Abstract
The function of the vast majority of mammalian long noncoding (lnc) RNAs remains unknown. Here, analysis of a highly abundant mammalian lncRNA, OIP5-AS1, known as cyrano in zebrafish, revealed that OIP5-AS1 reduces cell proliferation. In human cervical carcinoma HeLa cells, the RNA-binding protein HuR, which enhances cell proliferation, associated with OIP5-AS1 and stabilized it. Tagging OIP5-AS1 with MS2 hairpins to identify associated microRNAs revealed that miR-424 interacted with OIP5-AS1 and competed with HuR for binding to OIP5-AS1. We further identified a ‘sponge’ function for OIP5-AS1, as high levels of OIP5-AS1 increased HuR-OIP5-AS1 complexes and prevented HuR interaction with target mRNAs, including those that encoded proliferative proteins, while conversely, lowering OIP5-AS1 increased the abundance of HuR complexes with target mRNAs. We propose that OIP5-AS1 serves as a sponge or a competing endogenous (ce)RNA for HuR, restricting its availability to HuR target mRNAs and thereby repressing HuR-elicited proliferative phenotypes.
INTRODUCTION
Post-transcriptional regulatory processes critically influence eukaryotic gene expression programs. Pre-mRNA splicing and maturation, as well as mRNA transport, stability, storage, editing and translation are all subject to post-transcriptional gene regulation through the actions of noncoding RNAs (ncRNAs), including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), and RNA-binding proteins (RBPs) (1–5).
Despite the fact that ncRNAs were discovered decades ago (6–8), only with the recent advent of techniques for deep genomic analysis (e.g. tiling arrays, RNA sequencing) have we begun to appreciate that tens of thousands of ncRNAs are pervasively transcribed from >90% of genomes of eukaryotic species from yeast to human. In their mature form, some ncRNAs are small (e.g. miRNAs, small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs)) (9). Among these, miRNAs robustly regulate gene expression programs and have been studied in detail. They are transcribed as primary (pri-)miRNA transcripts and processed into miRNA precursors (pre-miRNAs) by the microprocessor complex, which includes the RNase Drosha. Following export to the cytoplasm, pre-miRNAs are cleaved by Dicer to form mature miRNAs, which are then loaded into the RNA-inducible silencing complex (RISC). MicroRNA-RISC complexes, which include the protein Argonaute 2 (AGO2), can target specific mRNAs, mainly through partial complementarity with the 3′-untranslated region (UTR) of the mRNA. In turn, this interaction leads to lower stability and/or translation of the target mRNA (10).
LncRNAs (>200 nt in length) also modulate gene expression programs through transcriptional and post-transcriptional mechanisms (5,11,12). Through their impact on gene expression patterns, lncRNAs are emerging as key regulators of cellular processes (proliferation, apoptosis, stress response, differentiation, senescence) as well as physiologic and pathologic processes (immune adaptation, cancer, neurodegeneration, cardiovascular disease and aging) (13–19). Akin to mRNAs, lncRNAs are transcribed as precursor transcripts and are subject to splicing and maturation in the nucleus, as well as cytoplasmic export, editing, transport and decay. In both the nucleus and the cytoplasm, lncRNAs are believed to control gene expression by interacting with chromatin regulators, transcriptional activators and repressors, chromosomal DNA, microRNAs, RBPs and mRNAs (5,15,20). However, the full spectrum of functions for the vast class of lncRNAs is poorly understood.
Ulitsky et al. (21) found evolutionarily conserved lncRNAs with biological significance in vertebrates. Transcriptomic analyses from zebrafish to human identified OIP5-AS1 (OIP5 antisense transcript 1) as the mammalian homolog of the zebrafish transcript cyrano. OIP5-AS1 is highly expressed in the nervous system and is important for controlling neurogenesis during development (21). While the sequence conservation for OIP5-AS1 is limited among the genomes from vertebrates examined, its gene structure and localization between the CHP1 and OIP5 genes (OIP5-AS1 is transcriptionally divergent from gene OIP5) are highly conserved (21,22).
To investigate in molecular detail the function of OIP5-AS1, we examined its interaction partners in HeLa (human cervical carcinoma) cells. Along with triggering suppression of HeLa cell proliferation, OIP5-AS1 associated with HuR, an interaction that rendered OIP5-AS1 stable. HuR is the ubiquitous member of the Hu/ELAV (human/embryonic lethal abnormal vision) RBP family and is predominantly nuclear, but its export to the cytoplasm is linked to the stabilization and/or translation of many target mRNAs, which typically bear U-rich 3′ UTRs (23–25). HuR is highly abundant in cancer and numerous HuR target mRNAs encode proteins that promote different aspects of tumorigenesis, such as cell proliferation, angiogenesis, cell survival, invasion and metastasis (26–29). Importantly, some of HuR actions depend on its interaction with miRNAs (30). For example, HuR can compete with miRNA-RISC for binding and regulation of targets (e.g. CAT1 and TOP2A mRNAs (31,32)), but it can also recruit miRNAs to certain target transcripts, as shown for MYC mRNA and for LINCRNAP21 (33,34). Our findings reveal that the interaction of OIP5-AS1 with HuR was competed by miRNA miR-424, thereby shifting HuR availability from OIP5-AS1 to target mRNAs. The interaction between OIP5-AS1 and HuR was found to sequester HuR away from target mRNAs, leading us to propose that OIP5-AS1 was a ‘sponge’ for HuR. Accordingly, lowering HuR enhanced miR-424 binding to OIP5-AS1, while overexpression of miR-424 reduced HuR binding to OIP5-AS1, in turn, freeing up HuR for binding to target mRNAs encoding proliferative proteins. We propose that two trans-acting regulators, HuR and miR-424, compete for binding lncRNA OIP5-AS1, and the balance of this interaction directly influences cell phenotypes controlled by HuR.
MATERIALS AND METHODS
Cell culture, transfection, analysis and fractionation
Human cervical carcinoma HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) and antibiotics. siRNA duplexes in Table 1 were transfected at 50 nM final concentration using Lipofectamine 2000 (Invitrogen). Cells were counted using an automatic cell counter (Bio-Rad) after staining with 0.4% Trypan Blue (Sigma). [3H]-thymidine (Amersham) incorporation and fluorescence-activated cell sorting (FACS) analyses were performed using standard procedures. Cellular fractionation was performed using the NE-PER nuclear and cytoplasmic extraction reagent (Thermo Fisher Scientific), following the manufacturer's protocol.
Table 1. siRNA duplexes.
| siRNA (Provider) | Sequence |
|---|---|
| Ctrl siRNA (Qiagen) | AATTCTCCGAACGTGTCACGT |
| HuR siRNA (Qiagen) | AATCTTAAGTTTCGTAAGTTA |
| TTCCTTTAAGATATATATTAA | |
| CGCAGAGATTCAGGTTCTCC | |
| OIP5-AS1 (IDT) | Sense1 rGrGrCrUrGrArGrUrUrUrCrArUrUrUrGrArArArCrArGrGTG |
| Antisense1 rCrArCrCrUrGrUrUrUrCrArArArUrGrArArArCrUrCrArGrCrCrUrU | |
| Sense2 rCrArUrGrCrArGrUrGrCrCrArUrCrUrGrArCrUrUrUrArUGG | |
| Antisense2 rCrCrArUrArArArGrUrCrArGrArUrGrGrCrArCrUrGrCrArUrGrArG | |
| Sense3 rCrArCrCrArArArCrArGrGrCrUrUrUrGrUrGrUrUrCrCrUTA | |
| Antisense3 rUrArArGrGrArArCrArCrArArArGrCrCrUrGrUrUrUrGrGrUrGrGrU | |
| miR-424 mimic (Qiagen) | CAGCAGCAAUUCAUGUUUUGAA (Cat. No. MSY0001341) |
RNA analysis
Total RNA was prepared using Trizol (Invitrogen) and analyzed by conventional reverse transcription (RT) using random hexamers and Maxima reverse transcriptase (Thermo Fisher Scientific) followed by real-time, quantitative PCR (qPCR) using target-specific primers (Table 2) and SYBR green master mix (Kapa Biosystems) in an Applied Biosystems 7300 instrument. For quantification of miRNAs (Table 3), RT was performed with Mir-XTM First-Strand synthesis kit (Clontech) and qPCR amplification was carried out using an Applied Biosystems 7900HT instrument and normalized to U6 RNA levels.
Table 2. mRNA qPCR primers.
| mRNA | Sequence |
|---|---|
| CCNA2-F | AACTTCAGCTTGTGGGCACT |
| CCNA2-R | AAACTCTGCTACTTCTGGGGG |
| CCNB1-F | CAAGCCCAATGGAAACATCT |
| CCNB1-R | GGATCAGCTCCATCTTCTGC |
| CCNB3-F | CCCATCTGAAAAGACGGGGG |
| CCNB3-R | GGGCTGGACAGGTTGACATT |
| CCND1-F | TGTTTGCAAGCAGGACTTTG |
| CCND1-R | TCATCCTGGCAATGTGAGAA |
| CCNE1-F | CCGCAGTATCCCCAGCAAAT |
| CCNE1-R | AAGGCCGAAGCAGCAAGTAT |
| CDK1-F | CGTAGCTGGGCTCTGATTGG |
| CDK1-R | TGGTAGATCCGCGCTAAAGG |
| CDK2-F | CCTGAAATCCTCCTGGGCTG |
| CDK2-R | CCCAGAGTCCGAAAGATCCG |
| HuR-F | CGCCAACTTGTACATCAGCG |
| HuR-R | TAAACGCAACCCCTCTGGAC |
| VHL-F | GACTGGACATCGTCAGGTCG |
| VHL-R | ATCCGTTGATGTGCAATGCG |
| SIRT1-F | TTGCAACAGCATCTTGCCTG |
| SIRT1-R | GTTCATCAGCTGGGCACCTA |
| PTMA-F | GAACCAAAACTTCCAAGGCCC |
| PTMA-R | GCTGGTTTGGTCATCCGAGA |
| TP53-F | AGGCCTTGGAACTCAAGGAT |
| TP53-R | TGAGTCAGGCCCTTCTGTCT |
| CDC25A-F | GAACAGCGAAGACAGCGTGA |
| CDC25A-R | AATCCAAACAAACGTGGCGG |
| WEE1-F | CGAGTGCGGCACCGATAA |
| WEE1-R | AAAGGCATCCTATGGCTCGG |
| OIP5-AS1-F | TGCGAAGATGGCGGAGTAAG |
| OIP5-AS1-R | TAGTTCCTCTCCTCTGGCCG |
| Mid-OIP5-AS1-F | TTCCAGTTTCAGCCACTACCA |
| Mid-OIP5-AS1-R | TCACAGGATGAGCCAGGATTT |
| 7SL-F | CAAAACTCCCGTGCTCATCA |
| 7SL-R | GGCTGGAGTGCAGTGGCTAT |
| 18S-F | CGAACGTCTGCCCTATCAACTT |
| 18S-R | ACCCGTGGTCACCATGGTA |
Table 3. miRNA qPCR primers.
| miRNA | Sequence |
|---|---|
| hsa-let-7b | TGAGGTAGTAGGTTGTGTGGTT |
| hsa-let-7c | TGAGGTAGTAGGTTGTATGGTT |
| hsa-let-7d | AGAGGTAGTAGGTTGCATAGTT |
| hsa-let-7e | TGAGGTAGGAGGTTGTATAGTT |
| hsa-let-7g | TGAGGTAGTAGTTTGTACAGTT |
| hsa-miR-16–1 | TAGCAGCACGTAAATATTGGCG |
| hsa-miR-16–2 | TAGCAGCACGTAAATATTGGCG |
| hsa-miR-17 | CAAAGTGCTTACAGTGCAGGTAG |
| hsa-miR-17* | ACTGCAGTGAAGGCACTTGTAG |
| hsa-miR-18a | TAAGGTGCATCTAGTGCAGATAG |
| hsa-miR-126 | TCGTACCGTGAGTAATAATGCG |
| hsa-miR-140–5p | CAGTGGTTTTACCCTATGGTAG |
| hsa-miR-183 | TATGGCACTGGTAGAATTCACT |
| hsa-miR-20a | TAAAGTGCTTATAGTGCAGGTAG |
| hsa-miR-21* | CAACACCAGTCGATGGGCTGT |
| hsa-miR-25 | CATTGCACTTGTCTCGGTCTGA |
| hsa-miR-26b | TTCAAGTAATTCAGGATAGGT |
| hsa-miR-27b | TTCACAGTGGCTAAGTTCTGC |
| hsa-miR-29a | TAGCACCATCTGAAATCGGTTA |
| hsa-miR-30b | TGTAAACATCCTACACTCAGCT |
| hsa-miR-301a | CAGTGCAATAGTATTGTCAAAGC |
| hsa-miR-424 | CAGCAGCAATTCATGTTTTGAA |
| hsa-miR-425 | AATGACACGATCACTCCCGTTGA |
| hsa-miR-452 | AACTGTTTGCAGAGGAAACTGA |
| hsa-miR-497 | CAGCAGCACACTGTGGTTTGT |
| hsa-miR-671 | AGGAAGCCCTGGAGGGGCTGGAG |
| hsa-miR-96 | TTTGGCACTAGCACATTTTTGCT |
| hsa-miR-98 | TGAGGTAGTAAGTTGTATTGTT |
| U6 | CACCACGTTTATACGCCGGTG |
Protein analysis
Total protein lysates were prepared in RIPA buffer containing protease inhibitor and 1 mM dithiothreitol (DTT). Proteins were size-separated by SDS-PAGE and transferred onto nitrocellulose membrane (Invitrogen). For Western blot analysis, primary antibodies were used that recognized HuR (1:1000), CCNA2 (1:1000), SIRT1 (1:1000), α-tubulin (TUBA, 1:2000), HSP90 (1:20 000) (all from Santa Cruz Biotechnology) and CCND1 (1:2000; from Cell Signaling). After incubation with appropriate secondary antibodies, protein signals were developed using chemiluminescence.
GST pulldown and ribonucleoprotein immunoprecipitation (RIP) analyses
For GST pulldown analysis, glutathione magnetic beads (Pierce) were incubated with cell lysate for 1 h at 4°C. For RIP analysis, agarose beads coated with protein A were pre-incubated with 2 μg each of antibody and isotype IgG for 4 h at 4°C. Cell lysates prepared in NT2 buffer containing RNase inhibitor, protease inhibitor and 1 mM DTT were incubated with pre-incubated beads at 4°C. After incubation for 1 h and washes in NT2 buffer, protein and RNA were extracted from the beads using NuPAGE sample buffer (Invitrogen) with 10% β-mercaptoethanol and Trizol (Invitrogen), respectively.
Statistical analysis
For statistical analysis of signals on Western blots, the intensities of bands from three independent experiments were quantified using ImageJ, the means ±S.E.M. were calculated, and P-values were determined using Student's t-test in each comparison. For statistical analysis of RNA levels, data from three independent qPCR results were calculated by the 2−ΔΔCt method and represented as the means ±S.D. for steady-state RNA levels, or the means ±S.E.M. for the levels of RBP-bound RNAs. P-values were determined using Student's t-test in each comparison. Only P-values lower than 0.05 were considered to be significant.
RESULTS
lncRNA OIP5-AS1/cyrano reduces proliferation of human cervical cancer cells
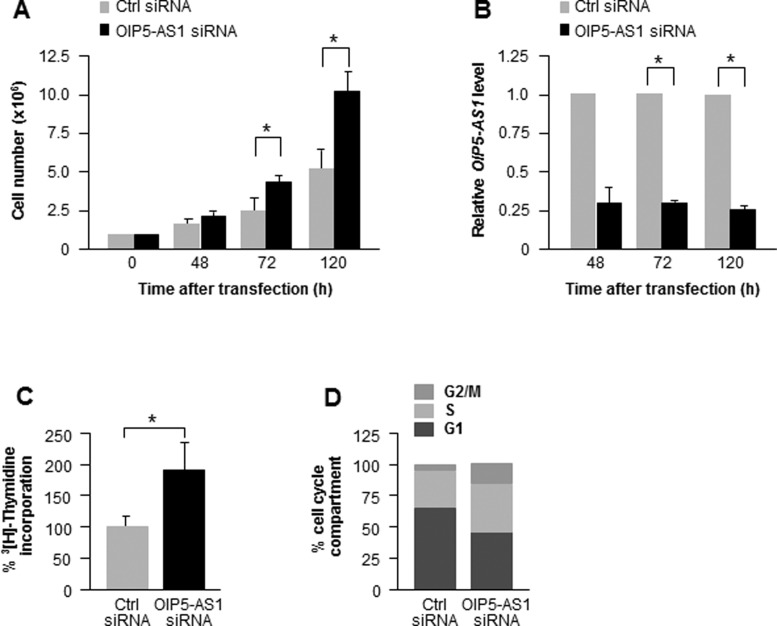
To begin examining the function of lncRNA OIP5-AS1, we silenced it in HeLa cells by transfecting a pool of siRNAs directed at OIP5-AS1 and studied the ensuing changes in cell numbers. As shown in Figure 1A, silencing OIP5-AS1 promoted cell proliferation, as assessed by counting cell numbers for up to 5 days; at the times examined, OIP5-AS1 levels were efficiently reduced, as determined by RT followed by qPCR analysis (Figure 1B). The individual siRNAs had a similar effect (Supplementary Figure S1). Analysis of [3H]-thymidine incorporation into replicating DNA confirmed that silencing OIP5-AS1 increased cell proliferation (Figure 1C), and FACS showed that silencing OIP5-AS1 increased the relative sizes of the S and G2/M cell compartments (Figure 1D). Taken together, these results indicate that lowering OIP5-AS1 enhances progression through the cell division cycle and suggest that OIP5-AS1 inhibits cell proliferation.
Figure 1.
Silencing OIP5-AS1 promotes cell proliferation. (A) HeLa cells were counted at the times indicated following transfection of siRNAs to silence OIP5-AS1. (B) Silencing efficiencies at each time point following transfection were examined by RT-qPCR analysis using total RNA; data were normalized to the levels of ACTN mRNA. (CandD) Seventy-two hours after transfection of HeLa cells with Ctrl or OIP5-AS1 siRNAs, [3H]-thymidine incorporation was measured during a 16-h period (C) and the relative percentages of HeLa cells in each cell cycle compartment were assessed by FACS analysis (D). *P< 0.05.
RNA-binding protein HuR binds to and stabilizes OIP5-AS1
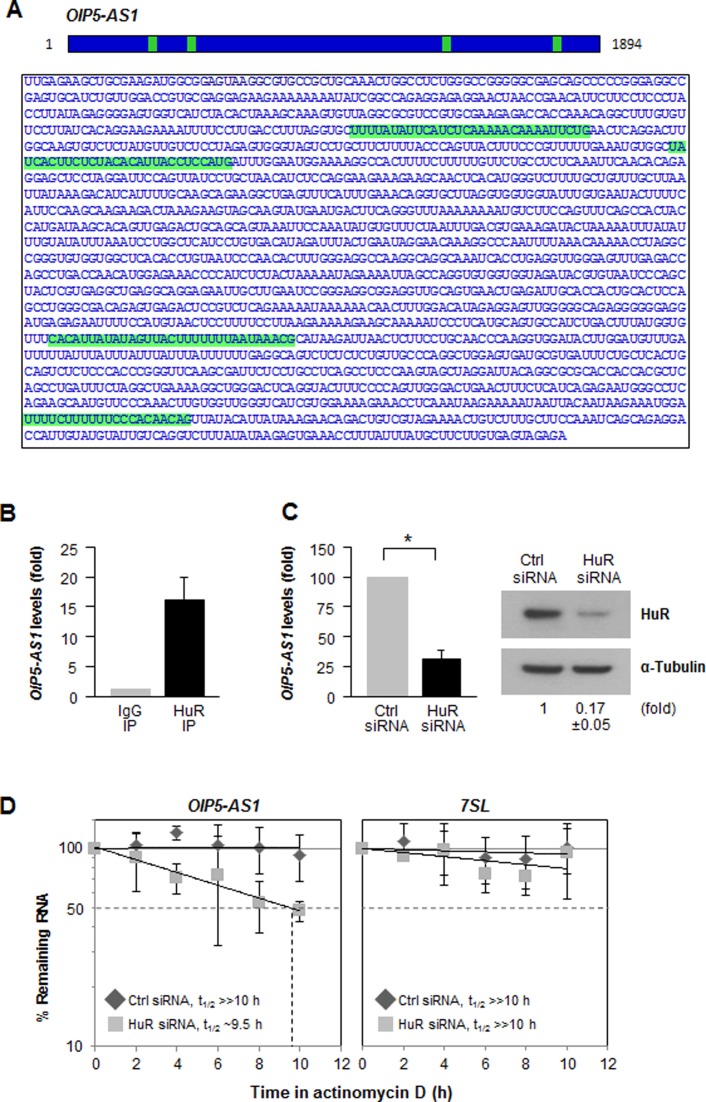
Among the vast collection of lncRNAs annotated for human sequences (9837 lncRNAs, Ensembl v72) (35,36), OIP5-AS1 was identified as the human homolog of cyrano, a lncRNA that plays a role in zebrafish development (21), and showed significant conservation in gene structure (22). OIP5-AS1 (15.6-kb long) comprises three short exons and a long fourth exon after the longest intron; the mature transcript (1.9-kb long) is shown in Figure 2A. A search for OIP5-AS1 interaction partners revealed that the RBP HuR associated with OIP5-AS1 at four different sites (24, GSM738185) (Figure 2A, green).
Figure 2.
HuR binds to lncRNA OIP5-AS1 and increases its stability. (A) HuR interaction sites (green) on OIP5-AS1, as identified using GSM738185.bed.gz. (B) RIP analysis of HuR interaction with OIP5-AS1 in HeLa cells. Following HuR immunoprecipitation (IP) or control using IgG IP, OIP5-AS1 levels were measured by RT-qPCR analysis and normalized to 7SL levels. (C) Seventy-two hours after transfecting Ctrl or HuR siRNAs, the levels of OIP5-AS1 (left) and HuR (right) were assessed by RT-qPCR and Western blot analyses, respectively. Data shown are the means and SEM from three independent experiments. (D) Cells transfected as in (C) were treated with actinomycin D to block de novo transcription and the levels of OIP5-AS1 and 7SL (a stable lncRNA) were assessed by RT-qPCR analysis and normalized to 18S rRNA levels, also quantified by RT-qPCR analysis; the half-life (t1/2) of OIP5-AS1 was measured as the time required for the levels of OIP5-AS1 to decline to 50% of their initial levels at 0 h.
We validated the interaction between HuR and OIP5-AS1 by ribonucleoprotein immunoprecipitation (RIP) analysis using anti-HuR and control IgG antibodies; after extracting RNA from the IP samples, RT-qPCR analysis was used to measure OIP5-AS1 levels, and normalized to GAPDH mRNA levels in each IP sample. OIP5-AS1 was found to be highly enriched (>15-fold) in HuR IP samples relative to IgG IP samples (Figure 2B), revealing that HuR selectively associates with OIP5-AS1. To investigate the biological significance of HuR binding to OIP5-AS1, we first examined if HuR regulates OIP5-AS1 stability. By 48 h after silencing HuR in HeLa cells, RT-qPCR analysis from total RNA revealed that OIP5-AS1 steady-state levels were reduced 3-fold (Figure 2C). To study if this reduction was due to increased OIP5-AS1 decay, HeLa cells were incubated in the presence of actinomycin D to block de novo transcription, and the half-life of OIP5-AS1 was measured as the time required to reach 50% of the RNA levels present at time 0 h (before adding actinomycin D); a very stable lncRNA, 7SL (37), was assessed side by side. Compared with control cells, where OIP5-AS1 RNA was quite stable, silencing HuR lowered OIP5-AS1 half-life to 9.5 h (Figure 2D), indicating that HuR contributed to stabilizing OIP5-AS1.
Identification of miRNAs with enhanced binding to OIP5-AS1 when HuR is silenced
To explore the mechanisms whereby HuR stabilizes OIP5-AS1, we investigated if other factors might be implicated in regulating OIP5-AS1 half-life. In silico analyses employing ‘RegRNA 2.0’ and ‘IntaRNA’ identified 30 miRNAs possibly interacting with OIP5-AS1. RIP analysis of Argonaute 2 (AGO2), an essential component of the miRNA-associated RISC, indicated that OIP5-AS1 was enriched in AGO2 complexes, as further evidence that OIP5-AS1 associated with miRNAs (Supplementary Figure S2A).
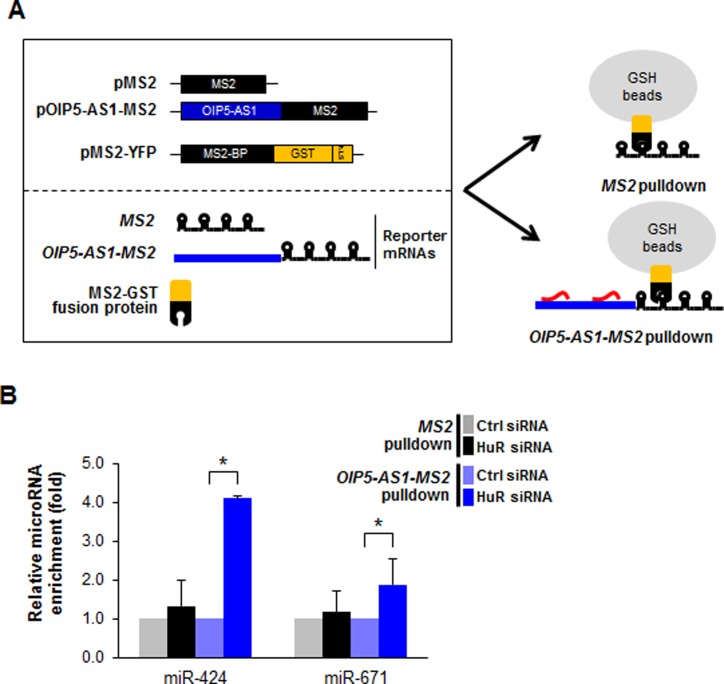
In order to study if OIP5-AS1 directly bound to these miRNAs in the cell, we prepared a vector that expressed OIP5-AS1 tagged with MS2 hairpins and coexpressed the chimeric RNA (OIP5-AS1-MS2) in HeLa cells along with a fusion protein that contained a glutathione-S-transferase domain fused to a domain that recognizes MS2 hairpins (MS2-GST). After OIP5-AS1-MS2 formed a complex with MS2-GST, miRNAs associated with the chimeric RNA were pulled down by using glutathione (GSH)-coated beads (Figure 3A). Following extraction of RNA from the beads, we employed RT-qPCR analysis to screen miRNAs predicted to have sites of interaction with OIP5-AS1, and further tested if silencing HuR (as in Figure 2C) changed the extent of interaction. Among these, nine miRNAs showed enhanced binding to OIP5-AS1-MS2 when HuR was silenced (Supplementary Figure S2B, red arrowheads). MiRNAs miR-424 and miR-671 were preferentially enriched in the OIP5-AS1-MS2 pulldown material over control transfected cells and over MS2 pulldown alone, although miR-671 was substantially less enriched in the MS2 pulldown than was miR-424 (Figure 3B, left). miR-7, a miRNA known to bind OIP5-AS1 (21) was included as a positive control and found to bind to OIP5-AS1-MS2 in pulldown experiments (data not shown). Collectively, these findings suggest that silencing HuR enhanced the access of certain miRNAs to OIP5-AS1, prompting us to hypothesize that miR-424 and HuR may compete for binding to OIP5-AS1.
Figure 3.
MiRNAs interacting with lncRNA OIP5-AS1/cyrano in the presence and the absence of HuR. (A) Schematic of constructs used for MS2 pulldown, including plasmids pMS2 (a control vector expressing MS2 RNA), pOIP5-AS1-MS2 (a vector expressing the chimeric RNA OIP5-AS1-MS2) and pMS2-GST, expressing a fusion protein (MS2-GST) that recognizes MS2 RNA tags and can be pulled down using glutathione (GSH) beads. (B) Forty-eight hours after transfection with either Ctrl or HuR siRNAs, cells were co-transfected with the plasmids in (A) and 24 h after that, cells were lysed and the lysates were analyzed by pulldown using GSH-conjugated beads. The relative interaction of miR-424 and miR-671 with OIP5-AS1-MS2 (selected among nine miRNAs that were chosen for further screening because they had predicted sites on OIP5-AS1 (Supplementary Figure S2B)) was examined in pulldown material and the impact of silencing HuR on the magnitude of the interactions was assessed. Fold changes were calculated over Ctrl siRNA group in each MS2 pulldown and normalized to U6 RNA levels. Error bars indicate SEM from three independent experiments in each pulldown.
miR-424 competes with HuR for binding OIP5-AS1
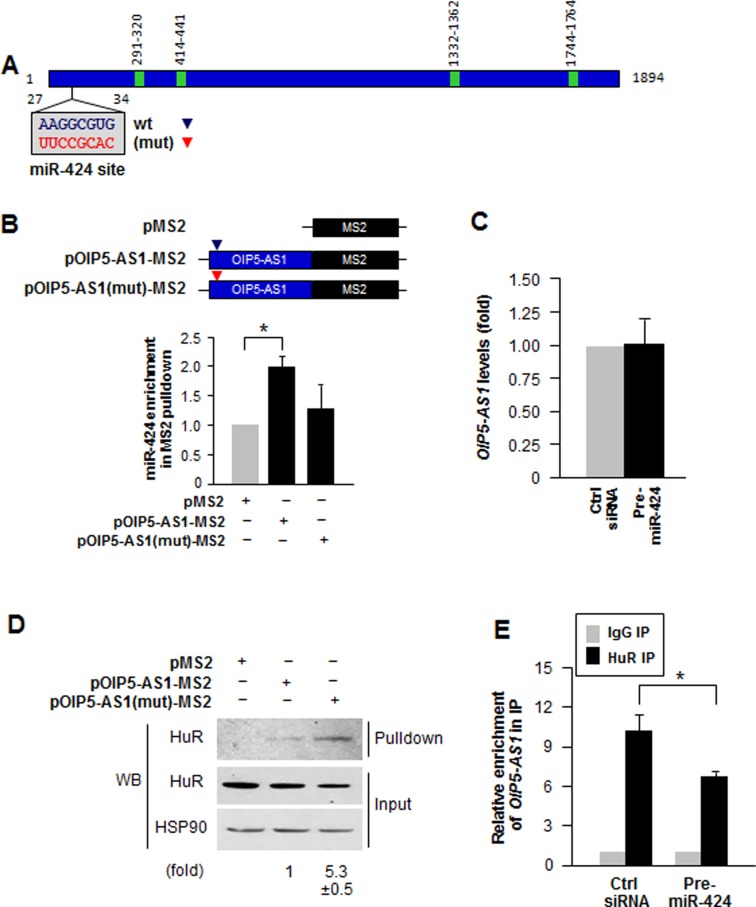
The miR-424 site on OIP5-AS1 is not located immediately adjacent to any of the HuR sites (Figure 4A). However, even though the nearest HuR site is >200 nucleotides away, folding of OIP5-AS1 could bring one or several HuR sites in proximity to the miR-424 site, such that the secondary structure of the RNA could lead to functional interactions among distal sites (31,33). To examine if miR-424 and HuR competed for binding to lncRNA OIP5-AS1, we mutagenized the miR-424 binding site on OIP5-AS1 at nucleotide positions 27–34 (Figure 4A). HeLa cells expressing the wild type (wt) OIP5-AS1-MS2 and mutated OIP5-AS1(mut)-MS2 were subjected to MS2 pulldown as described above (Figure 3A); miR-424 showed robust enrichment in wt OIP5-AS1 pulldown, but showed much less binding to OIP5-AS1(mut) (Figure 4B). Overexpression of miR-424 (Pre-miR-424) did not change OIP5-AS1 levels (Figure 4C), nor did it significantly change either OIP5-AS1 transcription rate, OIP5-AS1 interaction with AGO2 or OIP5-AS1 stability (Supplementary Figure S3).
Figure 4.
Mutation of miR-424 binding site enhances HuR binding to OIP5-AS1. (A) Schematic of the mutation of miR-424 binding site on OIP5-AS1. (B) HeLa cells transfected with plasmids pMS2, pOIP5-AS1-MS2 (wt) or pOIP5-AS1(mut)-MS2 were used for pulldown analysis using GSH beads to test the impact of mutating the OIP5-AS1 miR-424 site on miR-424 binding to the tagged OIP5-AS1. The level of miR-424 associated with each MS2-tagged RNA was measured by RT-qPCR analysis. (C) Steady-state levels of OIP5-AS1 in HeLa cells transfected with Pre-miR-424. Fold changes were relative to Ctrl siRNA transfection. (D) Cells transfected as in (B) were subjected to MS2 pulldown followed by detection of HuR levels associated with each MS2-tagged RNA by Western blot analysis; HuR and loading control HSP90 in the input material was also measured. (E) Twenty-four hours after transfecting HeLa cells with Ctrl siRNA or Pre-miR-424, the association of HuR with OIP5-AS1 was assessed by RIP analysis. The relative interaction levels were calculated by normalization with IgG IP. In B–E, errors indicate standard deviation (SD) from three independent experiments. *P < 0.05.
Conversely, analysis of the pulldown samples by Western blotting revealed that HuR was more abundant in OIP5-AS1(mut) pulldown samples (Figure 4D), suggesting that reduced miR-424 binding through mutation of the binding site on OIP5-AS1 permitted enhanced HuR binding to OIP5-AS1. To test this idea further, we overexpressed miR-424 by transfecting HeLa cells with Pre-miR-424, and measured HuR binding to OIP5-AS1 by RIP analysis. As shown, the 10-fold enrichment in HuR binding to OIP5-AS1 in control cells declined significantly to a 6-fold enrichment when miR-424 was overexpressed (Figure 4E). In summary, mutations which interfered with the access of miR-424 to OIP5-AS1 allowed enhanced HuR binding to OIP5-AS1, and conversely, when miR-424 was overexpressed, HuR binding to OIP5-AS1 was markedly reduced. These findings led us to conclude that miR-424 and HuR compete with each other for binding to OIP5-AS1.
Competitive binding of HuR and miR-424 to OIP5-AS1 affects HuR binding to target mRNAs, influences target mRNA fate
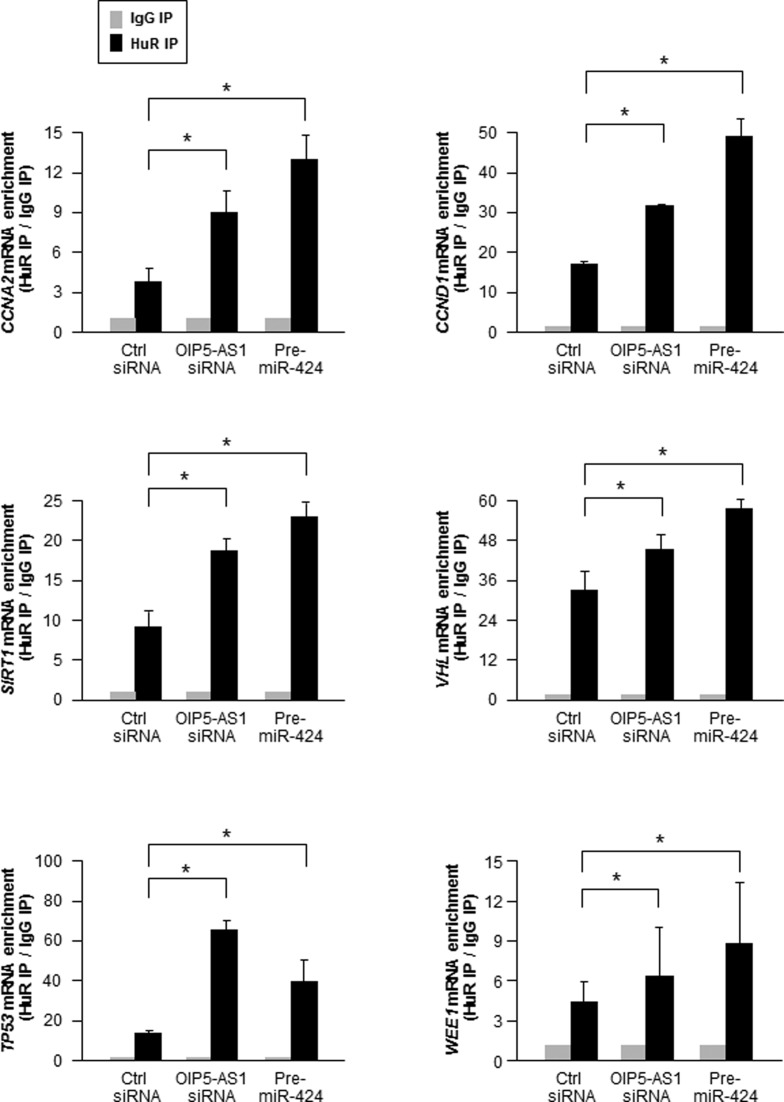
Next, we examined the consequences of HuR and miR-424 competing for binding to OIP5-AS1. Given that HuR binds to many mRNAs and that OIP5-AS1 is highly expressed in HeLa cells, we hypothesized that the interaction between OIP5-AS1 and HuR may influence the ability of HuR to bind other target mRNAs. To test this possibility, we silenced OIP5-AS1 in HeLa cells and performed RIP analysis of HuR interaction with target CCNA2 and CCND1 mRNAs (encoding cyclins A2 and D1, respectively), SIRT1 mRNA (encoding sirtuin 1), VHL and TP53 mRNAs (encoding the tumor suppressors VHL and p53), and WEE1 mRNA (encoding the cell cycle kinase WEE1). As shown in Figure 5, silencing OIP5-AS1 and overexpressing miR-424 each enhanced the interaction of HuR with target mRNAs relative to those seen in the corresponding control (Ctrl siRNA) transfections. The increased enrichments in HuR-mRNA complexes suggested that lowering OIP5-AS1 or increasing the abundance of competitor miR-424 ‘freed up’ HuR for binding to target mRNAs.
Figure 5.
Effect of silencing OIP5-AS1 or overexpression miR-424 on HuR binding to target mRNAs. By 72 h after transfecting Ctrl siRNA, OIP5-AS1 siRNA or Pre-miR-424, the relative binding of CCNA2, CCND1, SIRT1, VHL, TP53 and WEE1 mRNAs to HuR was calculated as ‘fold enrichment’ over IgG IP for each sample. Data are shown as the means and SEM from three independent experiments. *P < 0.05.
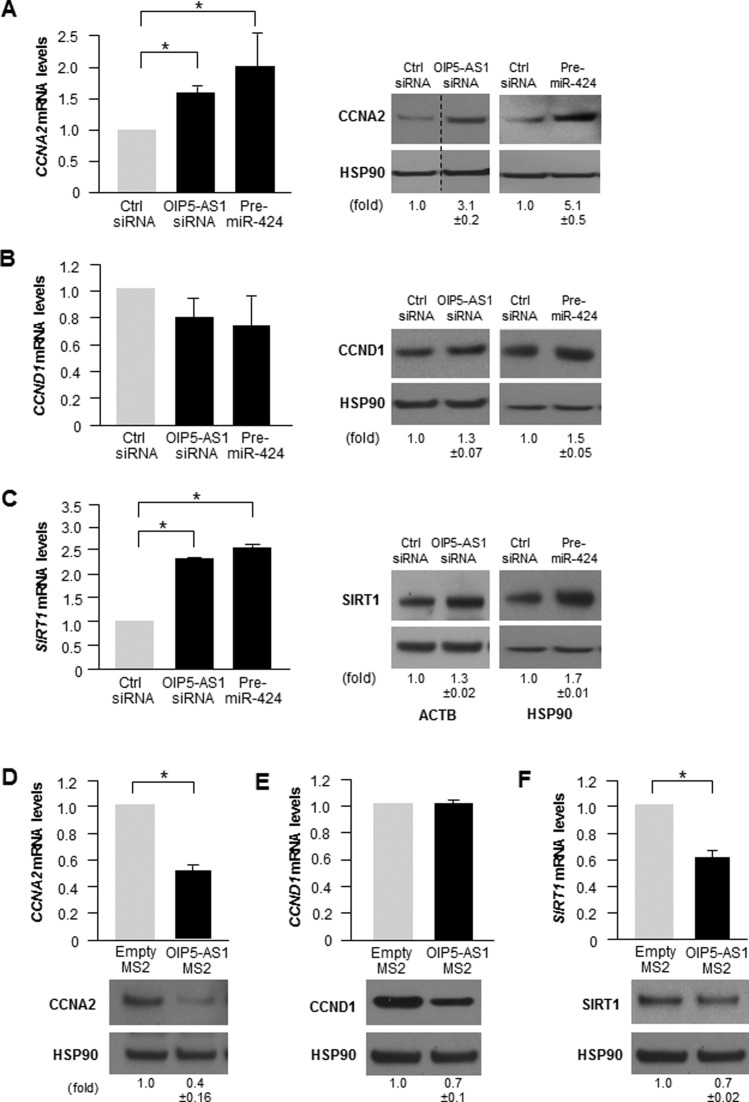
To investigate if the increased binding of HuR to target mRNAs due to miR-424 overexpression or OIP5-AS1 silencing influenced the levels of endogenous target mRNAs and proteins, we selected CCNA2, CCND1 and SIRT1 for further analysis. RT-qPCR and Western blot analyses after silencing OIP5-AS1 and after miR-424 overexpression (Figure 6A–C) revealed that the levels of CCNA2 mRNA and SIRT1 mRNA increased significantly, with a concomitant rise in CCNA2 and SIRT1 protein levels, while CCND1 mRNA levels did not change significantly, but CCND1 protein levels rose moderately. Conversely, overexpression of OIP5-AS1 as a fusion transcript (OIP5-AS1-MS2) reduced CCNA2 and SIRT1 mRNA levels and protein abundance, and also lowered CCND1 protein abundance but did not affect CCND1 mRNA levels (Figure 6D–F). Considering that HeLa cells have ∼1300 copies of HuR in the cytoplasm (17 200 total in the cell) (Supplementary Figure S4) and some 70 cytoplasmic copies of OIP5-AS1 (∼75 total in the cell (38–40), and that each individual OIP5-AS1 transcript has 4–6 binding sites for HuR (Supplementary Figure S5C), OIP5-AS1 could sponge 30% of cytoplasmic HuR. If one further considers that each site can accommodate multiple copies of HuR, as HuR can multimerize (41,42), and that some HuR is unable to bind RNA (for example, HuR phosphorylated at certain residues (43,44)), then OIP5-AS1 could well sequester the entire pool of cytoplasmic HuR. In this regard, the levels of HuR-OIP5-AS1 complexes in HeLa cytoplasm were far greater than those in the nucleus (Supplementary Figure S4E). In sum, these data support the notion that OIP5-AS1 can control the availability of HuR to bind and modulate the post-transcriptional fate of target mRNAs. Accordingly, we propose that OIP5-AS1 functions as a sponge for HuR.
Figure 6.
Impact of silencing OIP5-AS1 or overexpressing miR-424 on the expression of HuR target mRNAs. (A–C) Steady-state levels of CCNA2 mRNA (A), CCND1 mRNA (B) and SIRT1 mRNA (C), as measured by RT-qPCR analysis, as well as the encoded proteins, as measured by Western blot analysis, in cells that had been transfected 72 h earlier with Ctrl siRNA, OIP5-AS1 siRNA or Pre-miR-424. 18S rRNA was used to normalize mRNA levels (left), and HSP90 or ACTB was used to normalize protein input (right). In A, discontinuous lines flank one lane in order to exclude irrelevant lanes from that blot. (D) Steady-state levels of CCNA2, CCND1 and SIRT1 mRNAs and corresponding proteins in HeLa cells 72 h after transfection with empty pMS2 and pOIP5-AS1-MS2. Data are the means and SD from three independent experiments. Protein levels were quantified using ImageJ. *P< 0.05.
OIP5-AS1 HuR sites linked to regulation of HuR availability for binding mRNA
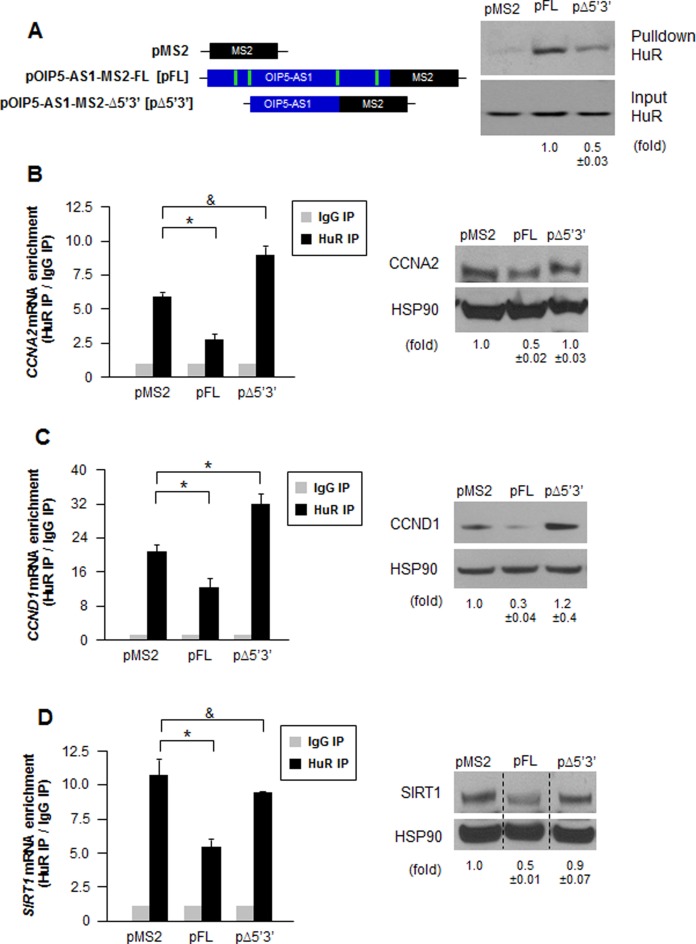
To solidify evidence for the notion that OIP5-AS1 can sponge HuR away from mRNAs, we sought to delete HuR binding sequences from OIP5-AS1. With knowledge of HuR binding sequences obtained from PAR-CLIP analysis (Figure 2A), we designed constructs in which the HuR sites were deleted at the 3′ and the 5′ ends (pOIP5-AS1-MS2-Δ5′3′ [pΔ5′3′]) (Figure 7A, left). This construct was used to evaluate the function of OIP5-AS1 as a sponge for HuR, compared to full-length (FL) OIP5-AS1 (pOIP5-AS1-MS2-FL [pFL]). First, we examined the efficiency of HuR binding to MS2-tagged RNAs (Figure 7A, right). As anticipated, FL RNA expressed from pOIP5-AS1-MS2 showed robust binding to HuR, but RNA expressed from pΔ5′3′ (pOIP5-AS1-MS2-Δ5′3′), pulled down substantially less HuR, suggesting that the deletion of the HuR binding sites strongly reduced the physical interaction between HuR and OIP5-AS1. Analysis of the residual binding of HuR to the OIP5-AS1-MS2-Δ5′3′ RNA using a biotinylated partial RNA confirmed that HuR was capable of binding modestly to this region. Further examination of this segment revealed that Kishore et al. (45) had identified two additional putative HuR sites within this region (Supplementary Figure S5C).
Figure 7.
OIP5-AS1 having deletions of HuR binding sites promotes HuR to bind with target mRNAs. (A) Left, plasmids pMS2 (parental vector), pOIP5-AS1-MS2-FL (pFL, expressing wild type, full-length OIP5-AS1) and pOIP5-AS1-MS2-Δ5′3′ (pΔ5′3′, expressing deletions on the HuR binding sites at the 5′ and 3′ ends) that were transfected into HeLa cells. The extent of HuR interaction with the RNAs expressed from each vector was assessed 72 h later by MS2 pulldown followed by Western blot analysis (right); HuR signals were quantified using ImageJ and indicated below. (B–D) HuR RIP analysis to quantify the interaction of HuR with target CCNA2 mRNA (B), CCND1 mRNA (C) and SIRT1 mRNA (D), each detected by RT-qPCR analysis (left), was performed 72 h after transfection of the different constructs shown in (A) into HeLa cells. After each transfection, Western blot analysis was used to determine the level of target protein expressed (right). In B–D, 18S rRNA was used for normalization of RNA and HSP90 for normalization of protein. In D, discontinuous lines flank one lane from a blot that was moved to exclude irrelevant lanes from that blot. Data are shown as the means and SEM from three independent experiments; *P< 0.05 and &, not significant. Images are a representative from three independent experiments.
Using this set of expression constructs, we tested whether HuR binding to target mRNAs was differentially influenced by OIP5-AS1 transcripts containing or lacking HuR binding sites. HuR IP was performed 72 h after overexpressing each construct in HeLa cells; after extracting RNA, RT-qPCR was used to quantify the levels of target CCNA2, CCND1 and SIRT1 mRNAs and represented as fold enrichment in HuR IP relative to control IgG IP unless. When we overexpressed FL OIP5-AS1, binding of HuR to target mRNAs was significantly suppressed compared to cells expressing empty pMS2 (a plasmid that does not contain OIP5-AS1 segments); however, this suppression disappeared when OIP5-AS1-MS2-Δ5′3′ was expressed instead, restoring the levels of HuR binding to target mRNAs to the levels seen when empty pMS2 was expressed (Figure 7B–D). The pattern of expression of proteins CCNA2, CCND1 and SIRT1 generally followed the pattern of binding, with proteins showing rescue after expressing the deletion mutant OIP5-AS1-MS2-Δ5′3′ compared to OIP5-AS1-MS2-FL. Overexpression of OIP5-AS1-MS2-Δ5′3′ caused HuR to bind more to CCNA2 and CCND1 mRNAs than other groups (Figure 7B and C, left) and led to higher production of CCND1 (Figure 7C, right), suggesting that the deletion mutant may selectively influence regulatory factors that affect adversely these mRNAs.
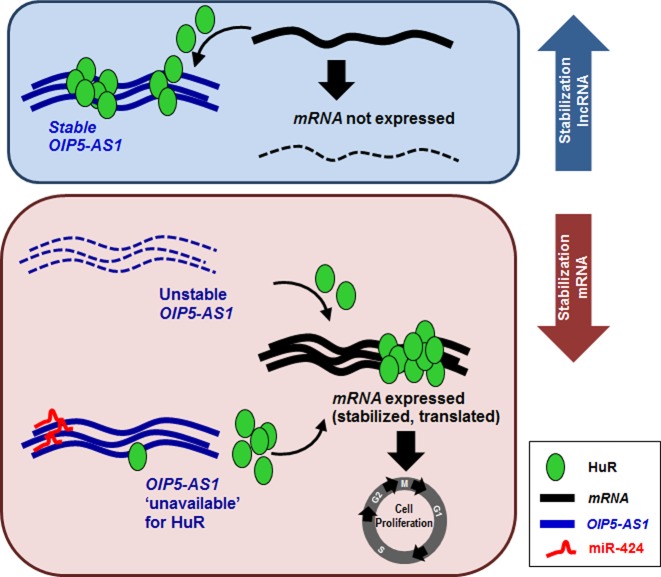
In sum, HuR binds OIP5-AS1 in competition with miR-424; given that HuR binding to OIP5-AS1 reduced HuR availability to target mRNAs, OIP5-AS1 mutants with impaired ability to bind HuR were unable to sponge HuR, permitting HuR to bind target mRNAs and enhance their expression. Taken together, our findings support a model whereby the accumulation of OIP5-AS1 can lead to sponging of HuR, sequestering it away from target mRNAs. Conversely, when OIP5-AS1 abundance declines or it becomes unavailable due to binding by miR-424, HuR availability increases, permitting HuR to elicit its post-transcriptional function on target mRNAs (Figure 8).
Figure 8.
OIP5-AS1 sponges HuR, preventing its interaction with target mRNAs, miR-424 binds OIP5-AS1 and releases HuR to coding mRNAs. Model proposed to explain our findings. When HuR binds OIP5-AS1, it does not bind target coding mRNAs and they are not expressed (HuR may stabilize them and/or enhance their translation). When HuR dissociates from OIP5-AS1 (including via displacement by miR-424), HuR becomes available to target mRNAs and promotes their expression, an effect linked to cell proliferation. HuR, OIP5-AS1, miR-424 and mRNA are represented in the box. Solid line, stable RNA; dashed line, unstable RNA (or untranslated mRNA).
DISCUSSION
We have identified a novel transcript interacting with HuR, the lncRNA OIP5-AS1, and have characterized the consequences of this interaction. Given that OIP5-AS1 reduced cell division (Figure 1), we examined the possibility that the interaction or OIP5-AS1 with HuR might impact the pro-proliferative phenotype attributed to HuR. After establishing that HuR binding OIP5-AS1 stabilized this RNA and increased its abundance, and that HuR competed with miR-424 for binding OIP5-AS1, we found that the availability of HuR for binding target mRNAs (e.g. those encoding CCNA2, CCND1, SIRT1, VHL, TP53, WEE1), increased when HuR interaction with OIP5-AS1 declined, whether it was by increasing miR-424 levels or by silencing OIP5-AS1. We propose that HuR stabilizes lncRNA OIP5-AS1, and in turn, OIP5-AS1 associates with and sponges HuR. This positive feedback loop is interrupted when miR-424 competes with HuR for binding to OIP5-AS1; loss of interaction with OIP5-AS1 releases HuR for binding to target mRNAs relevant for other cellular functions including proliferation. In this manner, OIP5-AS1 and miR-424 jointly work to fine-tune HuR binding to mRNAs (Figure 8).
Regulation of HuR function
As mentioned above, HuR binds to hundreds of cellular mRNAs and is best known for stabilizing and/or regulating their translation, although it has also been implicated in splicing and in nuclear export of mRNAs. HuR can achieve these actions in different ways, typically by interfering with or by recruiting different RNA-binding factors to target RNAs. For example, by modulating the associations of miRNAs and RBPs with subsets of HuR target RNAs, HuR can influence their splicing, transport, turnover and translation (30,32,46). The function of HuR has been found to be regulated in three main ways: by controlling its abundance, its export to the cytoplasm, and its interaction with mRNAs (29,47). HuR export to the cytosol requires a specialized HuR nucleocytoplasmic shuttling domain (HNS) and several proteins, including transportins 1 and 2, the chromosome region maintenance 1 (CRM1), and importin-1α; this process is controlled by cyclin-dependent kinase 1 (Cdk1), AMP-activated protein kinase (AMPK), protein kinase C (PKC) and mitogen-activated protein kinase (MAPK) p38, which phosphorylate HuR and HuR transport proteins (48–52). HuR abundance is regulated transcriptionally by NF-kB and by SMADs (53,54), and post-transcriptionally via miRNAs (including miR-125 and miR-519), and also through HuR ubiquitination and cleavage via caspases (55,56). HuR binding to mRNAs is likewise regulated via phosphorylation. Phosphorylation by the checkpoint kinase Chk2 at HuR residues S88, S100 and T118 (located between and within the HuR RNA-recognition motifs RRM1 and RRM2) modulates HuR binding to several target mRNAs (43,44,47). Phosphorylation by PKC at HuR S158, S221 and S318 promoted HuR binding to mRNAs. HuR methylation at N271 by CARM1 (coactivator-associated arginine methyltransferase 1) in response to lipopolysaccharide stimulation promoted HuR binding to and stabilization of a target mRNA (TNF mRNA) (57).
The regulation of HuR function reported here has some similarities with the function previously proposed in a study by Barnhart et al. (58), which showed that mRNAs encoded by the Sindbis virus contain 3′ UTRs that bind HuR robustly. This interaction excluded HuR from target mRNAs and caused alternative polyadenylation, splicing and degradation of cellular HuR target mRNAs. In the report by Barnhart, HuR sequestration by viral RNA helped elicit a gene expression program that favored viral infection. In our study, HuR is bound instead to a noncoding transcript, OIP5-AS1, and sequestration by OIP5-AS1, appeared to contribute to the growth inhibitory phenotype attributed to OIP5-AS1.
LncRNAs can sponge RBPs, not only miRNAs
Some lncRNAs have been reported to function as competing endogenous (ce)RNAs by serving as sponges that bind and sequester away miRNAs (59–66). For instance, this function was shown for lncRNA linc-MD1, which sponged miRNAs miR-133 and miR-135 (67), for lncRNA HOTAIR and miR-331–3p (68), and for lncRNA H19 and miRNAs miR-138 and miR-200a (69).
Based on our findings, we posit that lncRNAs may also function as endogenous competing RNAs for RBPs, sponging their activity away from target mRNAs. In the present report, the abundant lncRNA OIP5-AS1 sponged HuR away from HuR target mRNAs, generally lowering expression of these target transcripts. However, similar functions may be discovered for other highly expressed lncRNAs that interact with other RBPs. Like OIP5-AS1 and HuR, such sponge lncRNAs are predicted to be abundant and to have long half-lives and multiple sites of interaction with RBPs.
miR-424
It is possible that other lncRNAs that sponge RBPs also involve the action of miRNAs, as shown here for miR-424. Besides a physical competition, miR-424 appears to compete with HuR functionally, as miR-424 has a tumor suppressive function in many cancer types (70–74). In our study, miR-424 competed with HuR for binding OIP5-AS1 and hence counteracted the stabilizing effects of HuR on target mRNAs, which included proliferative and cancer-associated proteins. In this regard, it will be interesting to investigate in the future if miR-424 counteracts the growth suppressive phenotype of OIP5-AS1. HuR function is also closely linked to the stress response. While the impact of OIP5-AS1 was not studied in the context of cell damage, it is interesting to note that some stresses (e.g. ischemia/reperfusion (I/R) injury) elevated miR-424 expression in the brain. If this rise in miR-424 after stresses occurs broadly, miR-424 could mobilize HuR away from OIP5-AS1, enabling HuR actions in the stress response. Unexpectedly, miR-424 did not promote the decay of OIP5-AS1 and thus perhaps miR-424 serves primarily as an endogenous competitor, not a destabilizer of OIP5-AS1. In support of this notion was evidence that AGO2 interaction with OIP5-AS1 rose minimally after overexpressing miR-424 (Supplementary Figure S3A), and that while miR-424 overexpression displaced HuR, two-thirds of the cellular HuR-OIP5-AS1 complex was still detectable (Figure 4E), suggesting that much HuR is still bound and hence protecting OIP5-AS1. Further studies should test if OIP5-AS1 or miR-424 modulates the tumorigenic influence of HuR in vivo.
Concluding remarks
In closing, our findings indicate that HuR and miR-424 bind to OIP5-AS1 competitively and that the net impact of these interactions determine the level of HuR bound to OIP5-AS1, and hence the concentration of HuR available for other target mRNAs. The cytoplasmic-to-nuclear OIP5-AS1 ratio is close to 3:1 (Supplementary Figure S2C). The sponging action of OIP5-AS1 is expected to be particularly effective in the cytoplasm, since HuR abundance in this compartment is relatively low (Supplementary Figure S4B and D). Since PAR-CLIP analysis indicates that there are other abundant HuR-interacting lncRNAs (e.g. SNHG1, SNHG15 and MIR17HG), it is likely that there are additional RNA sponges for HuR in the cell. Our findings pave the way for future studies of other RBPs controlled via lncRNA sponges, as we endeavor to elucidate how RBPs and noncoding RNAs coordinately regulate gene expression programs.
Supplementary Material
Acknowledgments
This work was supported by the National Institute on Aging-Intramural Research Program, National Institutes of Health. We thank J.L. Martindale and J.H. Yoon for insightful discussions.
Authors’ contributions: J.K., K.A., X.Y., I.G. and J.H.N. performed experiments; J.K., K.A. and M.G. analyzed the data; K.A., S.D. and M.G. contributed reagents and expertise; and J.K. and M.G. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: NIA Intramural Research Program, NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Moore M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 2.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 3.Morris A.R., Mukherjee N., Keene J.D. Systematic analysis of posttranscriptional gene expression. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010;2:162–180. doi: 10.1002/wsbm.54. [DOI] [PubMed] [Google Scholar]
- 4.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M., et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 5.Yoon J.H., Abdelmohsen K., Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberg R.A., Penman S. Small molecular weight monodisperse nuclear RNA. J. Mol. Biol. 1968;38:289–304. doi: 10.1016/0022-2836(68)90387-2. [DOI] [PubMed] [Google Scholar]
- 7.Paul J., Duerksen J.D. Chromatin-associated RNA content of heterochromatin and euchromatin. Mol. Cell. Biochem. 1975;9:9–16. doi: 10.1007/BF01731728. [DOI] [PubMed] [Google Scholar]
- 8.Salditt-Georgieff M., Darnell J.E., Jr Further evidence that the majority of primary nuclear RNA transcripts in mammalian cells do not contribute to mRNA. Mol Cell. Biol. 1982;2:701–707. doi: 10.1128/mcb.2.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patil V.S., Zhou R., Rana T.M. Gene regulation by non-coding RNAs. Crit. Rev. Biochem. Mol. Biol. 2014;49:16–32. doi: 10.3109/10409238.2013.844092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 11.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Ann. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonasio R., Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qureshi I.A., Mattick J.S., Mehler M.F. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark M.B., Mattick J.S. Long noncoding RNAs in cell biology. Semin. Cell Dev. Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M. Non-coding RNAS in human diseases. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 16.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdelmohsen K., Panda A., Kang M.J., Xu J., Selimyan R., Yoon J.H., Martindale J.L., De S., Wood W.H., 3rd, Becker K.G., et al. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell. 2013;12:890–900. doi: 10.1111/acel.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grammatikakis I., Panda A.C., Abdelmohsen K., Gorospe M. Long noncoding RNAs (lncRNAs) and the molecular hallmarks of aging. Aging. 2014;6:992–1009. doi: 10.18632/aging.100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greco S., Gorospe M., Martelli F. Noncoding RNA in age-related cardiovascular diseases. J. Mol. Cell. Cardiol. 2015;83:142–155. doi: 10.1016/j.yjmcc.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Wu Z., Fu X., Han W. lncRNA: insights into their function and mechanics in underlying disorders. Mutat. Res. Rev. 2014;762:1–21. doi: 10.1016/j.mrrev.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebedeva S., Jens M., Theil K., Schwanhäusser B., Selbach M., Landthaler M., Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M., Jr, Tuschl T., Ohler U., Keene J.D. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srikantan S., Gorospe M. HuR function in disease. Front. Biosci. 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nabors L.B., Gillespie G.Y., Harkins L., King P.H. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001;61:2154–2161. [PubMed] [Google Scholar]
- 27.Dixon D.A., Tolley N.D., King P.H., Nabors L.B., McIntyre T.M., Zimmerman G.A., Prescott S.M. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan Z., Sanders A.J., Ye L., Jiang W.G. HuR, a key post-transcriptional regulator, and its implication in progression of breast cancer. Histol. Histopathol. 2010;25:1331–1340. doi: 10.14670/HH-25.1331. [DOI] [PubMed] [Google Scholar]
- 29.Abdelmohsen K., Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srikantan S., Tominaga K., Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 2012;13:372–379. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 32.Srikantan S., Abdelmohsen K., Lee E.K., Tominaga K., Subaran S.S., Kuwano Y., Kulshrestha R., Panchakshari R., Kim H.H., Yang X., et al. Translational control of TOP2A influences doxorubicin efficacy. Mol. Cell. Biol. 2011;31:3790–3801. doi: 10.1128/MCB.05639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H.H., Kuwano Y., Srikantan S., Lee E.K., Martindale J.L., Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon J.H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S., Huarte M., Zhan M., Becker K.G., Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang P., Ning S., Zhang Y., Li R., Ye J., Zhao Z., Zhi H., Wang T., Guo Z., Li X. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 2015;43:3478–3489. doi: 10.1093/nar/gkv233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdelmohsen K., Panda A.C., Kang M.J., Guo R., Kim J., Grammatikakis I., Yoon J.H., Dudekula D.B., Noh J.H., Yang X., et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 2014;42:10099–10111. doi: 10.1093/nar/gku686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Heesch S., van Iterson M., Jacobi J., Boymans S., Essers B.P., de Bruijn E., Hao W., MacInnes A.W., Cuppen E., Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lennox A.K., Behlke A.M. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2015;6:22513–22525. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabili M.N., Dunagin M.C., McClanahan P.D., Biaesch A., Padovan-Merhar O., Regev A., Rinn J.L., Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toba G., White K. The third RNA recognition motif of Drosophila ELAV protein has a role in multimerization. Nucleic Acids Res. 2008;36:1390–1399. doi: 10.1093/nar/gkm1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheiba R.M., Ibáñez de Opakua A., Díaz-Quintana A., Cruz-Gallardo I., Martínez-Cruz L.A., Martínez-Chantar M.L., Blanco F.J., Díaz-Moreno I. The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets. RNA Biol. 2014;11:1250–1261. doi: 10.1080/15476286.2014.996069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdelmohsen K., Pullmann R., Jr, Lal A., Kim H.H., Galban S., Yang X., Blethrow J.D., Walker M., Shubert J., Gillespie D.A., et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masuda K., Abdelmohsen K., Kim M.M., Srikantan S., Lee E.K., Tominaga K., Selimyan R., Martindale J.L., Yang X., Lehrmann E., et al. Global dissociation of HuR-mRNA complexes promotes cell survival after ionizing radiation. EMBO J. 2011;30:1040–1053. doi: 10.1038/emboj.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kishore S., Jaskiewicz L., Burger L., Hausser J., Khorshid M., Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 46.Hinman M.N., Lou H. Diverse molecular functions of Hu proteins. Cell Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eberhardt W., Doller A., Pfeilschifter J. Regulation of the mRNA-binding protein HuR by posttranslational modification: spotlight on phosphorylation. Curr. Protein Pept. Sci. 2012;13:380–390. doi: 10.2174/138920312801619439. [DOI] [PubMed] [Google Scholar]
- 48.Wang W., Fan J., Yang X., Fürer-Galban S., López de Silanes I., von Kobbe C., Guo J., Georas S.N., Foufelle F., et al. AMP-activated kinase regulates cytoplasmic HuR. Mol. Cell. Biol. 2002;22:3425–3436. doi: 10.1128/MCB.22.10.3425-3436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doller A., Huwiler A., Müller R., Radeke H.H., Pfeilschifter J., Eberhardt W. Protein kinase Cα-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doller A., Akool El-S., Huwiler A., Müller R., Radeke H.H., Pfeilschifter J., Eberhardt W. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cδ elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol. Cell. Biol. 2008;28:2608–2625. doi: 10.1128/MCB.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim H.H., Yang X., Kuwano Y., Gorospe M. Modification at HuR(S242) alters HuR localization and proliferative influence. Cell Cycle. 2008;7:3371–3377. doi: 10.4161/cc.7.21.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim H.H., Gorospe M. Phosphorylated HuR shuttles in cycles. Cell Cycle. 2008;7:3124–3126. doi: 10.4161/cc.7.20.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang M.J., Ryu B.K., Lee M.G., Han J., Lee J.H., Ha T.K., Byun D.S., Chae K.S., Lee B.H., Chun H.S., et al. NF-κB activates transcription of the RNA-binding factor HuR, via PI3K-AKT signaling, to promote gastric tumorigenesis. Gastroenterology. 2008;135:2030–2042. doi: 10.1053/j.gastro.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 54.Jeyaraj S.C., Singh M., Ayupova D.A., Govindaraju S., Lee B.S. Transcriptional control of human antigen R by bone morphogenetic protein. J. Biol. Chem. 2010;285:4432–4440. doi: 10.1074/jbc.M109.062216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mazroui R., Di Marco S., Clair E., von Roretz C., Tenenbaum S.A., Keene J.D., Saleh M., Gallouzi I.E. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J. Cell Biol. 2008;180:113–127. doi: 10.1083/jcb.200709030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdelmohsen K., Srikantan S., Yang X., Lal A., Kim H.H., Kuwano Y., Galban S., Becker K.G., Kamara D., de Cabo R., et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009;28:1271–1282. doi: 10.1038/emboj.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H., Park S., Kilburn B., Jelinek M.A., Henschen-Edman A., Aswad D.W., Stallcup M.R., Laird-Offringa I.A. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J. Biol. Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- 58.Barnhart M.D., Moon S.L., Emch A.W., Wilusz C.J., Wilusz J. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep. 2013;5:909–917. doi: 10.1016/j.celrep.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee D.Y., Jeyapalan Z., Fang L., Yang J., Zhang Y., Yee A.Y., Li M., Du W.W., Shatseva T., Yang B.B. Expression of versican 3′-untranslated region modulates endogenous microRNA functions. PLoS One. 2010;5:e13599. doi: 10.1371/journal.pone.0013599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y., Chen N., Sun F., Fan Q. CREB up-regulates long non-coding RNA HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeyapalan Z., Deng Z., Shatseva T., Fang L., He C., Yang B.B. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011;39:3026–3041. doi: 10.1093/nar/gkq1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X.H., Sun M., Nie F.Q., Ge Y.B., Zhang E.B., Yin D.D., Kong R., Xia R., Lu K.H., Li J.H., et al. LncRNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331–3p in gastric cancer. Mol. Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang W.C., Fu W.M., Wong C.W., Wang Y., Wang W.M., Hu G.X., Zhang L., Xiao L.J., Wan D.C., Zhang J.F., et al. The LncRNA H19 promotes epithelial to mesenchymal transition by functioning as MiRNA sponges in colorectal cancer. Oncotarget. 2015 doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu J., Li Y., Wang F., Wang X., Cheng B., Ye F., Xie X., Zhou C., Lu W. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2013;32:976–987. doi: 10.1038/onc.2012.121. [DOI] [PubMed] [Google Scholar]
- 71.Wu K., Hu G., He X., Zhou P., Li J., He B., Sun W. MicroRNA-424–5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol. Oncol. Res. 2013;19:739–748. doi: 10.1007/s12253-013-9637-x. [DOI] [PubMed] [Google Scholar]
- 72.Yu L., Ding G.F., He C., Sun L., Jiang Y., Zhu L. MicroRNA-424 is down-regulated in hepatocellular carcinoma and suppresses cell migration and invasion through c-Myb. PLoS One. 2014;9:e91661. doi: 10.1371/journal.pone.0091661. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Banno K., Iida M., Yanokura M., Kisu I., Iwata T., Tominaga E., Tanaka K., Aoki D. MicroRNA in cervical cancer: OncomiRs and tumor suppressor miRs in diagnosis and treatment. ScientificWorldJournal. 2014;2014:178075. doi: 10.1155/2014/178075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Q., Qiu X.M., Li Q.H., Wang X.Y., Li L., Xu M., Dong M., Xiao Y.B. MicroRNA-424 may function as a tumor suppressor in endometrial carcinoma cells by targeting E2F7. Oncol. Rep. 2015;33:2354–2360. doi: 10.3892/or.2015.3812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.