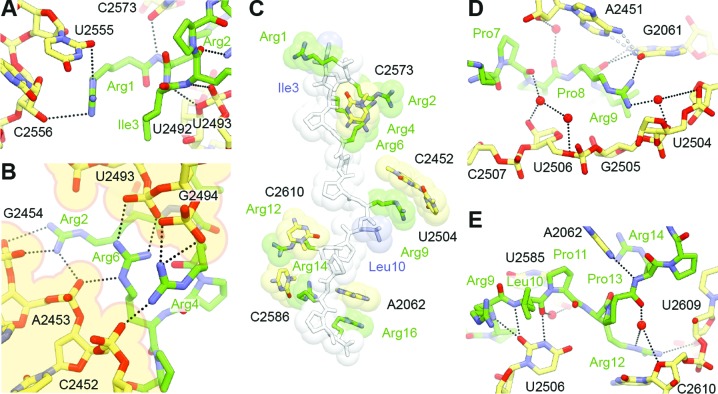
As with Onc112 (20,21), binding of Bac7(1–16) to the ribosome is accompanied by an induced fit involving 23S rRNA residues A2062, U2506 and U2585 (Supplementary Figure S3A; E. coli numbering is used throughout this work for the 23S rRNA), such that the base of this last nucleotide occupies a position that would normally clash with the formyl-methionyl moiety of fMet-tRNAiMet bound to the P-site of an initiation complex (Supplementary Figure S3B). Three modes of interaction are observed between Bac7(1–16) and the large 50S ribosomal subunit (Figure 2A–E).
Figure 2.
Interactions between Bac7(1–16) and the ribosome. (A) Bac7(1–16) (green) makes extensive contacts with the A-site tRNA binding region of the ribosome, in particular (B) electrostatic interactions between its N-terminal arginine stack and a deep groove lined by phosphate groups from the 23S rRNA (B). (C) π-stacking interactions between arginine side chains (green) of Bac7(1–16) and 23S rRNA bases contribute to much of the binding and are reinforced through further packing against aliphatic side chains (blue). (D and E) Water-mediated contacts between the peptide and the ribosome are also proposed to occur further down the exit tunnel, in addition to direct hydrogen bonding interactions between the two.
First, the N-terminal region of Bac7(1–16) forms multiple hydrogen bonds and salt bridges with the A-tRNA binding pocket of the ribosome (Figure 2A and B). In particular, the compact structure formed by Arg2, Arg4 and Arg6 provides a positively charged N-terminal anchor that displaces two magnesium ions from a deep groove lined by 23S rRNA residues C2452, A2453 and G2454 on one side, and residues U2493 and G2494 on the other (Figure 2B). This groove differs from the standard A-form RNA major groove in that it occurs between two unpaired, antiparallel strands of the 23S rRNA. Consequently, the compact arginine structure at the N-terminus of Bac7(1–16) is ideally sized and shaped to fit into this groove and the resulting interaction is likely to be specific in spite of its simple electrostatic nature. Further contacts in this region are likely to increase the specificity of Bac7(1–16) for the ribosome, such as the two hydrogen bonds between the side chain of Arg1 and 23S rRNA residues U2555 and C2556, and four hydrogen bonds between the backbone of Bac7(1–16) residues Arg2-Arg4 and 23 rRNA residues U2492, U2493 and C2573 (Figure 2A).
Second, the unusually high arginine (50%) and proline (37.5%) content of Bac7(1–16) restricts the types of contacts that this peptide can establish with the ribosome. This results in π-stacking interactions between the side chains of Arg2, Arg9, Arg12, Arg14 and Arg16 and exposed bases of 23S rRNA residues C2573, C2452/U2504, C2610, C2586 and A2062, respectively. Additional rigidity within the peptide is provided through the packing of Arg1 against Ile3 and Arg9 against Leu10, and through the compact arginine stack described above (Figure 2C).
Third, numerous possible hydrogen bonds can be established between the backbone of Bac7(1–16) and the ribosome (Figure 2A, D and E), including many indirect interactions via ordered solvent molecules (Figure 2D and E). Many of the water-mediated contacts suggested for Tth70S-Bac7 are likely to occur with oncocin, even though the lower resolution of the earlier Tth70S-Onc112 structures precluded the modelling of any water molecules (20,21). In addition, interactions such as those between 23S rRNA residue U2506 and the backbone of Bac7(1–16) residues Arg9 and Leu10 were also proposed to occur between the Onc112 peptide and the ribosome (20,21).
Bac7 and Onc112 compete with erythromycin for ribosome binding
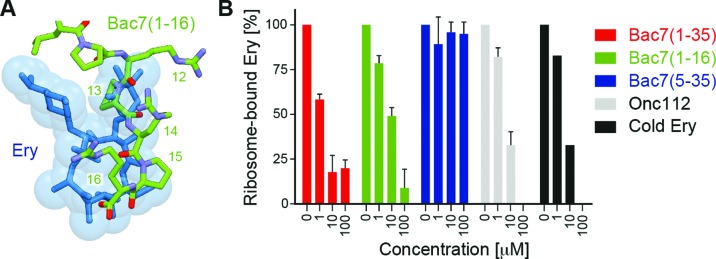
The C-terminal residues 12–16 of Bac7(1–16) overlap with the binding site of the macrolide antibiotic erythromycin on the bacterial ribosome (40,41), in particular with the region occupied by the cladinose sugar and part of the lactone ring (Figure 3A). Consistently, we could demonstrate that Bac7(1–16) and Bac7(1–35) efficiently compete with the binding of radiolabelled erythromycin to the 70S ribosome (Figure 3B). Similarly, Onc112 also efficiently competed with erythromycin (Figure 3B), as expected based on the similarity in binding mode with the ribosome for these regions of Onc112 and Bac7 (Figure 1B). In contrast, Bac7(5–35) was a poor competitor of erythromycin (Figure 3B), indicating that the highly cationic N-terminus of Bac7 and its interaction with the A-tRNA binding pocket (Figure 2B) are important for high affinity binding of Bac7 to the ribosome. Indeed, Bac7 derivatives lacking the first four N-terminal residues (RRIR), Bac7(5–35) and Bac7(5–23), exhibit dramatically reduced minimal inhibitory concentrations (MIC) against Gram-negative strains, such as E. coli, as well as Salmonella typhimurium (6). We note, however, that the internalization of Bac7(5–35) into bacteria is reduced, indicating that the N-terminal RRIR motif also plays an important role for cell penetration (11).
Figure 3.
Competition between Bac7 derivatives and erythromycin. (A) Superimposition of the binding site of erythromycin (blue) (40,41) with residues 11–16 of Bac7(1–16) (green). (B) A filter binding assay was used to monitor competition between radiolabelled [14C]-erythromycin and increasing concentrations (1–100 μM) of Bac7(1–35) (red), Bac7(1–16) (green), Bac7(5–35) (blue), Onc112 (grey) and cold (non-radioactive) erythromycin (ery, black).
Bac7 allows initiation, but prevents translation elongation
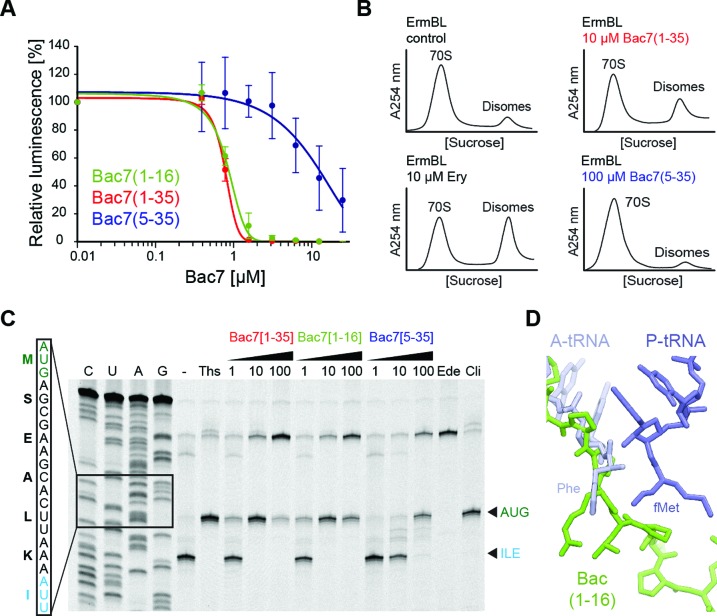
Consistent with the erythromycin binding assays and in agreement with previous results (Figure 4A) (19), we observed that Bac7(1–35) inhibits the production of luciferase with an IC50 of 1 μM in an E. coli in vitro translation system, similar to MetI and Pyr (Supplementary Figure S1), as well as that observed previously for Onc112 (20,21). Bac7(1–16) was an equally potent inhibitor as Bac7(1–35), consistent with the similar MICs observed for these two derivatives (6,10,11). In contrast, Bac7(5–35) inhibited in vitro translation with an IC50 of 10 μM, i.e. 10-fold higher than observed for Bac7(1–16) or Bac7(1–35), indicating that the reduced affinity for the ribosome, together with reduced cellular uptake (11), results in the higher MIC of the Bac7(5–35) derivative (6,42).
Figure 4.
Mechanism of action of Bac7 on the ribosome. (A) Effects of increasing concentrations of Bac7 derivatives Bac7(1–16) (green), Bac7(1–35) (red) and Bac7(5–35) (blue) on the luminescence resulting from the in vitro translation of firefly luciferase (Fluc) using an Escherichia coli lysate-based system. The error bars represent the standard deviation from the mean for triplicate experiments and the luminescence is normalized relative to that measured in the absence of peptide, which was assigned as 100%. (B) Sucrose gradient profiles to monitor disome formation from in vitro translation of the 2XErmBL mRNA in the absence (control) or presence of 20 μM erythromycin (Ery), 10 μM Bac7(1–35) (red) or 100 μM Bac7(5–35) (blue). (C) Toe-printing assay performed in the absence (−) or presence of increasing concentrations (1, 10, 100 μM) of Bac7(1–35), Bac7(1–16) or Bac7(5–35), or 100 μM thiostrepton (Ths), 50 μM edeine (Ede) or 50 μM clindamycin (Cli). Sequencing lanes for C, U, A and G and the sequence surrounding the toe-print bands (arrowed) when ribosomes accumulate at the AUG start codon (green, initiation complex) or the isoleucine codon (blue, stalled elongation complex) are included for reference. (D) Structural comparison of Phe-tRNAPhe (slate) in the A-site and fMet-tRNAiMet in the P-site (blue) (26) with the binding site of Bac7(1–16) (green).
Next we investigated the mechanism of inhibition by Bac7 using two in vitro translation assays. First, we compared the effect of Bac7(1–35) and Bac7(5–35) on the stabilization of disomes formed upon the stalling of ribosomes on a dicistronic mRNA (in this case 2XErmBL mRNA), as measured by sucrose gradient centrifugation (21,38,39). In the absence of inhibitor, the majority of ribosomes are present as 70S monosomes (control in Figure 4B), whereas the presence of erythromycin leads to translational arrest of the ribosomes on both cistrons of the 2XErmBL mRNA, thereby generating the expected disome peaks (Ery in Figure 4B). Consistent with the in vitro translation assays (Figure 4A), translation inhibition and thus disome formation was observed in the presence of 10 μM Bac7(1–35), whereas even 100 μM of Bac7(5–35) did not produce significant disomes (Figure 4B). These findings suggest that Bac7(1–35) but not Bac7(5–35) stabilizes an arrested ribosome complex, as observed previously for Onc112 (21).
Second, to monitor the exact site of translation inhibition of the Bac7 derivatives, we employed a toeprinting assay, which uses reverse transcription from the 3′ end of an mRNA to determine the exact location of the ribosomes that are translating it (35). In the absence of inhibitor, ribosomes initiated at the AUG start codon of the mRNA, translated through the open reading frame and ultimately became stalled on an isoleucine codon (Figure 4C) due to the omission of isoleucine from the translation mix. In the presence of thiostrepton or clindamycin, ribosomes accumulated at the AUG codon (Figure 4C), since these antibiotics prevent delivery and/or accommodation of aminoacyl-tRNA at the A-site directly following initiation (43). Similar results were observed when using the Bac7(1–35) and Bac7(1–16) derivatives, such that complete inhibition of translation elongation was observed at a peptide concentration of 10 μM (Figure 4C). These findings suggest that like Onc112 (21), Bac7 allows subunit joining and fMet-tRNAiMet binding, but prevents accommodation of the first aminoacyl-tRNA at the A-site, as suggested by the overlap in the binding site of Bac7 and the CCA-end of an A-tRNA (Figure 4D). Curiously, the toeprint for ribosomes stalled during initiation became weaker at 100 μM of Bac7(1–16) and Bac7(1–35) and the signal for the full-length mRNA became stronger, similar to the effect observed when the antibiotic edeine was used (Figure 4C). Edeine prevents 70S initiation complex formation by destabilizing fMet-tRNAiMet binding to the 30S subunit (43). Thus, Bac7 may have a similar effect when high cytosolic concentrations are achieved through active uptake into the cell, possibly due to the presence of non-specific interactions with the ribosome. In contrast to Bac7(1–16) and Bac7(1–35), Bac7(5–35) only stabilized the initiation complex at a much higher concentration (100 μM) (Figure 4C). This is consistent with a reduced affinity of Bac7(5–35) for the ribosome and reinforces the critical role played by the first four residues of Bac7 in its inhibitory activity (Figure 1A) (6,42).
Bac7 inhibits eukaryotic translation in vitro
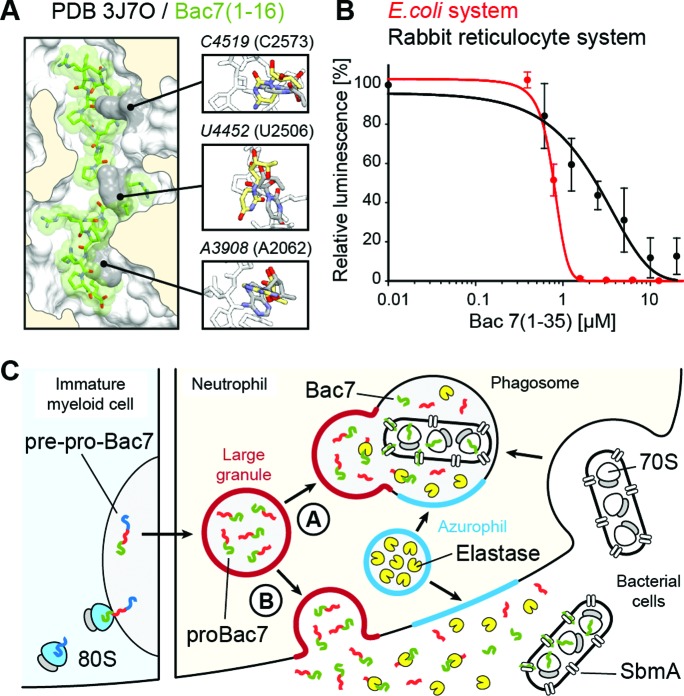
Bac7(1–35) is internalized by mammalian cells (42,44), yet no toxicity has been observed, even at concentrations well above those effective against microbes (12,13,42), raising the question as to whether Bac7 binds to eukaryotic cytosolic ribosomes. A comparison of the binding site of Bac7(1–16) on the bacterial 70S ribosome with the equivalent region of a mammalian 80S ribosome reveals that the rRNA nucleotide sequence is highly conserved. Structurally, the conformation of three 25S rRNA nucleotides, C4519 (C2573), U4452 (U2506) and A3908 (A2602), would be expected to preclude Bac7(1–16) from binding to the mammalian ribosome (Figure 5A). Nevertheless, these nucleotides are highly mobile and adopt different conformations depending on the functional state of the ribosome (26,39,45,46), suggesting that conformational rearrangements of these nucleotides could allow Bac7(1–16) binding. Indeed, we observed that increasing concentrations of Bac7(1–35) inhibited in vitro translation using a rabbit reticulocyte system (Figure 5B). Bac7(1–35) exhibited an IC50 of 2.5 μM, only 2.5-fold higher than that observed in the E. coli in vitro translation system (Figure 5B). The excellent inhibitory activity of Bac7(1–35) on mammalian ribosomes, combined with its lack of toxicity on mammalian cells (42), would be consistent with a mechanism of internalization via an endocytotic process (42) to ensure that Bac7 minimizes contact with the mammalian cytosolic ribosomes.
Figure 5.
Specificity of Bac7 for bacterial and eukaryotic ribosomes. (A) Superimposition of Bac7(1–16) (green) onto a mammalian 80S ribosome (PDB ID: 3J7O) (47) on the basis of the 23S and 25S rRNA chains in the corresponding structures, with inset illustrating three rRNA nucleotides whose conformation differs in the 80S (grey) and Tth70S-Bac7 (yellow) structures. (B) Effect of increasing concentrations of Bac7(1–35) on the luminescence resulting from the in vitro translation of firefly luciferase (Fluc) using an Escherichia coli lysate-based system (red) or rabbit reticulocyte-based system (black). The error bars represent the standard deviation from the mean for triplicate experiments and the fluorescence is normalized relative to that measured in the absence of peptide, which was assigned as 100%. (C) Model for the targeting of proBac7 to large granules and its processing by elastase to yield active Bac7 peptide. The latter is transported through the bacterial inner membrane by the SbmA transporter and binds within the tunnel of bacterial ribosomes to inhibit translation.
DISCUSSION
Our finding that Bac7 is active against eukaryotic translation, together with the current literature, allows us to present a model that explains how and why the mammalian cell prevents the active Bac7 peptide from being present in the cytoplasm (Figure 5C). Bac7 is produced by immature myeloid cells as a pre-pro-Bac7 precursor that is targeted to large granules, where it is stored as pro-Bac7 in differentiated neutrophils (48). The inactive proBac7 is cleaved by elastase, a serine protease that is present in azurophil granules, either upon (A) fusion with the phagosome, or (B) exocytosis and release into the extracellular matrix (Figure 5C) (48,49). The resulting activated Bac7 peptide can then enter into the bacterial cell through the SbmA transporter (10), where it subsequently binds to the ribosome to inhibit translation (Figure 5C) (19). Our structure of the Tth70S–Bac7 complex reveals specifically how Bac7 interacts with the bacterial ribosome (Figures 1 and 2) and inhibits translation by allowing initiation but preventing translation elongation (Figure 3). Although the overall mechanism of action of Bac7 is similar to that of insect-derived AMPs like oncocin (20,21), the high arginine content of Bac7 leads to a distinct mode of binding to the ribosome, namely through electrostatic and stacking interactions with the backbone and bases of 23S rRNA nucleotides, respectively (Figure 2C). It will be interesting to see whether such interactions are the basis for the translational arrest that has been observed when the ribosome translates a nascent polypeptide chain bearing positively charged arginine residues (50,51).
Supplementary Material
Acknowledgments
We thank the staff at the SOLEIL synchrotron (beamlines PROXIMA-2A and PROXIMA-1) for help during data collection and B. Kauffmann and S. Massip at the Institut Européen de Chimie et Biologie for help with crystal freezing and screening.
ACCESSION NUMBERS
PDB IDs: 5F8K, 5FDU, 5FDV.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale pour la Recherche [ANR-14-CE09–0001 to C.A.I., D.N.W.]; Conseil Régional d'Aquitaine [2012-13-01-009 to C.A.I.]; European Union [PCIG14-GA-2013-631479 to C.A.I.]; Deutsche Forschungsgemeinschaft [FOR1805, WI3285/4-1, GRK1721 to D.N.W.]; Fondo Ricerca di Ateneo [FRA2014 to M.S.]; Institut National de la Santé et de la Recherche Médicale Pre-doctoral Fellowship (to A.C.S.); Conseil Régional d'Aquitaine Pre-doctoral Fellowship (to A.C.S.). Funding for open access charge: Institut National de la Santé et de la Recherche Médicale [PCIG14-GA-2013-631479 to C.A.I.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 2.Scocchi M., Tossi A., Gennaro R. Proline-rich antimicrobial peptides: converging to a non-lytic mechanism of action. Cell. Mol. Life Sci. 2011;68:2317–2330. doi: 10.1007/s00018-011-0721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W., Tailhades J., O'Brien-Simpson N.M., Separovic F., Otvos L., Jr, Hossain M.A., Wade J.D. Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. Amino Acids. 2014;46:2287–2294. doi: 10.1007/s00726-014-1820-1. [DOI] [PubMed] [Google Scholar]
- 4.Ebbensgaard A., Mordhorst H., Overgaard M.T., Nielsen C.G., Aarestrup F.M., Hansen E.B. Comparative evaluation of the antimicrobial activity of different antimicrobial peptides against a range of pathogenic bacteria. PLoS One. 2015;10:e0144611. doi: 10.1371/journal.pone.0144611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernysh S., Cociancich S., Briand J.P., Hetru C., Bulet P. The inducible antibacterial peptides of the Hemipteran insect Palomena prasina: identification of a unique family of proline rich peptides and of a novel insect defensin. J. Insect. Physiol. 1996;42:81–89. [Google Scholar]
- 6.Benincasa M., Scocchi M., Podda E., Skerlavaj B., Dolzani L., Gennaro R. Antimicrobial activity of Bac7 fragments against drug-resistant clinical isolates. Peptides. 2004;25:2055–2061. doi: 10.1016/j.peptides.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Chan Y.R., Zanetti M., Gennaro R., Gallo R.L. Anti-microbial activity and cell binding are controlled by sequence determinants in the anti-microbial peptide PR-39. J. Invest. Dermatol. 2001;116:230–235. doi: 10.1046/j.1523-1747.2001.01231.x. [DOI] [PubMed] [Google Scholar]
- 8.Frank R.W., Gennaro R., Schneider K., Przybylski M., Romeo D. Amino acid sequences of two proline-rich bactenecins. Antimicrobial peptides of bovine neutrophils. J. Biol. Chem. 1990;265:18871–18874. [PubMed] [Google Scholar]
- 9.Tokunaga Y., Niidome T., Hatakeyama T., Aoyagi H. Antibacterial activity of bactenecin 5 fragments and their interaction with phospholipid membranes. J. Pept. Sci. 2001;7:297–304. doi: 10.1002/psc.317. [DOI] [PubMed] [Google Scholar]
- 10.Mattiuzzo M., Bandiera A., Gennaro R., Benincasa M., Pacor S., Antcheva N., Scocchi M. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 2007;66:151–163. doi: 10.1111/j.1365-2958.2007.05903.x. [DOI] [PubMed] [Google Scholar]
- 11.Podda E., Benincasa M., Pacor S., Micali F., Mattiuzzo M., Gennaro R., Scocchi M. Dual mode of action of Bac7, a proline-rich antibacterial peptide. Biochim. Biophys. Acta. 2006;1760:1732–1740. doi: 10.1016/j.bbagen.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Benincasa M., Pelillo C., Zorzet S., Garrovo C., Biffi S., Gennaro R., Scocchi M. The proline-rich peptide Bac7 (1–35) reduces mortality from Salmonella typhimurium in a mouse model of infection. BMC Microbiol. 2010;10:178. doi: 10.1186/1471-2180-10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghiselli R., Giacometti A., Cirioni O., Circo R., Mocchegiani F., Skerlavaj B., D'Amato G., Scalise G., Zanetti M., Saba V. Neutralization of endotoxin in vitro and in vivo by Bac7(1–35), a proline-rich antibacterial peptide. Shock. 2003;19:577–581. doi: 10.1097/01.shk.0000055236.26446.c9. [DOI] [PubMed] [Google Scholar]
- 14.Krizsan A., Knappe D., Hoffmann R. Influence of yjiL and upstream genes on the antibacterial activity of proline-rich antimicrobial peptides overcoming Escherichia coli resistance induced by the missing SbmA transporter system. Antimicrob. Agents Chemother. 2015;59:5992–5998. doi: 10.1128/AAC.01307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otvos L., O I., Rogers M.E., Consolvo P.J., Condie B.A., Lovas S., Bulet P., Blaszczyk-Thurin M. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry. 2000;39:14150–14159. doi: 10.1021/bi0012843. [DOI] [PubMed] [Google Scholar]
- 16.Czihal P., Knappe D., Fritsche S., Zahn M., Berthold N., Piantavigna S., Müller U., Van Dorpe S., Herth N., Binas A., et al. Api88 is a novel antibacterial designer peptide to treat systemic infections with multidrug-resistant gram-negative pathogens. ACS Chem. Biol. 2012;7:1281–1291. doi: 10.1021/cb300063v. [DOI] [PubMed] [Google Scholar]
- 17.Krizsan A., Volke D., Weinert S., Sträter N., Knappe D., Hoffmann R. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70 S ribosome. Angew. Chem. Int. Ed. 2014;53:12236–12239. doi: 10.1002/anie.201407145. [DOI] [PubMed] [Google Scholar]
- 18.Scocchi M., Lüthy C., Decarli P., Mignogna G., Christen P., Gennaro R. The proline-rich antibacterial peptide Bac7 binds to and inhibits in vitro the molecular chaperone DnaK. Int. J. Pept. Res. Therapeut. 2009;15:147–155. [Google Scholar]
- 19.Mardirossian M., Grzela R., Giglione C., Meinnel T., Gennaro R., Mergaert P., Scocchi M. The host antimicrobial peptide Bac71–35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem. Biol. 2014;21:1639–1647. doi: 10.1016/j.chembiol.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Roy R.N., Lomakin I.B., Gagnon M.G., Steitz T.A. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat. Struct. Mol. Biol. 2015;22:466–469. doi: 10.1038/nsmb.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seefeldt A.C., Nguyen F., Antunes S., Perebaskine N., Graf M., Arenz S., Inampudi K.K., Douat C., Guichard G., Wilson D.N., et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Mol. Biol. 2015;22:470–475. doi: 10.1038/nsmb.3034. [DOI] [PubMed] [Google Scholar]
- 22.Guida F., Benincasa M., Zahariev S., Scocchi M., Berti F., Gennaro R., Tossi A. Effect of size and N-terminal residue characteristics on bacterial cell penetration and antibacterial activity of the proline-rich peptide Bac7. J. Med. Chem. 2015;58:1195–1204. doi: 10.1021/jm501367p. [DOI] [PubMed] [Google Scholar]
- 23.Selmer M., Dunham C.M., Murphy F.V., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt E., Blanquet S., Mechulam Y. Crystallization and preliminary X-ray analysis of Escherichia coli methionyl-tRNAMetf formyltransferase complexed with formyl-methionyl-tRNAMetf. Acta Crystallogr. D. 1999;55:332–334. doi: 10.1107/S0907444998011780. [DOI] [PubMed] [Google Scholar]
- 25.Polikanov Y.S., Blaha G.M., Steitz T.A. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polikanov Y.S., Steitz T.A., Innis C.A. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014;21:787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polikanov Y.S., Melnikov S.V., Soll D., Steitz T.A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015;22:342–344. doi: 10.1038/nsmb.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou F.C., Echols N., Terwilliger T.C., Das R. RNA structure refinement using the ERRASER-phenix pipeline. Methods Mol. Biol. 2016;1320:269–282. doi: 10.1007/978-1-4939-2763-0_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starosta A.L., Karpenko V.V., Shishkina A.V., Mikolajka A., Sumbatyan N.V., Schluenzen F., Korshunova G.A., Bogdanov A.A., Wilson D.N. Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem. Biol. 2010;17:504–514. doi: 10.1016/j.chembiol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Hartz D., McPheeters D.S., Traut R., Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 36.Starosta A.L., Lassak J., Peil L., Atkinson G.C., Virumäe K., Tenson T., Remme J., Jung K., Wilson D.N. Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Res. 2014;42:10711–10719. doi: 10.1093/nar/gku768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polikanov Y.S., Starosta A.L., Juette M.F., Altman R.B., Terry D.S., Lu W., Burnett B.J., Dinos G., Reynolds K.A., Blanchard S.C., et al. Distinct tRNA accommodation intermediates observed on the ribosome with the antibiotics hygromycin A and A201A. Mol. Cell. 2015;58:832–844. doi: 10.1016/j.molcel.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arenz S., Meydan S., Starosta A.L., Berninghausen O., Beckmann R., Vazquez-Laslop N., Wilson D.N. Drug sensing by the ribosome induces translational arrest via active site perturbation. Mol. Cell. 2014;56:446–452. doi: 10.1016/j.molcel.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arenz S., Ramu H., Gupta P., Berninghausen O., Beckmann R., Vazquez-Laslop N., Mankin A.S., Wilson D.N. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat. Commun. 2014;5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulkley D., Innis C.A., Blaha G., Steitz T.A. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. 2010;107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunkle J.A., Xiong L., Mankin A.S., Cate J.H. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomasinsig L., Skerlavaj B., Papo N., Giabbai B., Shai Y., Zanetti M. Mechanistic and functional studies of the interaction of a proline-rich antimicrobial peptide with mammalian cells. J. Biol. Chem. 2006;281:383–391. doi: 10.1074/jbc.M510354200. [DOI] [PubMed] [Google Scholar]
- 43.Wilson D.N. The A-Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 44.Sadler K., Eom K.D., Yang J.L., Dimitrova Y., Tam J.P. Translocating proline-rich peptides from the antimicrobial peptide bactenecin 7. Biochemistry. 2002;41:14150–14157. doi: 10.1021/bi026661l. [DOI] [PubMed] [Google Scholar]
- 45.Schmeing T.M., Huang K.S., Strobel S.A., Steitz T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 46.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 47.Voorhees R.M., Fernandez I.S., Scheres S.H., Hegde R.S. Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zanetti M., Litteri L., Gennaro R., Horstmann H., Romeo D. Bactenecins, defense polypeptides of bovine neutrophils, are generated from precursor molecules stored in the large granules. J. Cell Biol. 1990;111:1363–1371. doi: 10.1083/jcb.111.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scocchi M., Skerlavaj B., Romeo D., Gennaro R. Proteolytic cleavage by neutrophil elastase converts inactive storage proforms to antibacterial bactenecins. Eur. J. Biochem. 1992;209:589–595. doi: 10.1111/j.1432-1033.1992.tb17324.x. [DOI] [PubMed] [Google Scholar]
- 50.Dimitrova L.N., Kuroha K., Tatematsu T., Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu J., Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.