Abstract
Skeletal muscle contains long multinucleated and contractile structures known as muscle fibers, which arise from the fusion of myoblasts into multinucleated myotubes during myogenesis. The myogenic regulatory factor (MRF) MYF5 is the earliest to be expressed during myogenesis and functions as a transcription factor in muscle progenitor cells (satellite cells) and myocytes. In mouse C2C12 myocytes, MYF5 is implicated in the initial steps of myoblast differentiation into myotubes. Here, using ribonucleoprotein immunoprecipitation (RIP) analysis, we discovered a novel function for MYF5 as an RNA-binding protein which associated with a subset of myoblast mRNAs. One prominent MYF5 target was Ccnd1 mRNA, which encodes the key cell cycle regulator CCND1 (Cyclin D1). Biotin-RNA pulldown, UV-crosslinking and gel shift experiments indicated that MYF5 was capable of binding the 3′ untranslated region (UTR) and the coding region (CR) of Ccnd1 mRNA. Silencing MYF5 expression in proliferating myoblasts revealed that MYF5 promoted CCND1 translation and modestly increased transcription of Ccnd1 mRNA. Accordingly, overexpressing MYF5 in C2C12 cells upregulated CCND1 expression while silencing MYF5 reduced myoblast proliferation as well as differentiation of myoblasts into myotubes. Moreover, MYF5 silencing reduced myogenesis, while ectopically restoring CCND1 abundance partially rescued the decrease in myogenesis seen after MYF5 silencing. We propose that MYF5 enhances early myogenesis in part by coordinately elevating Ccnd1 transcription and Ccnd1 mRNA translation.
INTRODUCTION
Skeletal muscle biogenesis during development and during regeneration following injury is tightly regulated. The regeneration of myofibers during muscle repair depends on quiescent mononucleated muscle stem cells known as satellite cells, which reside between the basal lamina and the fibers (1,2). Activated satellite cells start to proliferate into myoblasts, which further proliferate and fuse together to form multinucleated myotubes (3,4). Indeed, the process of adult muscle regeneration from satellite cells recapitulates the process of myogenesis during embryonic development. Like most developmental processes, myogenesis is regulated via changes in gene expression programs that drive activation of satellite cells followed by inhibition of myoblast growth and fusion into myotubes (5). The changes in expressed proteins during myogenesis are controlled both transcriptionally and post-transcriptionally. During myogenesis, transcription is mainly controlled by myogenic regulatory factors (MRFs) (6). Post-transcriptional regulators include RNA-binding proteins (RBPs) (7), a large class of proteins that modulate pre-mRNA splicing, maturation, transport, storage, translation and degradation (8), and regulatory noncoding RNAs like microRNAs and long noncoding (lnc)RNAs (9,10).
The four main MRFs driving myogenesis are MYF5, MYOD, MYOG (myogenin) and MRF4, each of them expressed with a distinct dynamic pattern (11–17). These factors can auto- and cross-regulate expression of each other and interact with the myocyte enhancer factor-2 (MEF2) family of transcription factors to activate the transcription of muscle-specific genes (18). MYF5 and MYOD are considered to be ‘commitment MRFs’, acting early in differentiation, while MYOG and MRF4 act later in differentiation and are considered ‘differentiation MRFs’ (19–21). During mouse embryogenesis, MYF5 is the first of the myogenic transcription factor to be expressed and defines the onset of myogenic development in mammals (22). Although MYOD and MYF5 play some redundant roles in myogenesis, they have distinct roles in the development of muscles from different parts of the body, and MYOD induction is delayed relative to that of MYF5 during adult skeletal muscle regeneration (19,23–25). Importantly, MYF5-deficient skeletal muscle shows a significant delay in regeneration after injury, as reported by Ustanina and colleagues (24). The reduced transition from proliferation to differentiation into myotubes in MYF5-deficient muscle was attributed to the delayed proliferation rate of MYF5-deficient myoblasts, indicating that MYF5 enhances myogenesis by promoting myoblast proliferation (24–26).
Our recent studies investigating the mechanisms that govern expression levels of MRFs during myogenesis using the mouse myoblast C2C12 line [an established cell model for skeletal muscle differentiation studies (27)] revealed that MEF2C expression was subject to post-transcriptional regulation by the RBP AUF1 (28). During the course of this investigation, we came upon the unexpected observation that MYF5 appeared to function as an RNA-binding protein and set out to investigate this possibility in molecular detail. Here, we present evidence that MYF5 also has affinity for mRNAs. We analyze in depth the interaction of MYF5 with one specific target transcript, Ccnd1 mRNA, which encodes the growth regulator Cyclin D1 (CCND1) that functions during the brief proliferative burst that initiates myogenesis. MYF5 was found to bind the coding region and 3′-untranslated region (UTR) of Ccnd1 mRNA, and further analysis of endogenous Ccnd1 mRNA and reporter constructs bearing segments of Ccnd1 mRNA revealed that MYF5 selectively enhances CCND1 translation. Accordingly, silencing MYF5 reduced CCND1 abundance, while overexpressing MYF5 elevated CCND1 levels, in turn impacting upon the rate of myotube formation. Interestingly, MYF5 also moderately upregulated Ccnd1 mRNA transcription. Together, we propose that MYF5 contributes to myoblast growth and differentiation at least in part by enhancing both the transcription and the translation of Ccnd1 mRNA and hence increasing Cyclin D1/CCND1 production.
MATERIALS AND METHODS
Plasmid constructs and reporter analysis
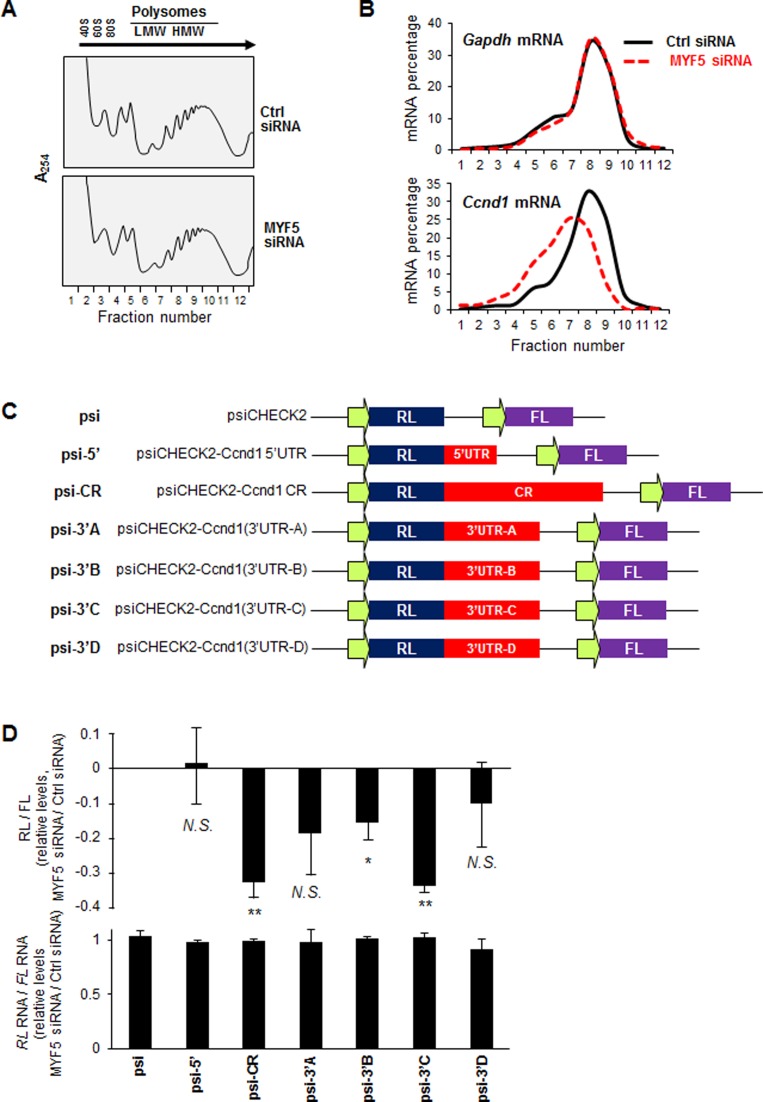
To prepare plasmid pcDNA3-FLAG3-MYF5, the MYF5 coding sequence (NM_008656.5) was amplified by polymerase chain reaction (PCR) from cDNA prepared from proliferating C2C12 cells. In order to assay the contribution of different regions of Ccnd1 mRNA (NM_007631.2) to the regulation by MYF5, several constructs were engineered by PCR amplification of different Ccnd1 mRNA segments (5′UTR, CR, 3′UTR-A, 3′UTR-B, 3′UTR-C and 3′UTR-D) and inserted into plasmid psiCHECK2 (Promega). Ccnd1 PCR products were inserted downstream of the Renilla luciferase (RL) open reading frame in psiCHECK2; Firefly luciferase (FL) was measured as internal control. Plasmids expressing GST-tagged MYF5 were constructed after amplifying Domain 1 (D1), Domain 2 (D2), Domain 3 (D3), or combinations of these domains and inserting them in frame downstream of GST.
For overexpression experiments, 1 μg of plasmids pcDNA3-FLAG3-MYF5 (constructed in-house), pcDNA3-FLAG, or pCCND1-Myc-DDK (Origene, MR226906) were transfected into growing C2C12 cells using Lipofectamine 2000 (Invitrogen). For reporter analyses, proliferating C2C12 cells were transfected with Ctrl siRNA or MYF5 siRNA using Lipofectamine 2000 and 24 h later with 50 ng of psiCHECK2 reporter plasmids containing Ccnd1 fragments. Sixteen hours after transfection of reporter plasmids, RL and FL activities were measured using Dual-Glo Luciferase Assay System (Promega) and RL mRNA and FL mRNA were measured by RT-qPCR analysis after elimination of residual plasmid using DNase I.
C2C12 cell culture, differentiation and creatine kinase assay
Proliferating C2C12 myoblasts cells were cultured in growth medium [(GM), Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Gibco), antibiotics and antimycotics (Life Technologies)]. Growing, subconfluent C2C12 cultures were induced to differentiate by replacing the GM with differentiation medium [(DM) DMEM with 2% horse serum]. For silencing experiments, control small interfering RNA (Ctrl siRNA) or MYF5 siRNA (sc-35989, Santa Cruz Biotech.) was transfected twice, 36 and 12 h before inducing differentiation. CCND1 siRNA (SR408533, Origene) was transfected 48 h before measuring cell number. Creatine kinase (CK) activity was determined using the EnzyChromTM Creatine Kinase Assay Kit (BioAssay Systems) following the manufacturer's protocol, and CK activity was calculated as arbitrary units per μg of total protein.
Ribonucleoprotein immunoprecipitation (RIP) and western blot analysis
The association of MYF5 with endogenous mRNAs in growing and differentiated C2C12 cells were analyzed by immunoprecipitation (IP) of endogenous ribonucleoprotein (RNP) complexes as described previously (28). Briefly, cytoplasmic extracts from growing (GM) C2C12 cells were prepared by lysing the cells in polysome extraction buffer (PEB; 20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2 and 0.5% NP-40) containing protease and RNase inhibitors for 10 min on ice and the supernatant was collected by centrifugation (15 000 × g for 10 min at 4°C). The supernatants were incubated with protein A sepharose beads coated with anti-MYF5 (sc-302, Santa Cruz Biotech.) or control IgG (Santa Cruz Biotech.) antibodies for 2 h at 4°C. After washes with NT2 buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40), bound RNA was extracted from the beads using TRIzol and subjected to reverse transcription (RT) followed by real-time, quantitative (q)PCR analysis using the primer pairs listed below. For detection of MYF5 in the MYF5 IP samples, pellets were incubated with elution buffer (0.2% SDS, 0.1% Tween-20, 50 mM Tris-HCl, pH 8.0) for 5 min at 25ºC with shaking; after a short spin, the supernatant was collected and further analyzed for the presence of MYF5 by western blot analysis.
Nuclear and cytoplasmic fractions of proliferating C2C12 cells were prepared using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Life Technologies) following the manufacturer's protocol. C2C12 whole-cell lysates were prepared using RIPA buffer, and western blot analysis was performed with 10 μg of protein fractionated by electrophoresis through 4–20% Mini-PROTEAN® TGX™ Gel (Bio-Rad) and transferred to nitrocellulose membrane using Trans-Blot® Turbo™ Transfer System (Bio-Rad). Primary antibodies recognized Cyclin D1 (92G2) (mAb #2978, Cell Signaling), MYF5 (sc-302, Santa Cruz Biotech.), the GST tag (sc-138), HSP90 (sc-13119), GAPDH (sc-32233), MYOG (sc-12732) and Lamin B (sc-6216). Following incubation with the appropriate secondary antibodies, signals were developed using Enhanced Chemiluminescence (ECL).
Production and purification of full-length GST-MYF5 and GST-MYF5 deletion products
GST-tagged full-length MYF5 and partial MYF5 domains were cloned in plasmid vector pGEX4T2, and transformed into competent BL21 E. coli. After overnight culture, cells were treated with 0.5 mM IPTG to induce GST-MYF5 production. Cells were then lysed in GST lysis buffer (50 mM Tris-HCl [pH 8.8], 200 mM NaCl, 1 mM EDTA) containing protease inhibitor using sonication and the lysate was cleared by centrifugation. The supernatant was supplemented with Triton X-100 (3%), and then incubated with Glutathione Sepharose 4B beads (GE Healthcare Life Sciences) to allow binding. Following washes with GST lysis buffer containing 1% Triton X-100 and elution with a buffer containing 300 mM Tris-HCl [pH 8.8], 120 mM NaCl and 20 mM Glutathione, GST-tagged proteins were dialyzed using the YM-10 centricon columns (Millipore) and washed with TBS buffer before use in binding assays.
Immunostaining
Growing or differentiated C2C12 cells were fixed with chilled acetone-methanol mix (50% each) for 10 min at RT, incubated with BSA-containing PBS (PBS-BSA) for 15 min and blocked with PBS-BSA, whereupon the cells were incubated for 1 h with anti-MYF5 (Santa Cruz Biotech.) or anti-MYH (H-300) (sc-20641, Santa Cruz Biotech.) antibodies. Following washes with PBS and incubation for 30 min with Donkey anti-Rabbit IgG (H+L) secondary antibody, Alexa Fluor® 488 conjugate (A-21206, Life Technologies) was used to visualize the signals using fluorescence microscopy. Nuclei were visualized by staining with DAPI.
Microarray analyses
RNA present in the MYF5 RIP samples was isolated using TRIzol extraction and amplified and labeled using the Illumina TotalPrep RNA Amplification Kit following the manufacturer's protocol (Thermo-Fisher, Waltham, MA). Biotinylated RNA hybridized to MouseRef-8 v2.0 Expression BeadChips (Illumina, San Diego, CA) was visualized using streptavidin-conjugated Cy3 staining and scanned at 0.53-μm resolution on an Illumina iScan Beadarray scanner. Raw microarray data were filtered by detection P-values of <0.02, normalized by Z-score transformation and tested for significant differences in signal intensity by Z-test and multiple comparison correction by false discovery rate, with additional sample group ANOVA test to control overall sample error. The sample quality was analyzed by scatter plot, principal component analysis and gene sample Z-score-based hierarchy clustering to exclude possible outliers. Transcripts were considered to be significantly changed when they had Z-test P-values of ≤0.05, Z-ratio absolute values of Z-ratio ≥1.5, a multiple-comparison correction false-discovery rate of ≤0.30, non-negative average Z-score signal of the comparison group samples, and an independent one-way analysis of variance (ANOVA) on sample group analysis P-value of ≤0.05.
RNA isolation and RT-qPCR analysis
RNA was isolated from cell extracts or from RIP samples using TRIzol (15596–026, Thermo Fisher Scientific) following the manufacturer's procedure. Reverse transcription (RT) was performed using 150 ng of random hexamers (11034731001, Roche) and Maxima reverse transcriptase (EP0741, Thermo Fisher Scientific) and incubated for 10 min at room temperature followed by 30 min incubation at 50°C using a Thermomixer. The RT enzyme was inactivated by heating at 85°C for 5 min before using the reaction products for real-time quantitative (q)PCR analysis. For qPCR analysis, KAPA SYBR® FAST qPCR Kits (ABI Prism) (KK4605, KAPA Biosystems) was used with 250 nM gene-specific primers (forward and reverse, respectively) GTAACCCGTTGAACCCCATT and CCATCCAATCGGTAGTAGCG for 18S rRNA [NR_003278.3], AACTTTGGCATTGTGGAAGG and GGATGCAGGGATGATGTTCT for Gapdh mRNA [NM_001289726.1], TATTACAGCCTGCCGGGACA and CTGCTGTTCTTTCGGGACCA for Myf5 mRNA [NM_008656.5], CTTCTACGCACCTGGACCG and ACTGTAGTAGGCGGTGTCGT for MyoD mRNA [NM_010866.2], CGGTGGAACTTTGACTTCGT and CAGGGCAGAGGAAGTACTGG for Cdkn1a mRNA [NM_007669.5], AGCCTCCAGAGGGCTGTCGG and TGGGGAGGGCTGTGGTCTCG for Ccnd1 mRNA [NM_007631.2], CCTCACGCGCGTCGCTTCTC and GGCACGTAGCGCTCCTCCAG for Ccnd3 mRNA [NM_007632.2], TCTGTGCATTCTAGCCATCG and ACAAAAGGCACCATCCAGTC for Ccne2 mRNA [NM_001037134.2], TCTACGTGGAACCTTTGCCG and GCTGCACACACTCTTTACCG for Atp2a2 mRNA [NM_001110140.3], CGGTGACTTTCCCTCCTTGTC and AAGCTGTAGCTGCACCACTCT for Atp1b1 mRNA [NM_009721.6], TACAAGTACCTCACCGCTTGGT and TGATCTTGTCTTGGTGCTCGTA for RL mRNA, CTAAGAAGGGCCTGCAGAAGAT and AAGCCCTGGTAGTCGGTCTTAG for FL mRNA, and CTGGAGCCCTTGAAGAAGAG and GGTGGTGGGTTACGTGGTTA for Ccnd1 pre-mRNA. RT-qPCR analysis was performed on Applied Biosystems 7300 and 7900 instruments with a cycle set up consisting of 3 min at 95°C and 40 cycles of 5 s at 95°C plus 20 s at 60°C; dissociation curves were routinely analyzed at a minimum when testing new primers for the first time. Relative expression levels were calculated after normalization to 18S rRNA or to Gapdh mRNA (29). Student's t-test was used for assessing significance (P-values) throughout the report.
Biotin pull down analysis
The primers used to prepare biotinylated RNA fragments spanning the Ccnd1 mRNA are listed below. To synthesize biotinylated transcripts, PCR fragments were prepared using forward primers that contained the T7 RNA polymerase promoter sequence (T7): AGTAATACGACTCACTATAGGG. The PCR template of Ccnd1 5′UTR was generated using primers (forward and reverse, respectively) (T7)TTTTCTCTGCCCGGCTTTG and GGCGCGGCCGTCTGGGGAGG, and primers to synthesize the Ccnd1 coding region (CR) were (T7)ATGGAACACCAGCTCCTGTG and TCAGATGTCCACATCTCGCA. PCR templates to synthesize the Ccnd1 3′UTR fragments were prepared using primers (T7)GGGCCACCGGGCAGGCGGGA and GCAAAGCAGAGTACATTTCTC for fragment A, (T7)GAGAAATGTACTCTGCTTTG and TCCATTCCATTAGAACCCCTC for fragment B, (T7)GAGGGGTTCTAATGGAATGGA and CCAATGGGGCCAATTGGGTT for fragment C, and (T7)AACCCAATTGGCCCCATTGG and GGTATTGTGAACAGGAACCTG for fragment D. After purification of the template PCR products, biotinylated transcripts were synthesized using the MegaScript T7 kit (Ambion) followed by purification using NucAway Spin Columns (AM10070, Applied Biosystems). The partially double-stranded template for the biotin-RNA preparation was generated from annealing the T7 promoter oligonucleotide and oligonucleotides corresponding to the complementary sequence of the 3′UTR-C of Ccnd1 mRNA with complementary T7 promoter sequence (CCCTATAGTGAGTCGTATTACT, referred to as T7AS) at the 3′ end. The complementary oligomers were ATCTGAATGCGTGTGTGGACATCCCCATCCATTCCATTAGAACCCCTC-T7AS for 3′C1, ACTTCCCAAGCACCTCATACTACCAGCCCTACAACCTGTTGTACAGCCATCTG-T7AS for 3′C2, AGATCCCGGTGGTGCGAGAACAGAGTTCTCTCTTCTTGACCCAACAAAACTTC-T7AS for 3′C3, CTTCCAAACACCAGCTGGCACCAAAGGATCCCTTCAACTTTGCAGGACAGATC-T7AS for 3′C4, AGAGTTGTCCCCAATCTCCTTGTCCAGGTAATGCCATCATGGTTCCTACTTCC-T7AS for 3′C5, GTATAAATTAGACATTTTAGTGTTTAAAAGCCTCCTGTGTGAGACTTAAGAGT-T7AS for 3′C6, CACTGGTCATGGGCAGCCTTTCCCATAAATACTCTTCTGTAGCCTTAAGTATA-T7AS for 3′C7, GGCAGAGGTGTGCGTTTGAATCAAGGGAGATCACATTGCTTTGAGTCACACTG-T7AS for 3′C8, CAATGAAAGACCAATCTCTCAGACATGGCCCTAAACCTTCTCCAGCAGGGCAG-T7AS for 3′C9, CCCCCCCCCCCCCCCCGTTGCCCAATGAAAGACCAATCTCTCAGACATGG-T7AS for 3′C9–10, TCTTTGTGGTTTTTTTTTTTTAAGGACCCCCCCCCCCCCCCCGTTGCCCAATG-T7AS for 3′C10, AAGGACCCCCCCCCCCCCCCCGTTGCCCAATG-T7AS for 3′C10–1, AAAGTCAAGCAGACCAAATCTCTGTCTTTGTGGTTTTTTTTTTTTAAGG-T7AS for 3′C10–11, and CCAATGGGGCCAATTGGGTTGGGAAAGTCAAGCAGACCAAATCTCTGTCTTT-T7AS for 3′C11 were used to generate the template for fragments corresponding to Ccnd1 3′UTR-C. The partial double-stranded templates were used to prepare biotinylated transcripts using the MegaShortscript T7 kit (Ambion) followed by purification using NucAway Spin Columns (AM10070, Applied Biosystems). Cytoplasmic C2C12 cell lysates (500 μg) prepared using PEB or 25 ng of recombinant His-MYF5 (TP760391, Origene) were incubated with 1 μg of purified biotinylated transcripts in 1X TENT buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 250 mM NaCl, 0.5% [v/v] Triton X-100), and 1× protease and 200 U/reaction RNase inhibitors for 30 min at room temperature, and then complexes were isolated with 50 μl of streptavidin-coupled Dynabeads (11206D, Invitrogen).
For pulldown using recombinant MYF5, 25–100 ng of His-MYF5 or GST-MYF5 were incubated with 1 μg of Ccnd1 RNA fragments followed by pulldown of MYF5 as described above. MYF5 levels in the pulldown material were studied by western blot analysis as described previously.
RNA electrophoretic mobility shift assay (EMSA) and crosslinking assay
Radiolabeled 3′C10–1 RNA was synthesized from semi-double stranded template (described above) and the annealed double stranded oligonucleotide was transcribed in presence of 50 μCi [α-32P]-CTP (PerkinElmer) at 37°C for 4 h using a MegaShortcript T7 kit (Ambion) and purified using NucAway Spin Columns. The 32P-CTP-labeled RNA was incubated with gel-shift mix (50 ng/μl BSA, 0.1 μg/μl heparin, 0.5 mM rATP, 75 mM KCl, 1× protease inhibitor, and 20 U Ribolock) to a final concentration of 500 nM GST or GST-MYF5 for 15 min on ice and the RNA-protein complex was resolved on a 8% native gel with 10% glycerol. For UV crosslinking assay, after performing the binding reaction as described above, the samples were irradiated with 400 mJ/cm2 at 254 nm using a Stratalinker 2400 and resolved by SDS-PAGE (4–20% polyacrylamide) after boiling in loading dye. RNA-protein complexes on the dried gel were visualized by autoradiography.
Thymidine incorporation
Growing C2C12 cells were transfected with Ctrl siRNA or MYF5 siRNA using Lipofectamine 2000 and 24 h later, cells were treated with [Methyl-3H]-Thymidine (NET027250UC, Perkin Elmer) for 16 h, whereupon cells were washed with PBS and collected. The incorporated radioactivity was measured in each sample using liquid scintillation counting, and the data were normalized to total protein amount.
Polysome analysis
For the analysis of polyribosomes, 48 h after transfection with Ctrl siRNA or MYF5 siRNA, C2C12 cells were incubated with cycloheximide (Calbiochem; 100 μg/ml, 15 min) followed by preparation of cytoplasmic lysates using PEB. Lysates were size-fractionated by centrifugation through 10–50% linear sucrose gradients and 12 fractions were collected for further analysis. RNA in each fraction was monitored by measuring absorbance at 254 nm using a spectrophotometer, and the distribution of Gapdh and Ccnd1 mRNAs over the gradient was analyzed by RT-qPCR analysis and plotted as a percentage of the specific mRNA in each fraction relative to the total amount of that mRNA in the gradient.
RESULTS
MYF5 expression is required for C2C12 myoblast growth and differentiation
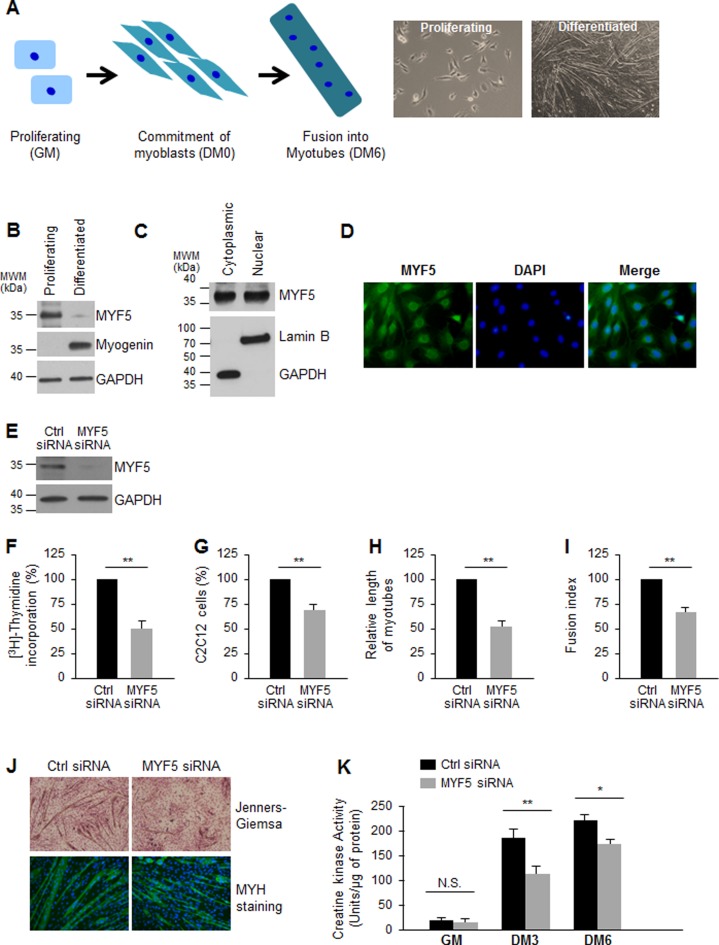
After proliferating C2C12 myoblasts cultured in growth medium (GM) are cultured with differentiation medium (DM), they undergo one or two final rounds of replication and subsequently begin to fuse and form multinucleated myotubes; this process, which resembles in vivo myogenesis, is completed by day 6 in DM (DM6, Figure 1A). The transcription factors MYF5 and MYOD are implicated in and serve as markers of early differentiation, while MYOG and MRF4 drive and define late differentiation stages. Western blot analysis indicated that MYF5 was downregulated while MYOG was upregulated in differentiated C2C12 myotubes, suggesting a role for MYF5 in proliferating myoblasts or in early differentiation (Figure 1B). In C2C12 myoblasts growing in GM conditions, MYF5 was localized in the nucleus as previously reported (30,31), where it functions as a transcription factor for myogenic gene expression (24–26), but unexpectedly it was also found in the cytoplasm (Figure 1C). Analysis of MYF5 by immunofluorescence confirmed the nuclear and cytoplasmic distribution of MYF5 in proliferating C2C12 myoblasts (Figure 1D).
Figure 1.
MYF5 is necessary for C2C12 myoblast growth and differentiation: (A)Left, Schematic of skeletal muscle differentiation. Proliferating myoblasts become committed to differentiation, exit the cell division cycle and begin to elongate, subsequently fusing together to form multinucleated myotubes. Right, phase-contrast images of proliferating C2C12 myoblasts cultured in growth medium (GM) and C2C12 cultures displaying myotubes after culture in differentiation medium [(DM) DMEM with 2% horse serum] for 6 days. (B) Western blot analysis of the levels of MYF5 (and loading control GAPDH) in proliferating and differentiated C2C12 myoblasts. (C) Western blot analysis of MYF5 localization in proliferating C2C12 myoblasts; GAPDH and Lamin B were assessed as cytoplasmic and nuclear fractionation markers, respectively. (D) Immunofluorescence detection of MYF5 in growing C2C12 myoblasts; nuclei were visualized by staining with DAPI. (E) Western blot analysis of MYF5 and GAPDH expression levels 48 h after transfection of proliferating C2C12 myoblasts with Ctrl siRNA or MYF5 siRNA. (F,G) Forty-eight hours after transfection of C2C12 myoblasts with Ctrl siRNA or MYF5 siRNA, measurements were taken for [3H]-thymidine incorporation (F) and cell numbers (using a TC10 automated cell counter, BioRad) (G). (H,I) After differentiation of C2C12 cells in which MYF5 levels were normal (Ctrl siRNA) or was reduced (MYF5 siRNA) through DM6, the length of myotubes (H) and the fusion index (fraction of total nuclei present myotube) (I) were assessed. (J) Micrographs of Jenners-Giemsa stain (a dye that stains myotubes and nuclei) (top) and fluorescence micrographs to detect MYH (present in differentiated myotubes) in C2C12 cells that had been transfected with MYF5 siRNA or Ctrl siRNA and placed in differentiation medium for 6 days (DM6). (K) C2C12 differentiation was monitored using creatine kinase activity at different time points of differentiation: cells in proliferation (growth) medium (GM), and cells in differentiation medium for 3 or 6 days (DM3 and DM6). In (F-I,K), data are the means and S.E.M. from three independent experiments. N.S., not significant; *, P < 0.05; **, P < 0.01 (Student's t-test).
Silencing of MYF5 (Figure 1E) significantly reduced the incorporation of 3H-thymidine in growing cells (Figure 1F), indicating that myoblast replication was diminished. Further analysis indicated a decrease in C2C12 cell numbers after MYF5 silencing, supporting the notion that MYF5 promoted cell proliferation (Figure 1G). Furthermore, C2C12 myoblasts expressing lower levels of MYF5 differentiated less efficiently than control C2C12 myoblasts, as determined on differentiation day 6 (DM6), where the relative length of myotubes (Figure 1H) and the fusion index (the fraction of total nuclei present inside myotubes) were lower in MYF5-silenced C2C12 cells (Figure 1I). Additional evidence that differentiation was impaired in MYF5-silenced C2C12 cells was obtained using microscopy after Jenners-Giemsa staining and assessing for the presence of MYH, a marker of differentiated myotubes. As shown (Figure 1J), MYF5 silencing robustly lowered the number, length and diameter of myotubes. In addition, silencing MYF5 in C2C12 cells decreased the enzymatic activity of creatine kinase, a marker of differentiated myoblasts, on differentiation days 3 and 6 (DM3 and DM6, respectively, Figure 1K). Taken together, these data confirm earlier reports (22,24) that MYF5 controls myogenic differentiation and suggest that this effect is in part elicited by promoting myoblast proliferation.
MYF5 binds myogenic mRNAs
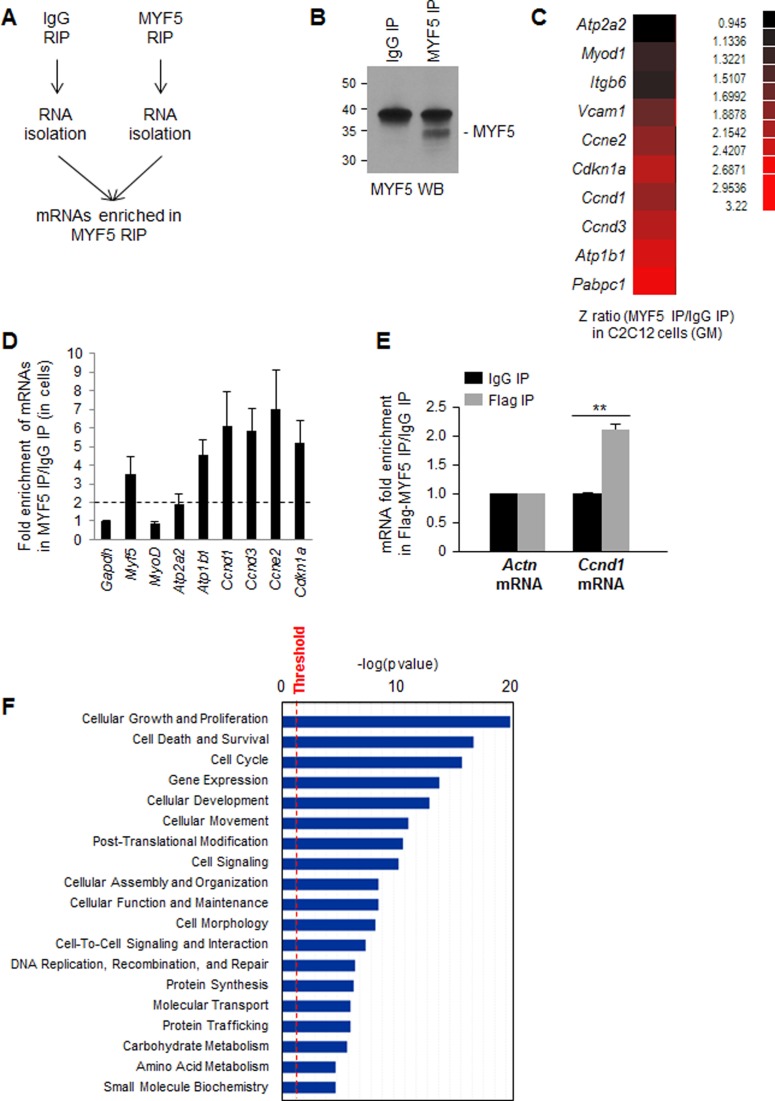
The first evidence that MYF5 might bind to RNA came from ribonucleoprotein immunoprecipitation (RNP IP or RIP) analysis (Materials and Methods). Following IP using anti-MYF5 antibody under conditions that preserved endogenous RNPs, mRNAs interacting with MYF5 in growing (GM) C2C12 myoblasts were investigated (Figure 2A,B). Microarray analysis was performed to compare mRNAs present in MYF5 RIP samples with those present at background levels in control IgG RIP samples, in order to identify the mRNAs preferentially interacting with MYF5 (GSE73704). Interestingly, in the MYF5 IP samples, a number of enriched mRNAs were identified which encoded proteins implicated in myoblast growth and differentiation (Figure 2C). Out of hundreds of MYF5-associated mRNAs, several were found to encode proteins involved in muscle development, including ion transport ATPases (e.g. Atp1b1 and Atp2a2 mRNAs), cell cycle regulators (e.g. Ccnd1, Ccnd3, Ccne2 and Cdkn1a mRNAs), cellular adhesion proteins (e.g. Vcam1 and Itgb6 mRNAs), translation regulators (e.g. Pabpc1 mRNA) and myogenic transcription factors (Myod mRNA). RT-qPCR analysis of MYF5 RIP samples verified the enrichment of transcripts encoding myogenic and proliferative factors in GM cells (Figure 2D), even though MYF5 silencing did not enrich cells in any specific cell cycle compartment (not shown). To further validate these interactions, C2C12 cells were transfected with a vector that expressed Flag-tagged MYF5 and the enrichment of Ccnd1 mRNA was analyzed by RIP analysis using anti-Flag antibody. This result confirmed the specific interaction of MYF5 with Ccnd1 mRNA in C2C12 cells (Figure 2E). The coordinate expression of MYF5 and Ccnd1 mRNA was also observed in primary myoblasts prepared from skeletal muscle from newborn mice (Supplementary Figure S1). Ingenuity pathway analysis (IPA) of the transcripts enriched in MYF5 RIP suggested that the most significantly enriched mRNAs encode proteins involved in cell growth and proliferation (Figure 2F).
Figure 2.
MYF5 target transcripts include mRNAs that encode proteins involved in myoblast proliferation and differentiation. (A) RIP assay using cytoplasmic lysates prepared from C2C12 cells using either anti-MYF5 antibody or IgG under conditions that preserved mRNA-RBP (mRNP) complexes. (B) Western blot analysis of MYF5 recovered in IP samples. (C) Following MYF5 RIP, MYF5-bound mRNAs were identified by microarray (RIP-chip) analysis in growing (GM) C2C12 cells. Data represent the Z-ratio of mRNAs in MYF5 RIP relative to IgG RIP. (D) RIP followed by RT-qPCR analysis to validate the association of MYF5 with mRNAs encoding myogenic proteins in proliferating C2C12 myoblasts; the levels of mRNAs in MYF5 IP were normalized to the levels of Gapdh mRNA and plotted as fold enrichment relative to the levels seen in control IgG IP samples. Discontinuous gray line: twofold enrichment in mRNAs bound to MYF5. (E) RIP analysis of Flag-MYF5 interaction with Ccnd1 mRNA. Forty-eight hours after C2C12 transfection with Flag-MYF5, RIP analysis was carried out using IgG or anti-Flag antibodies. Ccnd1 mRNA was detected by RT-qPCR analysis and its levels in Flag IP were compared with those in control IgG IP samples; Actn mRNA (encoding the housekeeping protein β-Actin) was measured to normalize sample input. Data in (D,E) represent the means and S.E.M. from three or more independent experiments. (F) Ingenuity pathway analysis (IPA) of mRNAs enriched in MYF5 IP relative to IgG in C2C12 growing myoblasts. *, P < 0.05 and **, P < 0.01 (Student's t-test).
The state-of-the-art method to identify the precise RNA sequences that interact with an RBP such as MYF5 involves crosslinking immunoprecipitation (CLIP) analysis transcriptome-wide followed by digestion of the bound mRNA and sequencing of the protected RNA fragments. Unfortunately, we could not carry out different types of CLIP analysis (e.g. specifically HITS-CLIP, PAR-CLIP and iCLIP) for MYF5 due to the limited uptake of plasmids by C2C12 cells (not shown). Instead, we analyzed the MYF5-bound mRNAs identified on microarrays following methods described earlier (supplementary text). Some of the top sequences among MYF5 targets identified by this method included several G-rich RNAs (Supplementary Figure S2A,B); among them, MYF5 showed preferential binding to G-rich RNAs (Supplementary Figure S2C).
MYF5 binds to sequences in the Ccnd1 coding region and 3′UTR
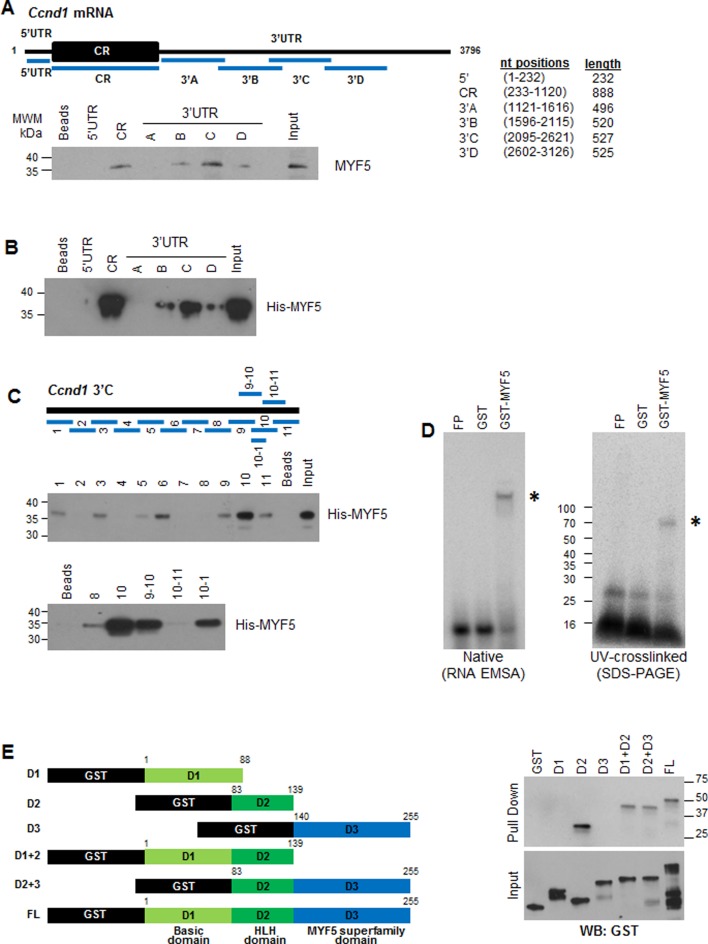
To identify the region(s) of Ccnd1 mRNA with which MYF5 associated, partial biotinylated fragments of the 5′UTR, the coding region (CR) and the 3′UTR of Ccnd1 mRNA were prepared (Figure 3A, schematic). The biotinylated RNA fragments were allowed to bind proteins present in cytoplasmic lysates prepared from growing C2C12 cells; after pulldown using streptavidin beads, the presence of MYF5 in the pulldown material was assessed by western blot analysis (Figure 3A). These data revealed that biotinylated Ccnd1 CR and 3′UTR-C fragments showed the strongest binding to MYF5 while the other fragments showed little or no binding to MYF5. We used recombinant His-MYF5 (Supplementary Figure S3A) to carry out similar RNA-protein affinity assays to analyze the interaction of recombinant His-MYF5 to different regions of Ccnd1 mRNA. CR and 3′UTR-C fragments (3′C, Figure 3B, Supplementary Figure S3B) were also found to be the best targets of interaction with His-MYF5, in agreement with the data in Figure 3A. We could not identify sequences that were clearly shared between CR and 3′UTR-C, although this finding was not unexpected, given that RBPs generally do not bind to strict RNA sequences and instead have degenerate requirements for RNA binding sites (as illustrated for MYF5 in Supplementary Figure S2) that are often complicated by secondary structures that present certain RNA sequences in double- or single-stranded conformations.
Figure 3.
MYF5 binds to specific sequences on the Ccnd1 mRNA. (A)Top, schematic showing biotinylated RNA fragments spanning the 5′UTR, coding region (CR), and 3′UTR of Ccnd1 mRNA used for pulldown. Bottom, biotinylated RNA fragments were incubated with cytoplasmic lysates from C2C12 cells (GM); after pulldown using streptavidin beads, the levels of MYF5 bound to the biotinylated RNA segments were detected by western blot analysis. (B) Recombinant purified His-MYF5 was incubated with biotinylated Ccnd1 RNA fragments followed by pulldown and detection of MYF5 by western blot analysis using anti-MYF5 antibody. (C) Schematic of biotinylated RNA fragments spanning the Ccnd1 3′UTR-C transcript (top), were tested for binding to His-MYF5 after pull-down using streptavidin beads; His-MYF5 interaction with RNA segments of fragment C (middle) and smaller RNAs after closer subdivision of fragments 9 through 11 (bottom) were assessed by western blot analysis using anti-MYF5 antibody. (D) GST or GST-MYF5 were incubated with radiolabeled Ccnd1 3′UTR-C10–1, then either resolved on native acrylamide gels (left) by RNA electrophoretic mobility shift assay (EMSA), or crosslinked by UV irradiation and resolved by SDS-PAGE (right). (E) The domain of MYF5 that interacts with the Ccnd1 3′-C fragment was mapped by creating GST-tagged truncations of MYF5 (left) and testing their interaction by biotin pulldown and western blot analysis using anti-GST antibody (right).
To narrow down the site of MYF5 association with Ccnd1 mRNA, we prepared smaller biotinylated fragments spanning the 3′UTR-C region (Figure 3C, schematic) and tested their interaction with His-MYF5 by biotin-RNA pulldown analysis. We found that MYF5 was most strongly recovered in the 3′UTR-C10 (3′-C10) fragment (Figure 3C) and further fragmentation of 3′C10 followed by pulldown revealed that MYF5 specifically binds to the 3′C10–1 sequence in the 3′UTR of Ccnd1 mRNA (Figure 3C; Supplementary Figure S4A). To test this interaction in a different way, we prepared radiolabeled 3′C10–1, incubated it with GST-MYF5 and then resolved the complexes by RNA electrophoretic mobility shift assay (EMSA) (Figure 3D, left) or by SDS-PAGE after UV-crosslinking (Figure 3D, right). In agreement with the high presence of G residues in the 3′C10–1 sequence (Supplementary Figure S4A), MYF5 was found to bind preferentially to a biotinylated G-rich oligomer relative to U- or A-rich ligands (Supplementary Figure S2C). This preference was further examined using GST-MYF5 and RNA segments spanning the segments of most binding: MYF5 CR, 3′B, 3′C and 3′D. After incubation in vitro, the complexes were subjected to crosslinking and immunoprecipitation (CLIP) analysis, followed by digestion using RNase T1, then isolation, cDNA preparation, cloning and sequencing of protected fragments. As shown in Supplementary Figure S4B, the protected fragments spanned G-rich stretches present primarily in fragment MYF5 3′C, but also in other regions such as the CR.
To identify the RNA-binding domain(s) of MYF5, we prepared GST-tagged recombinant MYF5 protein, either full-length (FL) or partial length containing the basic domain (D1), the helix-loop-helix (HLH) domain (D2), the MYF5 superfamily domain (D3) or combinations of these (Figure 3E; Supplementary Figure S4C). Equimolar concentration of the various GST-MYF5 proteins shown in (Figure 3E) were incubated with biotinylated 3′C fragment of Ccnd1 mRNA; after pulldown using streptavidin beads, bound proteins (top) and all proteins in the binding reactions (bottom) were detected by western blot analysis using an anti-GST antibody. Interestingly, all chimeric proteins bearing the helix-loop-helix (HLH) domain interacted specifically with the Ccnd1 3′C RNA (Figure 3E), as well as with the biotinylated Ccnd1 CR RNA (Supplementary Figure S4D). These findings suggested that the same HLH domain (D2), known to be responsible for MYF5 dimerization as well as binding to DNA with few amino acids from the basic domain (D1) might also be necessary for the binding of MYF5 to Ccnd1 mRNA.
MYF5 regulates CCND1 expression in C2C12 myoblasts
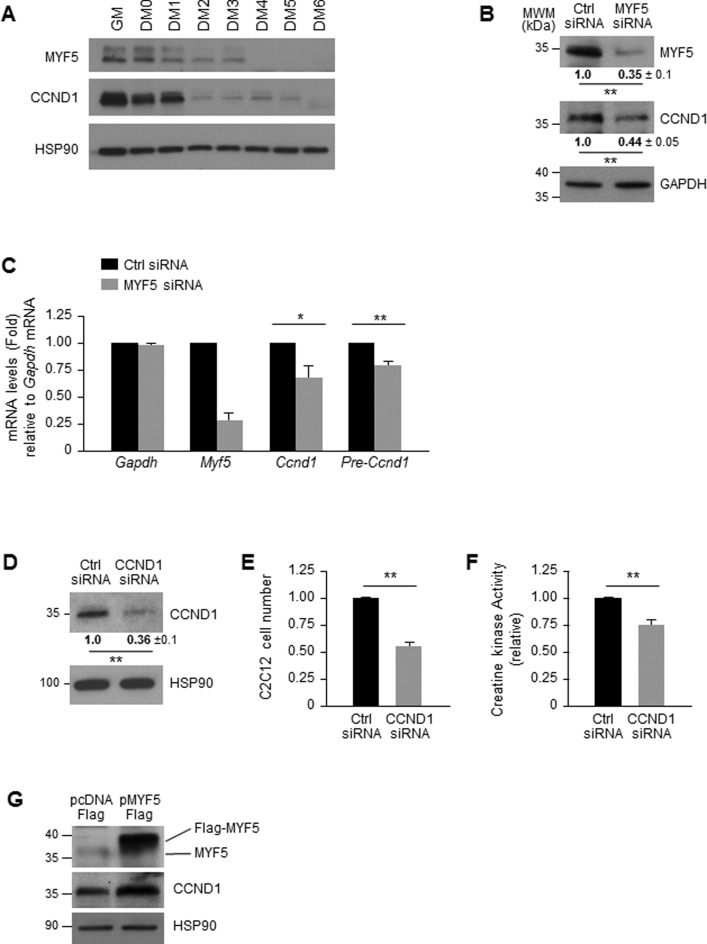
The interaction of MYF5 with Ccnd1 mRNA identified by RIP and in vitro binding analyses prompted us to investigate whether the levels of MYF5 affected CCND1 expression in C2C12 myoblasts. The first evidence in support of this possibility was obtained from experiments examining the correlation between MYF5 and CCND1 expression as a function of C2C12 myoblast differentiation. As shown in Figure 4A, CCND1 expression correlated closely with MYF5 expression levels during differentiation, suggesting that MYF5 might indeed control CCND1 expression in myoblasts. These findings were in keeping with the fact that the expression levels of CCND1 and MYF5 are regulated in a cell cycle-dependent manner (32). Moreover, MYF5 silencing in proliferating C2C12 cells led to a reduction in CCND1 levels (Figure 4B), which could result from reduced transcription, mRNA stability, or translation.
Figure 4.
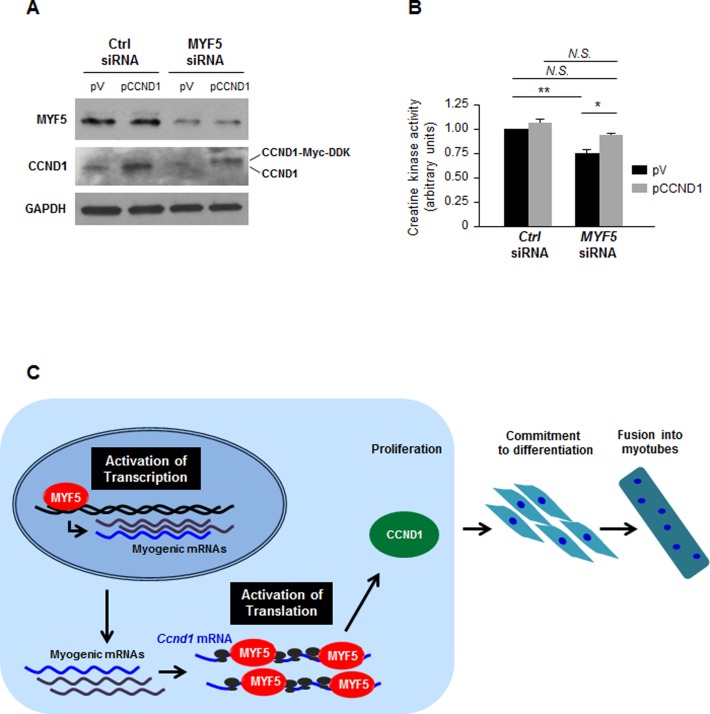
MYF5 regulates CCND1 expression in C2C12 myoblasts. (A) Western blot analysis of MYF5 and CCND1 expression during C2C12 differentiation; heat shock protein 90 (HSP90) was included as loading control. (B,C) Forty-eight hours after transfecting C2C12 cells with MYF5 siRNA or Ctrl siRNA, the levels of MYF5, CCND1 and loading control GAPDH were analyzed by western blot analysis (B) and the levels of Ccnd1 pre-mRNA and mRNA by RT-qPCR analysis (C). (D–F) Forty-eight hours after transfection of proliferating C2C12 myoblasts with Ctrl or CCND1 siRNAs, the levels of CCND1 and HSP90 were assessed by western blot analysis (D) and cell numbers were measured using a TC10 automated cell counter (BioRad) and represented as fold change in cell number after CCND1 silencing relative to those in the Ctrl siRNA group (E). At day 6 in differentiation medium (DM6) C2C12 differentiation was monitored by measuring creatine kinase activity (F). (G) Forty-eight hours after MYF5 was overexpressed using pFlag-MYF5 pcDNA3, the levels of MYF5, CCND1 and loading control HSP90 were studied by western blot analysis. Data in (B-F) represent the means and S.E.M. from three or four independent experiments. *, P < 0.05 and **, P < 0.01 (Student's t-test).
To assess if MYF5 influenced Ccnd1 transcription, the levels of Ccnd1 pre-mRNA were quantified as a surrogate measure of de novo transcription; as shown in Figure 4C, MYF5-silenced C2C12 myoblasts showed modestly reduced Ccnd1 pre-mRNA levels, suggesting a role of MYF5 in Ccnd1 transcription. A role for MYF5 in Ccnd1 gene transcription is certainly plausible, as several MYF5 target sites are present on the Ccnd1 promoter region (Supplementary Figure S5A) and most of them show interaction with MYF5 as assessed by chromatin IP (ChIP) analysis (Supplementary Figure S5B). However, given that a 20% reduction in Ccnd1 pre-mRNA following MYF5 silencing did not appear sufficient to account for a 65% reduction in CCND1 protein levels, we measured the stability of Ccnd1 mRNA in control and MYF5-silenced C2C12. After treatment with actinomycin D to block nascent mRNA transcription by inhibiting RNA polymerase II, measurement of the mRNA levels at subsequent times was used to determine the half-life of Ccnd1 mRNA, as well as that of a stable transcript, Gapdh mRNA (encoding a housekeeping protein). As shown, Ccnd1 mRNA was very stable in these cells, never reaching a reduction to 50% of its original abundance (discontinuous line), even after 8 h in actinomycin D (Supplementary Figure S5C). In addition, silencing of CCND1 (Figure 4D) significantly reduced the proliferation rate of C2C12 cells suggesting a role of CCND1 in myoblast proliferation (Figure 4E). Further analysis indicated that CCND1 silencing also lead to a decrease in C2C12 differentiation as measured by creatine kinase activity (Figure 4F), indicating that CCND1 regulates myogenesis by controling cell proliferation. Conversely, overexpression of MYF5 in growing C2C12 myoblasts using a Flag-MYF5 expression vector led to increased abundance of CCND1 in the MYF5-overexpressing cells compared to control cells (Figure 4G); unfortunately, extensive additional analysis after overexpressing MYF5 was not possible, as elevating MYF5 levels through transfection was toxic (not shown). In summary, MYF5 enhanced CCND1 expression, at least in part by elevating Ccnd1 mRNA transcription modestly. However, given the proportionally larger change in CCND1 protein levels, we set out to test the hypothesis that MYF5 might influence the translation of Ccnd1 mRNA.
MYF5 promotes Ccnd1 mRNA translation and hence myogenic differentiation
We first investigated if MYF5 regulated Ccnd1 mRNA translation by fractionating polysomes and studying if the relative size of Ccnd1 mRNA polysomes differed after modulating MYF5 levels. Two days after transfecting C2C12 cells (GM) with Ctrl siRNA or MYF5 siRNA, cytoplasmic lysates were fractionated through sucrose gradients and the distribution of Ccnd1 mRNA across the gradient fractions was examined. The overall distribution of polysomes in C2C12 cells, including the relative levels of 40S, 60S, 80S (monosomes), and low- and high-molecular-weight (LMW and HMW) polysomes, did not appear to vary as a function of MYF5 abundance (Figure 5A). From the 12 fractions in each gradient, RNA was isolated and subjected to RT-qPCR analysis, after which the relative abundance of Ccnd1 mRNA in each fraction was calculated as a percentage of the total Ccnd1 mRNA in the entire gradient; the distribution of Gapdh mRNA, which encodes a housekeeping protein and is not generally regulated by altered translation, was studied as a control. Interestingly, silencing MYF5 in C2C12 myoblasts shifted the relative size of Ccnd1 mRNA polysomes leftward, consistent with a reduction in the relative size of Ccnd1 mRNA polysomes; by contrast, the distribution of Gapdh mRNA was unaffected, supporting the view that Ccnd1 mRNA polyribosomes were selectively reduced in MYF5-silenced cells (Figure 5B). Control gradients were prepared in the presence of EDTA, in order to determine if these mRNAs simply cosedimented with polysomes or were true components of polyribosomes (Supplementary Figure S6A,B). Moreover, MYF5 was found to be associated with heavy translating polysomes in C2C12 cells (Supplementary Figure S6C); while the significance of this distribution pattern is unknown, it may be linked to MYF5-regulated translation, as shown for other RBPs (HuR and RBP42) interacting with target mRNAs (33–35).
Figure 5.
MYF5 promotes translation of Ccnd1 mRNA. (A,B) Forty-eight hours after Ctrl or MYF5 siRNA transfection of C2C12 cells, cytoplasmic extracts were fractionated through sucrose gradients to obtain cytoplasmic components of progressively larger weight: ribosomal subunits (40S, 60S), monosomes (80S) and low-molecular-weight (LMW) and high-molecular-weight (HMW) polysomes (A). The relative distribution of Gapdh mRNA, encoding a housekeeping protein, and Ccnd1 mRNA were measured by RT-qPCR analysis of RNA in each of the gradient fractions and represented as percentage of total RNA in the gradient (B). (C) Schematic of the dual luciferase reporter plasmids derived from the parent vector psiCHECK2 (psi), which expresses renilla luciferase (RL) and the internal control firefly luciferase (FL), and psiCHECK2-derived plasmids bearing the Ccnd1 fragments downstream of the RL coding region. (D)Top, Influence of MYF5 silencing on the expression of the reporter constructs. Twenty-four h after transfection of C2C12 cells with either MYF5 siRNA or Ctrl siRNA, each reporter plasmid was transfected, and 16 h later the ratio of RL activity to FL activity was measured. The decrease in relative RL/FL ratio of MYF5 siRNA-transfected cells relative to the RL/FL ratio of Ctrl siRNA-transfected cells is indicated. Bottom, RT-qPCR analysis of RL mRNA levels normalized to FL mRNA levels in each transfection group. Data represent the means and S.E.M. from 3 independent experiments. N.S., not significant; *, P < 0.05; **, P < 0.01 (Student's t-test).
To further study if MYF5 influenced CCND1 translation via its interaction with specific segments of Ccnd1 mRNA, we prepared luciferase reporter vectors derived from the parent plasmid psiCHECK2. Different fragments of Ccnd1 mRNA were cloned downstream of the renilla luciferase (RL) coding sequence, and firefly luciferase (FL) expressed from the same construct served as internal normalization control (Figure 5C). Twenty-four hours after transfecting C2C12 cells with MYF5 siRNA or Ctrl siRNA, cells were transfected with the vector control (psiCHECK2) or with constructs containing different fragments of Ccnd1 mRNA (as shown in Figure 5C). RL and FL activities were measured 16 h later, and the ratio of RL activity to FL activity (RL/FL) was measured for each construct in cells transfected with Ctrl siRNA and with MYF5 siRNA. The ratio of RL/FL activity in MYF5-silenced cells vs control cells for each construct was calculated using empty vector (psiCHECK2) as reference (Figure 5D, top). In this study, silencing MYF5 significantly reduced the relative expression of reporter constructs bearing Ccnd1 CR and Ccnd1 3′UTR-C (the segments interacting with MYF5, Figure 3) without significant changes in corresponding reporter RNA levels (Figure 5D, bottom), suggesting that the association of MYF5 to these regions of Ccnd1 mRNA is required for efficient translation. Taken together, these results suggest that MYF5 binds Ccnd1 mRNA and promotes the translation of CCND1 in proliferating C2C12 myoblasts.
In light of the findings that silencing MYF5 reduced CCND1 translation and hence CCND1 expression, we sought to test directly if the reduction in CCND1 abundance when MYF5 was silenced contributed to an impairment in myogenesis. In C2C12 cells in which MYF5 was either kept unchanged or silenced, pCCND1 or a vector control plasmid (pV) was ectopically overexpressed (Figure 6A). As shown, silencing MYF5 reduced myogenesis by day 6, as assessed by measuring creatine kinase activity; importantly, restoring CCND1 production by expression of a MYC-DDK-tagged CCND1 protein rescued significantly this loss of differentiation (Figure 6B). In closing, CCND1 is required to promote the progression of dividing cells through the G1 phase and through the G1/S transition point. Given that MYF5 promotes CCND1 expression partly via transcriptional induction and more robustly via translational upregulation, we propose that CCND1/Cyclin D1 is induced at multiple levels by MYF5 in order to elicit the early proliferative burst that initiates myogenic differentiation. The coordinated mechanisms by which MYF5 influences CCND1 expression are shown schematically (Figure 6C).
Figure 6.
Influence of MYF5 on myogenesis via regulation of CCND1 expression. (A,B) C2C12 cells were transfected with MYF5 siRNA or Ctrl siRNA, along with a control vector [pcDNA3-Flag (pV)] or a plasmid vector that expressed Myc-tagged CCND1. Forty-eight hours later, the levels of CCND1 and MYF5 were analyzed by western blot analysis (A) and the degree of differentiation was analyzed by measuring creatine kinase activity at day 6 into differentiation (B). Data presented are the means and S.E.M. from four independent experiments; significance (P) is indicated. (C) Proposed model whereby MYF5 modulates myogenesis by acting upon CCND1 expression on two levels: first, MYF5 activates Ccnd1 transcription moderately, and second, MYF5 binds the Ccnd1 mRNA at the CR and 3′UTR, promoting Ccnd1 mRNA translation. The net effect is a CCND1-mediated increase in myoblast proliferation necessary at the initiation of myogenesis. N.S., not significant; *, P < 0.05; **, P < 0.01 (Student's t-test).
DISCUSSION
Our results in the C2C12 myoblast model of myogenesis support the idea that besides modulating transcription, MYF5 can also bind to mRNAs and regulate the expression of myogenic proteins at the post-transcriptional level. The abundance of MYF5 was high in proliferating C2C12 cells and gradually declined as myoblasts differentiated into myotubes, in agreement with its role in myoblast proliferation (Figure 1). Although MYF5 was previously considered to be a nuclear protein (30,31), its discovery of in the cytoplasm of proliferating C2C12 cells was the first indication that MYF5 could have a non-nuclear role. A survey of MYF5-interacting mRNAs identified the key target Ccnd1 mRNA, which encodes Cyclin D1/CCND1, a protein required for the brief proliferative burst that precedes the onset of myoblast growth cessation and differentiation into myotubes. MYF5 specifically regulated Ccnd1 mRNA translation; accordingly, silencing MYF5 lowered Ccnd1 mRNA translation and CCND1 production, while overexpressing MYF5 overexpression elevated CCND1 levels. Moreover, while MYF5 silencing inhibited the early steps of C2C12 cell proliferation and its subsequent differentiation into myotubes, this inhibition was partially rescued when CCND1 was ectopically overexpressed (Figure 6). These findings are in agreement with earlier evidence indicating that initial cell division was important for establishing cell-cell contacts and for the fusion of myoblasts (36). Interestingly, Myf5 mRNA was stored in ribonucleoprotein granules in quiescent satellite cells, but was released and quickly translated upon myogenic stimulation (37), in turn promoting proliferation, differentiation and muscle fiber growth (24–26). In keeping with these previous observations, we propose that MYF5 is required for controlling CCND1 expression during myogenesis, particularly during the early proliferative stages of myoblast differentiation.
Although modestly, MYF5 promoted transcription of the Ccnd1 gene, as determined by the reduction in nascent Ccnd1 mRNA (Ccnd1 pre-mRNA) after silencing MYF5. Indeed, the binding sites identified on the Ccnd1 promoter (CAGCTG, Supplementary Figure S5A) are potential specific DNA target sequences shared by both MYF5 and MYOD. Both MYOD and MYF5 are myogenic transcription factors with largely redundant functions in myoblast differentiation; however, recent studies suggest that MYF5 regulates cell division in proliferating myoblasts, while MYOD functions later in the myogenic program (23,24). Supporting this idea are the findings that MYF5 levels are high in proliferative stages of the cell division cycle (S, G2 and early G1), while MYOD levels peak at the G1 phase, when cells exit the division cycle and enter growth arrest and differentiation (38–40). In fact, while MYOD and MYF5 can activate similar subsets of myoblast target genes transcriptionally, they follow distinct schedules of activation and tissue specificity (19,23,40–44). Structurally, both MYF5 and MYOD have central bHLH domains required for DNA binding and dimerization; differences in their N- and C-terminal segments are believed to be essential for their distinct kinetics and function.
MYF5 had a more substantial impact on translation of the Ccnd1 mRNA (Figure 5). The finding that the relative size of polysomes decreased after silencing MYF5 indicated that MYF5 promoted the initiation of translation of Ccnd1 mRNA. The exact mechanisms whereby MYF5 elevated CCND1 translation were not elucidated here, and there are no previous examples of MYF5 affecting translation of any mRNA. As we expand on this investigation, we will test if MYF5 promoted CCND1 translation by competing for binding with translational repressors, including other RBPs, microRNAs or other noncoding RNAs.
Discovering that MYF5 promoted Ccnd1 expression transcriptionally (as a nuclear DNA-binding protein) and post-transcriptionally (as a cytoplasmic RNA-binding protein) was unexpected. However, a rising number of RBPs are found which also bind DNA and regulate transcription. For example, the RBP NF90 [also named interleukin (IL) enhancer-binding factor 3 (ILF3)] binds the IL2 promoter and regulates transcription, while it binds numerous mRNAs via its two double-stranded (ds)RNA-binding domains (45,46). NF90 binds with high affinity and stabilizes several mRNAs containing AU-rich 3′UTRs, such as mRNAs that encode IL2, MKP-1, VEGF, the CDK inhibitor p21 and MYOD itself (45–47), while it represses the translation of other targets, such as the β-glucosidase mRNA (48). Another example is AUF1, an RBP best known for promoting target mRNA decay, although it can also have other post-transcriptional functions; AUF1 can also bind DNA and transcriptionally regulate the expression of MYC, CD21, telomerase (TERT) and MEF2C genes (28,49–51). Numerous other proteins bind both DNA and RNA, including NCL [nucleolin, (52,53)] and TP53 [the tumor suppressor p53 (54,55)]. In some cases, these dual interactions by a single protein have been mapped to DNA and RNA quadruplexes of different types (56). On a broader scale, it will be interesting to examine systematically if proteins capable of recognizing both RNA and DNA elements within a given gene employ this mechanism in order to coordinate the transcription and post-transcriptional fate of the same mRNA.
In closing, myogenic transcription factors (MRFs) govern myogenesis through their sequential, coordinated influence on the transcriptional production of myogenic genes (as reviewed in Ref. 57). Accordingly, MYF5, MYOD, myogenin and MRF4 function successively to assist quiescent satellite cells as they progress to activated, dividing myoblast, and then to growth-arrested myogenic cells that eventually fuse into differentiated myotubes. In an analogous manner, RBPs function sequentially to regulate myogenic gene expression at post-transcriptional levels to support myogenesis (7). According to this notion, MYF5 appears to be among the earliest RBPs necessary for activating proliferation in myoblasts through binding to Ccnd1 mRNA leading to a rise in CCND1 production, as reported here. The RBP HuR might be envisioned to function next, through binding and stabilizing the mRNAs that encode MRFs myogenin and MYOD, as well as the CDK inhibitor CDKN1A/p21. The ensuing increase in p21 establishes growth arrest in preparation for differentiation, while the elevation in myogenin and MYOD would further prepare myoblasts for differentiation (58). A subsequent association of RBPs HuR and KSRP, through their binding and destabilization of Npm mRNA [encoding NPM (nucleophosmin)] could solidify the growth inhibited state of myoblasts (59). Finally, the RBP AUF1 would function later in this temporal progression, assisting with advanced stages of myogenic differentiation by binding the Mef2c mRNA and promoting MEF2C production (28).
While this report focused on Ccnd1 mRNA, our analysis also identified other MYF5 target mRNAs. It will be important to investigate in depth the complete collection of MYF5 target mRNAs and assess whether MYF5 influences their expression transcriptionally as well as post-transcriptionally, and whether MYF5 interacts with similar RNA elements on Ccnd1 mRNA as on other targets mRNAs. Given that modulating MYF5 function may be a promising intervention strategy in diseases with poor muscle regeneration, it will be critical to gain a more complete understanding of MYF5 ribonucleoprotein complexes, particularly those that impact upon the expression of myogenic mRNAs.
Supplementary Material
Acknowledgments
This work was supported by the National Institute on Aging Intramural Research Program, National Institutes of Health. The authors thank J.Curtis and S.J.Mitchel (NIA, NIH) for assistance with this project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The National Institute on Aging Intramural Research Program, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Collins C.A., Olsen I., Zammit P.S., Heslop L., Petrie A., Partridge T.A., Morgan J.E. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Moss F.P., Leblond C.P. Satellite cells as the source of nuclei in muscles of growing rats. Anat. Rec. 1971;170:421–435. doi: 10.1002/ar.1091700405. [DOI] [PubMed] [Google Scholar]
- 3.Tedesco F.S., Dellavalle A., Diaz-Manera J., Messina G., Cossu G. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J. Clin. Invest. 2010;120:11–19. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chargé S.B., Rudnicki M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 5.Hawke T.J., Garry D.J. Myogenic satellite cells: physiology to molecular biology. J. Appl. Physiol. 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- 6.Carvajal J.J., Rigby P.W. Regulation of gene expression in vertebrate skeletal muscle. Exp. Cell Res. 2010;316:3014–3018. doi: 10.1016/j.yexcr.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Apponi L.H., Corbett A.H., Pavlath G.K. RNA-binding proteins and gene regulation in myogenesis. Trends Pharmacol. Sci. 2011;32:652–658. doi: 10.1016/j.tips.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glisovic T., Bachorik J.L., Yong J., Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovanda A., Režen T., Rogelj B. MicroRNA in skeletal muscle development, growth, atrophy, and disease. Wiley Interdiscip. Rev. RNA. 2014;5:509–525. doi: 10.1002/wrna.1227. [DOI] [PubMed] [Google Scholar]
- 10.Sohi G., Dilworth F.J. Noncoding RNAs as epigenetic mediators of skeletal muscle regeneration. FEBS J. 2015;282:1630–1646. doi: 10.1111/febs.13170. [DOI] [PubMed] [Google Scholar]
- 11.Berkes C.A., Tapscott S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Braun T., Buschhausen D.G., Bober E., Tannich E., Arnold H.H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversionin 10T1/2 fibroblasts. EMBO J. 1989;8:701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis R.L., Weintraub H., Lassar A.B. Expression of a single transfected eDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 14.Edmondson D.G., Olson E.N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989;3:628–640. doi: 10.1101/gad.3.5.628. [DOI] [PubMed] [Google Scholar]
- 15.Wright W.E., Sassoon D.A., Lin V.K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 16.Rhodes S.J., Konieczny S.F. Identification of MRF4: A new member of the muscle regulatory factor gene family. Genes Dev. 1989;3:2050–2061. doi: 10.1101/gad.3.12b.2050. [DOI] [PubMed] [Google Scholar]
- 17.Moncaut N., Rigby P.W., Carvajal J.J. Dial M(RF) for myogenesis. FEBS J. 2013;280:3980–3990. doi: 10.1111/febs.12379. [DOI] [PubMed] [Google Scholar]
- 18.Molkentin J.D., Black B.L., Martin J.F., Olson E.N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 19.Rudnicki M.A., Schnegelsberg P.N., Stead R.H., Braun T., Arnold H.H., Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 20.Rawls A., Morris J.H., Rudnicki M., Braun T., Arnold H.H., Klein W.H., Olson E.N. Myogenin's functions do not overlap with those of MyoD or Myf-5 during mouse embryogenesis. Dev. Biol. 1995;172:37–50. doi: 10.1006/dbio.1995.0004. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W., Behringer R.R., Olson E.N. Inactivation of the myogenic bHLH gene MRF4 results in upregulation of myogenin and rib anomalies. Genes Dev. 1995;9:1388–1399. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
- 22.Ott M.O., Bober E., Lyons G.E., Arnold H.H., Buckingham M. Early expression of the myogenic regulatory gene myf5 in precursor cells of skeletal muscle in the mouse embryo. Development. 1991;11:1097–1107. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- 23.Kablar B., Krastel K., Ying C., Asakura A., Tapscott S.J., Rudnicki M.A. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997;124:4729–4738. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- 24.Ustanina S., Carvajal J., Rigby P., Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- 25.Gayraud-Morel B., Chrétien F., Flamant P., Gomès D., Zammit P.S., Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev. Biol. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- 26.Montarras D., Lindon C., Pinset C., Domeyne P. Cultured myf5 null and myoD null muscle precursor cells display distinct growth defects. Biol. Cell. 2000;92:565–572. doi: 10.1016/s0248-4900(00)01110-2. [DOI] [PubMed] [Google Scholar]
- 27.Burattini S., Ferri P., Battistelli M., Curci R., Luchetti F., Falcieri E. C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur. J. Histochem. 2004;48:223–233. [PubMed] [Google Scholar]
- 28.Panda A.C., Abdelmohsen K., Yoon J.H., Martindale J.L., Yang X., Curtis J., Mercken E.M., Chenette D.M., Zhang Y., Schneider R.J., et al. RNA-binding protein AUF1 promotes myogenesis by regulating MEF2C expression levels. Mol. Cell. Biol. 2014;34:3106–3119. doi: 10.1128/MCB.00423-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y.H., Chen Y.H., Lu J.H., Tsai H.J. A 23-amino acid motif spanning the basic domain targets zebrafish myogenic regulatory factor myf5 into nucleolus. DNA Cell Biol. 2005;24:651–660. doi: 10.1089/dna.2005.24.651. [DOI] [PubMed] [Google Scholar]
- 31.Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T.K., Turner D., Rupp R., Hollenberg S., et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 32.Kitzmann M., Fernandez A. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell Mol. Life Sci. 2001;58:571–579. doi: 10.1007/PL00000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filippova N., Yang X., Wang Y., Gillespie G.Y., Langford C., King P.H., Wheeler C., Nabors L.B. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol. Cancer Res. 2011;9:648–659. doi: 10.1158/1541-7786.MCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraushar M.L., Thompson K., Wijeratne H.R., Viljetic B., Sakers K., Marson J.W., Kontoyiannis D.L., Buyske S., Hart R.P., Rasin M.R. Temporally defined neocortical translation and polysome assembly are determined by the RNA-binding protein Hu antigen R. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3815–E3824. doi: 10.1073/pnas.1408305111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das A., Morales R., Banday M., Garcia S., Hao L., Cross G.A., Estevez A.M., Bellofatto V. The essential polysome-associated RNA-binding protein RBP42 targets mRNAs involved in Trypanosoma brucei energy metabolism. RNA. 2012;18:1968–1983. doi: 10.1261/rna.033829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka K., Sato K., Yoshida T., Fukuda T., Hanamura K., Kojima N., Shirao T., Yanagawa T., Watanabe H. Evidence for cell density affecting C2C12 myogenesis: possible regulation of myogenesis by cell-cell communication. Muscle Nerve. 2011;44:968–977. doi: 10.1002/mus.22224. [DOI] [PubMed] [Google Scholar]
- 37.Crist C.G., Montarras D., Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Megeney L.A., Kablar B., Garrett K., Anderson J.E., Rudnicki M.A. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 39.Sabourin L.A., Girgis-Gabardo A., Seale P., Asakura A., Rudnicki M.A. Reduced differentiation potential of primary MyoD/ myogenic cells derived from adult skeletal muscle. J. Cell Biol. 1999;144:631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitzmann M., Carnac G., Vandromme M., Primig M., Lamb N.J., Fernandez A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J. Cell Biol. 1998;142:1447–1459. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin C.Y., Yung R.F., Lee H.C., Chen W.T., Chen Y.H., Tsai H.J. Myogenic regulatory factors Myf5 and Myod function distinctly during craniofacial myogenesis of zebrafish. Dev Biol. 2006;299:594–608. doi: 10.1016/j.ydbio.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 42.Kablar B., Krastel K., Tajbakhsh S., Rudnicki M.A. Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Dev. Biol. 2003;2:307–318. doi: 10.1016/s0012-1606(03)00139-8. [DOI] [PubMed] [Google Scholar]
- 43.Rudnicki M.A., Braun T., Hinuma S., Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH geneMyf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 44.Kaul A., Koster M., Neuhaus H., Braun T. Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell. 2000;102:17–19. doi: 10.1016/s0092-8674(00)00006-4. [DOI] [PubMed] [Google Scholar]
- 45.Masuda K., Kuwano Y., Nishida K., Rokutan K., Imoto I. NF90 in posttranscriptional gene regulation and microRNA biogenesis. Int. J. Mol. Sci. 2013;14:17111–17121. doi: 10.3390/ijms140817111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buaas F.W., Lee K., Edelhoff S., Disteche C., Braun R.E. Cloning and characterization of the mouse interleukin enhancer binding factor 3 (Ilf3) homolog in a screen for RNA binding proteins. Mamm. Genome. 1999;10:451–456. doi: 10.1007/s003359901022. [DOI] [PubMed] [Google Scholar]
- 47.Kuwano Y., Pullmann R., Jr, Marasa B.S., Abdelmohsen K., Lee E.K., Yang X., Martindale J.L., Zhan M., Gorospe M. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2010;38:225–238. doi: 10.1093/nar/gkp861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y.H., Grabowski G.A. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid beta-glucosidase and other mRNAs. Mol. Genet. Metab. 1999;68:441–454. doi: 10.1006/mgme.1999.2934. [DOI] [PubMed] [Google Scholar]
- 49.White E.J., Brewer G., Wilson G.M. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochim. Biophys. Acta. 2013;1829:680–688. doi: 10.1016/j.bbagrm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pont A.R., Sadri N., Hsiao S.J., Smith S., Schneider R.J. mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol. Cell. 2012;47:5–15. doi: 10.1016/j.molcel.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dempsey L.A., Hanakahi L.A., Maizels N. A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J. Biol. Chem. 1998;273:29224–29229. doi: 10.1074/jbc.273.44.29224. [DOI] [PubMed] [Google Scholar]
- 52.Brázda V., Hároníková L., Liao J.C., Fojta M. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014;15:17493–17517. doi: 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdelmohsen K., Gorospe M. RNA-binding protein nucleolin in disease. RNA Biol. 2012;9:799–808. doi: 10.4161/rna.19718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riley K.J., Ramirez-Alvarado M., Maher L.J., 3rd RNA-p53 interactions in vitro. Biochemistry. 2007;46:2480–2487. doi: 10.1021/bi061480v. [DOI] [PubMed] [Google Scholar]
- 55.Beckerman R., Prives C. Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cassiday L.A., Maher L.J., 3rd Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pownall M.E., Gustafsson M.K., Emerson C.P., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 58.van der Giessen K., Di-Marco S., Clair E., Gallouzi I.E. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J. Biol. Chem. 2003;278:47119–47128. doi: 10.1074/jbc.M308889200. [DOI] [PubMed] [Google Scholar]
- 59.Cammas A., Sanchez B.J., Lian X.J., Dormoy-Raclet V., van der Giessen K., López de Silanes I., Ma J., Wilusz C., Richardson J., Gorospe M., et al. Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation. Nat. Commun. 2014;5:4190. doi: 10.1038/ncomms5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.